Abstract
Newly designed primers for [Fe-Fe]-hydrogenases indicated that (i) fermenters, acetogens, and undefined species in a fen harbor hitherto unknown hydrogenases and (ii) Clostridium- and Thermosinus-related primary fermenters, as well as secondary fermenters related to sulfate or iron reducers might be responsible for hydrogen production in the fen. Comparative analysis of [Fe-Fe]-hydrogenase and 16S rRNA gene-based phylogenies indicated the presence of homologous multiple hydrogenases per organism and inconsistencies between 16S rRNA gene- and [Fe-Fe]-hydrogenase-based phylogenies, necessitating appropriate qualification of [Fe-Fe]-hydrogenase gene data for diversity analyses.
Molecular hydrogen (H2) is important in intermediary ecosystem metabolism (i.e., processes that link input to output) in wetlands (7, 11, 12, 33) and other anoxic habitats like sewage sludges (34) and the intestinal tracts of animals (9, 37). H2-producing fermenters have been postulated to form trophic links to H2-consuming methanogens, acetogens (i.e., organisms capable of using the acetyl-coenzyme A [CoA] pathway for acetate synthesis) (7), Fe(III) reducers (17), and sulfate reducers in a well-studied moderately acidic fen in Germany (11, 12, 16, 18, 22, 33). 16S rRNA gene analysis revealed the presence of Clostridium spp. and Syntrophobacter spp., which represent possible primary and secondary fermenters, as well as H2 producers in this fen (11, 18, 33). However, H2-producing bacteria are polyphyletic (30, 31, 29). Thus, a structural marker gene is required to target this functional group by molecular methods. [Fe-Fe]-hydrogenases catalyze H2 production in fermenters (19, 25, 29, 30, 31), and genes encoding [Fe-Fe]-hydrogenases represent such a marker gene. The objectives of this study were to (i) develop primers specific for highly diverse [Fe-Fe]-hydrogenase genes, (ii) analyze [Fe-Fe]-hydrogenase genes in pure cultures of fermenters, acetogens, and a sulfate reducer, (iii) assess [Fe-Fe]-hydrogenase gene diversity in H2-producing fen soil enrichments, and (iv) evaluate the limitations of the amplified [Fe-Fe]-hydrogenase fragment as a phylogenetic marker.
Design of [Fe-Fe]-hydrogenase-specific primers.
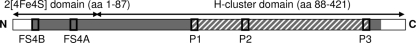
Degenerate PCR primers corresponding to the signatures FeFe_P1 (forward primer HydH1f) and FeFe_P3 (reverse primer HydH3r) were designed according to the consensus degenerate hybrid oligonucleotide primer (CODEHOP) strategy (Table 1 and Fig. 1) (23). Signatures FeFe_P1 and FeFe_P3 represent two of the three most conserved segments within the catalytic domains (H-cluster domains) of [Fe-Fe]-hydrogenases (30).
TABLE 1.
Sequences and properties of [Fe-Fe]-hydrogenase-specific primers
| Primer | Sequencea | Source or reference | Pos.b | Deg.c | No. of sequences with indicated no. of primer-template mismatchesd |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | ≥5 | |||||
| HydH1f | TTIACITSITGYWSYCCIGSHTGG | This study | 523 | 192 | 85 | 71 | 28 | 3 | 1 | 0 |
| HydH3r | CAICCIYMIGGRCAISNCAT | This study | 1126 | 64 | 138 | 16 | 26 | 8 | 0 | 0 |
| hydF1 | GCCGACCTKACMATMATGGA | 34 | 445 | 8 | 8 | 19 | 29 | 43 | 43 | 46 |
| hydR1 | ATRCARCCRCCSGGRCAGGCCAT | 34 | 1126 | 32 | 11 | 20 | 39 | 40 | 32 | 46 |
| FeFe-272F | GCHGAYMTBACHATWATGGARGA | 3 | 445 | 432 | 39 | 47 | 52 | 21 | 10 | 19 |
| FeFe-427R | GCNGCYTCCATDACDCCDCCNGT | 3 | 895 | 864 | 82 | 48 | 20 | 12 | 7 | 19 |
I, inosine. IUPAC nomenclature was used for mixed bases (5).
