Abstract
Nuclear receptor subfamily 1, group D member 1 (Nr1d1), also known as Rev-erb-α, belongs to the family of “orphan receptors” and functions as a member of clock gene family. In addition to being an important member of clock circuitry, Nr1d1, also regulates cell proliferation, lipid metabolism, and inflammation and is also touted as a tumor suppressor. Our focus on Nr1d1 was stimulated by data from a genome-wide search for mRNA correlates of cigarette smoke (CS) sensitive—whole smoke (WS) and filtered smoke (FS)—lung transcriptomes in tumor-resistant C57BL6 and tumor-susceptible AJ mice strains. Differential analysis of ~15 000 genes using Affymetrix 430A 2.0 high-density oligonucleotide arrays identified modulation of genes related to circadian pathways by CS in lungs of both mouse strains. Nr1d1 expression was downregulated by both WS and FS irrespective of mouse strain as compared to respective air-breathing controls. WS was more effective than FS on decreasing Nr1d1 expression. The present data suggest that transcriptional regulation of Nr1d1 by CS may affect circadian rhythmicity and thus may play a complementary role in CS-induced lung respiratory tract pathobiology and/or lung tumorigenesis.
Keywords: Nr1d1, Rev-erb-α, C57BL6 mice strain, AJ mice strain, lungs, cigarette smoke, gene expression profiling, chronobiology
Introduction
Nuclear receptor subfamily 1, group D member 1 (Nr1d1), also known as, Rev-erb-α, is a member of “orphan receptors”1 and is encoded on the noncoding strand of the thyroid hormone receptor gene.2 Orphan receptors are proteins lacking in, as yet, well-defined ligands or particular physiological or biological functions.3 However, recent studies have shown that heme may function as a ligand for Nr1d1 and its closely related receptor—Nr1d2 (Rev-erbβ).4 Nr1d1 was reported to act as a major molecular link through which proteins related to circadian regulation drive the circadian processes.5,6 These proteins are encoded by “clock genes” such as period homologs 1 and 2 (Per1 and Per2), cryptochromes 1 and 2 (Cry 1 and Cry2), aryl hydrocarbon receptor nuclear translocator-like (Arntl; also known as Bmal1), circadian locomotor output cycles kaput (Clock) and neuronal PAS domain protein 2 (Npas2).7 The “clock circuitry” is composed of interacting feedback loops that regulate the transcription of the clock genes and are highly conserved among animals.8 The circadian clock is controlled by clock genes and is synchronized to the external time cues by input signals and regulate peripheral circadian rhythms via output signals.9 Studies in Nr1d1 null mice (Nr1d1−/−) had shown that the expression of suprachiasmatic nucleus (SCN) and liver Nr1d1 in mice is negatively regulated by Per and Cry proteins but positively regulated by Clock and Arntl. The cyclic accumulation of Nr1d1 then imposes circadian regulation of Arntl transcription.5 Apart from its circadian regulatory function, Nr1d1 was also reported to modulate cell proliferation/differentiation,10,11 lipid metabolism,12,13 and NF-κB pathways.14 Altered circadian rhythms may contribute to sleep disorders, psychotic disorders, and tumorigenesis.15–17
Cigarette smoke (CS) exposure is known to alter normal circadian rhythm in lungs by disrupting lung function18 or by modulating the expression of genes related to circadian circuitry.19 The present study was carried out to identify circadian gene expression changes in lung tissue in two strains of mice—tumor-resistant C57BL6 mice and tumor-susceptible AJ mice—by exposing them to whole smoke (WS) and to filtered smoke (FS).
Materials and Methods
Animals
The protocols for the care and use of animals were approved by the Institutional Care and Use Committee at the University of California, Davis. Male C57BL/6 and AJ mice (3 months old) were purchased from Jackson Laboratories, Bar Harbor, ME. Mice were housed in polycarbonate cages and maintained at 21° to 23°C and 60% to 70% humidity on a 12-hour light/dark schedule and with ad libitum access to water and food. After a 2-week acclimatization period, both strains of mice were assigned at random to different groups (n = 5 per group) and exposed to filtered air, WS, or FS. Mice had unrestricted access to water and diet, and body weight was measured before and after respective exposures. Immediately after the last filtered air or CS exposures, the animals were euthanized by pentobarbital overdose (120 mg/kg body weight, i.p.). Lung parenchymal tissue was dissected away from extraparenchymal airways and blood vessels and stored at −80°C until further analyses.
Cigarette Smoke Exposure
The CS exposure studies were carried out at the University of California, Davis CS Exposure Facility (Director: Dr Kent Pinkerton). Kentucky 2R4F research cigarettes obtained from Tobacco Research Institute, University of Kentucky, Lexington, were used. Three-month-old male mice from both strains (C57BL6 and AJ) were exposed to WS, 6 hours a day for a period of 10 days at 90 mg/m3 of total suspended particles (TSP), and then increased to 125 mg/m3 over 5 days, and continued for 3 additional days at 125 mg/m3.20,21. For FS exposure, WS was drawn through a high-efficiency particulate air (HEPA) filter generating FS before entering the exposure chamber.21,22 The chamber atmosphere was controlled with relative humidity of 40% ± 8% and temperature 75°F ± 3°F. Chamber atmospheres were also monitored daily for carbon monoxide (CO), nicotine, and TSP. Animals breathing filtered air served as controls for the study.
