Abstract
RNase III proteins play vital roles in processing of several types of RNA molecules and gene silencing. Recently, it has been discovered that some plant and animal viruses encode RNase III-like proteins as well. Genome sequencing of four virus species belonging to the Ascoviridae family has revealed sequence conservation of an RNase III open reading frame among the viruses. These have not been explored in ascoviruses, and therefore their role in host-virus interaction is unknown. Here, we confirmed expression of Heliothis virescens ascovirus (HvAV-3e) open reading frame 27 (orf27) that encodes an RNase III-like protein after infection and demonstrated dsRNA specific endoribonuclease activity of the encoded protein. Analysis of the expression patterns of orf27 in virus-infected insect cells and a bacterial expression system revealed autoregulation of this protein over time. Moreover, HvAV-3e RNase III was found essential for virus DNA replication and infection using RNA interference (RNAi)-mediated gene silencing. In addition, using green fluorescent protein gene as a marker, we provide evidence that RNase III is involved in the suppression of gene silencing. To our knowledge, this is the first insect virus-encoded RNase III described and shown to suppress host cell RNAi defense mechanism.
RNases III are double-stranded RNA (dsRNA)-specific processing enzymes present in prokaryotes, eukaryotes, and some viruses. Their functions are well documented in the processing and maturation of precursors of rRNA (pre-rRNA) (14), tRNA (26), small nuclear RNA, small nucleolar RNA (28), mRNA decay (13) and, above all, gene silencing (22). RNase III proteins all contain an RNase signature motif but, based on structure, are divided into four classes. Class I enzymes (in bacteria and bacteriophages) contain only one RNase domain (endoND) and a dsRNA-binding domain (dsRBD) (25). Class II riboendonucleases (in yeast) are highly variable and contain an additional 100-amino-acid N-terminal domain (10). Class III enzymes possess a dsRBD and two endoND domains. Drosha is one of the most extensively studied member of this class that guides microRNA (miRNA) biogenesis by cleaving primary miRNA (pri-miRNA) transcripts into precursor miRNA (pre-miRNA) (17). The largest class, which includes Dicer, is class IV, in which members contain a dsRBD, two endoNDs, a helicase, and a Piwi Agronaute Zwille (PAZ) domain (24). Dicers process small RNAs of multiple types, including short interfering RNAs (siRNAs) and miRNA.
Recently, viruses (either with RNA or DNA genomes) infecting a wide range of organisms have been reported to encode RNase III-like proteins. Thus far, well-studied examples of viral RNase III proteins among RNA viruses is from sweet potato chlorotic stunt virus (SPCSV) (20), while in DNA viruses are from chlorella virus (PBCV-1) (34) and rock bream iridovirus (33). Genome annotations of other DNA viruses belonging to Ascoviridae and Iridoviridae indicate the presence of open reading frames (ORFs) that putatively encode RNase III proteins. The molecular masses of viral RNase III proteins vary from 20 to 80 kDa and can be grouped into the class I category since they all possess a single endoND with a conserved RNase signature motif and a dsRBD domain. The role of these proteins in virus biology is still largely unknown. It was earlier found that SPCSV RNase III enhances RNA interference (RNAi) suppression induced by another virus-encoded protein (p22) (20). Recently, the same protein was reported to mediate synergism with other unrelated viruses to break down antiviral resistance in plants (see the Discussion) (12).
Ascoviruses (AVs) are pathogenic to lepidopteran insects at larval and pupal stages (6). After infection, the host cell morphology resembles that of apoptotic cells producing vesicles that look like apoptotic bodies through which virions are disseminated (5). The virions can be transmitted mechanically by the ovipositor of female parasitic wasps between caterpillars (15). Genome sequences (119- to 187-kbp dsDNA) of four recognized AV species, Spodoptera frugiperda AV (SfAV-1), Trichoplusia ni AV (TnAV-2), Heliothis virescens AV (HvAV-3), and Diadromus pulchellus AV (DpAV-4) are now available (1, 4, 6, 30). Phylogenetic studies suggest that AVs may have evolved from iridoviruses that infect lepidopteran larvae (15, 29). There are 180 potential ORFs in the HvAV-3e genome, most of which are homologs to genes involved in nucleic acid metabolism, virus replication, apoptosis, and lipid metabolism (1). In the present study, we analyzed an HvAV-3e-encoded RNase III protein (from orf27) at a functional level by exploring its differential expression after virus infection, catalytic activity for dsRNA and autoregulation. Our RNAi-mediated knockdown experiment for orf27 suggested its vital role in virus DNA replication and infection. Moreover, we demonstrated that the protein is a suppressor of gene silencing by degrading siRNAs.
MATERIALS AND METHODS
Cell cultures and virus infection.
A Helicoverpa zea fat body cell line (HzFB) (23) and a Spodoptera frugiperda cell line (Sf9) were maintained in SF900-II serum-free medium (Invitrogen) as monolayers in cell culture flasks at 27°C.
