Abstract
The receptor for parathyroid hormone (PTHR) is a main regulator of calcium homeostasis and bone maintenance. As a member of class B of G protein-coupled receptors, it harbors a large extracellular domain, which is required for ligand binding. Here, we demonstrate that the PTHR extracellular domain is cleaved by a protease belonging to the family of extracellular metalloproteinases. We show that the cleavage takes place in a region of the extracellular domain that belongs to an unstructured loop connecting the ligand-binding parts and that the N-terminal 10-kDa fragment is connected to the receptor core by a disulfide bond. Cleaved receptor revealed reduced protein stability compared with noncleaved receptor, suggesting degradation of the whole receptor. In the presence of the agonistic peptides PTH(1–34), PTH(1–14), or PTH(1–31), the processing of the PTHR extracellular domain was inhibited, and receptor protein levels were stabilized. A processed form of the PTHR was also detected in human kidney. These findings suggest a new model of PTHR processing and regulation of its stability.
Keywords: G Proteins/Coupled Receptors (GPCR), Hormones/Peptide, Proteases/Metalloprotease, Protein/Degradation, Protein/Post-translational Modification, Protein/Stability, Receptors/Modification, Receptors/Regulation, Parathyroid Hormone Receptor Type 1
Introduction
Parathyroid hormone (PTH),2 an 84-amino acid polypeptide, is a key regulator of blood calcium levels controlling mineral ion homeostasis and bone metabolism. In response to low calcium or high phosphate plasma levels, PTH is synthesized in and secreted from the parathyroid glands. The primary effector organs for PTH are kidney and bone. In the kidney, PTH stimulates reabsorption of calcium from renal tubules, stimulates 1,25-dihydroxyvitamine D synthesis, and prevents reabsorption of phosphate. In bone, PTH mediates bone resorption by osteoclasts and reduces osteoblast proliferation, resulting in calcium liberation and decreased bone mass (1). However, intermittent administration of full-length PTH or a N-terminal fragment of PTH, PTH(1–34), results in enhanced bone formation and increased bone density, providing a valuable option for the treatment of severe osteoporosis (2).
The multiple effects of PTH are mainly mediated by the activation of the PTH receptor type 1 (PTHR), which belongs to the class B G protein-coupled receptor family (3). Upon activation by an agonist, PTHR triggers at least two signaling pathways: Gq/11-mediated phospholipase Cβ stimulation, leading to inositol 1,4,5-trisphosphate production, calcium mobilization, and protein kinase C activation, and Gs-mediated activation of adenylyl cyclase, resulting in cAMP production and cAMP-dependent protein kinase activation (4–6).
Like other members of the class B receptor family, the PTHR has a relatively long N-terminal extracellular domain of ∼200 amino acids. Significant efforts have been undertaken to reveal the mechanisms of ligand binding and activation of the PTHR. The N-terminal 34-residue fragment of PTH (PTH(1–34)) is capable of fully activating the receptor to the same degree as full-length PTH (7). Although the first 14 amino acids of PTH were found to directly bind to the transmembrane domain of the PTHR and activate the receptor, amino acids 15–34 bind to the extracellular domain (8–10). From these data, a two-step activation model was derived where PTH first binds to the extracellular domain, resulting in a conformational change of the extracellular domain, and a subsequent binding of the N-terminal parts of PTH to the receptor core leading to activation of the receptor (11–13). Structural analysis revealed the extracellular domain to be consisting of a three-layer α-β-β-α fold forming a hydrophobic groove into which parts of PTH can bind (10). Three conserved disulfide bonds are required for proper folding of the extracellular domain and for ligand binding (10, 14, 15). Although the N-terminal and the juxtamembrane parts of the receptor N terminus are structured, x-ray crystallography as well as studies investigating the functional role of segments of the extracellular domain demonstrated that an intermediate stretch of 44 residues within the extracellular domain, corresponding to exon E2 of the PTHR gene, is not required for ligand binding (10, 16).
Here we reveal a new post-translational modification of the PTHR. We demonstrate that the PTHR ectodomain is subject to proteolytic cleavage within a region that most likely corresponds to exon E2. We further show that this cleavage can be inhibited by PTH.
EXPERIMENTAL PROCEDURES
Materials
Lipofectamine 2000 was from Invitrogen. Human PTH (1–31), [Nle8,18,Tyr34]PTH (1–34), and [d-Trp12,Tyr34]PTH (7–34) were from Bachem, and human [Aib1,3,M]PTH (1–14) was a kind gift from Dr. Thomas Gardella (Massachusetts General Hospital, Boston, MA). Monoclonal HA.11 anti-hemagglutinin (HA) antibody was purchased from Covance, anti-HA, and anti-FLAG affinity agarose, monoclonal anti-β-actin, and monoclonal anti-FLAG M2 antibody were from Sigma-Aldrich. Anti-mouse and anti-rabbit peroxidase-conjugated secondary antibodies were obtained from Dianova. Cy2-conjugated anti-mouse antibody was from Jackson Immuno Research Lab. Endoglycosidase H (Endo H) and neuraminidase were from New England Biolabs. N-Glycosidase F (PNGase F) was purchased from Roche Applied Science. EXPRE [35S]protein labeling mix was from PerkinElmer Life Sciences. All of the cell culture media were obtained from PAN Biotech. GM6001, MMP3 inhibitor 2, and BB2516 (Marimastat) were obtained from Calbiochem. TIMP-1 and TIMP-2 were from Enzo Life Sciences. TNF-484 was a kind gift from Dr. Olga Dolnik (Institute of Virology, Philipps-University Marburg). All other reagents were from Sigma-Aldrich or Applichem.
cDNA Constructs
Human PTHR harboring a HA epitope in exon E2 (17) was subcloned into pcDNA3 (Invitrogen) plasmid using the restriction sites EcoRI and SalI. To generate a receptor mutant with two epitope tags in the extracellular domain, a FLAG epitope was inserted after the signal peptide into the HA-PTHR by PCR using the sense primer GAT TAT AAA GAT GAT GAT GAT AAA TAC GCG CTG GTG GAT GCA and the antisense primer CGG CTT ATC ATC ATC ATC TTT ATA ATC CGC GGA GCT GAG CAC GGG. All of the constructs were confirmed by sequencing.
Cell Culture and Transfection
Chinese hamster ovary (CHO) cells and rat osteosarcoma ROS 17.2/8 cells were maintained in 1:1 Dulbecco's modified Eagle's medium/Ham's F-12 medium containing 10% (v/v) fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. The cells were maintained at 37 °C in a humidified atmosphere of 5% CO2, 95% air. To generate stable cell lines, the cells were transfected with PTHR, HA-PTHR, or FLAG-HA-PTHR in pcDNA3 using Lipofectamine 2000 according to the manufacturer's instructions. 48 h after transfection, the cells were selected in culture medium supplemented with 1 mg/ml G418 for approximately 2 weeks. Clonal cell lines were derived from limited dilution series and screened for expression of the desired protein by Western blot.
Antibody Generation
We previously generated polyclonal antisera against the C-terminal tail of the human PTHR (18). The identity of the peptide used for the immunization of the rabbits was EEASGPERPPALLQEEWETVM. The specificity of the antisera was initially tested using immunodot blot analysis. For subsequent analysis, antibodies were affinity-purified against their immunizing peptide using the Pierce SulfoLink Immobilization Kit for Peptides (Thermo Scientific) according to the instructions of the manufacturer. In initial dot blot analyses and preliminary immunohistochemical investigations, the antiserum (antiserum 1781) displayed high affinity along with strong and specific immunostaining.