RNA Extraction and Biotin-Labeled RNA for GeneChip Analysis
RNA from lung tissue was extracted and processed for GeneChip analysis as previously described.23 Briefly, RNA from lung tissues were extracted with Trizol reagent and purified and quantified according to the manufacturer’s (Invitrogen, Carlsbad, CA) protocol. Five replicates, each from a mouse lung, from each of the 3 groups (n = 5 per group) exposed to filtered air, WS, or FS, were further processed as follows for GeneChip analysis.
The methods used to prepare the eukaryotic target preparation were followed according to the manufacturer’s protocol (Affymetrix, Inc.) for expression arrays. An aliquot (10 μg) of RNA solution was used for preparation of one-cycle cDNA synthesis (first-strand and second-strand cDNA synthesis) followed by cleanup of double-stranded cDNA and synthesis of biotin-labeled cRNA. The biotin-labeled cRNA (40 μg) was used for fragmenting for target preparation. Fragmented cRNA samples were hybridized to high density oligonucleotide Affymetrix Mouse 430A 2.0 GeneChips (Santa Clara, CA), which contains 22 600 probe sets representing transcripts and variants from more than 14 000 well-characterized mouse genes. For hybridization, gene expression was assessed using one chip per mouse lung.
Oligonucleotide Microarray Analysis and Statistics
The scanned images of hybridization signals were analyzed with the Affymetrix GeneChip Operating Software (GCOS 1.4), including the GeneChip scanner 3000 High-Resolution Scanning Patch and DNA-D chip analyzer (d-Chip), a software package implementing model-based expression analysis of oligonucleotide arrays at http://www.dchip.org.24 The absolute mRNA expression (present or absent) and differential mRNA expression data were obtained from the pivot data. When the P value for detection signal was <.049 (range of P values .0002 to .0490), the expression of the mRNA was classified as present (P). All mRNAs with the P value for detection >.05 were considered absent (A). The signal intensities for transcripts classified as present ranged from 5 to 7000 U. We also performed Gene Ontology (GO) (www.geneontology.org) and PANTHER (Protein ANalysis THrough Evolutionary Relationships)25 analysis for classification of differentially expressed genes for characterizing the biological properties and pathways involved.
Statistical Analyses
Statistical evaluation of analytical data was done by Student’s t test using the statistical software GraphPad Prism 4.0 (San Diego, CA). In all comparisons P < .05 was considered as significant. Results are expressed as mean ± standard error of the mean (mean ± SEM).
Results
Body Weight and CS Exposure Data
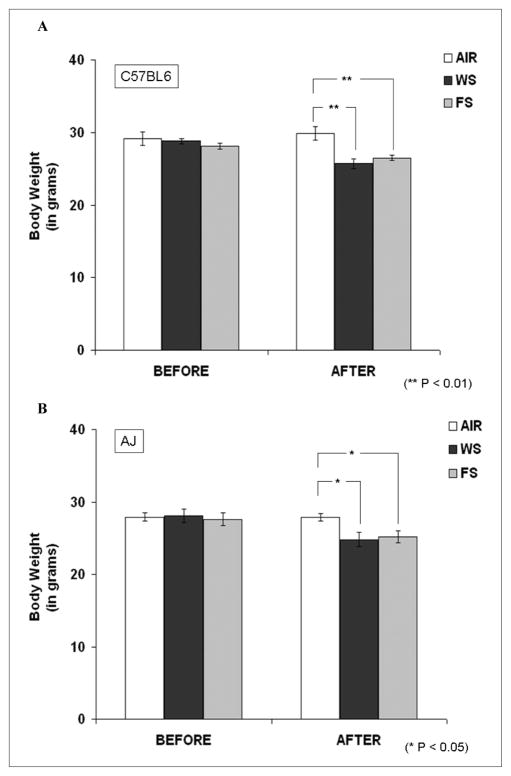
As displayed in Figures 1A and 1B, CS, both WS and FS, exposures caused a significant decrease in the body weights of the 2 mouse strains as compared with their respective filtered air controls (P < .01 and P < .05, respectively). There was no significant difference in the effects of WS and FS on the reduction in body weight in both the mouse strains.
Figure 1. Body weight data before and after cigarette smoke (CS) exposure in C57BL6 and AJ mice strains.
The mice were exposed to filtered air, incremental whole smoke (WS; 90 to 125 mg/m3), or filtered smoke (FS) for 8 days (6 h/d). (A) C57BL6 mice exposed to WS and FS showed a significant reduction in body weight (P < .01). Similarly, (B) AJ mice also showed a significant reduction in body weight post exposure (P < .05). However, there was no significant change in body weight levels comparing C57BL6 and AJ mice in any of the exposure groups. Data are represented as mean ± standard error of the mean, n = 5 to 6.
Analyses of CS exposure chambers (based on observations from 8 different exposure days), revealed that TSP, CO, and nicotine levels were 129.5 ± 11.6 mg/m3, 291 ± 21.9 ppm, and 12.8 ± 0.19 mg/m3, respectively, in WS chambers. By introducing HEPA filters to generate FS, TSP, and nicotine levels were effectively lowered to 1.0 ± 0.15 mg/m3 and 2.6 ± 0.08 mg/m3 respectively, with a 15% drop in CO levels (245 ± 19.6 ppm).