5′ end in the nucleic acid sequence of the D. vulgaris hydrogenase (GenBank accession no. AAS96246) gene.
Degeneracy, i.e., the number of combinations for the degenerate primers.
The total number of sequences was 188 (see Table S1 in the supplemental material for the number of mismatches per sequence). Inosine positions were not counted as mismatches (32).
FIG. 1.
Distribution of conserved amino acid (aa) motifs (black rectangles) in bacterial [Fe-Fe]-hydrogenases. FS4B and FS4A are [4Fe4S]-cluster binding sites (13). P1, P2, and P3 are highly conserved motifs coordinating the H-cluster (30). Amino acid positions are given in parentheses and are according to amino acid residues of the D. vulgaris hydrogenase (GenBank accession no. AAS96246). Abbreviations: N, amino terminal; C, carboxy terminal. The fragments used for the [Fe-Fe]-hydrogenase trees in Fig. 3 and Fig. S1 in the supplemental material and in Fig. 2 and Fig. S2 in the supplemental material are shaded gray and hatched, respectively.
The primer pair HydH1f/HydH3r is characterized by a clearly lower number of primer-template mismatches to publicly available [Fe-Fe]-hydrogenase genes than the [Fe-Fe]-hydrogenase-specific primer pair hydF1/hydR1 (34) (Table 1). The fragment length amplified with FeFe-272F/FeFe-427R, another [Fe-Fe]-hydrogenase-specific primer pair (3), was 66% of that amplified by HydH1f/HydH3r, indicating that the latter primers are better suited for phylogenetic analysis (Table 1). The degeneracy of HydH1f/HydH3r is five- to sevenfold lower than that of FeFe-272F/FeFe-427R (Table 1), suggesting higher amplification efficiencies (20). Eighty-one percent of the available 188 [Fe-Fe]-hydrogenase gene sequences had ≤2 mismatches to HydH1f and HydH3r (see Table S1 in the supplemental material), indicating that these primers target a large portion of the presently known [Fe-Fe]-hydrogenase diversity.
PCR conditions were optimized with DNA of Desulfovibrio vulgaris DSM 644 (see materials and methods in the supplemental material). HydH1f/HydH3r amplified approximately 620-bp fragments. The optimized PCR conditions were as follows: 2 μM each primer, 200 μM deoxynucleoside triphosphates (dNTPs), PCR buffer (1× concentrated), 3 mM magnesium acetate, TaqMaster PCR enhancer (1× concentrated), 0.12 μg bovine serum albumin ml−1, 0.02 U Taq DNA polymerase μl−1 (all from Eppendorf, Hamburg, Germany), and approximately 2 ng DNA μl−1. PCR consisted of an initial denaturation at 95°C for 10 min; 40 cycles consisting of sequential temperature regimens of 94°C for 45 s, 55°C for 45 s, and 72°C for 90 s; and a final extension at 72°C for 5 min. Amplified D. vulgaris [Fe-Fe]-hydrogenase gene fragments were 98% identical (translated amino acid sequence) to the published [Fe-Fe]-hydrogenase (GenBank accession no. AAS96246).
Phylogeny of pure-culture [Fe-Fe]-hydrogenase gene fragments.
DNA samples of the H2-producing aerotolerant fermenter Clostridium intestinale strain RC DSM 16614 (10) and the acetogens Clostridium magnum DSM 2767, Clostridium glycolicum strain RD-1 DSM 13865, and Thermoanaerobacter kivui DSM 2030, which are likewise all capable of H2 production during fermentation (2, 6, 15), were chosen to further evaluate the designed primers. With the choice of these species, including the sulfate reducer D. vulgaris, which is capable of secondary fermentation (4, 21, 28), most functional groups that are known to contain [Fe-Fe]-hydrogenases and that are potentially linked to H2 production or consumption in the moderately acidic fen (see above) were covered.