Ligand Stimulation Experiments
Subconfluently grown cells in 12-well dishes were stimulated with different PTH peptides as indicated. For time series, the ligands were added in a reversed temporal order starting with the longest incubation time to guarantee the same control point for all of the samples. Subsequently, the cells were lysed in SDS sample buffer, heated for 20 min at 60 °C, and subjected to SDS-PAGE and Western blot analysis.
Immunoprecipitation Assays and Western Blotting
Immunoprecipitation and Western blots were performed as described elsewhere (19). Briefly, the cells were lysed in ice-cold radioimmune precipitation assay buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% (w/v) Nonidet P-40, 0.1% (w/v) SDS, 0.5% (w/v) sodium deoxycholate, 5 mm EDTA) supplemented with a mix of protease inhibitors (10 μg/ml soybean trypsin inhibitor, 30 μg/ml benzamidine, 1 mg/ml leupeptin, 100 μm phenylmethylsulfonyl fluoride). The lysates were cleared by centrifugation at 20,000 × g for 30 min at 4 °C, and the supernatant was incubated for 2 h with anti-HA affinity agarose or anti-FLAG affinity agarose. The precipitates were collected by gentle centrifugation and washed three times in cold radioimmune precipitation assay buffer. The proteins were eluted with SDS sample buffer, subjected to SDS-PAGE, and the proteins were transferred onto polyvinylidene difluoride membranes (Millipore). The blots were incubated with primary antibodies as indicated, and bound antibodies were visualized with secondary antibodies and ECL Plus Western blotting detection reagent (GE Healthcare) in accordance with the manufacturer's instructions.
Metabolic Labeling of the PTHR
Pulse-chase experiments were essentially done as described elsewhere (19). Briefly, the cells stably expressing HA-PTHR were starved for 1 h in Met/Cys-depleted Dulbecco's modified Eagle's medium (PAN Biotech) and labeled for 1 h in Dulbecco's modified Eagle's medium containing [35S]Met/Cys (150 μCi/ml). The cells were washed twice in phosphate-buffered saline and chased with complete culture medium supplemented with 2 mm methionine and 2 mm cysteine for 1–12 h in the absence or presence of 100 nm PTH(1–34). Subsequently, the cells were lysed, PTHR was precipitated with anti-HA affinity agarose as described above, and the precipitates were separated by SDS-PAGE. The gels were Coomassie-stained, soaked with Amplify fluorographic solution (GE Healthcare), and dried. Radiolabeled HA-PTHR was visualized and quantified by autoradiography on a PMI phosphorimager (Bio-Rad).
Receptor Deglycosylation
PTHR was precipitated with anti-HA affinity agarose as described above. The precipitates were eluted from the affinity resin and denatured with 1% (w/v) SDS, 50 mm sodium phosphate, pH 7.5, for 40 min at room temperature. Before the enzyme reaction, the eluates were diluted 10-fold with either buffer E (50 mm sodium phosphate, pH 5.5, 50 mm EDTA, 0.5% (w/v) dodecylmaltoside, 1% 2-mercaptoethanol; Endo H), buffer P (50 mm sodium phosphate, pH 7.5, 50 mm EDTA, 1% Triton X-100, 1% 2-mercaptoethanol; PNGase F), or buffer N (50 mm sodium phosphate, pH 6.0; Neuraminidase). All of the buffers were supplemented with a mix of protease inhibitors (10 μg/ml soybean trypsin inhibitor, 30 μg/ml benzamidine, 1 mg/ml leupeptin, 100 μm phenylmethylsulfonyl fluoride). The enzymes were added at final concentrations of 5 units/ml PNGase F, 250 units/ml Endo H, or 50 units/ml neuraminidase. The samples were incubated at 37 °C for 16 h, and the reaction was terminated by adding SDS sample buffer.
Receptor Enrichment from Kidney Tissue
Fresh tissue from human kidney was homogenized in ice-cold 50 mm Tris (pH 7.4) supplemented with a mix of protease inhibitors (10 μg/ml soybean trypsin inhibitor, 30 μg/ml benzamidine, 1 mg/ml leupeptin, 100 μm phenylmethylsulfonyl fluoride) using an Ultra-Turrax tissue homogenizer followed by brief sonification. Homogenates were diluted 1:1 with radioimmune precipitation assay buffer, incubated on a rotary wheel for 1 h at 4 °C, and cleared by centrifugation at 20,000 × g for 30 min at 4 °C. The supernatant was incubated for 3 h with wheat germ agglutinin-agarose. The precipitates were collected by gentle centrifugation and washed three times in ice-cold radioimmune precipitation assay buffer. The proteins were eluted with elution buffer (50 mm NaPO4, pH 7.5, 0.5 m N-acetylglucosamine, 10 mm EDTA, 1% Nonidet P-40 (w/v), 0.1% (w/v) SDS) and concentrated with a Microcon centrifugal filter device (10,000 molecular weight cut-off; Millipore).
Immunocytochemistry and Confocal Imaging
CHO cells stably expressing HA-PTHR were grown on coverslips overnight. The cells were then exposed to different ligands for 30 min as indicated. The cells were fixed with 4% paraformaldehyde and 0.2% picric acid in 0.1 m phosphate buffer, pH 6.9, for 30 min at room temperature and washed five times in phosphate-buffered saline. For permeabilization the cells were incubated for 5 min in methanol. After 10 min of preincubation in phosphate-buffered saline containing 0.35% bovine serum albumin, the cells were incubated with anti-HA antibody at a dilution of 1:1000 in phosphate-buffered saline containing 0.35% bovine serum albumin for 1 h at 37 °C. Bound primary antibody was detected with Cy2-labeled goat anti-mouse IgG (1:200). The specimens were examined using a Leica SP2 laser scanning confocal microscope.
Generation of Dynamin Wild Type and K44A Recombinant Adenoviruses
Wild type dynamin (DynWT) and dynamin K44A (DynK44A) cDNAs in the pCB1 vector were kindly provided by Dr. Marc Caron (Duke University, Durham, NC). Dynamin cDNA fragments were excised from pCB1 plasmids and cloned between the EcoRI and XmaI sites of vector pIRES2-AcGFP1-Nuc (Clontech). Thereafter, the entire cassettes containing dynamin, IRES, and nuclear green fluorescent protein sequences were removed and cloned into the EcoRI/NotI sites of pENTR vector (Invitrogen) and subsequently introduced into adenoviral vectors (pAD/CMV/V5-DEST; Invitrogen) by homologous recombination. Recombinant adenoviral vectors were generated by standard procedures following the manufacturer's instructions. Adenovirus titers were determined by measuring absorbance at 260 nm with a spectrophotometer.