GeneChip Analysis
The microarray analysis by Affymetrix GeneChips detected ~16 000 genes in lung tissues from the 2 mouse strains exposed to either filtered air or CS. A report of comprehensive analysis and interpretations of the transcriptomic data is under preparation. Here we give a summary of the data and focus on the expression of Nr1d1 and other genes related to circadian circuitry.
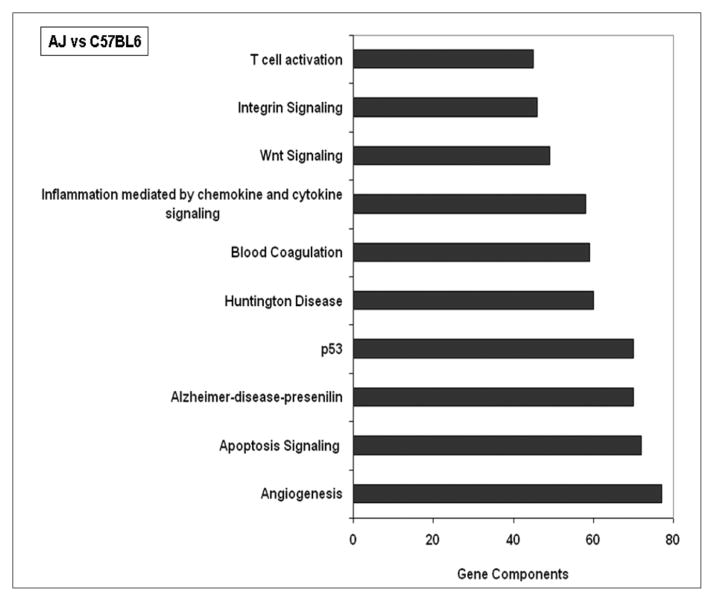
Comparative analysis of the data from air-exposed, tumor-susceptible AJ and tumor-resistant C57BL6 mice identified 468 differentially expressed genes in lung tissue. Of these 468 genes, 232 were overexpressed and 236 genes were underexpressed in C57BL6 lungs compared with those in AJ lungs. Functional classification of the differentially expressed genes was obtained by GO ontology (www.geneontology.org) using dChip software and related pathways analyzed by PANTHER analysis. Pathways related to angiogenesis, apoptosis signaling, p53, and inflammatory-immune responses were seen to be largely affected in AJ lung tissue basally as compared with lungs in C57BL6 mice strain (Figure 2).
Figure 2. Major pathways affected in lungs of AJ mice as compared with lungs of C57BL6 mice.
Differentially expressed lung genes in air-breathing AJ mice and C57BL6 mice were subjected to pathway analysis by PANTHER software. Out of 85 pathways identified, the top 10 pathways, with the maximum gene component “hits” are shown (N = 5/group). Circadian clock system was one of the pathways (44th in the list of 85 pathways) identified with 14 gene component hits.
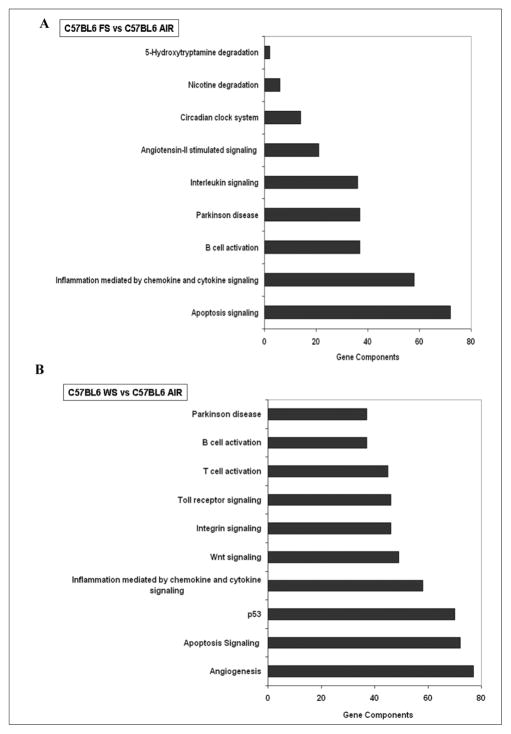
The lungs of the 2 mouse strains also showed distinct responses to CS exposures. Furthermore, the analysis also revealed distinct transcriptomic signatures of WS and FS. WS exposures changed the expression of 134 genes (37 upregulated and 97 downregulated) in lung tissues of C57BL6 mice. In contrast, FS exposure affected the expression of 32 genes (17 genes were induced and 15 were repressed). As seen in Figures 3A and 3B, FS and WS exposure to C57BL6 mice affected genes related to multiple pathways in lung tissue.
Figure 3. Major pathways affected in lungs of C57BL6 mice exposed to (A) filtered smoke (FS) or (B) whole smoke (WS) as compared with respective air-breathing controls.