An HvAV isolate (HvAV-3e) was used throughout the experiments (1). For virus infection, cells were transferred into 35-by-10-mm petri dishes (Falcon) and allowed to settle for 1 h. The medium was removed, and the cells were inoculated with 106 HvAV-3e PFU, determined by plaque assay, at a multiplicity of infection of 5 (19). An hour after incubation at room temperature with rocking, fresh medium was added to the cells, followed by further incubation at 27°C.
Cloning and expression of HvAV-3e RNase III gene.
For the bacterial expression of RNase III, three gene constructs were produced. A pair of specific primers was used to amplify full-length RNase III gene from genomic DNA extracted from HvAV-3e-infected HzFB cells. The PCR-amplified product (2,176 bp) was cloned into pQE30 bacterial expression vector (Qiagen) at the BamHI and HindIII sites, yielding pQE/RNaseIII. Similarly, a truncated gene construct containing the first 1,800 bp was cloned at the same sites resulting in pQE/RNaseIII-1800 clone. Another construct with a deleted RNase domain was produced after SalI restriction digestion and self-ligation of the pQE/RNaseIII clone. This clone was designated pQE/RNaseΔrnd. Escherichia coli M15(pREP4) competent cells were used for protein expression of these three constructs. For protein expression, cultures harboring these clones were grown overnight at 37°C in LB medium containing ampicillin (100 μg/ml) and kanamycin (50 μg/ml), and cells were collected at different times after IPTG (isopropyl-β-d-thiogalactopyranoside) induction (0.5 mM).
Construction of recombinant baculovirus and protein purification.
We used the Bac-to-Bac expression system for the overexpression and purification of RNase III protein. Full-length RNase III PCR product was cloned into pFastBac-Hta vector at the BamHI and HindIII restriction sites. All of the procedures from transfection to purification of the fusion protein were conducted according to the manufacturer's instructions (Invitrogen). Expression of the recombinant protein was confirmed by Western blotting, using an alkaline phosphatase-conjugated monoclonal antibody to His residues (1:5,000; Sigma). The fusion protein was purified under native conditions using Ni-NTA beads (Invitrogen) and finally eluted in 200 and 300 mM imidazole, which were mixed together.
Northern hybridization.
Gene transcripts were detected by Northern blot analyses of total RNA (10 μg) run on 1.2% agarose formaldehyde gels as described previously (27). DNA probes were labeled with [32P]dCTP using a random primer DNA labeling kit (GE Healthcare), and all hybridization and washing steps were carried out at 65°C. Blots were then exposed to a phosphorimager screen for 2 h, and radioactive signals were detected by using a phosphorimager scanner.
qPCR.
Total genomic DNA was extracted from cells and subjected to quantitative PCR (qPCR) using specific primers to HvAV-3e orf19 (orf19-For, 5′-ATCTCGACTGGCATACGC-3′; orf19-Rev, 5′-CGCAAAGTCCGTGAGTAGC-3′). DNA concentrations were measured with a Nanodrop, and 50 ng of total genomic DNA was used for each qPCR using SYBR green (Invitrogen) with a Rotor-Gene 6000. The real-time PCR conditions were as follows: 50°C for 2 min and 95°C for 2 min, followed by 40 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 20 s, with a final extension of 72°C for 20 s. Actin was used for normalizing the data with the primers Act-For (5′-ATGGAGAAGATCTGGCAC-3′) and Act-Rev (5′-GTTGGCCTTGGGGTTGAG-3′). Relative DNA levels from each sample were compared in the QGene template program. Reactions were repeated three times.
RNAi-mediated gene silencing.
For RNAi, we used dsRNA synthesized in vitro by using a MegaScript transcription kit according to the manufacturer's instructions (Ambion). T7 promoter sequences were incorporated in both forward and reverse primers designed to amplify a 450-bp DNA fragment of RNase III gene. For dsRNA synthesis, 1 μg of PCR product was used during a 16-h incubation period at 37°C, DNase treated, and precipitated by the lithium chloride method (Ambion). We used 0.5 μg of dsRNA for transfection into HzFB cells (106 cells) at 24 h prior to virus inoculation. The cells were collected for further analyses at 48 h postinfection (hpi).
Suppression of gene silencing.
To investigate the RNAi suppressor activity of HvAV-3e RNase III protein, we used green fluorescent protein (GFP) as a reporter gene. We cloned the full-length GFP orf (700 bp) into pIZ vector at the HindIII and XbaI restriction sites, resulting in a pIZ/GFP construct. dsRNA (500 bp) specific to GFP was synthesized in vitro over the region between nucleotides 101 and 600. We transfected 2 μg of pIZ/GFP with or without 0.5 μg of GFP dsRNA into Sf9 cells. After 48 h, GFP-silenced cells were mock inoculated or inoculated separately with HvAV-3e, RNase III recombinant baculovirus, and HvAV-3e DNA polymerase (orf1) recombinant baculovirus as a control. Total RNA was extracted at 48 hpi and subjected to qRT-PCR for GFP transcript levels by using a primer pair designed to amplify the first 100 bp of the GFP sequence. After DNase I treatment, total RNA was reverse transcribed into cDNA by using Superscript III (Invitrogen) and subsequently subjected to qPCR as described above.