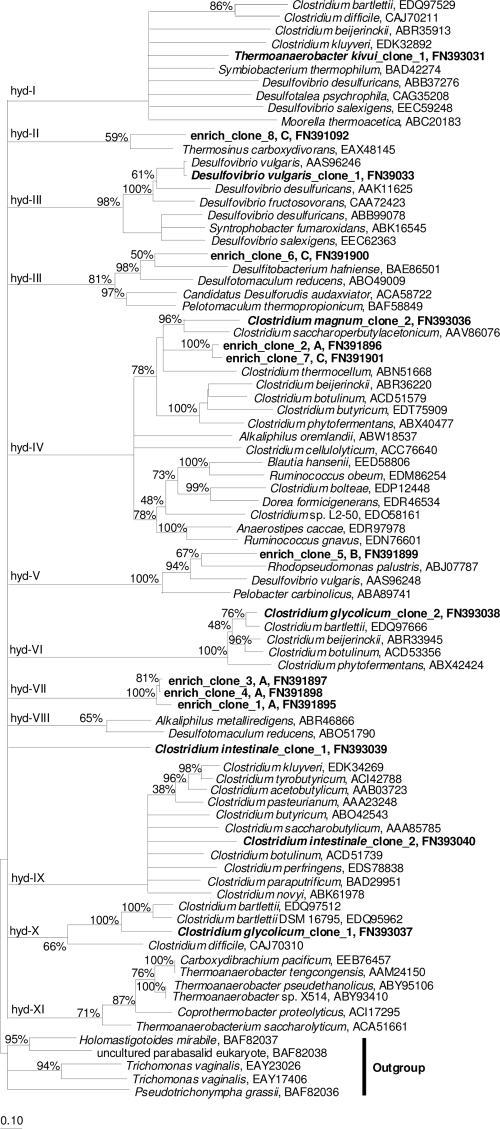
Both C. glycolicum and C. intestinale yielded two different [Fe-Fe]-hydrogenase gene fragments, indicating that these clostridia contain homologous hydrogenase genes (Fig. 2). Only one [Fe-Fe]-hydrogenase gene fragment was retrieved from C. magnum. Four of the clostridial [Fe-Fe]-hydrogenase sequences clustered with known clostridial [Fe-Fe]-hydrogenases (Fig. 2). However, [Fe-Fe]-hydrogenase 1 of C. intestinale was not closely affiliated to any cluster (Fig. 2). Amino acid identities of in silico-translated [Fe-Fe]-hydrogenase genes 1 and 2 of C. glycolicum to those of Clostridium bartlettii were 75% and 92%, respectively. Identities of [Fe-Fe]-hydrogenases 1 and 2 from C. intestinale to those of Clostridium botulinum were 56% and 76%, respectively. The [Fe-Fe]-hydrogenase from C. magnum was most similar to the [Fe-Fe]-hydrogenase of Clostridium saccharoperbutylacetonicum (71% amino acid identity). The [Fe-Fe]-hydrogenase of T. kivui was most similar to the [Fe-Fe]-hydrogenase of Symbiobacterium thermophilum (73% amino acid identity) and did not cluster with those of other Thermoanaerobacter species, indicating that species within a 16S rRNA gene-defined monophyletic genus can have distantly related [Fe-Fe]-hydrogenase genes (see the discussion below). These data indicate that fermenters and acetogens host phylogenetically closely related as well as distinct and hitherto undetected [Fe-Fe]-hydrogenase gene sequences.
FIG. 2.
Phylogenetic tree of amplified [Fe-Fe]-hydrogenase genes (boldface) and closely related sequences (residues 183 to 375 of the D. vulgaris hydrogenase [see Fig. 1]). The origin of enrichment-derived sequences (enrich_clone) is indicated by the following capital letters: A, 10−4 dilution of fen soil; B, 10−7 dilution of preincubated, nonsupplemented fen soil slurry; C, 10−2 dilution of preincubated, TSB (0.3 g liter−1)-supplemented fen soil slurry (for details see materials and methods in the supplemental data). The consensus tree was drawn based on neighbor-joining, maximum-parsimony, and maximum-likelihood trees. Incongruencies of tree topologies between the three methods were indicated by multifurcations. Branch lengths and bootstrap values (500 resamplings) were based on the maximum-parsimony analysis. Bootstrap values are displayed for nodes supported by all three analyses. The bar indicates 0.1 estimated change per amino acid. See Fig. S2 in the supplemental material for a tree that includes all presently available pure-culture [Fe-Fe]-hydrogenases.
