Abstract
Human enterovirus 71 (EV-71) is one of the major etiologic causes of hand, foot, and mouth disease (HFMD) among young children worldwide, with fatal instances of neurological complications becoming increasingly common. Global VP1 capsid sequences (n = 628) sampled over 4 decades were collected and subjected to comprehensive evolutionary analysis using a suite of phylogenetic and population genetic methods. We estimated that the common ancestor of human EV-71 likely emerged around 1941 (95% confidence interval [CI], 1929 to 1952), subsequently diverging into three genogroups: B, C, and the now extinct genogroup A. Genealogical analysis revealed that diverse lineages of genogroup B and C (subgenogroups B1 to B5 and C1 to C5) have each circulated cryptically in the human population for up to 5 years before causing large HFMD outbreaks, indicating the quiescent persistence of EV-71 in human populations. Estimated phylogenies showed a complex pattern of spatial structure within well-sampled subgenogroups, suggesting endemicity with occasional lineage migration among locations, such that past HFMD epidemics are unlikely to be linked to continuous transmission of a single strain of virus. In addition, rises in genetic diversity are correlated with the onset of epidemics, driven in part by the emergence of novel EV-71 subgenogroups. Using subgenogroup C1 as a model, we observe temporal strain replacement through time, and we investigate the evidence for positive selection at VP1 immunogenic sites. We discuss the consequences of the evolutionary dynamics of EV-71 for vaccine design and compare its phylodynamic behavior with that of influenza virus.
Enterovirus 71 (EV-71) is a member of the genus Enterovirus in the family Picornaviridae. Classified as human enterovirus species A (HEV-A) along with some group A coxsackieviruses (CV-A), EV-71 is a small, nonenveloped, positive-stranded RNA virus with a genome approximately 7,400 bases long and is genetically most related to CV-A16. EV-71 is divided into three major genogroups (denoted A, B, and C), and various subgenogroups within genogroups B and C.
Since its first isolation in the United States in 1969 (71), EV-71 has been identified worldwide as a common cause of hand, foot, and mouth disease (HFMD) in young children and infants. Large EV-71-associated HFMD outbreaks have been reported in the United States, Europe, Australia, and Asia and constitute a significant and emerging threat to global public health (9, 50, 62, 63). Although EV-71 infection manifests most frequently as a mild, self-limited febrile illness characterized by papulovesicular lesions on the hands, feet, oropharyngeal mucosa, and buttocks, a small proportion of acute infections are associated with fatal neurological symptoms, including brain stem encephalitis, aseptic meningitis, and poliomyelitis-like paralysis (4, 28, 47). Such cases of neurological disease with a high case fatality rate were first reported in Bulgaria in 1975 (21) and Hungary in 1978 (52). However, large HFMD epidemics with high mortality rates resurfaced 2 decades later, in Malaysia in 1997 (2, 13, 16, 43) and Taiwan in 1998 (33, 42). Following these outbreaks, the Asia-Pacific region has experienced more frequent large-scale EV-71-associated HFMD epidemics—most with a high incidence of neurotropic infections and significant case fatality rates—and the virus has attracted global attention (3, 5, 14, 15, 18, 37, 46, 48, 55, 57, 74, 81, 82). Intriguingly, almost all outbreaks reported in the Asia-Pacific region during the last decade were caused by previously undefined EV-71 subgenogroups, raising questions about their origin, genetic complexity, and epidemiological behavior.
The icosahedral particles of EV-71, which are structurally similar to those of other members of the Picornaviridae, consist of structural proteins (capsid proteins VP1 to VP4) assembled as pentameric subunits (66). The VP1 protein is highly exposed and usually targeted by host neutralizing antibodies, predisposing the VP1 gene to constant immune selective pressure. This selection may drive the adaptive evolution of the capsid region of many enteroviruses, possibly resulting in amino acid fixations in virus populations (19, 45, 79). Because the VP1 gene of enteroviruses is thought to play an important role in viral pathogenesis and virulence (10, 12, 30), understanding the tempo and mode of evolution of the capsid protein can provide new insights into the epidemiological dynamics of EV-71 that may be useful in predicting the genetic basis and periodicity of future EV-71 epidemics and in facilitating the development of an effective EV-71 vaccine candidate.
In this study, we investigated the evolutionary dynamics and genetic history of EV-71. We estimate the dates of emergence of various subgenogroups identified in recent HFMD outbreaks. Using recently developed Bayesian methods of evolutionary analysis, we estimate the divergence time of EV-71 from its closely related ancestor CV-A16, thereby providing a date of origin for EV-71. We also reconstruct the global population dynamics of EV-71 over the past 40 years, revealing temporal trends in genetic diversity within and between major epidemics. Finally, despite finding little evidence of positive selection in the VP1 capsid protein, we observed a pattern of continuous strain and lineage replacement through time, with strong selective pressure detected at several potentially immunogenic sites. The impact of EV-71 evolution on the development of an EV-71 vaccine is also discussed.
MATERIALS AND METHODS
Enterovirus 71 sequence collection and phylogenetic analyses.
A total of 628 complete VP1 gene sequences with known collection dates between 1970 and 2008 were retrieved from GenBank (www.ncbi.nlm.nih.gov), corresponding to virtually all reported EV-71 VP1 genes (as of mid-2008). The nucleotide sequences were isolated mainly from large HFMD outbreaks that occurred in the last 4 decades in Asia, Europe, and America. The data set includes recently described full-length EV-71 genomes sampled from Malaysia (85) and data derived from the first EV-71 surveillance study in the United Kingdom (8). EV-71 strains known to be associated with severe or fatal neurological complications in Europe, Malaysia, Taiwan, Vietnam, and China were also included. Genogroup A was represented by the BrCr-CA-70 isolate, an early prototype strain collected in 1970. Phylogenetic estimates of EV-71 genogroups B and C and their respective subgenogroups were consistent among different phylogenetic approaches, specifically, maximum-likelihood (ML), neighbor-joining (using PAUP*, version 4.0 beta [78]) and the Bayesian Markov chain Monte Carlo (MCMC) method (using BEAST [24], described below). All accession numbers for the VP1 sequences are provided in Table S1 in the supplemental material. To test for the presence of recombination in the VP1 gene, sequences were screened using the Recombination Detection Program (RDP) version 3.27, under both the default and triplet settings. RDP implements a combination of methods including RDP, CHIMAERA, GENECONV, MAXIMUM χ2, and 3Seq for recombination detection (we also used BOOTSCAN and SISTER SCANNING for secondary scanning) (44). Potential recombinant sequences were suggested when more than three methods showed significant support for recombination with a Bonferroni-corrected P value cutoff of 0.05.
Bayesian MCMC evolutionary analyses.
Rates of evolution, molecular clock phylogenies, divergence times, demographic histories, and other evolutionary parameters were jointly estimated from heterochronous VP1 gene sequences of genogroups B and C (with sampling date ranges of 32 and 26 years, respectively) using the Bayesian MCMC method implemented in BEAST, version 1.4.8 (24). To reduce excessive computational load, closely related sequences collected at the same location and time point (i.e., from a single outbreak/source) were manually removed without compromising the genetic or geographical heterogeneity of each alignment, resulting in a down-sampling of the genogroup B and C data sets to 229 and 251 sequences, respectively. Bayesian MCMC analyses (36) were performed using a relaxed molecular clock model (the uncorrelated lognormal-distributed model [UCLD]) (22). Analyses were independently performed using the Hasegawa-Kishino-Yano (HKY) (32) and general time-reversible (GTR) (68) nucleotide substitution models, with a gamma-distributed among-site rate variation with four rate categories (γ4) (84). Bayesian MCMC analyses were repeated using the constant size and exponential growth models in order to investigate the degree to which dating estimates are affected by the demographic model chosen (23). Each Bayesian MCMC analysis was run for 20 million states and sampled every 10,000 states. Posterior probabilities were calculated with a burn-in of 2 million states and checked for convergence using Tracer, version 1.4 (64). The posterior distribution of the substitution rate obtained from the heterochronous sequences was subsequently incorporated as a prior distribution for the evolutionary rate of EV-71 genogroups B and C, thereby adding a timescale to the phylogenetic histories of these strains and enabling the times of their most recent common ancestors (tMRCA) to be calculated (60). Bayesian skyline plots (25), which depict the relative viral genetic diversity (g) through time, were estimated for genogroups B and C. Finally, to investigate the phylogenetic relationships of EV-71 and the closely related CV-A16, the divergence time of EV-71 from reference CV-A16 strains (56) was estimated using a coalescent approach similar to that described above.