RNA cleavage assays.
In order to study RNA cleavage in vitro, we produced dsRNA specific to the HvAV-3e Bro11 (orf98; 350 bp) and GFP (500 bp) genes. The dsRNA substrates were incubated with 0.3 pmol of purified RNase III and Vn50 (control) proteins expressed by recombinant baculoviruses. Vn50 is a venom protein from the Cotesia rubecula parasitoid wasp with similarity to serine protease homologs, but it lacks any enzyme activity (2). A cleavage assay was performed at 37°C for 0, 10, 20, 30, 45, and 60 min of incubation in cleavage buffer (100 mM NaCl, 10 mM MgCl2, 20 mM HEPES [pH 8.0], 5 mM dithiothreitol, 1 mM EDTA, 0.1% Triton X-100, 20% glycerol). The reactions were then stopped by adding loading buffer containing 20 mM EDTA and run on 1% agarose gel.
For small RNA cleavage, we used synthesized siRNA duplexes 21 nucleotides in length (GenePharma). The sequences of the oligonucleotides were the siRNA duplex-25 GUCCGGAUACUCUUUGCGGAC and siRNA duplex-11 GGAGGAAGAAAGGAGAAAGGA. These were incubated with purified RNase III and Vn50 proteins for 60 min at 37°C in cleavage buffer (30 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 5% glycerol [vol/vol]). The samples were then run on a 15% urea-polyacrylamide gel and subsequently stained with ethidium bromide.
RESULTS
HvAV-3e orf27 encodes an RNase III protein.
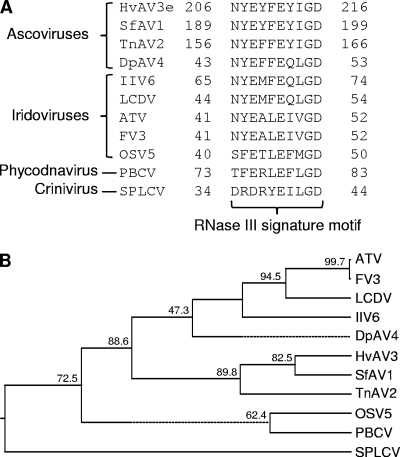
Several viruses with DNA or RNA genomes have been reported to encode RNase III-like proteins. Genome annotation of HvAV-3e revealed that orf27 (nuclear coordinates 32493 to 34679) encodes an RNase-like protein of 728 amino acids (1), which is significantly larger than those encoded by other viruses (average size, 280 amino acids). The RNase-specific domain (Rnc) and the RNase III C-terminal domain (RIBOc) are located in between amino acids 183 and 442 and amino acids 191 and 336, respectively. These domains are present in eukaryotic, bacterial, and archaeal RNase III proteins. Amino acid sequence alignments of HvAV-3e RNase III domain with those from other viruses indicated conserved RNase III signature motifs (Fig. 1A). orf27 shows 51.9, 35.9, and 17.2% amino acid identity to SfAV-1a orf22, TnAV-2c orf8, and DpAV-4a orf3, respectively, and clustered closely with SfAV-1a and TnAV-2c (Fig. 1B).
FIG. 1.
Different viruses encode class I RNase III-like proteins. (A) Only RNase III signature motifs in amino acid alignments that are necessary for dsRNA cleavage are shown. The positions of these motifs with respect to amino acid coordinates are variable in different viruses. (B) The full-length amino acid sequences of RNase III from viruses used in panel A were aligned by the CLUSTAL W method based on which a phylogenetic tree was constructed (DNASTAR; MegAlign). A dotted line on the phenogram indicates a negative branch length, a common result of averaging. HvAV-3e (Heliothis virescens AV), SfAV-1 (Spodoptera frugiperda AV), TnAV-2 (Trichoplusia ni AV), DpAV-4 (Diadromus pulchellus AV), IIV-6 (Invertebrate iridescent virus 6), LCDV (Lymphocystis disease virus 1), ATV (Ambystoma tigrinum virus), FV3 (Frog virus 3), OSV 5 (Ostreococcus virus 5), PBCV (Paramecium bursaria chlorella virus), and an RNase III sequence from the RNA virus SPCSV are included in the phenogram. Bootstrap values are shown on the tree.
Differential expression of RNase III.
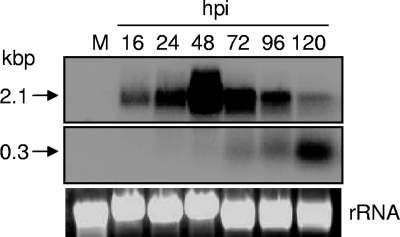
Northern blot analyses was used to study transcript levels of HvAV-3e RNase III at different times after virus infection in the Sf9 and HzFB cell lines (Fig. 2; only Sf9 data are shown since the HzFB results were similar). A full-length RNase III probe produced by PCR was used in the hybridization. A transcript band of 2.1 kb was detected at 16 hpi with maximum intensity at 48 hpi, which then decreased significantly at later times of up to 120 hpi. At 48 hpi the highest level of expression was detected and later, from 72 to 120 hpi, we observed a decrease in the 2.1-kb transcript levels with an increase in another smaller, ∼300-nucleotide fragment (Fig. 2). These results suggested cleavage of RNase III transcript later in infection which prompted us to investigate the mechanism.