Diversity of [Fe-Fe]-hydrogenases in H2-producing fen enrichments.
Soil of a moderately acidic methane-emitting fen (25) and preincubated fen soil slurries were serially diluted in anoxic tryptic soy broth (TSB) containing glucose (3 g liter−1, pH 5.5; for details see materials and methods in the supplemental material). Growth and glucose consumption occurred at 30°C in 10−1 to 10−7 dilutions. Up to 11.0 mM acetate, 3.2 mM formate, 2.9 mM propionate, 1.8 mM butyrate, and 1.3 mM H2 were found in enrichments, underscoring the importance of acid-tolerant fermenters for the intermediary ecosystem metabolism of this fen (8, 25).
Diverse [Fe-Fe]-hydrogenase gene fragments from five distantly related clusters were amplified from fen enrichments (Fig. 2). Most hydrogenase gene fragments originated from 10−4 to 10−7 dilutions of fen soil (Fig. 2) and were only distantly related to those previously amplified from a saline microbial mat or domestic sewage (data not shown) (3, 34). Hydrogenases were grouped into five distinct clusters and were affiliated with hydrogenases of species from the cellulolytic fermenter Clostridium thermocellum (68 to 69% amino acid sequence identity to enrich_clones 2 and 7 [36]), the sulfate and metal reducer Desulfotomaculum reducens (71% identity to enrich_clone 6 [27]), the primary fermenter and carboxydotroph Thermosinus carboxydivorans (70% identity to enrich_clone 8 [26]), and a sulfate reducer capable of syntrophic H2 production, D. vulgaris (21, 28) (64% identity to enrich_clone 5). Enrich_clones 1, 3, and 4 showed highest identities (70 to 71%) to a [Fe-Fe]-hydrogenase from the metal reducer Alkaliphilus metalliredigens (35).
Specificity of [Fe-Fe]-hydrogenase primer.
Ninety-eight percent of 74 sequences retrieved from pure cultures and 87% of 90 sequences from fen soil enrichments were related to [Fe-Fe]-hydrogenase genes available in GenBank (1). Such specificity is above or in the range of values reported for other well-established structural marker genes. For example, for dsrAB, which encodes the dissimilatory bisulfite reductase, only 60 to 70% of all clones from a moderately acidic fen contained dsrAB-related inserts (18). Thus, HydH1f/HydH3r facilitated efficient amplification of diverse [Fe-Fe]-hydrogenases.
Limitations of [Fe-Fe]-hydrogenase gene fragment-based analyses.
[Fe-Fe]-hydrogenase trees based on either the structural framework (residues 35 to 389 of the D. vulgaris hydrogenase) (Fig. 1) (13, 14, 19, 31) or the amplified fragment (residues 183 to 375) showed similar topologies (see Fig. S1 and S2 in the supplemental material). Thus, the amplified fragment contained sufficient information for reliable phylogenetic analysis of most presently known [Fe-Fe]-hydrogenase genes.