Natural selection and adaptation.
Selection pressure on the EV-71 VP1 gene was investigated by estimating the ratio of nonsynonymous substitutions to synonymous substitutions (dN/dS) using the codon-based phylogenetic method implemented in CODEML (distributed in the PAML, version 4, package). To investigate the difference in dN/dS on the trunk and terminal branches (see Results section for definitions), we employed a branch-specific codon model that defines two independent dN/dS ratios, one for the trunk and one for the terminal branches. This lineage-specific codon model was estimated using maximum likelihood and was compared, using a likelihood ratio test (LRT) with one degree of freedom, to a null model that assumes a uniform dN/dS ratio across all branches.
To chronologically trace the evolution of nonsynonymous changes throughout the evolutionary history of EV-71 subgenogroup C1 (i.e., 1986 to 2006), we reconstructed nonsynonymous changes from the heterochronous virus sequences using a joint maximum-likelihood method (59), as implemented in HyPhy, version 0.99 (58). Since the phylogeny used in this approach was the maximum clade credibility (MCC) tree from the Bayesian MCMC molecular clock analysis, we were able to estimate the time when each VP1 nonsynonymous substitution occurred.
RESULTS
Phylogenetic relationships of EV-71 subgenogroups.
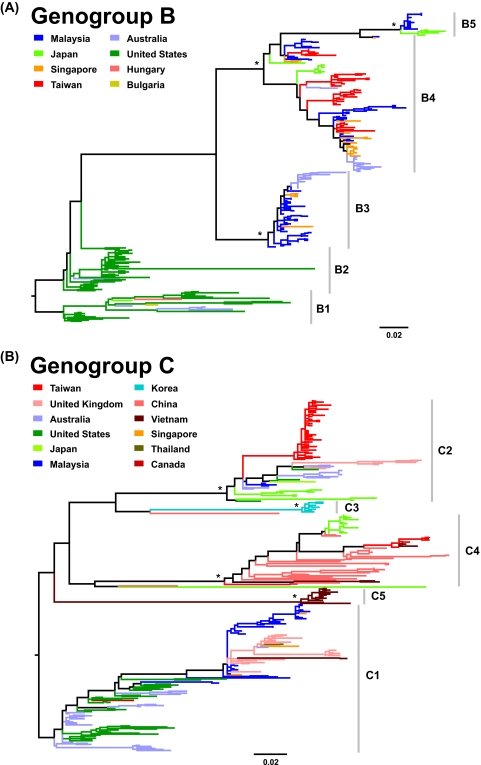
Maximum-likelihood phylogenetic reconstructions of 628 VP1 gene sequences, sampled between 1970 and 2008, classified the isolates into three major genogroups, denoted A, B, and C. Genogroups B and C were each composed of distinct subgenogroups—B1 to B5 and C1 to C5—consistent with previously assigned nomenclatures (Fig. 1) (1, 11, 14, 33, 38, 46, 48, 81). Genogroup A (71), which no longer plays a role in contemporary epidemics, was represented by a single prototype isolate (BrCr-CA-70) located outside genogroups B and C (data not shown). Phylogeographic analysis revealed that subgenogroups B1 to B5, C1, C2, and C4 exhibit a global, cosmopolitan distribution, whereas the C3 and C5 lineages are restricted to Korea and Vietnam, respectively (Fig. 1). Cocirculation of multiple subgenogroups has been observed in some countries, for example, Malaysia, the United States, Taiwan, and Japan. Early subgenogroups (B1 and B2) that caused major HFMD epidemics in the 1970s and 1980s (4, 21, 52, 69) appear to have been replaced (11) by new subgenogroups (B3, B4, and B5) that have since become predominant and circulate endemically in the Asia-Pacific region, generating major EV-71-associated HFMD outbreaks (Fig. 1A), including those with fatal neurological cases in Malaysia and Taiwan (1, 48, 82). In contrast, subgenogroup C1, which was first described in Australia and the United States in the mid-1980s (Fig. 1B) (4, 70), has continuously been identified in various countries since its initial detection. Later in the 1990s, the diversity of genogroup C expanded significantly due to the emergence of a number of novel subgenogroups in the Asia-Pacific region, including subgenogroups C2, C4, and C5 that have been associated with fatal HFMD cases in Taiwan (1998) (33), China (2008) (5, 18), and Vietnam (2005) (81), respectively. Expanded phylogenies showing the isolate name, country of origin, and sampling time for each sequence are provided in Fig. S1 and S2 in the supplemental material. Additionally, rigorous recombination analysis showed a lack of recombination in the capsid-encoding VP1 region, consistent with previous analyses on human enterovirus species (73).
FIG. 1.
Phylogenies of human enterovirus 71 (EV-71) VP1 genes. The maximum-likelihood phylogenetic trees of EV-71 VP1 genes from genogroup B and C are shown. Complete VP1 genes of genogroup B (n = 283) and C (n = 344) with known sampling dates (1972 to 2008) were used. Each genogroup is classified into five subgenogroups, denoted B1 to B5 and C1 to C5, respectively. The trees are midpoint rooted, and significant bootstrap support values (≥80%; 1,000 bootstrap replicates) are indicated by asterisks at major nodes. Scale bars signify a genetic distance of 0.02 nucleotide substitutions per site. For clarity, the year of isolation of each sequence is not shown in this figure but is listed in Fig. S1 and S2 in the supplemental material. Several C3- and C4-like isolates are also shown at the base of the C3 and C4 clusters, respectively.
Of note, EV-71 isolates belonging to the same subgenogroup but sampled from different locations showed a complex pattern of relatedness (except for subgenogroups C3 and C5, which were sampled from only a single country). Within a subgenogroup, clusters of sequences sampled from the same location were observed, but these clusters were interspersed with those from other locations. This suggests an intermediate level of geographic gene flow, such that a particular subgenogroup may endemically persist in diverse parts of the world, with occasional movement of viral lineages among locations. This also suggests that contemporary HFMD outbreaks in different countries caused by the same EV-71 subgenogroup may be the result of different lineages and are less likely to have been caused by the sequential transmission of a single lineage between countries (72). In addition, a ladder-like tree structure was observed within subgenogroups that were sufficiently sampled in both time and space (B1 to B4, C1, C2, and C4) (Fig. 1). This characterizes trees in which viral sequences isolated at early time points are located toward the tree root, while sequences collected later are found further from the root, together with limited genetic diversity observed at any one time. Such phylogenetic patterns may suggest a process of temporal strain replacement occurring on a global scale.
Evolutionary rate, time of subgenogroup emergence, and origin of EV-71.
Since the 1990s, seven novel subgenogroups have been discovered in the Asia-Pacific region. To understand the evolutionary behavior of EV-71, we estimated the dates of origin of each subgenogroup using a Bayesian relaxed molecular clock method, a newly developed approach that assumes no a priori correlation between a lineage's rate of evolution and that of its ancestor (22). The evolutionary rate of the VP1 gene was estimated to be 4.5 × 10−3 to 4.6 × 10−3 and 4.2 × 10−3 substitutions/site/year for genogroups B and C, respectively (Table 1). The estimated substitution rates for genogroups B and C are similar and higher than those estimated using the less statistically appropriate linear regression method previously used (11). Our analyses also indicated substantial heterogeneity in evolutionary rates among viral lineages, with an estimated coefficient of variation of 0.44 (95% credible region [CR], 0.28 to 0.57) and 0.39 (95% CR, 0.25 to 0.53) for genogroups B and C, respectively. Using the estimated molecular clock, the common ancestor of subgenogroup B1 (first identified in 1972 in the United States) was dated to around 1967 (Table 1), that is, before the recognition of EV-71 as a human pathogen. Other novel subgenogroups have since emerged, i.e., in the late 1970s (B2), early to mid-1990s (B3 and B4), and early 2000s (B5). When our estimates of origin are compared with the dates of first detection of each subgenogroup, we find that the divergent B subgenogroups typically circulated for approximately 2 to 5 years before causing large HFMD outbreaks (Table 1).