RESULTS
Prolonged Activation of the PTHR Results in Increased Molecular Mass Forms of the Receptor
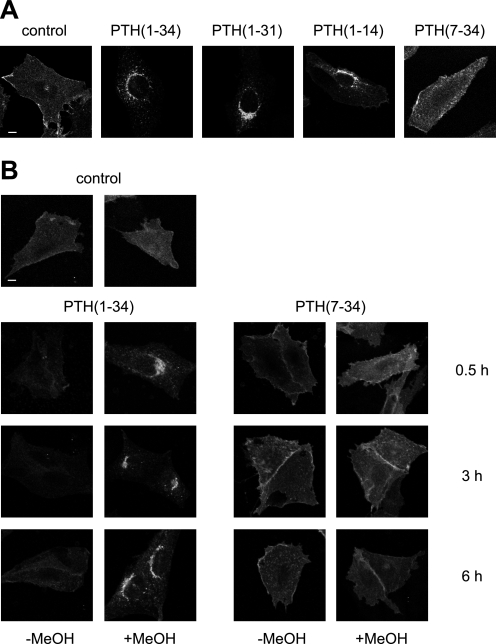
Although PTH(1–34) and PTH(1–14) have been described to fully activate the PTHR with almost equal potency compared with full-length PTH, PTH(1–31) has been reported to stimulate adenylyl cyclase activity but not to activate protein kinase C (20, 21). PTH(7–34) is a competitive antagonist to PTH(1–34) and is believed to have no intrinsic activity (22). First, we investigated the capacity of these four PTH peptides to induce PTHR internalization. CHO cells stably expressing HA-tagged PTHR were incubated with saturating concentrations of PTH(1–34), PTH(1–31), PTH(1–14), or PTH(7–34) for 30 min. To monitor PTHR trafficking, the cells were fixed, PTHR was stained with an anti-HA antibody followed by a Cy2-coupled secondary antibody, and confocal images of each condition were taken. Although in unstimulated cells the receptor was mainly distributed at the cell surface, treatment with the agonistic peptides PTH(1–34), PTH(1–31), or PTH(1–14) resulted in a perinuclear accumulation of immunoreactive vesicles indicating a rapid internalization of PTHR. In contrast, the antagonistic peptide PTH(7–34) did not significantly internalize the receptor (Fig. 1A). To assess how prolonged stimulation affected PTHR internalization and to assess the extent of internalization, CHO cells stably expressing HA-tagged PTHR were incubated with 100 nm PTH(1–34) or 1 μm PTH(7–34) for 30 min, 3 h, or 6 h. The cells were fixed and either permeabilized with methanol or left unpermeabilized to detect receptor at the cell surface only. PTHR was visualized as described above. As shown in Fig. 1B, PTH(1–34) almost completely internalized the receptor from the cell surface. Also, continuous application of PTH(1–34) for up to 6 h did not lead to a significant recovery of the receptor levels at the cell surface. In contrast, neither given as a short pulse nor applied for up to 6 h did PTH(7–34) have an effect on PTHR internalization.
FIGURE 1.
Ligand-induced internalization of the PTHR. A, CHO cells stably expressing HA-PTHR were treated with 100 nm PTH(1–34), 1 μm PTH(1–31), 3 μm PTH(1–14), or 1 μm PTH(7–34) for 30 min. Subsequently, the cells were fixed, permeabilized, and stained with mouse anti-HA antibody followed by a Cy2-labeled anti-mouse antibody. HA-PTHR was visualized by confocal microscopy. B, CHO cells stably expressing HA-PTHR were left untreated (control) or were treated with 100 nm PTH(1–34) or 1 μm PTH(7–34) for 30 min, 3 h, or 6 h. The cells were fixed and either permeabilized with MeOH or left unpermeabilized. White scale bars represent 5 μm. PTHR was visualized as described above.
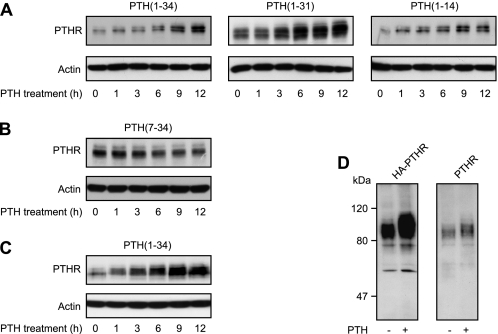
Next, we investigated the effect of prolonged application of PTH on the PTHR protein. CHO cells stably expressing HA-tagged PTHR were incubated with saturating concentrations of different PTH peptide fragments for 30 min to 12 h. PTHR expression was monitored in Western blots from whole cell lysates and was detected as a broad band with an approximate molecular mass of 90 kDa. Interestingly, with the progressive presence of agonistic peptides an increasing fraction of the PTHR appeared to have a slower electrophoretic mobility compared with unstimulated receptors (Fig. 2A). Moreover, in PTH(1–34)-, PTH(1–31)-, and PTH(1–14)-treated cells, an increased immunoreactive signal was detected, suggesting an increase of PTHR protein levels. Repeating these experiments in the presence of cycloheximide to block protein synthesis did not diminish the PTH(1–34) evoked increase in protein levels (supplemental Fig. S1), thus suggesting that PTH(1–34) rather reduces the degradation of PTHR than stimulates protein expression. In contrast, cells treated with the antagonistic peptide PTH(7–34) exhibited major changes neither of PTHR mass nor of protein levels (Fig. 2B). To confirm these observations in a more physiological setting, we used rat osteosarcoma cells that stably expressed HA-PTHR. Again, a shift in the electrophoretic mobility of the PTHR was observed after prolonged stimulation with PTH(1–34) along with an increasing signal strength (Fig. 2C).
FIGURE 2.
Prolonged agonist stimulation results in reduced electrophoretic mobility of the PTHR. A and B, CHO cells stably expressing HA-PTHR were treated with 100 nm PTH(1–34), 1 μm PTH(1–31), 3 μm PTH(1–14), or 1 μm PTH(7–34) for the indicated times. Subsequently, the cells were lysed, and the proteins were separated by SDS-PAGE and blotted to polyvinylidene difluoride membranes. PTHR was detected in Western blots with an anti-HA antibody. C, ROS 17/2.8 cells stably expressing HA-PTHR were treated with 100 nm PTH(1–34) for the indicated times. The cells were processed as described above. D, CHO cells stably expressing HA-PTHR or wild type PTHR were treated with 100 nm PTH(1–34) as indicated for 12 h and processed as above. PTHR was detected in Western blot using antibodies detecting an epitope at the PTHR C terminus. The positions of molecular weight standards are marked on the left.
The HA-tagged PTHR, which was used in the above experiments, has been shown to have similar properties to wild type PTHR in terms of ligand binding and signaling (16, 17). However, to exclude that the observed effects of PTH were caused by the modification of the PTHR with an HA epitope, we compared CHO cells stably expressing either HA-tagged PTHR or wild type human PTHR. To visualize wild type PTHR, we generated an antiserum against the final 21 amino acids in the C terminus of the human PTHR; this antiserum detected human HA-PTHR as well as human wild type PTHR with high sensitivity and specificity (18). After 12 h of incubation with 100 nm PTH(1–34), an increased molecular mass was detected for HA-PTHR (Fig. 2D, left panel) as well as for wild type PTHR (Fig. 2D, right panel) compared with unstimulated receptor. This indicated that the observed difference of molecular mass was not affected by the HA epitope and suggests that the stimulation of PTHR does indeed change its electrophoretic mobility and increase its levels.
PTHR Shift Is Not Mediated by Changes in Glycosylation
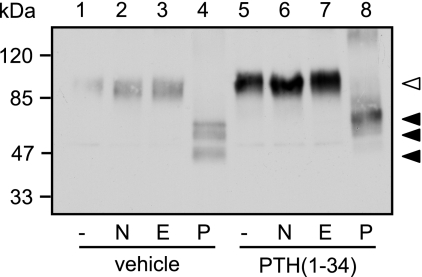
Previous studies revealed that some ligands may facilitate the maturation of vasopressin 2 receptor mutants and the δ opioid receptor by acting as molecular chaperones. These ligands promoted glycosylation of immature receptors, which are retained in the endoplasmic reticulum. Therefore in these two examples mature and immature receptors differ in their glycosylation pattern. Although immature receptors were susceptible to deglycosylation by Endo H and by PNGase F, mature receptors were resistant to Endo H (23, 24). PTHR is highly glycosylated at four N-glycosylation sites constituting ∼30% of the apparent molecular mass of the mature receptor (25). To test whether a similar mechanism of molecular chaperoning could underlie our findings for the PTHR, we assessed the glycosylation pattern of unstimulated and PTH-treated PTHR. CHO cells expressing HA-PTHR were stimulated with PTH(1–34) for 12 h, HA-PTHR was immunoprecipitated using anti-HA agarose, and precipitates were treated with neuraminidase, Endo H, or PNGase F. As can be seen in Fig. 3, neither neuraminidase nor Endoglycosidase H affected the mobility of the PTHR without or with prior PTH treatment (lanes 2 and 3 versus lanes 6 and 7). PNGase F treatment, however, resulted in three different bands with approximate Mr values of 50,000, 57,000, and 60,000 (lane 4). In cells that had been treated with PTH, only a single band with a Mr of 60,000 was observed after PNGase F deglycosylation (lane 8). Thus, the change in mobility of the PTHR after prolonged PTH treatment does not appear to be due to altered glycosylation because it was also seen after PNGase F-induced deglycosylation. Moreover, complete deglycosylation with PNGase F revealed an apparent difference of ∼10 kDa between the smallest and the largest receptor proteins.