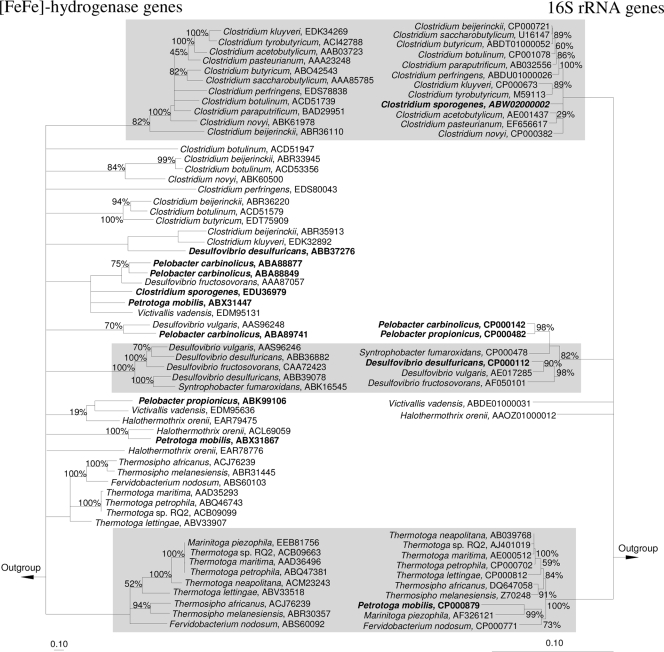
Topologies of phylogenetic trees based on either the structural framework of [Fe-Fe]-hydrogenase genes or the 16S rRNA genes were compared to evaluate whether [Fe-Fe]-hydrogenase phylogeny reflects the evolutionary history of its host or not (Fig. 3; see Fig. S1 in the supplemental material). The hydrogenase genes of bacteria from one 16S rRNA cluster formed similar clusters in the hydrogenase tree for Clostridia, Thermotoga, Gamma- and Deltaproteobacteria, Dehalococcoides, and Bacteroidetes (Fig. 3; see Fig. S1 in the supplemental material). For these organisms, hydrogenase clusters often had more multifurcations and lower bootstrap values than did 16S rRNA gene clusters, indicating that the hydrogenase fragment tree had a lower resolution than did the 16S rRNA gene tree. [Fe-Fe]-hydrogenases of species within monophyletic 16S rRNA-defined clusters also grouped with hydrogenases of distantly related species (e.g., Pelobacter propionicus hydrogenase clustered next to a hydrogenase of Victivallis vadensis) (Fig. 3). Several homologs of [Fe-Fe]-hydrogenases have been found in a number of bacterial genomes (19) (Fig. 3) and also in C. intestinale and C. glycolicum in this study (Fig. 2). Such homologs can originate from gene duplication and diversification or lateral gene transfer (19) and may be stabilized in the genome by performing different functions in the cell (24). The presence of homologs had negligible effects on phylogenetic inferences for [Fe-Fe]-hydrogenases ABB36882 and ABB39078 of Desulfovibrio desulfuricans (Fig. 3). In contrast, a third [Fe-Fe]-hydrogenase of D. desulfuricans (GenBank accession no. ABB37276) had a common root with hydrogenases from Clostridium beijerinckii and Clostridium kluyveri, resulting in phylogenetic inconsistencies. Thus, species level diversity of hydrogen producers might be overestimated, and identification of 16S rRNA gene-defined species based on hydrogenase gene fragment analysis is not always possible, necessitating appropriate qualification of [Fe-Fe]-hydrogenase gene data for diversity analyses.
FIG. 3.
Comparison of [Fe-Fe]-hydrogenase- and 16S rRNA gene-based phylogenetic trees (residues 35 to 389 of the D. vulgaris hydrogenase) (see Fig. 1). Both trees are consensus trees calculated and displayed as described for Fig. 2. Consistent monophyletic groups between both trees are shaded in gray. Examples of hydrogenases and hosts with inconsistent phylogenies are in boldface. Bars indicates 0.1 estimated change per amino acid or nucleotide. See Fig. S1 in the supplemental material for trees that include all presently available pure-culture [Fe-Fe]-hydrogenases and corresponding 16S rRNA genes.
Conclusions.
These collective data demonstrate that diverse H2 producers are abundant in the fen and that the newly designed primer pair for [Fe-Fe]-hydrogenases is an efficient tool for molecular detection of H2 producers.
Nucleotide sequence accession numbers.
Sequences obtained in this study were deposited in the EMBL nucleotide sequence database (http://www.ebi.ac.uk) under accession numbers FN391895 to FN391902 and FN393031 to FN393040.
Supplementary Material
Acknowledgments
Support for this study was provided by the Deutsche Forschungsgemeinschaft (DFG DR 310/3-3 and HO 4020/2-2) and the University of Bayreuth.