TABLE 1.
Evolutionary characteristics of EV-71 genogroups B and C and their respective subgenogroups based on the VP1 gene
| EV-71 genogroup and subgenogroup | Year first isolated (country) | Divergence according to the indicated evolutionary modela |
|||
|---|---|---|---|---|---|
| Exponential growth (GTR+γ4) |
Constant size (GTR+γ4) |
||||
| Substitution rate (CR)b | Time of emergence (yr [CR]) | Substitution rate (CR)b | Time of emergence (yr [CR]) | ||
| Genogroup B | 4.5 (4.3-4.7) | 4.6 (4.4-4.8) | |||
| B1 | 1972 (United States) | 1966.8 (1964.7-1968.8) | 1967.0 (1964.9-1968.9) | ||
| B2 | 1981 (United States) | 1978.8 (1977.9-1979.8) | 1978.9 (1977.9-1979.8) | ||
| B3 | 1997 (Malaysia) | 1994.8 (1993.9-1995.7) | 1994.7 (1993.6-1995.5) | ||
| B4 | 1997 (Malaysia) | 1993.2 (1991.8-1994.6) | 1993.2 (1991.7-1994.6) | ||
| B5 | 2003 (Japan) | 2001.2 (2000.6-2001.8) | 2001.2 (2000.6-2001.8) | ||
| Genogroup C | 4.2 (4.0-4.4) | 4.2 (4.0-4.4) | |||
| C1 | 1986 (Australia) | 1983.3 (1982.3-1984.2) | 1983.3 (1982.2-1984.2) | ||
| C2 | 1995 (Australia) | 1992.9 (1991.5-1994.0) | 1992.7 (1991.3-1993.9) | ||
| C3 | 2000 (Korea) | 1998.8 (1998.2-1999.4) | 1998.8 (1998.1-1999.2) | ||
| C4 | 1998 (Taiwan) | 1992.8 (1991.0-1994.5) | 1992.8 (1991.0-1994.6) | ||
| C5 | 2005 (Vietnam) | 2002.8 (2001.9-2003.6) | 2002.7 (2001.8-2003.6) | ||
Based on relaxed (uncorrelated lognormal) coalescent inference. The 95% highest posterior density CRs are given in parentheses. See Materials and Methods for further details of the estimation procedure.
Substitution rates are expressed as 10−3 substitutions per site per year.
Similar results were obtained for genogroup C lineages; the various subgenogroups within genogroup C were identified only 1 to 5 years after their dates of common ancestry (Table 1). The evolutionary models employed in the coalescent analyses had no significant effect on estimated dates (Table 1; see also Table S2 in the supplemental material, where divergence times obtained under two further models are provided). The cryptic circulation of each EV-71 subgenogroup for years prior to its detection suggests that (i) the virus was circulating at low prevalence, such that the number of HFMD cases was small and occasional or no deaths were reported, and/or (ii) infection symptoms were not severe during the course of infection (17), thus attracting little or no attention.
Using the VP1 sequences from all genogroups, the time of origin of EV-71—a probable descendant of the closely related CV-A16—was estimated to be 1941.0 (95% CR, 1928.8 to 1952.2) (Fig. 2). Our analyses suggest that EV-71 is a relatively recent human pathogen that emerged in the mid-20th century (51). Marginal-likelihood analysis (77) showed that a demographic model of exponential growth fits the VP1 data set better than a model of constant population size (log10 Bayes factor, 7.6).
FIG. 2.
Origin of human enterovirus 71 (EV-71). (A) Phylogenetic relationships of EV-71 genogroups with coxsackievirus A16. Coxsackievirus A16 sequences shown in the cladogram were previously reported by Perera et al. (56). (B) The date of the MRCA of EV-71 was estimated to be 1941.0 (95% CR, 1928.8 to 1952.2) using a relaxed molecular clock and an exponential population growth model (22), as implemented in BEAST (24).
Dynamics of population growth.
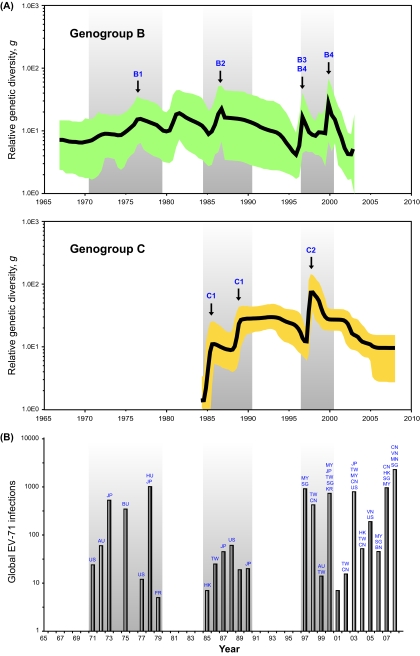
Bayesian skyline plot analyses (25) were performed to reconstruct the past population history of EV-71 by measuring the dynamics of VP1 genetic diversity over time (Fig. 3). Sharp but transient increases in relative genetic diversity (g) were observed for genogroup B in the late 1990s. These correspond to the period when large-scale HFMD outbreaks were reported in Malaysia, Taiwan, and other countries in the Asia-Pacific region, during which subgenogroups B3 and B4 were originally described (1, 82). Prior to this, irregular cycles in genetic diversity can be observed between early 1970s and mid-1980s, perhaps reflecting the swift but sporadic occurrence of EV-71 outbreaks in various parts of the world, which were mainly caused by subgenogroups B1 or B2 although these earlier trends are less statistically significant. Crucially, the dynamics of the skyline plot are largely in agreement with the global surveillance data of laboratory-confirmed EV-71 cases (Fig. 3B) longitudinally recorded over the past 4 decades (9), suggesting that elevation in genetic diversity is correlated with EV-71 epidemics, as has been previously observed for the human influenza A virus (65). Likewise, skyline plot estimation based on the VP1 gene of genogroup C revealed a significant rise in genetic diversity around 1985, coinciding with the emergence of C1 (Fig. 3). Interepidemic viral diversity of genogroup C was maintained until major HFMD outbreaks (driven primarily by subgenogroup C2) occurred in Taiwan in 1998 (33).
FIG. 3.
The genetic diversity dynamics of EV-71 genogroups B and C. (A) Bayesian skyline plot estimates depicting the past genetic diversity dynamics through time of EV-71 genogroups B and C. The plot for genogroup B shows a sharp rise in relative genetic diversity in the late 1990s. Other increases can be seen in the early 1970s and mid-1980s. The putative EV-71 subgenogroups that are proposed to be implicated in escalating genetic diversity are indicated by arrows. Similarly, the plot for genogroup C shows sharp rises in genetic diversity in the mid-to-late 1980s and late 1990s. (B) The global reporting of EV-71 infections from the 1970s to 2008, compiled from the published literature (see Table S4 in the supplemental material), suggests that at least three independent waves of major outbreaks—one in each decade—have occurred worldwide since 1970. These episodes of outbreaks are indicated by gray shading in all plots. Country names are abbreviated as follows: US, United States; AU, Australia; JP, Japan; BU, Bulgaria; FR, France; HK, Hong Kong; TW, Taiwan; MY, Malaysia; SG, Singapore; CN, China; KR, Republic of Korea; VN, Viet Nam; HU, Hungary; MN, Mongolia; BN, Brunei.