FIGURE 3.
Glycosylation does not contribute to the PTH-induced changes in molecular mass of the PTHR. CHO cells stably expressing HA-PTHR were treated with 100 nm PTH(1–34) for 12 h as indicated. The cells were lysed, and PTHR was precipitated using anti-HA affinity beads. Precipitated proteins were incubated with 50 units/ml of neuraminidase (N), 250 units/ml of Endo H (E), or 5 units/ml of PNGase F (P) for 16 h. Enzyme reactions were stopped by the addition of SDS sample buffer, and proteins were separated by SDS-PAGE. HA-PTHR was detected in Western blot analysis. The empty arrowhead indicates a fully glycosylated receptor form. The closed arrowheads indicate the different receptor forms after PNGase treatment. The positions of molecular weight standards are marked on the left.
PTH Prevents Degradation of the PTHR
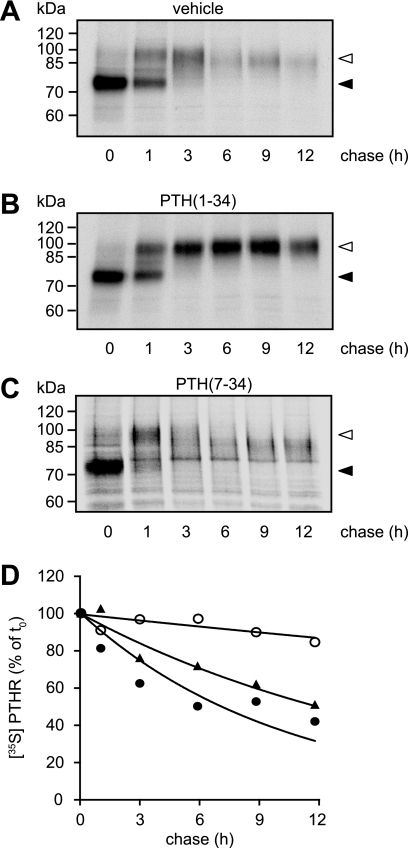
Because glycosylation seemed not to be responsible for the different molecular weight species of the PTHR, we set out to monitor PTHR maturation in the absence or presence of PTH by metabolic labeling. CHO cells stably expressing HA-PTHR were metabolically labeled with [35S]methionine/cysteine and then chased in medium containing unlabeled amino acids. The chase medium was supplemented with PTH(1–34) or PTH(7–34) or without any ligand. Subsequently, the cells were lysed, and the receptors were isolated by immunoprecipitation and detected by SDS-PAGE and autoradiography. Two major forms of the PTHR were detected: a Mr 75,000 species, which was present directly after the pulse and most probably represents core-glycosylated receptor, and a Mr 100,000 species, which appeared after 1 h of chase and was the predominant form after 3 h, therefore presumably representing fully glycosylated PTHR (Fig. 4, A–C). Interestingly, in the course of chase in the absence of PTH, the Mr 100,000 form progressively lost mass, resulting in an effective molecular weight of ∼90,000 after 12 h of chase (Fig. 4A). In contrast, when PTH(1–34) was present during the chase, the molecular weight of the mature PTHR remained constant at 100,000 (Fig. 4B). When we applied the antagonist PTH(7–34) to the chase medium, no stabilization of the PTHR protein was detectable, comparable with the situation without any ligand (Fig. 4C). Moreover, when comparing the stability of the radiolabeled PTHR over time, a strong difference in protein half-life was observed: although in the absence of ligand and in the presence of PTH(7–34) PTHR had half-lives of 12 and 7 h, respectively, PTH(1–34) led to a dramatic stabilization resulting in a half-life estimated at >60 h (Fig. 4D).
FIGURE 4.
PTH stabilizes the high molecular mass form of the PTHR. A–C, CHO cells stably expressing HA-PTHR were pulse-labeled with [35S]methionine/cysteine-containing medium (150 μCi/ml) and then chased for the indicated times with medium containing nonradioactive methionine and cysteine in the absence (A) or presence of 100 nm PTH(1–34) (B) or 1 μm PTH(7–34) (C). HA-PTHR was precipitated from cell lysates using HA antibody. The samples were analyzed by SDS-PAGE and fluorography of the dried gels with a Bio-Rad phosphorimager. The closed arrowheads indicate newly synthesized PTHR; the open arrowheads indicate fully mature PTHR. D, decay of radiolabeled PTHR was quantified using a monoexponential decay function. ▴, untreated; ○, PTH(1–34); ●, PTH(7–34). The means of three independent experiments are shown.
Together with the above findings, these results indicate that the PTHR protein undergoes a degradative process during its lifetime, resulting in a reduced protein mass and ultimately leading to reduced protein stability. This process can be blocked by the presence of PTH(1–34) but not of PTH(7–34).
PTHR Is Cleaved within the Extracellular Domain
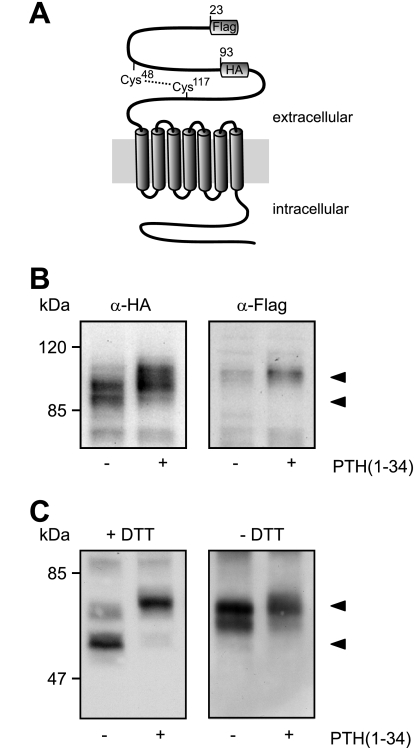
Taking into account that changes in receptor glycosylation were not responsible for the different electrophoretic mobilities of the PTHR and that the receptor mass decreased over time, we speculated whether a proteolytic event might be occurring. In such a case, the extracellular N terminus or the intracellular C terminus would most likely be affected. However, as we demonstrated that an antibody directed against the PTHR C terminus still was capable of detecting both the high and the low molecular weight PTHR form (Fig. 2D), a cleavage in the PTHR C terminus seemed unlikely. The HA epitope, which was used for detection of the PTHR, had been inserted into the extracellular domain in exon E2 of the PTHR (amino acids 93–101). Both the fully processed and the apparently degraded forms of the receptor were equally well detected with an anti-HA antibody (Fig. 2, A–C), giving rise to the assumption that a potential proteolytic cleavage should occur in the PTHR extracellular domain N-terminal from the HA epitope. To investigate this, we generated a PTHR mutant containing a FLAG epitope directly after the signal peptide and an HA epitope in exon E2 (Fig. 5A). CHO cells stably expressing this receptor construct were treated with PTH(1–34) for 12 h, and Western blots of cell lysates were probed with anti-HA and anti-FLAG antibodies. As expected, in the HA blots a Mr 90,000 band and a Mr 100,000 band were detectable in cells without and with PTH incubation, respectively (Fig. 5B, left panel). In contrast, the anti-FLAG antibody detected only a single mobility form of Mr 100,000, which was very faint in the unstimulated sample and more prominent in the PTH-treated sample (Fig. 5B, right panel). This strongly suggests that the N-terminal part of the PTHR ectodomain, harboring the FLAG epitope, is cleaved in untreated cells and thus cannot be detected in anti-FLAG Western blots.