Footnotes
Published ahead of print on 29 January 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, B. A. Rapp, and D. L. Wheeler. 2000. GenBank. Nucleic Acids Res. 28:15-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bomar, M., H. Hippe, and B. Schink. 1991. Lithotrophic growth and hydrogen metabolism. FEMS Micriobiol. Lett. 83:347-350. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, E. S., J. R. Spear, and J. W. Peters. 2009. [Fe-Fe]-hydrogenase genetic diversity provides insight into molecular adaption in a saline microbial mat community. Appl. Environ. Microbiol. 75:4620-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carepo, M., J. F. Baptista, A. Pamplona, G. Fauque, J. J. G. Moura, and M. A. M. Reis. 2002. Hydrogen metabolism in Desulfovibrio desulfuricans strain New Jersey (NCIMB 8313)—comparative study with D. vulgaris and D. gigas species. Anaerobe 8:325-332. [DOI] [PubMed] [Google Scholar]
- 5.Cornish-Bowden, A. 1985. IUPAC-IUB symbols for nucleotide nomenclature. Nucleic Acids Res. 13:3021-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniel, S. L., T. Hsu, S. I. Dean, and H. L. Drake. 1990. Characterization of the H2- and CO-dependent chemolithotrophic potentials of the acetogens Clostridium thermoaceticum and Acetogenium kivui. J. Bacteriol. 172:4464-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drake, H. L., K. Küsel, and C. Matthies. 2006. Acetogenic prokaryotes, p. 354-420. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 2. Springer-Verlag, New York, NY. [Google Scholar]
- 8.Drake, H. L., M. A. Horn, and P. K. Wüst. 2009. Intermediary ecosystem metabolism as a main driver of methanogenesis in acidic wetland soil. Environ. Microbiol. Rep. 1:307-318. [DOI] [PubMed] [Google Scholar]
- 9.Flint, H. J. 1997. The rumen microbial ecosystem-some recent developments. Trends Microbiol. 5:483-488. [DOI] [PubMed] [Google Scholar]
- 10.Gößner, A. S., K. Küsel, D. Schulz, S. Tenz, G. Acker, C. R. Lovell, and H. L. Drake. 2006. Trophic interactions of the aerotolerant anaerobe Clostridium intestinale and the acetogen Sporomusa rhizae sp. nov. isolated from roots of the black needlerush Juncus roemerianus. Microbiology 152:1209-1219. [DOI] [PubMed] [Google Scholar]
- 11.Hamberger, A., M. A. Horn, M. G. Dumont, J. C. Murrell, and H. L. Drake. 2008. Anaerobic consumers of monosaccharides in a moderately acidic fen. Appl. Environ. Microbiol. 74:3112-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horn, M. A., C. Matthies, K. Küsel, A. Schramm, and H. L. Drake. 2003. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 69:74-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horner, D. S., P. G. Foster, and T. M. Embley. 2000. Iron hydrogenases and the evolution of anaerobic eukaryotes. Mol. Biol. Evol. 17:1695-1709. [DOI] [PubMed] [Google Scholar]
- 14.Horner, D. S., B. Heil, T. Happe, and T. M. Embley. 2002. Iron hydrogenases—ancient enzymes in modern eukaryotes. Trends Biochem. Sci. 27:148-153. [DOI] [PubMed] [Google Scholar]
- 15.Küsel, K., A. Karnholz, T. Trinkwalter, R. Devereux, G. Acker, and H. L. Drake. 2001. Physiological ecology of Clostridium glycolicum RD-1, an aerotolerant acetogen isolated from sea grass roots. Appl. Environ. Microbiol. 67:4734-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Küsel, K., M. Blöthe, D. Schulz, M. Reiche, and H. L. Drake. 2008. Microbial reduction of iron and porewater biogeochemistry in acidic peatlands. Biogeosciences 5:1537-1549. [Google Scholar]