Natural selection and adaptation in VP1.
To investigate the extent of selective pressure on the VP1 gene of EV-71, we estimated the ratio of nonsynonymous to synonymous substitution using a codon-based method. Maximum-likelihood analyses showed that the evolution of EV-71 is driven by strong purifying selection, with estimates of mean dN/dS values ranging from 0.029 to 0.075 for all subgenogroups (except B3, for which the mean dN/dS is significantly higher) (Table 2). This strong signal of negative selection is perhaps surprising, given that estimates of EV-71 VP1 substitution rates are similar to those observed for the surface glycoproteins of the influenza virus (26) and HIV-1 (67), which show evidence of repeated positive selection. Nonetheless, some individual sites under positive selection were detected in well-sampled subgenogroups, including B2, B3, B4, C1, and C2, and are located at positions 145 (P < 0.05), 98 (P < 0.05), 237, and 241 (P < 0.05) (Table 2).
TABLE 2.
Selection analysis of the VP1 gene for various EV-71 subgenogroups
| Subgenogroup | Mean dN/dS (CI)a | Positive selection site datab |
||
|---|---|---|---|---|
| Position | Normalized dN − dS | P value | ||
| B1 | 0.036 (0.025-0.049) | NA | ||
| B2 | 0.063 (0.046-0.084) | 145 | 19.849 | 0.0078 |
| B3 | 0.172 (0.128-0.226) | 145 | 38.285 | 0.0112 |
| 237 | 23.138 | 0.0899 | ||
| B4 | 0.075 (0.059-0.094) | 98 | 3.616 | 0.0706 |
| 145 | 16.695 | <0.0001 | ||
| 241 | 4.260 | 0.0146 | ||
| B5 | 0.050 (0.023-0.093) | NA | ||
| C1 | 0.039 (0.031-0.048) | 98 | 2.980 | 0.0282 |
| 145 | 3.264 | 0.0248 | ||
| C2 | 0.054 (0.039-0.072) | 241 | 3.247 | 0.0979 |
| C3 | 0.074 (0.023-0.171) | NA | ||
| C4 | 0.029 (0.021-0.040) | NA | ||
| C5 | 0.047 (0.020-0.091) | NA | ||
CI, 95% confidence interval.
NA, not applicable.
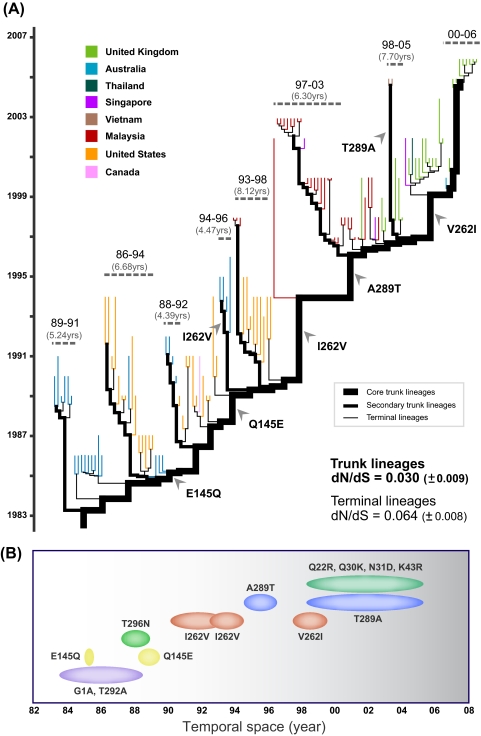
As mentioned above, maximum-likelihood phylogenies of EV-71 subgenogroups are characterized by a ladder-like structure resulting from the continual turnover of viral lineages through time (Fig. 1). Branches in such phylogenies can be distinguished as either trunk or terminal branches. Mutations that occur on trunk branches may contribute to the persistence and survival of viral lineages through time, whereas those on the terminal branches are transient and not observed at later time points. We therefore studied whether the pattern of adaptive evolution for subgenogroup C1 differed between these sets of branches. We define “core trunk” branches as those connecting the tree root to the MRCA of the most recently sampled isolates, and we defined terminal branches as those at the tree tips (Fig. 4A). In addition, we defined a set of secondary trunk lineages as those that immediately branch off the core trunk lineage and that persist for an intermediate period of time (∼4 to 9 years).
FIG. 4.
Evolutionary dynamics of EV-71 subgenogroup C1. (A) Molecular clock phylogeny of EV-71 subgenogroup C1, longitudinally sampled between 1986 and 2006. The phylogeny branch lengths are in units of time. The trunk lineages (which include both core and secondary trunk lineages) are drawn thicker than the terminal lineages; all tip branches are colored according to their countries of isolation. Clusters of secondary trunk and terminal lineages are annotated by dashed horizontal lines, and the ranges of sampling time for these isolates are shown, below which the duration in years of the secondary trunk lineage is shown in parentheses. The subgenogroup C1 phylogeny shows temporal strain replacement (a ladder-like topology; also seen in the ML phylogeny). (B) An illustration of the temporal amino acid evolution of the VP1 gene for subgenogroup C1. Observed amino acid substitutions are plotted against their estimated times of occurrence, as obtained from the molecular clock phylogeny in panel A using the joint ML method implemented in HyPhy.
Lineage-specific codon-based analyses of subgenogroup C1 VP1 genes revealed a significantly lower dN/dS ratio for the trunk lineages (0.030 ± 0.009) than for the terminal lineages (0.064 ± 0.008) (P = 0.055). This is best explained by a combination of strong purifying selection on all lineages, as proposed above, combined with a greater occurrence of transient deleterious mutations on terminal branches than on trunk branches. This is in agreement with the conclusions of a comparative study that found the same pattern in most RNA viruses (61). Interestingly, we found instances of repeated nonsynonymous substitution in the same codon on different terminal lineages (see Table S3 in the supplemental material). For instance, the VP1 position 98 underwent frequent amino acid replacements (e.g., E98K/G, n = 13; K98N, n = 1) on terminal lineages, but no changes at this site were found on the trunk lineages. These amino acid changes were unidirectional, from glutamic acid to lysine (or sometimes to glycine), which is unlikely to occur by chance. Positions 1, 145, and 289 also showed repeated amino acid replacements on terminal lineages, but the changes were less directional than those at position 98. The finding suggests that the maintenance of glutamic acid at position 98, which is completely exposed on the VP1 capsid surface and thus potentially immunogenic (66), might be important for the long-term persistence of subgenogroup C1. Two nonexclusive hypotheses might help to explain this observation: (i) the E98K/G substitution directly reduces viral fitness, leading to lineage extinction, or (ii) the E98K/G substitution is an adaptive/epistatic change associated with mutations elsewhere in the EV-71 genome or with infection conditions such as hyperimmune responses exerted by the host. Of note, amino acid substitutions observed in our analysis are unlikely to be a cell culture artifact because, in most laboratory settings, PCR and sequencing are usually performed directly on clinical specimens (75) or low-passage virus isolates (1, 2), limiting the possibility that cell culture-adapted mutations are generated (20). Furthermore, studies by Bible and colleagues have shown that virus cultivation in cell cultures may have negligible influence on the native sequence of the VP1 gene (8). Our results suggest the need for further studies to elucidate the biological properties of EV-71 immunogenic sites.
We also chronologically traced trunk lineage-specific nonsynonymous mutations in the VP1 gene of subgenogroup C1 from 1983 to 2006 (including mutations on core and secondary trunk branches) (Fig. 4; see also Table S3 in the supplemental material). Only 14 nonsynonymous substitutions are observed on the trunk branches, and they occur at 10 different sites. These mutations are candidates for the changes that may determine viral survival and persistence, possibly by generating antigenic novelty. Interestingly, we found reversed change, or “toggling,” of amino acids in the VP1 gene. For instance, glutamic acid at position 145 changed to glutamine in 1985 to 1986 and later reverted to glutamic acid in 1988 to 1989; isoleucine at position 262 was replaced by valine in 1990 to 1994 and returned to isoleucine in 1997 to 1999; alanine at position 289 changed to threonine in 1994 to 1996 and reverted to alanine in 1998 to 2005. Moreover, most of this amino acid toggling occurred on the core trunk branches, and changed residues persisted for about 3 to 5 years (Fig. 4). It is therefore essential to understand how the occurrence of these changes may be correlated with the epidemiology of EV-71 outbreaks, which typically occur in a periodic manner.