FIG. 2.
Northern blot analysis of HvAV-3e RNase III expression at different time intervals of virus infection in HzFB cells. A full-length RNase III probe was used for hybridization. A specific 2.1-kbp transcript was detected at 16 hpi that maximized at 48 h and then decreased at later times with an increase in the levels of an ∼300-nucleotide fragment. rRNA bands show equal loading of total RNA samples in the gel.
RNase III catalyzes dsRNA.
A baculovirus expression system was used for overexpression and purification of the RNase III protein. Western blot analysis with a specific monoclonal antibody to His residues detected an ∼80-kDa fusion protein in Sf9 cells infected with the recombinant virus (Fig. 3A). The band was not detected in mock-infected cells. The predicted size for HvAV-3e RNase III is 77.2 kDa, which is close to the detected band, considering the addition of six His residues. The purified protein was then tested for catalytic activity. We synthesized dsRNA specific to Bro11 (HvAV-3e orf98; 350 bp) and GFP (500 bp) sequences by in vitro transcription. In cleavage assays, 100 ng of Bro11 and GFP dsRNAs were incubated with 0.3 pmol of purified RNase III, as well as a control protein (Vn50) (2) in the presence of a catalysis buffer containing Mg2+. Vn50 also contained a His tag and was produced and purified from Sf9 cells. The reactions were incubated at 37°C; stopped after 0, 10, 20, 30, 45, and 60 min; and run on 1% agarose gels. We found significant reductions in the dsRNA concentration over time in the presence of RNase III, whereas in the absence of the enzyme or the presence of Vn50 protein the dsRNA concentration remained the same (Fig. 3B). These results showed the dsRNA catalytic activity of HvAV-3e RNase III. In another experiment, we tested dsRNA cleavage in vivo, as GFP dsRNA (0.5 μg) was transfected into Sf9 cells and 24 h after transfection, cells were inoculated with HvAV-3e, RNase III, and Vn50 recombinant baculoviruses or were mock inoculated (no virus). Total RNA was extracted 48 hpi, and a GFP probe was used in a Northern blot analysis. dsRNA detection was significantly lower in HvAV-3e and RNase III baculovirus-infected cells (Fig. 3C, lanes 3 and 4) compared to that in mock and Vn50 baculovirus infections (Fig. 3C, lanes 1 and 2).
FIG. 3.
HvAV-3e RNase III cleaves dsRNA. (A) Expression of RNase III protein in Sf9 cells infected with RNase III recombinant baculovirus. Samples were collected at 48 and 72 hpi and analyzed by Western blotting with an antiserum against the His tag. Lane M, mock infection. An ∼80-kDa fusion protein is indicated by an arrow. (B) In vitro catalysis of Bro11 and GFP dsRNAs in the presence of recombinant RNase III. No enzyme and the Vn50 protein were used as controls. (C) In vivo catalysis of GFP dsRNA in Sf9 cells. At 48 h after the transfection of 0.5 μg of dsRNA, Sf9 cells were inoculated with Vn50 baculovirus, mock inoculated, inoculated with HvAV-3e, and inoculated with RNase III recombinant baculovirus (lanes 1, 2, 3, and 4, respectively). rRNA bands show equal loading of the total RNA samples in the gel.
RNase III autoregulates its own expression.
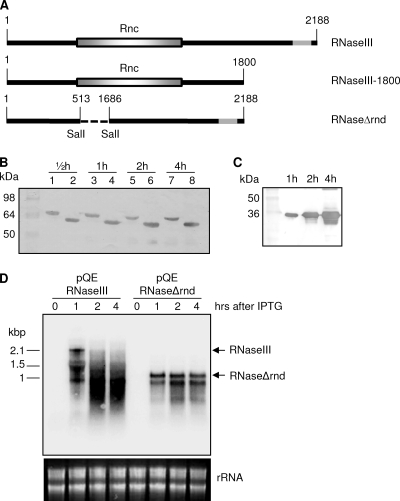
When we expressed the RNase III gene using the baculovirus expression system, we analyzed the pattern of transcript levels at different time intervals similar to that in the AV infection (Fig. 4). Similarly, we found the highest level of expression at 48 hpi with a significant decline in transcript levels from 72 hpi, which continued to decrease over time (Fig. 4). This clearly demonstrated that the stability of transcripts is independent of AV. This led us to explore the possibility of the autocatalytic activity of this protein. We tested the autocatalytic activity of RNase III in a bacterial expression system by producing three gene constructs of RNase III in pQE30 vector: a full-length construct (pQE/RNaseIII), a construct with a truncation at the 3′ end (pQE/RNaseIII-1800), and one without the endoribonuclease domain (pQE/RNaseΔrnd) (Fig. 5A). We carried out a Western blot analysis with antisera against the His tag in samples collected 30 min and 1, 2, and 4 h after IPTG induction. RNase III-associated protein expression after the addition of IPTG remained relatively unchanged over time in pQE/RNaseIII-1800 (Fig. 5B, lanes 2, 4, 6, and 8) and pQE/RNaseIII (Fig. 5B, lanes 1, 3, 5, and 7). However, overexpression of pQE/RNaseΔrnd showed significantly higher quantities of the protein produced, which increased over time (Fig. 5C). This suggested that the protein might be involved in autoregulation of RNase III transcripts. To explore this further, we found, using Northern blot analysis, cleavage products of RNase III transcripts in pQE/RNaseIII at 1 h post-IPTG induction, followed by excessive degradation at 2 and 4 h after induction (Fig. 5D), whereas the transcript levels remained the same in pQE/RNaseΔrnd (Fig. 5D). This finding further confirmed that HvAV-3e-encoded RNase III regulates its own expression by autocleaving its transcripts.