Differentially expressed lung genes in air, FS, and/or WS exposed C57BL6 mice were subjected to pathway analysis by PANTHER software. (A) In FS-exposed C57BL6 mice lungs, only 9 pathways were seen to be primarily affected of which one was that of circadian clock system. (B) In WS-exposed mice, out of 34 pathways identified, the top 10 pathways, with the maximum gene component “hits” are shown (N = 5/group). Circadian clock system being one of the pathways (21st in the list of 34 pathways) identified with 14 gene component hits.
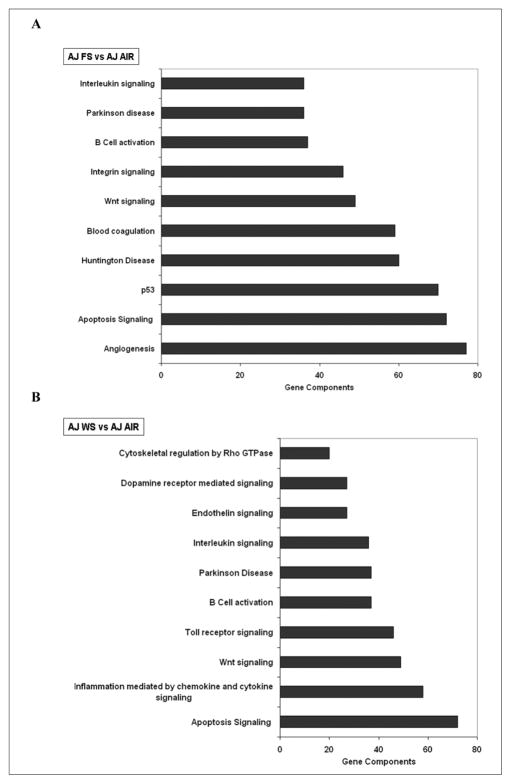
WS affected the expression of 87 lung genes in AJ mice (47 genes were induced and 40 were repressed). FS altered the expression of 62 lung genes (47 induced and 15 repressed) in lungs of AJ mice. FS exposure to AJ mice was as effective as WS exposure in modulating lung genes related to angiogenesis, apoptosis signaling, p53, and inflammatory-immune response pathways as compared with respective air-breathing control mice (Figures 4A and 4B).
Figure 4. Major pathways affected in lungs of AJ mice exposed to (A) filtered smoke (FS) or (B) whole smoke (WS) as compared with respective air-breathing controls.
Differentially expressed lung genes in air, FS, and/or WS exposed AJ mice were subjected to pathway analysis by PANTHER software. (A) In FS- exposed AJ mice lungs, out of 19 pathways identified, the top 10 pathways, with the maximum gene component “hits” is shown here. Circadian clock system pathway was listed as the 16th out of 19 pathways affected. (B) In WS-exposed mice, out of 20 pathways identified (circadian clock system pathway was listed at 12th), the top 10 pathways, with the maximum gene component “hits” is shown here (N = 5/group).
Nr1d1 Expression Suppressed by CS Exposure
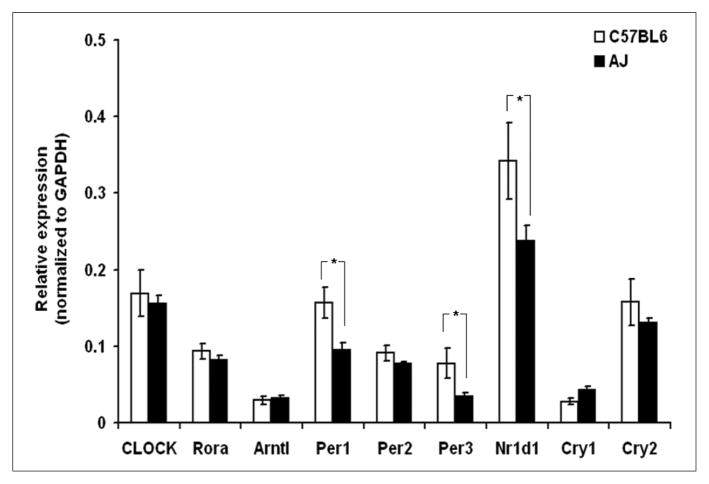
The present article is primarily focused on the CS-induced modulation of genes related to circadian circuitry. As compared to lungs of air-breathing C57BL6 mice strain, the basal transcriptomic expressions of circadian genes, period homolog 1 (Per1), period homolog 3 (Per3), and nuclear receptor subfamily 1, group D member 1 (Nr1d1), were significantly (P < .05) suppressed in AJ lungs (Figure 5). This is an interesting observation, as period holomogs and Nr1d1 also act as tumor suppressors26–28 and AJ mice strains are known to be “tumor-susceptible”29 as compared with “tumor-resistant” C57BL6 mice strains.
Figure 5. Basal expression of genes related to circadian circuitry in lungs of C57BL6 and AJ mice.
As compared with C57BL6 lungs, mRNA expression of Per1, Per3, and Nr1d1 were seen to be significantly downregulated (*P < .05) in lungs of AJ mice. Data are represented as mean ± standard error of the mean, n = 5 to 6.