DISCUSSION
This study establishes the early origin, spatiotemporal divergence, and epidemiological dynamics of EV-71 for the first time, providing new insights into virus-host interactions and disease periodicity. We reconstructed the epidemic history of EV-71 and found that the virus is a recently emerged pathogen that originated around the mid-20th century. Using the relaxed molecular clock and coalescent approaches, we conclude that EV-71 subgenogroups have been circulating cryptically in human populations for years before causing large-scale HFMD outbreaks. Despite our large sample size, we cannot at present exclude the possibility that currently available virus diversity may underestimate the date of EV-71 origin, a problem that can be resolved by sequencing of more archival samples (80). Thus, the estimated divergence times in this study could be more recent than the actual dates of outbreak origin. Fortunately, we found no evidence of homologous recombination within our data sets, suggesting that our evolutionary analyses (which do not explicitly incorporate recombination) are unlikely to be strongly affected in this instance. Furthermore, the correspondence between our inferred genetic diversity dynamics and known epidemiological trends is reassuring. Although potential biases in estimation can arise from model misspecification (34, 53), they have likely been avoided here by the use of highly flexible models such as the relaxed clock and skyline plot. However, since recombination outside the VP1 region has been reported (73), our divergence dates should be considered specific to VP1 and may not be applicable to other genome regions.
Persistence in genetic diversity between epidemic peaks, as illustrated by the Bayesian skyline plots (Fig. 3), suggests that human populations sustain EV-71 transmission at low levels without triggering huge epidemics (65). Further, the increase in genetic diversity—most evident in the late 1990s—is likely a consequence, at least in part, of the emergence of heterogeneous EV-71 subgenogroups. Together, our data show that the genetic diversity dynamics of EV-71 are associated with the emergence of new subgenogroups and can be used to reflect past HFMD outbreaks.
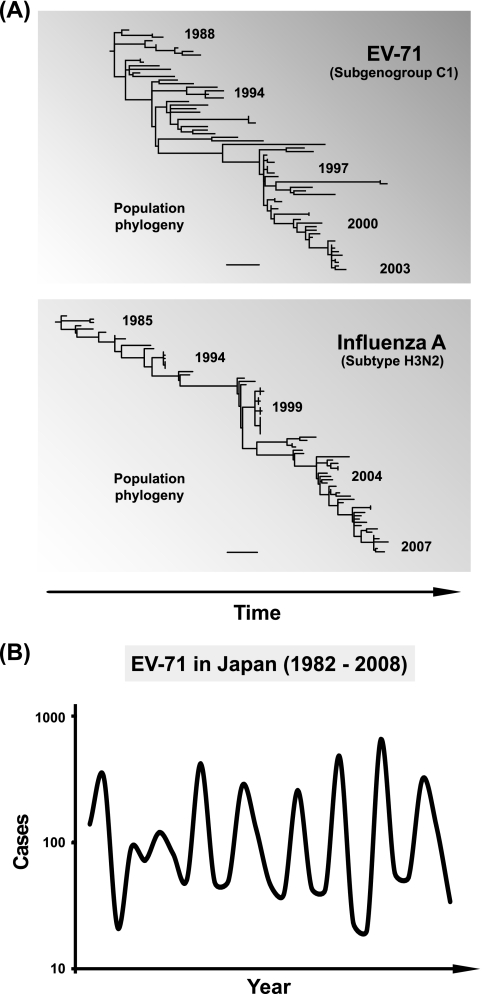
When a phylogeny of longitudinally sampled subgenogroup C1 isolates is compared with a comparable human influenza virus A phylogeny (subtype H3N2; hemagglutinin gene; isolates sampled between 1985 and 2007 from six Asian countries), we note some similarities between the evolutionary dynamics of the two viruses (Fig. 5A). Both phylogenies are characterized by temporal structure with continuous strain replacement through time although strain replacement is perhaps more vigorous for influenza virus. For influenza virus, the low level of host cross-immunity among antigenic types is thought to generate, at the population level, positive selection for immune escape variants (termed antigenic drift). This process has been linked to the strongly temporal shape of influenza virus phylogenies, with limited genetic diversity at any one time (29, 76).
FIG. 5.
Adaptive evolution and periodicity of EV-71. (A) Phylogeny of the longitudinally sampled EV-71 subgenogroup C1, together with a comparable phylogeny of the human influenza A virus. Population sequences from 73 individuals of the human influenza A virus (subtype H3N2) hemagglutinin (HA) gene (∼1.7 kb) isolated from six countries across Asia and sampled between 1985 and 2007 were downloaded from the NCBI's Influenza Virus Resource (7). Trees were plotted using PAUP*, version 4.0 beta (78), and scale bars represent 0.02 nucleotide substitutions per site. (B) The annual incidence of EV-71 infections in Japan from 1982 to 2008. EV-71 isolations/detections are reported by public health institutes and compiled by the Infectious Disease Surveillance Center (IDSC) of the National Institute of Infectious Diseases, Tokyo (idsc.nih.go.jp/iasr/index).
Although influenza virus-like antigenic evolution is one possible explanation of the observed EV-71 phylogeny shape, it is not without problems. Other enteroviruses, notably poliovirus, have remained antigenically stable for many years. Furthermore, it is possible that the temporal structure of the EV-71 phylogeny results from cyclical epidemic dynamics that do not depend on antigenic escape from host immunity. Specifically, gradual accumulation of susceptible hosts through birth (40) and low EV-71 seroprevalence or seroconversion rates at the population level (17, 42) may trigger large-scale viral transmission once a threshold density of susceptible hosts is reached. Intriguingly, EV-71 outbreaks do appear to show a cyclical pattern, occurring about every 2 to 3 years. In Japan, for instance, oscillation in EV-71 infections has been documented over the last 3 decades (Fig. 5B), and a similar trend has also been observed recently in Malaysia (57). Whether antigenic evolution is important in causing these cycles is a key area of uncertainty. Clearly, further research is needed to understand the dynamic relationships between the virus, its hosts, and the environment; this will require integration of population and exposure data before the onset of disease with comprehensive sequencing of pathogen and host genomes in large-scale prospective birth cohorts over an adequate period of time (41).
The selection for immune escape variants in influenza virus presents a major challenge to the development of an efficacious vaccine that elicits broadly neutralizing antibodies against the virus (35). Although current influenza virus vaccines can limit illness, they must be regularly updated to match the influenza virus variants that are “predicted” to constitute future global epidemics (31). Little is known about EV-71 genetic diversity; however, if it does exhibit influenza virus-like antigenic drift, then the strategies adopted in developing an influenza virus vaccine may be relevant for EV-71. Developing an EV-71 vaccine may present a daunting challenge if actively circulating subgenogroups are antigenically distinct. Multiple EV-71 subgenogroups can cocirculate during an outbreak (1, 46, 82), and rapid switching among subgenogroups has become increasingly common, perhaps caused by virus importation, evolution of novel lineages, or other factors (14, 39, 48, 57, 82). Although broadly neutralizing antibodies against selected EV-71 subgenogroups have been reported in animal models (6, 27, 49, 54, 83), the degree of cross-protection and the potential for EV-71 escape evolution in humans remain uninvestigated and unknown. Therefore, addressing the dynamic relationships between the rapidly evolving EV-71 and its human hosts is critical in gaining a better understanding of the disease.