FIG. 4.
Northern blot analysis to examine expression of RNase III expressed by recombinant baculoviruses in Sf9 cells at different time intervals. A full-length RNase III probe was used for hybridization. rRNA bands show equal loading of the total RNA samples in the gel.
FIG. 5.
RNase III autoregulation. (A) Cloning strategy of three versions of HvAV-3e RNase III in pQE30 vector (pQE/RNaseIII, full length; pQE/RNaseIII-1800, C terminus removed; and pQE/RNaseΔrnd, endoND removed). (B) Western blot analyses showing bacterial expression of pQE/RNaseIII (lanes 1, 3, 5, and 7) and pQE/RNaseIII-1800 (lanes 2, 4, 6, and 8) at various times after IPTG induction. (C) pQE/RNaseΔrnd expression at 1, 2, and 4 h after IPTG addition. A monoclonal antiserum against the His tag was used for the detection of the fusion proteins in panels B and C. (D) Northern blot analysis of RNA samples extracted from cells with pQE/RNaseIII and pQE/RNaseΔrnd plasmids after IPTG induction.
RNase III is essential for virus pathology and DNA replication.
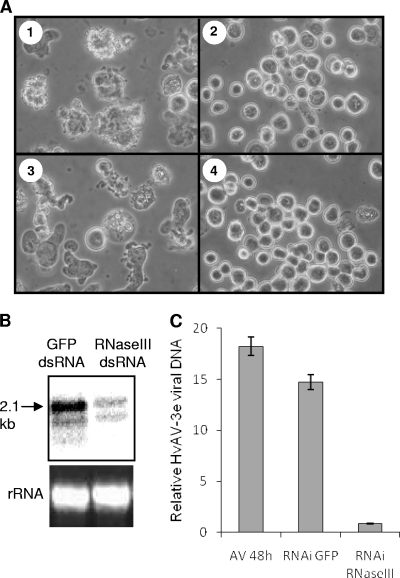
We applied RNAi to silence HvAV-3e RNase III to find out whether it is essential in virus infection. dsRNA specific to RNase III transcript was produced in vitro and transfected into HzFB cells. At 24 h after transfection, the cells were inoculated with HvAV-3e and collected at 24 and 48 hpi. HvAV-3e-associated pathology was observed in GFP dsRNA-transfected cells infected with AV; this pathology was inhibited in cells transfected with RNase III-specific dsRNA (Fig. 6A) . Downregulation of RNase III transcript levels in an RNase III knockdown treatment was confirmed by Northern hybridization (Fig. 6B). To find out whether knockdown of RNase III had any effect on virus DNA replication, we used qPCR to determine the relative quantities of viral DNA according to the treatments described above. In these analyses, we found a significant reduction in virus DNA replication in the presence of RNase III dsRNA compared to control cells that were transfected with GFP dsRNA (P = 0.003) or were mock transfected (P = 0.0005) prior to HvAV-3e infection (Fig. 6C). Moreover, in a recent study in which RNase III silencing was used as a positive control, it was shown that silencing of the gene leads to significant reductions in the detection of the major capsid protein compared to GFP dsRNA- or mock-transfected cells (see Fig. 7B in reference 18). These results suggest that RNase III is essential for HvAV-3e DNA replication and pathology.
FIG. 6.
RNase III is required for HvAV-3e pathology and DNA replication. (A) Phase-contrast images of HzFB cells at 48 hpi after mock transfection (subpanel 1) or transfection with RNase III (subpanel 2) and GFP dsRNA (subpanel 3). Subpanel 4 shows mock-transfected cells that were mock infected. (B) Northern hybridization showing downregulation of RNase III transcript levels in RNase III-specific dsRNA transfected cells after 48 h of virus infection. (C) qPCR analysis of total genomic DNA from cells treated as in panel A that shows the relative viral DNA levels in the treatments. Error bars represent the standard deviations of averages from three replicates.
FIG. 7.