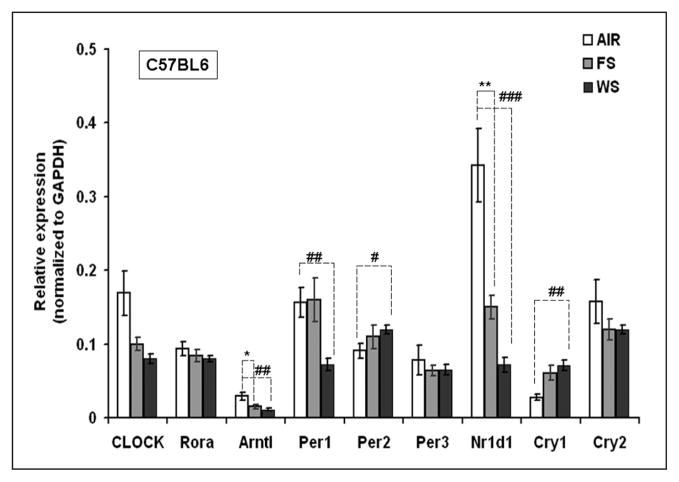
CS exposure to C57BL6 mice was seen to modulate the expression of circadian-related lung genes (Figure 6), of which aryl hydrocarbon receptor nuclear translocator-like (Arntl; also known as Bmal1) and Nr1d1 were seen to be significantly downregulated by both FS (P < .05 and P < .01, respectively) and WS (P < .01 and P < .001. respectively) exposure as compared to air-breathing controls. At the same time, the expression of circadian genes, Per2 and cryptochrome 1 (Cry1), were seen to be significantly upregulated by WS exposure (Figure 6).
Figure 6. Expression of genes related to circadian circuitry in cigarette smoke (CS)-exposed lungs of C57BL6 mice.
Three-month-old male mice were exposed to whole smoke (WS), 6 hours a day for a period of 10 days at 90 mg/m3 of total suspended particles (TSP), and then increased to 125 mg/m3 over 5 days, and continued for 3 additional days at 125 mg/m3. For filtered smoke (FS) exposure, WS was drawn through a high-efficiency particulate air (HEPA) filter generating FS before entering the exposure chamber. *P < .05 and **P < .01 represent level of significance in FS-exposed group as compared with the respective air-group. #P < .05, ##P < .01, and ###P < .001 represent level of significance in WS-exposed group as compared with respective air-breathing controls. Data are represented as mean ± standard error of the mean, n = 5 to 6.
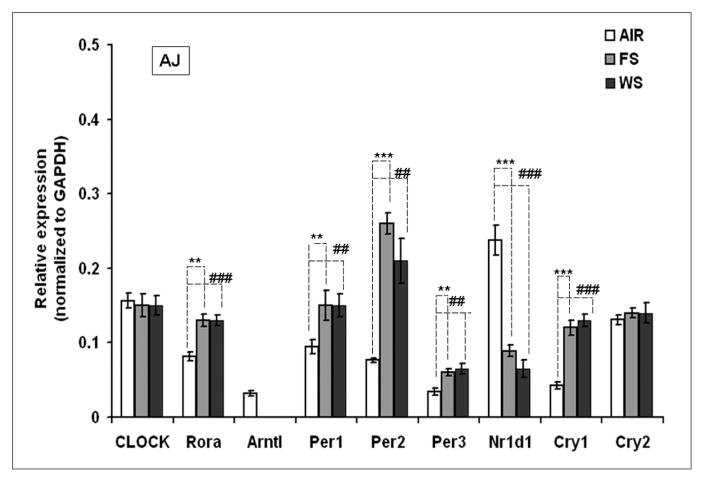
Similarly, FS and WS exposure to AJ mice was seen to further modulate the expression of circadian related genes in lungs as compared with air-breathing controls (Figure 7). Of note, Arntl expression was completely abolished by FS and WS exposure in AJ mice. Nr1d1 expression as compared with C57BL6 and AJ mice lungs (Figure 5), was seen to be further downregulated (P < .001) by FS and WS exposure (Figure 7).
Figure 7. Expression of genes related to circadian circuitry in cigarette smoke (CS)-exposed lungs of AJ mice.
Three-month-old male mice were exposed to whole smoke (WS), 6 hours a day for a period of 10 days at 90 mg/m3 of total suspended particles (TSP), and then increased to 125 mg/m3 over 5 days, and continued for 3 additional days at 125 mg/m3. *P < .05 and **P < .01 represent level of significance in FS-exposed group as compared with the respective air-group. #P < .05, ##P < .01, and ###P < .001 represent level of significance in WS-exposed group as compared with respective air-breathing controls. Data are represented as mean ± standard error of the mean, n = 5 to 6.