Supplementary Material
Acknowledgments
We gratefully acknowledge Naoki Yamamoto and Edward C. Holmes for support and the anonymous reviewers for their constructive comments and suggestions.
This study was supported in part by grants from the Ministry of Health, Labor and Welfare, Japan (H18-AIDS-General-016) to Y.T.; the Ministry of Science, Technology and Innovation, Malaysia (eScienceFund 02-01-03-SF0379) to A.K.; and a Royal Society International Project Fund to O.G.P. and Y.T. K.K.T. is a recipient of the Japanese Foundation for AIDS Prevention research resident fellowship.
We declare that no competing interests exist.
Footnotes
Published ahead of print on 20 January 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.AbuBakar, S., H. Y. Chee, M. F. Al-Kobaisi, J. Xiaoshan, K. B. Chua, and S. K. Lam. 1999. Identification of enterovirus 71 isolates from an outbreak of hand, foot and mouth disease (HFMD) with fatal cases of encephalomyelitis in Malaysia. Virus Res. 61:1-9. [DOI] [PubMed] [Google Scholar]
- 2.Abubakar, S., H. Y. Chee, N. Shafee, K. B. Chua, and S. K. Lam. 1999. Molecular detection of enteroviruses from an outbreak of hand, foot and mouth disease in Malaysia in 1997. Scand. J. Infect. Dis. 31:331-335. [DOI] [PubMed] [Google Scholar]
- 3.AbuBakar, S., I. C. Sam, J. Yusof, M. K. Lim, S. Misbah, N. MatRahim, and P. S. Hooi. 2009. Enterovirus 71 outbreak, Brunei. Emerg. Infect. Dis. 15:79-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander, J. P., Jr., L. Baden, M. A. Pallansch, and L. J. Anderson. 1994. Enterovirus 71 infections and neurologic disease-United States, 1977-1991. J. Infect. Dis. 169:905-908. [DOI] [PubMed] [Google Scholar]
- 5.Anonymous. 2008. Outbreak news. Enterovirus, China. Wkly. Epidemiol. Rec. 83:169-170. [PubMed] [Google Scholar]
- 6.Arita, M., N. Nagata, N. Iwata, Y. Ami, Y. Suzaki, K. Mizuta, T. Iwasaki, T. Sata, T. Wakita, and H. Shimizu. 2007. An attenuated strain of enterovirus 71 belonging to genotype a showed a broad spectrum of antigenicity with attenuated neurovirulence in cynomolgus monkeys. J. Virol. 81:9386-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao, Y., P. Bolotov, D. Dernovoy, B. Kiryutin, L. Zaslavsky, T. Tatusova, J. Ostell, and D. Lipman. 2008. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 82:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bible, J. M., M. Iturriza-Gomara, B. Megson, D. Brown, P. Pantelidis, P. Earl, J. Bendig, and C. Y. Tong. 2008. Molecular epidemiology of human enterovirus 71 in the United Kingdom from 1998 to 2006. J. Clin. Microbiol. 46:3192-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bible, J. M., P. Pantelidis, P. K. Chan, and C. Y. Tong. 2007. Genetic evolution of enterovirus 71: epidemiological and pathological implications. Rev. Med. Virol. 17:371-379. [DOI] [PubMed] [Google Scholar]
- 10.Bouchard, M. J., D. H. Lam, and V. R. Racaniello. 1995. Determinants of attenuation and temperature sensitivity in the type 1 poliovirus Sabin vaccine. J. Virol. 69:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, B. A., M. S. Oberste, J. P. Alexander, Jr., M. L. Kennett, and M. A. Pallansch. 1999. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 73:9969-9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cameron-Wilson, C. L., Y. A. Pandolfino, H. Y. Zhang, B. Pozzeto, and L. C. Archard. 1998. Nucleotide sequence of an attenuated mutant of coxsackievirus B3 compared with the cardiovirulent wildtype: assessment of candidate mutations by analysis of a revertant to cardiovirulence. Clin. Diagn. Virol. 9:99-105. [DOI] [PubMed] [Google Scholar]
- 13.Cardosa, M. J., S. Krishnan, P. H. Tio, D. Perera, and S. C. Wong. 1999. Isolation of subgenus B adenovirus during a fatal outbreak of enterovirus 71-associated hand, foot, and mouth disease in Sibu, Sarawak. Lancet 354:987-991. [DOI] [PubMed] [Google Scholar]
- 14.Cardosa, M. J., D. Perera, B. A. Brown, D. Cheon, H. M. Chan, K. P. Chan, H. Cho, and P. McMinn. 2003. Molecular epidemiology of human enterovirus 71 strains and recent outbreaks in the Asia-Pacific region: comparative analysis of the VP1 and VP4 genes. Emerg. Infect. Dis. 9:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan, K. P., K. T. Goh, C. Y. Chong, E. S. Teo, G. Lau, and A. E. Ling. 2003. Epidemic hand, foot and mouth disease caused by human enterovirus 71, Singapore. Emerg. Infect. Dis. 9:78-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan, L. G., U. D. Parashar, M. S. Lye, F. G. Ong, S. R. Zaki, J. P. Alexander, K. K. Ho, L. L. Han, M. A. Pallansch, A. B. Suleiman, M. Jegathesan, and L. J. Anderson for the Outbreak Study Group. 2000. Deaths of children during an outbreak of hand, foot, and mouth disease in Sarawak, Malaysia: clinical and pathological characteristics of the disease. Clin. Infect. Dis. 31:678-683. [DOI] [PubMed] [Google Scholar]
- 17.Chang, L. Y., C. C. King, K. H. Hsu, H. C. Ning, K. C. Tsao, C. C. Li, Y. C. Huang, S. R. Shih, S. T. Chiou, P. Y. Chen, H. J. Chang, and T. Y. Lin. 2002. Risk factors of enterovirus 71 infection and associated hand, foot, and mouth disease/herpangina in children during an epidemic in Taiwan. Pediatrics 109:e88. [DOI] [PubMed] [Google Scholar]
- 18.Chinese Center for Disease Control and Prevention, and Office of the World Health Organization in China. 2008. Report on the hand, foot and mouth disease outbreak in Fuyang City, Anhui Province and the prevention and control in China. World Health Organization, Beijing, China. http://www.wpro.who.int/china.