FIGURE 5.
Modification of the PTHR is restricted to the extracellular domain. A, schematic representation of the PTHR. A FLAG epitope was inserted after the signal sequence (amino acids 22 and 23), and an HA epitope was inserted replacing amino acids 93–101. A disulfide bridge between Cys48 and Cys117 stabilizing the extracellular domain is depicted by a dotted line. B, CHO cells stably expressing the FLAG-HA-PTHR were stimulated with PTH(1–34) for 12 h. The cells were lysed, and the samples were subjected to SDS-PAGE and subsequent Western blot analysis. PTHR was detected using an anti-HA antibody (left panel) or an anti-FLAG-M2 antibody (right panel). C, CHO cells stably expressing the HA-PTHR were stimulated with PTH(1–34) for 12 h. To gain better resolution of the proteins, the cells were treated 12 h prior to and throughout the experiment with 5 μg/ml tunicamycin to block N-glycosylation. The cells were lysed in SDS-buffer in the presence (left panel) or absence (right panel) of 100 mm dithiothreitol (DTT). The lysates were subjected to SDS-PAGE and subsequent Western blot analysis. PTHR was detected using an anti-HA antibody. The PTHR forms are indicated by arrowheads.
As mentioned above, the extracellular domain of the PTHR is structured by three disulfide bridges (10, 14). One of these disulfide bridges is formed between Cys48 and Cys117 (Fig. 5A); this bridge might stabilize the cleaved fragment under native conditions. To test this hypothesis, CHO cells expressing HA-tagged PTHR were stimulated for 12 h with PTH(1–34). To obtain better protein separation in the subsequent SDS-PAGE, the cells were treated for 12 h prior to and throughout the whole experiment with tunicamycin to block glycosylation of the PTHR. The cell lysates were prepared in the presence or absence of 100 mm dithiothreitol as a reducing agent, and PTHR was detected in Western blot using an anti-HA antibody. Under reducing conditions, a band at 55 kDa in unstimulated cells and a band at 65 kDa in PTH-treated cells were detected, most likely representing the cleaved and the uncleaved receptor forms, respectively (Fig. 5C, left panel). Under nonreducing conditions, in both unstimulated and stimulated cells only a major band of 65 kDa was detected. Taken together, these findings indicate that the PTHR is proteolytically cleaved within the N terminus. The cleaved fragment is apparently under native conditions tethered to the receptor core by a single disulfide bond.
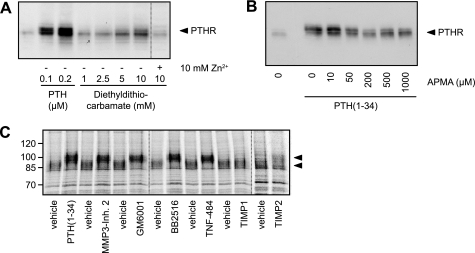
PTHR Cleavage Is Mediated by Zinc-dependent Metalloproteinases
Assuming that a proteolytic cleavage was responsible for PTHR cleavage, as suggested by our above findings, we searched for the responsible enzyme. Therefore, a number of known inhibitors of proteinases were tested. CHO cells expressing HA-tagged PTHR were incubated with medium containing protease inhibitors or PTH(1–34) for 6 h. The cells were lysed, and HA-PTHR was detected in Western blot. No reduced cleavage was observed with inhibitors of cysteine, serine, and aspartate proteases (10 μm pepstatin, 1 μm aprotinin, 100 μm leupeptin, 40 μm ALLM, 100 mm AEBSF, 50 mm N-ethylmaleimide, and 1 mg/ml α2-macroglobulin; data not shown). However, significant inhibition of PTHR cleavage was observed for metal ion-chelating inhibitors; diethyldithiocarbamate blocked PTHR cleavage at a concentration of 5–10 mm, and this blockage was reversed by the addition of equimolar amounts of zinc ions (Fig. 6A). Similarly, another ion chelator, 1,10-phenanthroline, also prevented PTHR cleavage (data not shown). The effect of both inhibitors was concentration-dependent. The organomercurial compound p-aminophenyl-mercuric acetate (APMA) is an effective activator of metalloproteinases (26, 27). To test whether APMA would provoke PTHR cleavage in the presence of PTH, CHO cells expressing HA-PTHR were incubated with PTH(1–34) for 12 h, and different concentrations of APMA were added to the medium thereafter and incubated for 6 h. The cells were lysed, and HA-PTHR was analyzed in Western blots. APMA overcame the PTH-induced inhibition of PTHR cleavage in a concentration-dependent manner, resulting in a PTHR size pattern comparable with unstimulated PTHR (Fig. 6B). A further characterization of the protease responsible for PTHR cleavage was done by testing various inhibitors of metalloproteinases. For this, the cells were pulse-labeled with [35S]methionine/cysteine as described above and chased for 8 h. During the chase MMP3 inhibitor 2, GM6001, Marimastat, TNF-484, TIMP-1, or TIMP-2 was added to the medium. PTH(1–34) served as a positive control. All of the pharmacological inhibitors of matrix metalloproteinases (MMPs) inhibited PTHR cleavage comparable with PTH(1–34), as was observed from the increased electrophoretic mobility of the labeled receptor. Also TIMP-2 prevented cleavage of the receptor to some extent. Only TIMP-1 did not reveal any inhibitory effect on the cleavage (Fig. 6C). Taken together, these data strongly indicate that the protease responsible for the cleavage of the PTHR extracellular domain belongs to the proteinase superfamily of zinc-dependent metalloproteinases.
FIGURE 6.
PTHR is cleaved by an extracellular, zinc-dependent protease. A, CHO cells stably expressing HA-PTHR were treated for 6 h with increasing concentrations of PTH(1–34) or diethyldithiocarbamate in the absence or presence of equimolar concentrations of zinc phosphate. The cells were lysed, and the lysates were subjected to SDS-PAGE and subsequent Western blot analysis. PTHR was detected using an anti-HA antibody. B, CHO cells stably expressing HA-PTHR were treated for 12 h with PTH(1–34). APMA was added to the medium in increasing concentrations, and the cells were incubated for 6 h. Subsequently, the cells were lysed, and the lysates were subjected to SDS-PAGE and subsequent Western blot analysis using an anti-HA antibody. C, CHO cells stably expressing HA-PTHR were pulse-labeled with [35S]methionine/cysteine-containing medium (150 μCi/ml) and then chased for 8 h with nonradioactive medium containing PTH(1–34) (100 nm), GM6001 (20 μm), MMP3 inhibitor 2 (20 μm), Marimastat/BB2515 (20 μm), TNF-484 (20 μm), TIMP-1 (40 nm), or TIMP-2 (40 nm). HA-PTHR was precipitated from cell lysates using HA antibody. The samples were analyzed by SDS-PAGE and fluorography of the dried gels with a Bio-Rad phosphorimager. The arrowheads indicate cleaved and uncleaved PTHR species.