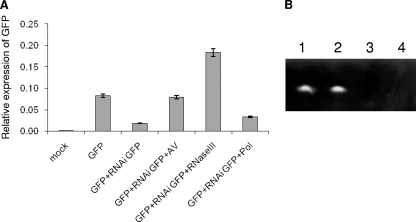
RNase III suppresses RNAi-mediated gene silencing. (A) qRT-PCR analysis of GFP expression in Sf9 cells. The cells were mock transfected (mock), transfected with pIZ/GFP (GFP), transfected with pIZ/GFP plus GFP-specific dsRNA (GFP+RNAiGFP), transfected with pIZ/GFP plus GFP-specific dsRNA and infected with HvAV-3e (GFP+RNAiGFP+AV), transfected with pIZ/GFP plus GFP-specific dsRNA and infected with RNase III recombinant baculovirus (GFP+RNAiGFP+RNaseIII), or transfected with pIZ/GFP plus GFP-specific dsRNA infected with HvAV-3e DNA polymerase (orf1) recombinant baculovirus (GFP+RNAi GFP+Pol [control]). The mRNA levels were determined and compared by setting the level observed in silenced cells to 1.0. Error bars represent the standard deviations of averages from three replicates. (B) siRNA duplex degradation by RNase III. Two different siRNA duplexes were incubated in the presence of a control protein (Vn50; lanes 1 and 2) and RNase III (lanes 3 and 4). Reactions were run on a 15% urea-PAGE gel and stained with ethidium bromide.
RNase III suppresses RNAi-mediated gene silencing.
A GFP-based silencing system was used to determine whether RNase III is involved in the suppression of RNAi-mediated gene silencing. Cotransfection of pIZ/GFP and GFP dsRNA in Sf9 cells resulted in a significant reduction in GFP transcript levels (Fig. 7A). However, when these cells were infected with HvAV-3e, the transcript levels of GFP were similar to those of pIZ/GFP. Interestingly, when GFP-silenced cells were infected with the RNase III recombinant baculovirus that overexpresses the protein, the GFP transcript levels were significantly higher than other treatments. However, a recombinant baculovirus expressing HvAV-3e orf1 (DNA polymerase), which was used as a control, did not suppress RNAi silencing of GFP (Fig. 7A). This clearly demonstrated that RNase III suppresses GFP silencing. siRNAs produced from degradation of dsRNAs mediate RNAi silencing of genes (9). To find out whether HvAV-3e RNase III degrades siRNAs, we used synthetic 21 nucleotide siRNA duplexes in enzyme assays applying purified RNase III and Vn50 (as control) proteins expressed in the baculovirus expression system. As expected, RNase III protein, but not the control protein, was able to degrade the siRNA duplexes (Fig. 7B).
DISCUSSION
Similar to other organisms, some viral genomes also encode RNase III proteins comparable to those of class I RNases containing a single endoND and a dsRBD domain. In the present study, we analyzed orf27 from HvAV-3e that encodes a functional RNase III and examined its roles in dsRNA processing, virus infection, and autoregulation.
Based on size, HvAV-3e RNase III (728 amino acids) is the largest among those reported from viruses and possesses a conserved RNase III signature motif (NYEYFEYIGD) at N terminus. Sequencing of four AV species suggests that RNase III proteins are conserved among all AVs with different percent homologies. These are closely related to RNase III from iridoviruses. Recently, it was shown that the RNase III encoded by rock bream iridovirus has activity against dsRNA (33). Our in vitro cleavage assays with the purified protein confirmed endoND activity of HvAV-3e RNase III against dsRNA, which is a characteristic feature of all RNases (11, 28). We tested two different types of dsRNAs of various lengths, one specific to Bro11 and the other to GFP. RNase III-mediated degradation of both were the same, which suggests size-independent RNase activity of the protein. The catalytic complex of class I RNase III and dsRNA is highly symmetric and contains four RNA-binding motifs (RBMs) in RNase III and three protein-interacting boxes in dsRNA. In dsRBD, RBM 1 and RBM 2 play a role in dsRNA recognition and binding, whereas in endoND, RBM 3 and RBM 4 function in substrate recognition and scissile-bond selection (16). Accordingly, dimerization of endoNDs is essential for RNase III function in that residues from one subunit (RBM 3 residues) are involved in the selection of the scissile bond, whereas those from the partner subunit are involved in the cleavage (16). Residues from each monomer catalyze two breaks in the phosphodiester backbone of dsRNA on each strand, resulting in an ∼11-nucleotide product with 2-nucleotide, 3′ overhanging ends.
Differential transcript levels of the RNase III gene were detected over different time intervals after HvAV-3e infection in HzFB cells. In our Northern blot analysis, the RNase III-specific transcript was found to be cleaved initially into two fragments at 48 hpi, followed by the appearance of a third, smaller fragment later in infection. A similar pattern was observed when we expressed this gene in a baculovirus expression system, independent of HvAV-3e. A chlorella virus (PBCV) RNase III gene was also shown to be expressed very early in infection (15 to 120 min postinfection), followed by the downregulation and disappearance of transcripts (34). Based on a previous report (32) demonstrating autoregulation of an RNase III, we investigated whether HvAV-3e RNase III protein is also able to autoregulate its own transcripts. We found several predicated stem-loop structures in orf27 that could provide possible cleavage sites for the RNase III protein. We hypothesized that orf27 regulates its own levels by cleaving hairpins in its coding region, thus destabilizing the mature transcript. The hypothesis was tested in a bacterial expression system using deletion mutations. The protein expression levels of the gene construct without the endoND (pQE/RNaseΔrnd) were significantly higher than those of the full-length or N-terminus-deleted constructs. This was found to be due to autoregulation of the transcripts in the constructs containing the endoND causing degradation of the transcripts.