- 19.Chua, B. H., P. C. McMinn, S. K. Lam, and K. B. Chua. 2001. Comparison of the complete nucleotide sequences of echovirus 7 strain UMMC and the prototype (Wallace) strain demonstrates significant genetic drift over time. J. Gen. Virol. 82:2629-2639. [DOI] [PubMed] [Google Scholar]
- 20.Chua, B. H., P. Phuektes, S. A. Sanders, P. K. Nicholls, and P. C. McMinn. 2008. The molecular basis of mouse adaptation by human enterovirus 71. J. Gen. Virol. 89:1622-1632. [DOI] [PubMed] [Google Scholar]
- 21.Chumakov, M., M. Voroshilova, L. Shindarov, I. Lavrova, L. Gracheva, G. Koroleva, S. Vasilenko, I. Brodvarova, M. Nikolova, S. Gyurova, M. Gacheva, G. Mitov, N. Ninov, E. Tsylka, I. Robinson, M. Frolova, V. Bashkirtsev, L. Martiyanova, and V. Rodin. 1979. Enterovirus 71 isolated from cases of epidemic poliomyelitis-like disease in Bulgaria. Arch. Virol. 60:329-340. [DOI] [PubMed] [Google Scholar]
- 22.Drummond, A. J., S. Y. Ho, M. J. Phillips, and A. Rambaut. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drummond, A. J., G. K. Nicholls, A. G. Rodrigo, and W. Solomon. 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161:1307-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drummond, A. J., A. Rambaut, B. Shapiro, and O. G. Pybus. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22:1185-1192. [DOI] [PubMed] [Google Scholar]
- 26.Fitch, W. M., R. M. Bush, C. A. Bender, and N. J. Cox. 1997. Long term trends in the evolution of H(3) HA1 human influenza type A. Proc. Natl. Acad. Sci. U. S. A. 94:7712-7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foo, D. G., S. Alonso, V. T. Chow, and C. L. Poh. 2007. Passive protection against lethal enterovirus 71 infection in newborn mice by neutralizing antibodies elicited by a synthetic peptide. Microbes Infect. 9:1299-1306. [DOI] [PubMed] [Google Scholar]
- 28.Gilbert, G. L., K. E. Dickson, M. J. Waters, M. L. Kennett, S. A. Land, and M. Sneddon. 1988. Outbreak of enterovirus 71 infection in Victoria, Australia, with a high incidence of neurologic involvement. Pediatr. Infect. Dis. J. 7:484-488. [DOI] [PubMed] [Google Scholar]
- 29.Grenfell, B. T., O. G. Pybus, J. R. Gog, J. L. Wood, J. M. Daly, J. A. Mumford, and E. C. Holmes. 2004. Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303:327-332. [DOI] [PubMed] [Google Scholar]
- 30.Halim, S., and A. I. Ramsingh. 2000. A point mutation in VP1 of coxsackievirus B4 alters antigenicity. Virology 269:86-94. [DOI] [PubMed] [Google Scholar]
- 31.Harper, S. A., K. Fukuda, T. M. Uyeki, N. J. Cox, and C. B. Bridges. 2004. Prevention and control of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recommend. Rep. 53:1-40. [PubMed] [Google Scholar]
- 32.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 33.Ho, M., E. R. Chen, K. H. Hsu, S. J. Twu, K. T. Chen, S. F. Tsai, J. R. Wang, and S. R. Shih, for the Taiwan Enterovirus Epidemic Working Group. 1999. An epidemic of enterovirus 71 infection in Taiwan. N. Engl. J. Med. 341:929-935. [DOI] [PubMed] [Google Scholar]
- 34.Jorba, J., R. Campagnoli, L. De, and O. Kew. 2008. Calibration of multiple poliovirus molecular clocks covering an extended evolutionary range. J. Virol. 82:4429-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karlsson Hedestam, G. B., R. A. Fouchier, S. Phogat, D. R. Burton, J. Sodroski, and R. T. Wyatt. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 6:143-155. [DOI] [PubMed] [Google Scholar]
- 36.Kingman, J. F. C. 1982. The coalescent. Stochastic Processes Appl. 13:235-248. [Google Scholar]
- 37.Komatsu, H., Y. Shimizu, Y. Takeuchi, H. Ishiko, and H. Takada. 1999. Outbreak of severe neurologic involvement associated with enterovirus 71 infection. Pediatr. Neurol. 20:17-23. [DOI] [PubMed] [Google Scholar]
- 38.Li, L., Y. He, H. Yang, J. Zhu, X. Xu, J. Dong, Y. Zhu, and Q. Jin. 2005. Genetic characteristics of human enterovirus 71 and coxsackievirus A16 circulating from 1999 to 2004 in Shenzhen, People's Republic of China. J. Clin. Microbiol. 43:3835-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, K. H., K. P. Hwang, G. M. Ke, C. F. Wang, L. Y. Ke, Y. T. Hsu, Y. C. Tung, P. Y. Chu, B. H. Chen, H. L. Chen, C. L. Kao, J. R. Wang, H. L. Eng. S. Y. Wang, L. C. Hsu, and H. Y. Chen. 2006. Evolution of EV71 genogroup in Taiwan from 1998 to 2005: an emerging of subgenogroup C4 of EV71. J. Med. Virol. 78:254-262. [DOI] [PubMed] [Google Scholar]
- 40.Lin, T. Y., S. J. Twu, M. S. Ho, L. Y. Chang, and C. Y. Lee. 2003. Enterovirus 71 outbreaks, Taiwan: occurrence and recognition. Emerg. Infect. Dis. 9:291-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipkin, W. I. 2009. Microbe hunting in the 21st century. Proc. Natl. Acad. Sci. U. S. A. 106:6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu, C. Y., C. Y. Lee, C. L. Kao, W. Y. Shao, P. I. Lee, S. J. Twu, C. C. Yeh, S. C. Lin, W. Y. Shih, S. I. Wu, and L. M. Huang. 2002. Incidence and case-fatality rates resulting from the 1998 enterovirus 71 outbreak in Taiwan. J. Med. Virol. 67:217-223. [DOI] [PubMed] [Google Scholar]
- 43.Lum, L. C., K. T. Wong, S. K. Lam, K. B. Chua, A. Y. Goh, W. L. Lim, B. B. Ong, G. Paul, S. AbuBakar, and M. Lambert. 1998. Fatal enterovirus 71 encephalomyelitis. J. Pediatr. 133:795-798. [DOI] [PubMed] [Google Scholar]
- 44.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 45.Martin, J., G. Dunn, R. Hull, V. Patel, and P. D. Minor. 2000. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J. Virol. 74:3001-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McMinn, P., K. Lindsay, D. Perera, H. M. Chan, K. P. Chan, and M. J. Cardosa. 2001. Phylogenetic analysis of enterovirus 71 strains isolated during linked epidemics in Malaysia, Singapore, and Western Australia. J. Virol. 75:7732-7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melnick, J. L. 1984. Enterovirus type 71 infections: a varied clinical pattern sometimes mimicking paralytic poliomyelitis. Rev. Infect. Dis. 6(Suppl. 2):S387-S390. [DOI] [PubMed] [Google Scholar]
- 48.Mizuta, K., C. Abiko, T. Murata, Y. Matsuzaki, T. Itagaki, K. Sanjoh, M. Sakamoto, S. Hongo, S. Murayama, and K. Hayasaka. 2005. Frequent importation of enterovirus 71 from surrounding countries into the local community of Yamagata, Japan, between 1998 and 2003. J. Clin. Microbiol. 43:6171-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizuta, K., Y. Aoki, A. Suto, K. Ootani, N. Katsushima, T. Itagaki, A. Ohmi, M. Okamoto, H. Nishimura, Y. Matsuzaki, S. Hongo, K. Sugawara, H. Shimizu, and T. Ahiko. 2009. Cross-antigenicity among EV71 strains from different genogroups isolated in Yamagata, Japan, between 1990 and 2007. Vaccine 27:3153-3158. [DOI] [PubMed] [Google Scholar]
- 50.Modlin, J. F. 2007. Enterovirus deja vu. N. Engl. J. Med. 356:1204-1205. [DOI] [PubMed] [Google Scholar]
- 51.Morens, D. M., G. K. Folkers, and A. S. Fauci. 2008. Emerging infections: a perpetual challenge. Lancet Infect. Dis. 8:710-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagy, G., S. Takatsy, E. Kukan, I. Mihaly, and I. Domok. 1982. Virological diagnosis of enterovirus type 71 infections: experiences gained during an epidemic of acute CNS diseases in Hungary in 1978. Arch. Virol. 71:217-227. [DOI] [PubMed] [Google Scholar]
- 53.Navascues, M., and B. C. Emerson. 2009. Elevated substitution rate estimates from ancient DNA: model violation and bias of Bayesian methods. Mol. Ecol. 18:4390-4397. [DOI] [PubMed] [Google Scholar]
- 54.Ong, K. C., S. Devi, M. J. Cardosa, and K. T. Wong. 2010. Formaldehyde-inactivated whole virus vaccine protects a murine model of enterovirus 71 encephalomyelitis against disease. J. Virol. 84:661-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ooi, M. H., S. C. Wong, Y. Podin, W. Akin, S. del Sel, A. Mohan, C. H. Chieng, D. Perera, D. Clear, D. Wong, E. Blake, J. Cardosa, and T. Solomon. 2007. Human enterovirus 71 disease in Sarawak, Malaysia: a prospective clinical, virological, and molecular epidemiological study. Clin. Infect. Dis. 44:646-656. [DOI] [PubMed] [Google Scholar]
- 56.Perera, D., M. A. Yusof, Y. Podin, M. H. Ooi, N. T. Thao, K. K. Wong, A. Zaki, K. B. Chua, Y. A. Malik, P. V. Tu, N. T. Tien, P. Puthavathana, P. C. McMinn, and M. J. Cardosa. 2007. Molecular phylogeny of modern coxsackievirus A16. Arch. Virol. 152:1201-1208. [DOI] [PubMed] [Google Scholar]
- 57.Podin, Y., E. L. Gias, F. Ong, Y. W. Leong, S. F. Yee, M. A. Yusof, D. Perera, B. Teo, T. Y. Wee, S. C. Yao, S. K. Yao, A. Kiyu, M. T. Arif, and M. J. Cardosa. 2006. Sentinel surveillance for human enterovirus 71 in Sarawak, Malaysia: lessons from the first 7 years. BMC Public Health 6:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pond, S. L., S. D. Frost, and S. V. Muse. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676-679. [DOI] [PubMed] [Google Scholar]
- 59.Pupko, T., I. Pe'er, R. Shamir, and D. Graur. 2000. A fast algorithm for joint reconstruction of ancestral amino acid sequences. Mol. Biol. Evol. 17:890-896. [DOI] [PubMed] [Google Scholar]
- 60.Pybus, O. G., A. J. Drummond, T. Nakano, B. H. Robertson, and A. Rambaut. 2003. The epidemiology and iatrogenic transmission of hepatitis C virus in Egypt: a Bayesian coalescent approach. Mol. Biol. Evol. 20:381-387. [DOI] [PubMed] [Google Scholar]
- 61.Pybus, O. G., A. Rambaut, R. Belshaw, R. P. Freckleton, A. J. Drummond, and E. C. Holmes. 2007. Phylogenetic evidence for deleterious mutation load in RNA viruses and its contribution to viral evolution. Mol. Biol. Evol. 24:845-852. [DOI] [PubMed] [Google Scholar]
- 62.Qiu, J. 2008. Enterovirus 71 infection: a new threat to global public health? Lancet Neurol. 7:868-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiu, J. 2009. Viral outbreak in China tests government efforts. Nature 458:554-555. [DOI] [PubMed] [Google Scholar]
- 64.Rambaut, A., and A. J. Drummond. 2007. Tracer v1.4. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, Scotland. http://tree.bio.ed.ac.uk.