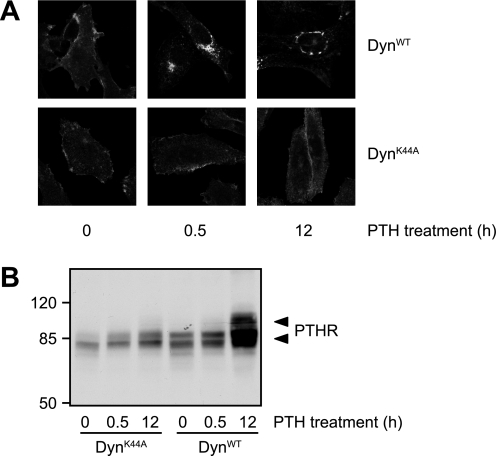
Agonists Prevent PTHR Cleavage by Internalizing the Receptor
As shown above, only agonists that internalized the PTHR were able to prevent its N-terminal cleavage, and the responsible protease most likely resides in the extracellular space. Thus, we rationalized that the mechanism through which agonists could prevent cleavage might involve the removal of receptor from the cell surface. To test this hypothesis, we blocked PTH-induced internalization of the PTHR and monitored receptor cleavage. For this, we took advantage of a dominant negative mutant of dynamin (DynK44A) that blocks clathrin-dependent internalization of membrane proteins (28). To obtain maximal expression of wild type and K44A dynamin, CHO cells stably expressing HA-PTHR were infected using an adenoviral vector. To monitor the extent of internalization suppression, infected cells were treated with PTH(1–34) for 30 min or 12 h, and HA-PTHR was detected by immunofluorescence. Although in cells expressing DynWT PTHR was internalized in a fashion similar to noninfected cells and was detected in intracellular vesicles after 30 min and 12 h of stimulation, in DynK44A-infected cells no significant internalization of the receptor occurred (Fig. 7A, compared with Fig. 1). PTHR cleavage of DynWT- or DynK44A-infected cells was assessed by Western blot analysis of the PTHR. Fig. 7B shows that although in DynWT-infected cells PTH(1–34) inhibited PTHR cleavage significantly after 12 h, no high molecular weight form of the receptor was detected in DynK44A-infected cells after PTH stimulation. Thus, PTH blocks receptor cleavage by removing the receptor from the cell surface, thereby making it inaccessible for extracellular proteases.
FIGURE 7.
PTH inhibits ectodomain cleavage by internalization of the PTHR. CHO cells stably expressing HA-PTHR were infected with adenovirus encoding for DynWT or dynamin K44A (DynK44A). 48 h after the infection, the cells were stimulated with 100 nm PTH(1–34) for 30 min or 12 h. A, cells were fixed, permeabilized, and stained with mouse anti-HA antibody followed by a Cy2-labeled anti-mouse antibody. HA-PTHR was visualized by confocal microscopy. B, cells were lysed, and the proteins were separated by SDS-PAGE and blotted to polyvinylidene difluoride membranes. PTHR was detected in Western blots with an anti-HA antibody. The positions of molecular weight standards are marked on the left. The arrowheads indicate cleaved and uncleaved PTHR species.
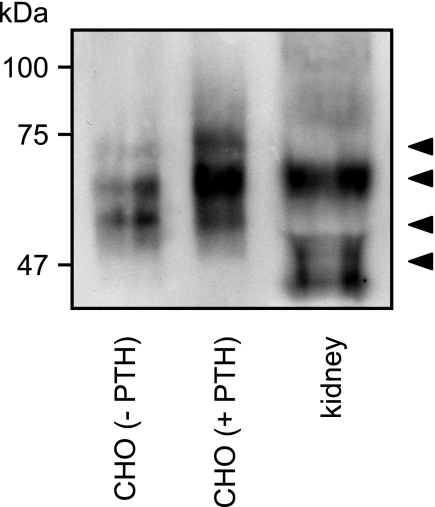
Detection of a Cleaved Form of the PTHR in Human Kidney
So far, all of the experiments were performed with human PTHR in cell culture. To investigate whether the cleavage of the PTHR ectodomain also occurs in vivo, we examined PTHR from human kidney. To be able to detect the PTHR, we enriched them using lectin affinity purification. To better assess the molecular weight of the receptors, the proteins were deglycosylated subsequently with PNGase F and separated by reducing SDS-PAGE. PTHR was detected by Western blot analysis using anti-PTHR-CT antibody. As a control, Fig. 8 shows that in CHO cells PTH(1–34) shifted the PTHR from a low molecular form of 60 kDa to a high molecular weight form of ∼70–75 kDa. In human kidney, the major fraction of the PTHR was similar to the major band of PTH-treated CHO cells, i.e. resembling uncleaved receptor. However, a minor fraction of the receptor from human kidney was detected at a lower molecular weight, which was comparable with the lower forms of cleaved PTHR from unstimulated CHO cells. Because the receptors were detected with antibodies directed against the PTHR-CT, this strongly suggests that also in vivo the PTHR is a target to cleavage of the N-terminal domain.
FIGURE 8.
Detection of a cleaved form of the PTHR in human kidney. Glycoproteins from human kidney tissue or from CHO cells stably expressing HA-PTHR, which had been treated for 12 h with 100 nm PTH(1–34), were enriched by wheat germ agglutinin affinity purification. Thereafter proteins were deglycosylated with PNGase F for 16 h. The enzyme reactions were stopped by the addition of SDS sample buffer, and the proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. PTHR was detected by Western blot using an anti-PTHR-CT antiserum. The arrowheads indicate cleaved and uncleaved PTHR species.
DISCUSSION
PTH has differential effects on bone metabolism. It has been long known that persistently elevated PTH levels lead to bone resorption, thereby liberating calcium and organic phosphate and elevating plasma calcium levels. In contrast, an intermittent application of exogenous PTH has been proven to stimulate bone apposition, thereby preventing bone loss. Thus, recombinant PTH(1–34) is being used for the treatment of severe osteoporosis. However, the exact underlying molecular mechanisms are still not fully understood. Here we investigated the direct effect of continuous application of PTH on the PTHR. We found that prolonged stimulation with agonists led to an apparently increased molecular weight and to increased protein levels of HA-tagged as well as wild type PTHR. For some G protein-coupled receptors, an accumulation of immature receptor species in the endoplasmic reticulum has been described. However, alterations in the glycosylation pattern of the receptor, which have been ascribed to immature receptor species, were not found for the PTHR. Therefore, our initial hypothesis that this phenomenon might be due to a chaperoning effect evoked by PTH on immature receptor trapped in the endoplasmic reticulum could not be held up. This is in line with a previous report documenting that impaired glycosylation of the PTHR does not fundamentally affect the functionality and localization of the receptor (29). Notably, other post-translational modifications, which can lead to an increased protein mass, such as SUMOylation, ubiquitination, or phosphorylation, were either not detectable or did not contribute to the PTH-induced molecular weight shift of the PTHR (data not shown). Interestingly, when investigating the steady-state levels of PTHR protein, we observed a progressive decrease of molecular weight of PTHR, which had not been activated by an agonist, from an initial Mr 100,000 form to a Mr 90,000 form. A similar mass change was observed when the receptor was treated with the antagonist PTH(7–34). PTH(1–34), however, prevented this effect and significantly reduced PTHR protein turnover. These findings would explain our initial observation in whole cell lysates of an apparent increase of molecular mass during the presence of PTH; while in the presence of PTH, the Mr 100,000 protein was stabilized and accumulated over time, whereas under unstimulated conditions only the Mr 90,000 form was present.