It is now well documented that RNAi is a natural antiviral defense in eukaryotes and, in return, viruses encode suppressor proteins to tackle this response. Several plant and animal viruses with DNA genomes, such as geminiviruses (7) and herpesviruses (31), and with RNA genomes, such as SPCSV (12), human immunodeficiency virus type 1 (3), and influenza virus (21), have been reported to encode RNAi suppressors. We also found that HvAV-3e RNase III possesses RNAi suppressor activity since GFP silencing was reversed in the presence of this protein. Degradation of duplex siRNAs by RNase III in our in vitro assay suggests that the mechanism of suppression lies downstream of the RNAi pathway. Similarly, it was recently demonstrated that the SPCSV-encoded RNase III eliminates the antiviral defense of the host plant (sweet potato; Ipomoea batatas) by degrading double-stranded siRNAs of 21 to 24 bp into smaller fragments that are inactive in RNAi (12). Most virus-encoded RNAi suppressors are pathogenicity determinants as well, since they suppress the host immune response that facilitates the rapid replication of virus genomes (8). In our RNAi experiment, specific silencing of RNase III led to significant reduction in virus DNA replication, as well as AV symptomology. Therefore, the role of RNase III encoded by AVs is clearly in suppression of antiviral immunity boosting viral DNA replication. A recent finding on the role of a silencing suppressor in a dsDNA virus (herpes simplex virus type 1) DNA replication supports our results (31). Nevertheless, other possible role(s) for the RNase III protein cannot be ruled out (e.g., interfering with microRNA biogenesis). Recently, we showed that expression of a subset of host cell (HzFB) microRNAs (miRNA) was downregulated after HvAV-3e infection at 24 and 48 hpi when RNase III expression is at its peak and later upregulated (18). If the protein is involved in the degradation of miRNA duplexes, then its function has to be specific against certain subset of miRNAs rather than blocking the process in general. This, however, requires further investigations. To our knowledge, this is the first insect virus-encoded RNase III described and shown to suppress the host cell RNAi defense mechanism.
Acknowledgments
This project was partly funded by Horticulture Australia, Ltd. (VG06044), to S.A. and a University of Queensland Ph.D. Scholarship to M.H.
Footnotes
Published ahead of print on 13 January 2010.
REFERENCES
- 1.Asgari, S., J. Davis, D. Wood, P. Wilson, and A. McGrath. 2007. Sequence and organization of the Heliothis virescens ascovirus genome. J. Gen. Virol. 88:1120-1132. [DOI] [PubMed] [Google Scholar]
- 2.Asgari, S., G. Zhang, R. Zareie, and O. Schmidt. 2003. A serine proteinase homolog venom protein from an endoparasitoid wasp inhibits melanization of the host hemolymph. Insect Biochem. Mol. Biol. 33:1017-1024. [DOI] [PubMed] [Google Scholar]
- 3.Bennasser, Y., S.-Y. Le, M. Benkirane, and K.-T. Jeang. 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22:607-619. [DOI] [PubMed] [Google Scholar]
- 4.Bideshi, D. K., M. V. Demattei, F. Rouleux-Bonnin, K. Stasiak, Y. Tan, S. Bigot, Y. Bigot, and B. A. Federici. 2006. Genomic sequence of Spodoptera frugiperda ascovirus 1a, an enveloped, double-stranded DNA insect virus that manipulates apoptosis for viral reproduction. J. Virol. 80:11791-11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bideshi, D. K., Y. Yeping Tan, Y. Bigot, and B. A. Federici. 2005. A viral caspase contributes to modified apoptosis for virus transmission. Gene Dev. 19:1416-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bigot, Y., S. Renault, J. Nicolas, C. Moundras, M. V. Demattei, S. Samain, D. K. Bideshi, and B. A. Federici. 2009. Symbiotic virus at the evolutionary intersection of three types of large DNA viruses; iridoviruses, ascoviruses, and ichnoviruses. PLoS One 4:e6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisaro, D. M. 2006. Silencing suppression by geminivirus proteins. Virology 344:158-168. [DOI] [PubMed] [Google Scholar]
- 8.Brigneti, G., O. Voinnet, W.-X. Li, L.-H. Ji, S.-W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Buchon, N., and C. Vaury. 2006. RNAi: a defensive RNA-silencing against viruses and transposable elements. Heredity 96:195-202. [DOI] [PubMed] [Google Scholar]
- 10.Catala, M., B. Lamontagne, S. Larose, G. Ghazal, and S. A. Elela. 2004. Cell cycle-dependent nuclear localization of yeast RNase III is required for efficient cell division. Mol. Biol. Cell 15:3015-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conrad, C., and R. Rauhut. 2002. Ribonuclease III: new sense from nuisance. Int. J. Biochem. Cell Biol. 34:116-129. [DOI] [PubMed] [Google Scholar]