Discussion
The significant and novel outcome of this study is the identification of a cluster of genes, encoding members of the transcription factor family that regulate circadian rhythm, which are differentially expressed between tumor-susceptible and tumor-resistance mice. The basal expression of the major circadian regulatory lung genes, transcriptomic expression of Nr1d1 (also known as Rev-erbα) was seen to be suppressed in AJ lungs as compared with lungs of C57BL6 mice (Figure 5). This observation is noteworthy, as Nr1d1, a “clock gene” has also been reported to act as a “tumor suppressor” gene regulating cell proliferation.28 Preitner et al5 showed in Nr1d1 null mice that the expression of SCN and liver Nr1d1 is negatively regulated by Per and Cry proteins but positively regulated by Clock and Arntl and the cyclic accumulation of Nr1d1 then imposes circadian regulation of Arntl transcription. Hence, suppressed Nr1d1 expression in AJ mice lungs may suggest that these mice may have dysregulated circadian functions and maybe one of the factors attributed to its tumor susceptibility. It must also be noted that these data were obtained from 3-month-old AJ mice and that no lung tumors were observed at this early time period. In this strain, sporadic lung tumors start to develop by 5 to 6 months.30 We speculate that Nr1d1 could be touted as one of the “early carcinogenic biomarkers (preneoplastic)” for early detection of cancer in lungs.
Disruption in “chronobiology” or the biological rhythm as represented by the circadian rhythm may result in abnormal homeostatic control of normal cellular proliferation leading to cancer as a result of environmental or genetic stimuli. Circadian clock gene mutations, repeat shifts in light–dark cycle, ablation of SCN in the hypothalamus, and so on have been reported to be the causative factors.31–35 However, there is a paucity of reports related to CS-induced dysregulation of genes related to circadian circuitry.19 Clegg et al36 have reported that nicotine administration (1 mg/kg body weight s.c.) to pregnant Sprague–Dawley rats induced c-fos (FBJ osteosarcoma oncogene) mRNA expression in maternal habenula and hypothalamic paraventricular nucleus. Similar induction was also seen in fetal brain in both habenula and hypothalamic paraventricular nucleus and also in the SCN.36 Though a direct link on the effect of nicotine on SCN is not clearly understood, nicotine-induced c-fos mRNA expression in SCN has been implicated as causing a phase-shift in circadian systems.36 However, reports by other workers had shown that nicotine may not significantly alter circadian activities.37,38
There is overwhelming evidence that CS plays a major role in the epidemiology of lung cancer39 and the effects of CS on tumor-susceptible mouse strain, AJ, have been extensively studied.21,22,29,30,40–42 Dysregulated xenobiotic metabolism, inflammatory-immune responses, and/or epigenetic modifications, with respect to methylation/acetylation of DNA binding sites/histones have all been thought to play contributing roles in CS-induced lung carcinogenesis.43 Rutter et al44 have reported that DNA binding activity of the circadian genes CLOCK:Arntl and neuronal PAS domain protein 2 (NPAS2:Arntl) heterodimers are influenced by the redox state of nicotinamide adenine dinucleotide (NAD).44 Thus, reduced forms of cofactors NAD(H) and NADP(H) enhanced DNA-binding, whereas oxidized forms inhibited it, suggesting that circadian systems may be modulated by the redox-status of the biological system.44 As seen in our observations, Nr1d1 expression was suppressed by FS and WS exposure in both the mice strains, where WS was seen to be more potent (Figures 6 and 7). Thus, Nr1d1 expression, which was suppressed in AJ lungs as compared with lungs of C57BL6 mice, and further downregulated by CS exposure, implicates that AJ mice are more susceptible to dysregulation of circadian rhythms in lung tissue as compared with C57BL6 mice. It must also be noted that we have been consistently observing suppression of Nr1d1 mRNA expression in lungs of CS-exposed mice in several of our studies in a different cohorts of mice.23,45 In a recently published report, we observed ozone (O3)-induced downregulation of Nr1d1 expression in lungs in a vitamin E deficient mice model though no changes were seen in O3-exposed vitamin E sufficient mice.46 These observations, along with the present data, support our hypothesis that Nr1d1 may represent a redox-sensitive gene. Nr1d1 were reported to be “orphan receptors” without any identified physiological ligands;1 however, recent studies have shown that “heme” may act as a ligand, thus regulating this receptor’s function.47,48 This does open the question as to whether there is a link to its ligand, heme and Nr1d1’s redox-sensitive status. Further studies that would resolve this possibility, along with characterizations of which of the lung’s major cell types predominantly express Nr1d1, are warranted.
Summary
To summarize, CS exposure to tumor-resistant C57BL6 mice and tumor-susceptible AJ mice revealed dysregulated expression of lung Nr1d1, a key circadian rhythm and tumor-suppressor gene. Apart from their circadian regulatory functions, Nr1d1 is also reported to regulate cell proliferation/differentiation,10,11 lipid metabolism,12,13 and NF-κB pathways.14 Hence, CS-induced dysregulation of Nr1d1 expression may affect several cellular processes that could affect CS-induced lung carcinogenic and non-carcinogenic (eg, chronic obstructive pulmonary disorder) pathobiologies. It must also be noted that the data presented here are of just one time-point and further studies need to be done to understand the light and night cycle lung gene expression studies at different time-points and its modulation by environmental perturbations such as CS. The present study reveals a “snapshot” of CS-modulated expression of genes related to circadian circuitry in lungs and the lesser studied interactions of CS and/or redox-sensitive circadian genes.
Acknowledgments
We gratefully acknowledge Professor Hanspeter Witschi for his advice and support for these studies and Dr Saji Oommen for his expertise in mouse studies and gene expression analysis.