- 17.Lovley, D. R., E. J. P. Philips, D. J. Lonergan, and P. K. Widman. 1995. Fe(III) and S0 reduction by Pelobacter carbinolicus. Appl. Environ. Microbiol. 61:2132-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loy, A., K. Küsel, A. Lehner, H. L. Drake, and M. Wagner. 2004. Microarray and functional gene analyses of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal co-occurrence of recognized genera and novel lineages. Appl. Environ. Microbiol. 70:6998-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer, J. 2007. [FeFe] hydrogenases and their evolution: a genomic perspective. Cell. Mol. Life Sci. 64:1063-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan, Z., R. Barry, A. Lipkin, and M. Soloviev. 2007. Selection strategy and the design of hybrid oligonucleotide primers for RACE-PCR: cloning a family of toxin-like sequences from Agelena orientalis. BMC Mol. Biol. 8:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereira, P. M., Q. He, F. M. A. Valente, A. V. Xavier, J. Zhou, I. A. C. Pereira, and R. O. Louro. 2008. Energy metabolism in Desulfovibrio vulgaris Hildenborough: insights from transcriptome analysis. Antonie Van Leeuwenhoek 93:347-362. [DOI] [PubMed] [Google Scholar]
- 22.Reiche, M., G. Torburg, and K. Küsel. 2008. Competition of Fe(III) reduction and methanogenesis in an acidic fen. FEMS Microbiol. Ecol. 65:88-101. [DOI] [PubMed] [Google Scholar]
- 23.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly-related sequences. Nucleic Acids Res. 26:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schut, G. I., and M. W. W. Adams. 2009. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J. Bacteriol. 191:4451-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz, E., and B. Friedrich. 2006. The H2-metabolizing prokaryotes, p. 496-563. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 3rd ed. Springer-Verlag, New York, NY.
- 26.Sokolova, T. G., J. M. Gonzalez, N. A. Kostrikina, N. A. Chernyh, T. V. Slepova, E. A. Bonch-Osmolovskaya, and F. T. Robb. 2004. Thermosinus carboxydivorans gen. nov., sp. nov., a new anaerobic, thermophilic, carbon-monoxide-oxidizing, hydrogenotrophic bacterium from a hot pool of Yellowstone National Park. Int. J. Syst. Evol. Microbiol. 54:2353-2359. [DOI] [PubMed] [Google Scholar]
- 27.Tebo, B. M., and A. Y. Obraztsova. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 162:193-198. [Google Scholar]
- 28.Traore, A. S., M.-L. Fardeau, C. E. Hatchikian, J. Le Gall, and J.-P. Belaich. 1983. Energetics of growth of a defined mixed culture of Desulfovibrio vulgaris and Methanosarcina barkeri: interspecies hydrogen transfer in batch and continuous cultures. Appl. Environ. Microbiol. 46:1152-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vignais, P. M. 2008. Hydrogenases and H+-reduction in primary energy conservation. Results Probl. Cell Differ. 45:223-252. [DOI] [PubMed] [Google Scholar]
- 30.Vignais, P. M., and B. Billoud. 2007. Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107:4206-4272. [DOI] [PubMed] [Google Scholar]
- 31.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 32.Watkins, N. E., and J. SantaLucia. 2005. Nearest-neighbour thermodynamics of deoxyinosine pairs in DNA duplexes. Nucleic Acids Res. 33:6258-6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wüst, P. K., M. A. Horn, and H. L. Drake. 2009. Trophic links between fermenters and methanogens in a moderately acidic fen soil. Environ. Microbiol. 11:1395-1409. [DOI] [PubMed] [Google Scholar]
- 34.Xing, D., N. Ren, and B. E. Rittmann. 2008. Genetic diversity of hydrogen-producing bacteria in an acidophilic ethanol-H2-coproducing system, analyzed using the [Fe]-hydrogenase gene. Appl. Environ. Microbiol. 74:1232-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye, Q., Y. Roh, S. L. Carroll, B. Blair, J. Zhou, C. L. Zhang, and M. W. Fields. 2004. Alkaline anaerobic respiration: isolation and characterization of a novel alkaliphilic and metal-reducing bacterium. Appl. Environ. Microbiol. 70:5595-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, Y.-H. P., and L. R. Lynd. 2005. Cellulose utilization by Clostridium thermocellum: bioenergetics and hydrolysis product assimilation. Proc. Natl. Acad. Sci. U. S. A. 102:7321-7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmermann, P. R., J. P. Greenberg, S. O. Wandiga, and P. J. Crutzen. 1982. Termites: a potentially large source of atmospheric methane, carbon dioxide, and molecular hydrogen. Science 218:563-565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.