Several lines of evidence suggest that the loss of molecular weight of the PTHR is caused by proteolytic cleavage occurring within the PTHR ectodomain; an antibody directed against the last 20 amino acids was able to detect the high and the low molecular form of PTHR, ruling out a cleavage in the C terminus of the receptor. Using a PTHR mutant harboring two epitope tags in the N-terminal domain, we showed that only the very N-terminal FLAG tag was lost from the receptor, whereas the HA tag more proximal to the receptor core remained. The loss of the N-terminal fragment of the PTHR extracellular domain was dependent on reducing conditions, indicating that the cleaved fragment is tethered in vivo to the receptor by a disulfide bridge, most likely between Cys48 and Cys117. Although the exact position of the cleavage could not be resolved, based on the size of the fragment of ∼10 kDa, the position of both epitope tags and the fact that cysteine residues stabilized the N-terminal fragment, we propose a cleavage occurring within residues 50–90 of the PTHR. This region was found to be within a flexible, unstructured loop of the PTHR extracellular domain, which would allow easy attachment of proteases (10). Interestingly, amino acid sequence comparison of the PTH/secretin/calcitonin receptor family revealed that residues 61–105 of the PTHR, which are encoded by exon E2, were not conserved in the other members of the class B G protein-coupled receptors (25). A similar finding has been described for the family of glycoprotein receptors, where the thyrotropin receptor was found to contain a motif in the extracellular domain not present in other family members (30). Most notably, it has long been known that the respective region in thyrotropin receptor is cleaved by a still not identified extracellular protease, resulting in a two-subunit receptor (31, 32). Based on our present findings, we propose a similar mechanism for the PTHR. This underlines the remarkable biochemical and evolutionary similarities between both receptor families.
Two possible explanations emerged for how ligands could prevent receptor cleavage. Agonist binding to the receptor might directly interfere with protease cleavage. However, because PTH(1–14), which also blocked PTHR cleavage, only binds to the transmembrane region of the PTHR, this explanation seems to be unlikely (9). All three agonists used in this study, but not the antagonist PTH(7–34), rapidly internalized the PTHR. Moreover, when we blocked agonist-evoked receptor internalization with dominant negative dynamin, the PTHR was cleaved irrespective of agonist binding. Thus, receptor internalization caused by these agonists appears to be involved in the inhibition of receptor cleavage, presumably by removing the receptor from the cell surface and restricting the access of extracellular proteases.
Notably, cleavage of the extracellular domain reduced the overall stability of the PTHR protein, and this was reversed by agonist application. The most likely explanation for this finding would be that the N-terminal domain is required for the stabilization of the whole receptor protein. Interestingly, these findings are in contrast to the common concept that prolonged agonist exposure generally reduces receptor numbers and thereby blunts long term signaling. Here, we rather propose that, depending on the surrounding environment, receptors that remain “unused” for a certain time are cleaved and then degraded, thereby driving cells into a state of unresponsiveness to PTH.
Because PTHR cleavage occurred with nonactivated PTHR at the cell surface, it was reasonable to assume that extracellular proteases would be responsible. No significant reduction of PTHR cleavage was achieved by classical inhibitors of cysteine, serine, and aspartate proteases. Instead, several lines of evidence indicate that a metalloproteinase is responsible for PTHR cleavage; the two metal ion chelators 1,10-phenanthroline and diethyldithiocarbamate, which inhibit a wide range of metalloproteinases by binding zinc ions, reduced PTHR cleavage to a significant extent. Moreover, the addition of APMA, an organo-mercuric compound acting as an activator of metalloproteinases (27), to PTH-treated cells provoked a rapid cleavage of PTHR.
More selective inhibitors of MMPs and ADAMs (a disintegrin and metalloprotease) also blocked PTHR cleavage. However, matrix metalloproteinases are a large family with overlapping functions. This makes the exact identification of the protease difficult. The hydroxamate-derivates GM6001, MMP3 inhibitor 2, and Marimastat possess inhibitory effects against several MMPs. However, at least Marimastat and GM6001 also block several members of the ADAM family. TNF-484 inhibits tumor necrosis factor-α converting enzyme (ADAM17) but can also inhibit some MMPs (33). TIMP-2, which is an inhibitor of almost all MMPs and of tumor necrosis factor-α converting enzyme, prevented PTHR cleavage. Only TIMP-1, which predominantly inhibits MMPs 1, 3, and 9, did not prevent PTHR cleavage. Thus, at present it remains to be seen whether only a single protease is responsible for PTHR cleavage or whether several members from the metalloproteinase family can substitute for each other.
More experiments will be needed to find out whether and how the cleavage is necessary for the biological activity of the PTHR. Our identification of cleaved receptor in human kidney shows that this process takes place in vivo. Moreover, several lines of evidence suggest that MMPs influence the effects of PTH on bone and vice versa; MMPs generally are involved in the degradation of extracellular matrix. In bone these proteases have been shown to be crucial for resorption and remodeling processes (34–36). PTH has been shown to positively regulate the expression of MMP9 and MMP13 (37–40). Conversely, inhibition of MMPs activity by TIMP1 increased the osteogenic effect of PTH mainly by reducing osteoclastic activity (41, 42). Thus, a direct influence of MMPs on PTHR biology seems plausible.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Gardella (Massachusetts General Hospital) for providing the human [Aib1,3,M]PTH (1–14). We are grateful to Michaela Hoffmann for excellent technical assistance and to Dr. Wolfgang Garten and Dr. Olga Dolnik (Institute of Virology, Philipps-University Marburg) for providing TNF-484 inhibitor and for helpful discussions on metalloproteases. We also want to thank Dr. Hubertus Riedmiller (Department of Urology, University of Würzburg) for providing the human kidney samples.
This work was supported by grants from the Deutsche Forschungsgemeinschaft and the European Research Council (to M. J. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PTH
- parathyroid hormone
- APMA
- p-aminophenyl-mercuric acetate
- HA
- hemagglutinin
- HA-PTHR
- HA-tagged PTHR
- MMP
- matrix metalloproteinase
- PTHR
- parathyroid hormone receptor type 1
- TIMP
- tissue inhibitor of matrix metalloproteinases
- Endo H
- endoglycosidase H
- PNGase
- F,N-glycosidase F
- CHO
- Chinese hamster ovary
- DynWT
- wild type dynamin
- DynK44A
- dynamin K44A.