Funding
The authors disclosed receipt of the following financial support for the research and/or authorship of this article:
National Institute of Health Sciences (NIH #ES011895) University of California Clinical Nutrition Research Unit and Center for Human Nutrition Research at Davis
Footnotes
Declaration of Conflicting Interests
The authors declared no conflicts of interest with respect to the authorship and/or publication of this article.
References
- 1.O’Malley BW, Conneely OM. Orphan receptors: in search of a unifying hypothesis for activation. Mol Endocrinol. 1992;6:1359–1361. doi: 10.1210/mend.6.9.1331771. [DOI] [PubMed] [Google Scholar]
- 2.Lazar MA, Hodin RA, Darling DS, Chin WW. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA alpha transcriptional unit. Mol Cell Biol. 1989;9:1128–1136. doi: 10.1128/mcb.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burris TP. Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock. Mol Endocrinol. 2008;22:1509–1520. doi: 10.1210/me.2007-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Preitner N, Damiola F, Lopez-Molina L, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 6.Triqueneaux G, Thenot S, Kakizawa T, et al. The orphan receptor Rev-erbalpha gene is a target of the circadian clock pacemaker. J Mol Endocrinol. 2004;33:585–608. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 8.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 10.Downes M, Carozzi AJ, Muscat GE. Constitutive expression of the orphan receptor, Rev-erbA alpha, inhibits muscle differentiation and abrogates the expression of the myoD gene family. Mol Endocrinol. 1995;9:1666–1678. doi: 10.1210/mend.9.12.8614403. [DOI] [PubMed] [Google Scholar]
- 11.Chawla A, Lazar MA. Induction of Rev-ErbA alpha, an orphan receptor encoded on the opposite strand of the alpha-thyroid hormone receptor gene, during adipocyte differentiation. J Biol Chem. 1993;268:16265–16269. [PubMed] [Google Scholar]
- 12.Gervois P, Chopin-Delannoy S, Fadel A, et al. Fibrates increase human REV-ERBalpha expression in liver via a novel peroxisome proliferator-activated receptor response element. Mol Endocrinol. 1999;13:400–409. doi: 10.1210/mend.13.3.0248. [DOI] [PubMed] [Google Scholar]
- 13.Raspe E, Duez H, Mansen A, et al. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. J Lipid Res. 2002;43:2172–2179. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- 14.Migita H, Morser J, Kawai K. Rev-erbalpha upregulates NF-kappaB-responsive genes in vascular smooth muscle cells. FEBS Lett. 2004;561:69–74. doi: 10.1016/S0014-5793(04)00118-8. [DOI] [PubMed] [Google Scholar]
- 15.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 16.Cermakian N, Boivin DB. A molecular perspective of human circadian rhythm disorders. Brain Res Brain Res Rev. 2003;42:204–220. doi: 10.1016/s0165-0173(03)00171-1. [DOI] [PubMed] [Google Scholar]
- 17.Shi J, Wittke-Thompson JK, Badner JA, et al. Clock genes may influence bipolar disorder susceptibility and dysfunctional circadian rhythm. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1047–1055. doi: 10.1002/ajmg.b.30714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casale R, Natali G, Colantonio D, Pasqualetti P. Circadian rhythm of peak expiratory flow in children passively exposed and not exposed to cigarette smoke. Thorax. 1992;47:801–803. doi: 10.1136/thx.47.10.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gebel S, Gerstmayer B, Kuhl P, Borlak J, Meurrens K, Muller T. The kinetics of transcriptomic changes induced by cigarette smoke in rat lungs reveals a specific program of defense, inflammation, and circadian clock gene expression. Toxicol Sci. 2006;93:422–431. doi: 10.1093/toxsci/kfl071. [DOI] [PubMed] [Google Scholar]
- 20.Witschi H, Espiritu I, Peake JL, Wu K, Maronpot RR, Pinkerton KE. The carcinogenicity of environmental tobacco smoke. Carcinogenesis. 1997;18:575–586. doi: 10.1093/carcin/18.3.575. [DOI] [PubMed] [Google Scholar]
- 21.Witschi H, Espiritu I, Maronpot RR, Pinkerton KE, Jones AD. The carcinogenic potential of the gas phase of environmental tobacco smoke. Carcinogenesis. 1997;18:2035–2042. doi: 10.1093/carcin/18.11.2035. [DOI] [PubMed] [Google Scholar]
- 22.Witschi H. Carcinogenic activity of cigarette smoke gas phase and its modulation by beta-carotene and N-acetylcysteine. Toxicol Sci. 2005;84:81–87. doi: 10.1093/toxsci/kfi043. [DOI] [PubMed] [Google Scholar]
- 23.Gohil K, Oommen S, Vasu VT, Aung HH, Cross CE. Tocopherol transfer protein deficiency modifies nuclear receptor transcriptional networks in lungs: modulation by cigarette smoke in vivo. Mol Aspects Med. 2007;28:453–480. doi: 10.1016/j.mam.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2(8):RESEARCH0032. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen-Goodspeed M, Lee CC. Tumor suppression and circadian function. J Biol Rhythms. 2007;22:291–298. doi: 10.1177/0748730407303387. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Wood PA, Ansell CM, et al. Beta-catenin induces beta-TrCP-mediated PER2 degradation altering circadian clock gene expression in intestinal mucosa of ApcMin/+ mice. J Biochem. 2009;145:289–297. doi: 10.1093/jb/mvn167. [DOI] [PubMed] [Google Scholar]