- 65.Rambaut, A., O. G. Pybus, M. I. Nelson, C. Viboud, J. K. Taubenberger, and E. C. Holmes. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ranganathan, S., S. Singh, C. L. Poh, and V. T. Chow. 2002. The hand, foot and mouth disease virus capsid: sequence analysis and prediction of antigenic sites from homology modelling. Appl. Bioinformatics 1:43-52. [PubMed] [Google Scholar]
- 67.Robbins, K. E., P. Lemey, O. G. Pybus, H. W. Jaffe, A. S. Youngpairoj, T. M. Brown, M. Salemi, A. M. Vandamme, and M. L. Kalish. 2003. U.S. Human immunodeficiency virus type 1 epidemic: date of origin, population history, and characterization of early strains. J. Virol. 77:6359-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodriguez, F., J. L. Oliver, A. Marin, and J. R. Medina. 1990. The general stochastic model of nucleotide substitution. J. Theor. Biol. 142:485-501. [DOI] [PubMed] [Google Scholar]
- 69.Samuda, G. M., W. K. Chang, C. Y. Yeung, and P. S. Tang. 1987. Monoplegia caused by enterovirus 71: an outbreak in Hong Kong. Pediatr. Infect. Dis. J. 6:206-208. [DOI] [PubMed] [Google Scholar]
- 70.Sanders, S. A., L. J. Herrero, K. McPhie, S. S. Chow, M. E. Craig, D. E. Dwyer, W. Rawlinson, and P. C. McMinn. 2006. Molecular epidemiology of enterovirus 71 over two decades in an Australian urban community. Arch. Virol. 151:1003-1013. [DOI] [PubMed] [Google Scholar]
- 71.Schmidt, N. J., E. H. Lennette, and H. H. Ho. 1974. An apparently new enterovirus isolated from patients with disease of the central nervous system. J. Infect. Dis. 129:304-309. [DOI] [PubMed] [Google Scholar]
- 72.Shimizu, H., A. Utama, K. Yoshii, H. Yoshida, T. Yoneyama, M. Sinniah, M. A. Yusof, Y. Okuno, N. Okabe, S. R. Shih, H. Y. Chen, G. R. Wang, C. L. Kao, K. S. Chang, T. Miyamura, and A. Hagiwara. 1999. Enterovirus 71 from fatal and nonfatal cases of hand, foot and mouth disease epidemics in Malaysia, Japan and Taiwan in 1997-1998. Jpn. J. Infect. Dis. 52:12-15. [PubMed] [Google Scholar]
- 73.Simmonds, P., and J. Welch. 2006. Frequency and dynamics of recombination within different species of human enteroviruses. J. Virol. 80:483-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Singh, S., V. T. Chow, K. P. Chan, A. E. Ling, and C. L. Poh. 2000. RT-PCR, nucleotide, amino acid and phylogenetic analyses of enterovirus type 71 strains from Asia. J. Virol. Methods 88:193-204. [DOI] [PubMed] [Google Scholar]
- 75.Singh, S., V. T. Chow, M. C. Phoon, K. P. Chan, and C. L. Poh. 2002. Direct detection of enterovirus 71 (EV71) in clinical specimens from a hand, foot, and mouth disease outbreak in Singapore by reverse transcription-PCR with universal enterovirus and EV71-specific primers. J. Clin. Microbiol. 40:2823-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith, D. J., A. S. Lapedes, J. C. de Jong, T. M. Bestebroer, G. F. Rimmelzwaan, A. D. Osterhaus, and R. A. Fouchier. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371-376. [DOI] [PubMed] [Google Scholar]
- 77.Suchard, M. A., R. E. Weiss, and J. S. Sinsheimer. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 18:1001-1013. [DOI] [PubMed] [Google Scholar]
- 78.Swofford, D. L. 2003. PAUP*, phylogenetic analysis using parsimony (*and other methods), 4.0 beta ed. Sinauer Associates, Sunderland, MA.
- 79.Takeda, N., M. Tanimura, and K. Miyamura. 1994. Molecular evolution of the major capsid protein VP1 of enterovirus 70. J. Virol. 68:854-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tee, K. K., O. G. Pybus, X. J. Li, X. Han, H. Shang, A. Kamarulzaman, and Y. Takebe. 2008. Temporal and spatial dynamics of human immunodeficiency virus type 1 circulating recombinant forms 08_BC and 07_BC in Asia. J. Virol. 82:9206-9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tu, P. V., N. T. T. Thao, D. Perera, T. H. Khanh, N. T. K. Tien, T. C. Thuong, O. M. How, M. J. Cardosa, and P. C. McMinn. 2007. Epidemiologic and virologic Invest. of hand, food, and mouth disease, southern Vietnam, 2005. Emerg. Infect. Dis. 13:1733-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang, J. R., Y. C. Tuan, H. P. Tsai, J. J. Yan, C. C. Liu, and I. J. Su. 2002. Change of major genotype of enterovirus 71 in outbreaks of hand-foot- and-mouth disease in Taiwan between 1998 and 2000. J. Clin. Microbiol. 40:10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu, T. C., Y. F. Wang, Y. P. Lee, J. R. Wang, C. C. Liu, S. M. Wang, H. Y. Lei, I. J. Su, and C. K. Yu. 2007. Immunity to avirulent enterovirus 71 and coxsackie A16 virus protects against enterovirus 71 infection in mice. J. Virol. 81:10310-10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang, Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39:306-314. [DOI] [PubMed] [Google Scholar]
- 85.Yoke-Fun, C., and S. AbuBakar. 2006. Phylogenetic evidence for intertypic recombination in the emergence of human enterovirus 71 subgenotypes. BMC Microbiol. 6:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.