REFERENCES
- 1.Potts J. T. (2005) J. Endocrinol. 187, 311–325 [DOI] [PubMed] [Google Scholar]
- 2.Neer R. M., Arnaud C. D., Zanchetta J. R., Prince R., Gaich G. A., Reginster J. Y., Hodsman A. B., Eriksen E. F., Ish-Shalom S., Genant H. K., Wang O., Mitlak B. H. (2001) N. Engl. J. Med. 344, 1434–1441 [DOI] [PubMed] [Google Scholar]
- 3.Foord S. M., Bonner T. I., Neubig R. R., Rosser E. M., Pin J. P., Davenport A. P., Spedding M., Harmar A. J. (2005) Pharmacol. Rev. 57, 279–288 [DOI] [PubMed] [Google Scholar]
- 4.Abou-Samra A. B., Jüppner H., Force T., Freeman M. W., Kong X. F., Schipani E., Urena P., Richards J., Bonventre J. V., Potts J. T., Jr. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 2732–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pines M., Fukayama S., Costas K., Meurer E., Goldsmith P. K., Xu X., Muallem S., Behar V., Chorev M., Rosenblatt M., Tashjian A. H., Jr., Suva L. J. (1996) Bone 18, 381–389 [DOI] [PubMed] [Google Scholar]
- 6.Offermanns S., Iida-Klein A., Segre G. V., Simon M. I. (1996) Mol. Endocrinol. 10, 566–574 [DOI] [PubMed] [Google Scholar]
- 7.Tregear G. W., Van Rietschoten J., Greene E., Keutmann H. T., Niall H. D., Reit B., Parsons J. A., Potts J. T., Jr. (1973) Endocrinology 93, 1349–1353 [DOI] [PubMed] [Google Scholar]
- 8.Gardella T. J., Jüppner H., Wilson A. K., Keutmann H. T., Abou-Samra A. B., Segre G. V., Bringhurst F. R., Potts J. T., Jr., Nussbaum S. R., Kronenberg H. M. (1994) Endocrinology 135, 1186–1194 [DOI] [PubMed] [Google Scholar]
- 9.Luck M. D., Carter P. H., Gardella T. J. (1999) Mol. Endocrinol. 13, 670–680 [DOI] [PubMed] [Google Scholar]
- 10.Pioszak A. A., Xu H. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5034–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castro M., Nikolaev V. O., Palm D., Lohse M. J., Vilardaga J. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16084–16089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vilardaga J. P., Bünemann M., Krasel C., Castro M., Lohse M. J. (2003) Nat. Biotechnol. 21, 807–812 [DOI] [PubMed] [Google Scholar]
- 13.Gensure R. C., Gardella T. J., Jüppner H. (2005) Biochem. Biophys. Res. Commun. 328, 666–678 [DOI] [PubMed] [Google Scholar]
- 14.Grauschopf U., Lilie H., Honold K., Wozny M., Reusch D., Esswein A., Schäfer W., Rücknagel K. P., Rudolph R. (2000) Biochemistry 39, 8878–8887 [DOI] [PubMed] [Google Scholar]
- 15.Karpf D. B., Arnaud C. D., Bambino T., Duffy D., King K. L., Winer J., Nissenson R. A. (1988) Endocrinology 123, 2611–2620 [DOI] [PubMed] [Google Scholar]
- 16.Lee C., Gardella T. J., Abou-Samra A. B., Nussbaum S. R., Segre G. V., Potts J. T., Kronenberg H. M., Jüppner H. (1994) Endocrinology 135, 1488–1495 [DOI] [PubMed] [Google Scholar]
- 17.Castro M., Dicker F., Vilardaga J. P., Krasel C., Bernhardt M., Lohse M. J. (2002) Endocrinology 143, 3854–3865 [DOI] [PubMed] [Google Scholar]
- 18.Lupp A., Klenk C., Röcken C., Evert M., Mawrin C., Schulz S. (2010) Eur. J. Endocrinol., in press [DOI] [PubMed] [Google Scholar]
- 19.Klenk C., Humrich J., Quitterer U., Lohse M. J. (2006) J. Biol. Chem. 281, 8357–8364 [DOI] [PubMed] [Google Scholar]
- 20.Jouishomme H., Whitfield J. F., Gagnon L., Maclean S., Isaacs R., Chakravarthy B., Durkin J., Neugebauer W., Willick G., Rixon R. H. (1994) J. Bone Miner Res. 9, 943–949 [DOI] [PubMed] [Google Scholar]
- 21.Shimizu N., Guo J., Gardella T. J. (2001) J. Biol. Chem. 276, 49003–49012 [DOI] [PubMed] [Google Scholar]
- 22.Goldring S. R., Roelke M. S., Bringhurst F. R., Rosenblatt M. (1985) Biochemistry 24, 513–518 [DOI] [PubMed] [Google Scholar]
- 23.Morello J. P., Salahpour A., Laperrière A., Bernier V., Arthus M. F., Lonergan M., Petäjä-Repo U., Angers S., Morin D., Bichet D. G., Bouvier M. (2000) J. Clin. Invest. 105, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petäjä-Repo U. E., Hogue M., Bhalla S., Laperrière A., Morello J. P., Bouvier M. (2002) EMBO J. 21, 1628–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schipani E., Karga H., Karaplis A. C., Potts J. T., Jr., Kronenberg H. M., Segre G. V., Abou-Samra A. B., Jüppner H. (1993) Endocrinology 132, 2157–2165 [DOI] [PubMed] [Google Scholar]
- 26.Santavicca M., Noel A., Angliker H., Stoll I., Segain J. P., Anglard P., Chretien M., Seidah N., Basset P. (1996) Biochem. J. 315, 953–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galazka G., Windsor L. J., Birkedal-Hansen H., Engler J. A. (1996) Biochemistry 35, 11221–11227 [DOI] [PubMed] [Google Scholar]
- 28.van der Bliek A. M., Redelmeier T. E., Damke H., Tisdale E. J., Meyerowitz E. M., Schmid S. L. (1993) J. Cell Biol. 122, 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bisello A., Greenberg Z., Behar V., Rosenblatt M., Suva L. J., Chorev M. (1996) Biochemistry 35, 15890–15895 [DOI] [PubMed] [Google Scholar]
- 30.Wadsworth H. L., Chazenbalk G. D., Nagayama Y., Russo D., Rapoport B. (1990) Science 249, 1423–1425 [DOI] [PubMed] [Google Scholar]
- 31.Loosfelt H., Pichon C., Jolivet A., Misrahi M., Caillou B., Jamous M., Vannier B., Milgrom E. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 3765–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka K., Chazenbalk G. D., McLachlan S. M., Rapoport B. (1999) J. Biol. Chem. 274, 33979–33984 [DOI] [PubMed] [Google Scholar]
- 33.Trifilieff A., Walker C., Keller T., Kottirsch G., Neumann U. (2002) Br. J. Pharmacol. 135, 1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaissé J. M., Engsig M. T., Everts V., del Carmen Ovejero M., Ferreras M., Lund L., Vu T. H., Werb Z., Winding B., Lochter A., Karsdal M. A., Troen T., Kirkegaard T., Lenhard T., Heegaard A. M., Neff L., Baron R., Foged N. T. (2000) Clin. Chim. Acta 291, 223–234 [DOI] [PubMed] [Google Scholar]
- 35.Delaissé J. M., Andersen T. L., Engsig M. T., Henriksen K., Troen T., Blavier L. (2003) Microsc. Res. Tech. 61, 504–513 [DOI] [PubMed] [Google Scholar]
- 36.Andersen T. L., del Carmen Ovejero M., Kirkegaard T., Lenhard T., Foged N. T., Delaissé J. M. (2004) Bone 35, 1107–1119 [DOI] [PubMed] [Google Scholar]
- 37.Luo X. H., Liao E. Y., Su X., Wu X. P. (2004) J. Bone Miner. Metab. 22, 19–25 [DOI] [PubMed] [Google Scholar]
- 38.Porte D., Tuckermann J., Becker M., Baumann B., Teurich S., Higgins T., Owen M. J., Schorpp-Kistner M., Angel P. (1999) Oncogene 18, 667–678 [DOI] [PubMed] [Google Scholar]
- 39.Peeters-Joris C., Hammani K., Singer C. F. (1998) Biochim. Biophys. Acta 1405, 14–28 [DOI] [PubMed] [Google Scholar]
- 40.Kawashima-Ohya Y., Satakeda H., Kuruta Y., Kawamoto T., Yan W., Akagawa Y., Hayakawa T., Noshiro M., Okada Y., Nakamura S., Kato Y. (1998) Endocrinology 139, 2120–2127 [DOI] [PubMed] [Google Scholar]
- 41.Geoffroy V., Marty-Morieux C., Le Goupil N., Clement-Lacroix P., Terraz C., Frain M., Roux S., Rossert J., de Vernejoul M. C. (2004) J. Bone Miner Res. 19, 811–822 [DOI] [PubMed] [Google Scholar]
- 42.Merciris D., Schiltz C., Legoupil N., Marty-Morieux C., de Vernejoul M. C., Geoffroy V. (2007) Bone 40, 75–83 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.