- 28.Filipski E, Innominato PF, Wu M, et al. Effects of light and food schedules on liver and tumor molecular clocks in mice. J Natl Cancer Inst. 2005;97:507–517. doi: 10.1093/jnci/dji083. [DOI] [PubMed] [Google Scholar]
- 29.Witschi H. A/J mouse as a model for lung tumorigenesis caused by tobacco smoke: strengths and weaknesses. Exp Lung Res. 2005;31:3–18. doi: 10.1080/01902140490494959. [DOI] [PubMed] [Google Scholar]
- 30.Witschi H, Espiritu I, Maronpot RR. Lung tumors in 2 year old strain A/J mice exposed for 6 months to tobacco smoke. Cancer Lett. 2006;241:64–68. doi: 10.1016/j.canlet.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Filipski E, King VM, Li X, et al. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002;94:690–697. doi: 10.1093/jnci/94.9.690. [DOI] [PubMed] [Google Scholar]
- 32.Wille JJ., Jr Circadian rhythm of tumor promotion in the two-stage model of mouse tumorigenesis. Cancer Lett. 2003;190:143–149. doi: 10.1016/s0304-3835(02)00594-3. [DOI] [PubMed] [Google Scholar]
- 33.Filipski E, Delaunay F, King VM, et al. Effects of chronic jet lag on tumor progression in mice. Cancer Res. 2004;64:7879–7885. doi: 10.1158/0008-5472.CAN-04-0674. [DOI] [PubMed] [Google Scholar]
- 34.Levin RD, Daehler MA, Grutsch JF, et al. Circadian function in patients with advanced non-small-cell lung cancer. Br J Cancer. 2005;93:1202–1208. doi: 10.1038/sj.bjc.6602859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood PA, Du-Quiton J, You S, Hrushesky WJ. Circadian clock coordinates cancer cell cycle progression, thymidylate synthase, and 5-fluorouracil therapeutic index. Mol Cancer Ther. 2006;5:2023–2033. doi: 10.1158/1535-7163.MCT-06-0177. [DOI] [PubMed] [Google Scholar]
- 36.Clegg DA, O’Hara BF, Heller HC, Kilduff TS. Nicotine administration differentially affects gene expression in the maternal and fetal circadian clock. Brain Res Dev Brain Res. 1995;84:46–54. doi: 10.1016/0165-3806(94)00152-p. [DOI] [PubMed] [Google Scholar]
- 37.Liu C, Gillette MU. Cholinergic regulation of the suprachiasmatic nucleus circadian rhythm via a muscarinic mechanism at night. J Neurosci. 1996;16(2):744–751. doi: 10.1523/JNEUROSCI.16-02-00744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benowitz NL, Hansson A, Jacob P., 3rd Cardiovascular effects of nasal and transdermal nicotine and cigarette smoking. Hypertension. 2002;39:1107–1112. doi: 10.1161/01.hyp.0000018825.76673.ea. [DOI] [PubMed] [Google Scholar]
- 39.De Flora S, Izzotti A, D’Agostini F, et al. Induction and modulation of lung tumors: genomic and transcriptional alterations in cigarette smoke-exposed mice. Exp Lung Res. 2005;31:19–35. doi: 10.1080/01902140490494986. [DOI] [PubMed] [Google Scholar]
- 40.Shimkin MB, Stoner GD. Lung tumors in mice: application to carcinogenesis bioassay. Adv Cancer Res. 1975;21:1–58. doi: 10.1016/s0065-230x(08)60970-7. [DOI] [PubMed] [Google Scholar]
- 41.Witschi H. The complexities of an apparently simple lung tumor model: the A/J mouse. Exp Toxicol Pathol. 2005;57(suppl 1):171–181. doi: 10.1016/j.etp.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Witschi H, Espiritu I, Ly M, Uyeminami D. The chemopreventive effects of orally administered dexamethasone in strain A/J mice following cessation of smoke exposure. Inhal Toxicol. 2005;17:119–122. doi: 10.1080/08958370590899712. [DOI] [PubMed] [Google Scholar]
- 43.Haussmann HJ. Smoking and lung cancer: future research directions. Int J Toxicol. 2007;26:353–364. doi: 10.1080/10915810701490463. [DOI] [PubMed] [Google Scholar]
- 44.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 45.Aung HH, Vasu VT, Valacchi G, et al. Effects of dietary carotenoids on mouse lung genomic profiles and their modulatory effects on short-term cigarette smoke exposures. Genes Nutr. 2009;4:23–39. doi: 10.1007/s12263-008-0108-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vasu VT, Oommen S, Lim Y, et al. Modulation of ozone-sensitive genes in alpha-tocopherol transfer protein null mice [published online ahead of print April 17, 2009] Inhal Toxicol. doi: 10.1080/08958370902838145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raghuram S, Stayrook KR, Huang P, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogers PM, Ying L, Burris TP. Relationship between circadian oscillations of Rev-erbalpha expression and intracellular levels of its ligand, heme. Biochem Biophys Res Commun. 2008;368:955–958. doi: 10.1016/j.bbrc.2008.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]