- 12.Cuellar, W. J., J. F. Kreuze, M.-L. Rajamäkia, K. R. Cruzado, M. Untiveros, and J. P. T. Valkonen. 2009. Elimination of antiviral defense by viral RNase III. Proc. Natl. Acad. Sci. U. S. A. 106:10354-10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danin-Kreiselman, M., C. Y. Lee, and G. Chanfreau. 2003. RNase III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell 11:1279-1289. [DOI] [PubMed] [Google Scholar]
- 14.Drider, D., and C. Condon. 2004. The continuing story of endoribonuclease III. J. Mol. Microbiol. Biotechnol. 8:195-200. [DOI] [PubMed] [Google Scholar]
- 15.Federici, B. A., D. K. Bideshi, Y. Tan, T. Spears, and Y. Bigot. 2009. Ascoviruses: superb manipulators of apoptosis for viral replication and transmission. Curr. Top. Microbiol. Immunol. 328:171-196. [DOI] [PubMed] [Google Scholar]
- 16.Gan, J., J. E. Tropea, B. P. Austin, D. L. Court, D. S. Waugh, and X. Ji. 2006. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell 124:355-366. [DOI] [PubMed] [Google Scholar]
- 17.Han, J., Y. Lee, K.-H. Yeom, Y.-K. Kim, H. Jin, and V. N. Kim. 2004. The Drosha-DGCR8 complex in primary microRNA processing. Gene Dev. 18:3016-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussain, M., and S. Asgari. 2010. Functional analysis of a cellular microRNA in insect host-ascovirus interaction. J. Virol. 84:612-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King, L. A., and R. D. Possee. 1992. The baculovirus expression system: a laboratory guide. Chapman & Hall, London, England.
- 20.Kreuze, J. F., E. I. Savenkov, W. J. Cuellar, X. Li, and J. P. T. Valkonen. 2005. Viral class 1 RNase III involved in suppression of RNA silencing. J. Virol. 79:7227-7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, W.-X., H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. A. Brydon, K. L. Johnson, A. Garcia-Sastre, L. A. Ball, P. Palese, and S.-W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. U. S. A. 101:1350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacRae, I. J., and J. A. Doudna. 2007. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 17:138-145. [DOI] [PubMed] [Google Scholar]
- 23.McIntosh, A. H., and C. M. Ignoffo. 1981. Replication and infectivity of the single-embedded nuclear polyhedrosis virus, Baculovirus heliothis, in homologous cell lines. J. Invertebr. Pathol. 37:258-264. [Google Scholar]
- 24.Meister, G., and T. Tuschl. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431:343-349. [DOI] [PubMed] [Google Scholar]
- 25.Nicholson, A. W. 1999. Function, mechanism, and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 23:371-390. [DOI] [PubMed] [Google Scholar]
- 26.Régnier, P., and M. Grunberg-Manago. 1989. Cleavage by RNase III in the transcripts of the metY-nusA-infB operon of Escherichia coli releases the tRNA and initiates the decay of the downstream mRNA. J. Mol. Biol. 210:293-302. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Saunders, L. R., and G. N. Barber. 2003. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB 17:961-983. [DOI] [PubMed] [Google Scholar]
- 29.Stasiak, K., S. Renault, M.-V. Demattei, Y. Bigot, and B. A. Federici. 2003. Evidence for the evolution of ascoviruses from iridoviruses. J. Gen. Virol. 84:2999-3009. [DOI] [PubMed] [Google Scholar]
- 30.Wang, L., J. Xue, C. P. Seaborn, B. M. Arif, and X.-W. Cheng. 2006. Sequence and organization of the Trichoplusia ni ascovirus 2C (Ascoviridae) genome. Virology 354:167-177. [DOI] [PubMed] [Google Scholar]
- 31.Wu, Z., Y. Zhu, D. M. Bisaro, and D. S. Parris. 2009. Herpes simplex virus type 1 suppresses RNA-induced gene silencing in mammalian cells. J. Virol. 83:6652-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu, W., J. Huang, and S. N. Cohen. 2008. Autoregulation of AbsB (RNase III) expression in Streptomyces coelicolor by endoribonucleolytic cleavage of absB operon transcripts. J. Bacteriol. 190:5526-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zenke, K., and K. H. Kim. 2008. Functional characterization of the RNase III gene of rock bream iridovirus. Arch. Virol. 153:1651-1656. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y., I. Calin-Jageman, J. R. Gurnon, T.-J. Choi, B. Adams, A. W. Nicholson, and J. L. Van Ettena. 2003. Characterization of a chlorella virus PBCV-1 encoded ribonuclease III. Virology 317:73-83. [DOI] [PubMed] [Google Scholar]