Abstract
Nuclear factor E2-related factor 2 (Nrf2) is a cap-n-collar basic leucine zipper (CNC-bZIP) transcription factor that is well established as a master regulator of phase II detoxification and antioxidant gene expression and is strongly expressed in tissues involved in xenobiotic metabolism including liver and kidney. Nrf2 is also abundantly expressed in adipose tissue; however, the exact function of Nrf2 in adipocyte biology is unclear. In the current study we show that targeted knock-out of Nrf2 in mice decreases adipose tissue mass, promotes formation of small adipocytes, and protects against weight gain and obesity otherwise induced by a high fat diet. In mouse embryonic fibroblasts, 3T3-L1 cells, and human subcutaneous preadipocytes, selective deficiency of Nrf2 impairs adipocyte differentiation. Deficiency of Nrf2 also leads to decreased expression of peroxisome proliferator-activated receptor γ (PPARγ), CCAAT enhancer-binding protein α (C/EBPα), and their downstream targets during adipocyte differentiation. Conversely, activation of Nrf2 in 3T3-L1 cells by stable knockdown of its negative regulator Keap1 enhances and accelerates hormone-induced adipocyte differentiation. Transfection of Nrf2 stimulates Pparγ promoter activity, and stable knockdown of Keap1 enhances PPARγ expression in 3T3-L1 cells. In addition, chromatin immunoprecipitation studies show that Nrf2 associates with consensus binding sites for Nrf2 in the Pparγ promoter. These findings demonstrate a novel biologic role for Nrf2 beyond its participation in detoxification and antioxidant pathways and place Nrf2 within the limited network of transcription factors that control adipocyte differentiation by regulating expression of PPARγ.
Keywords: Adipocyte, Chromatin Immunoprecipitation (ChIP), Differentiation, Gene Knockout, Gene Regulation, Obesity, Oxidative Stress, shRNA, NFE2L2, Antioxidant Response Element
Introduction
Fat cells play an important role in energy storage and metabolism and secrete a variety of factors that influence appetite, insulin sensitivity, inflammation, and many other pathways of biologic and clinical significance (1). Because of the significance of adipocyte biology in the pathogenesis of obesity and related metabolic, cardiovascular, and inflammatory disorders, there has been intense interest in defining the network of transcription factors that controls the expression of genes involved in fat cell development. Peroxisome proliferator-activated receptors (PPARs)4 are ligand-activated transcription factors that belong to a nuclear hormone receptor family including related molecules that function by forming heterodimers with retinoid X receptors (2). PPARγ is expressed abundantly in adipose tissue and is considered to be the dominant transcriptional regulator of adipogenic differentiation (3). Accordingly, much attention has been directed at identifying factors that control PPARγ expression during the process of adipogenesis.
CCAAT enhancer-binding proteins (C/EBPs) are leucine zipper transcription factors expressed in both white and brown adipose tissue and have been extensively studied for their roles in regulating PPARγ activity and adipogenesis (4, 5). C/EBPβ and C/EBPδ are transiently expressed at the onset of the adipose differentiation program (6, 7). This phase is then followed by induction of PPARγ and C/EBPα expression (8–10). In addition, PPARγ and C/EBPα form a positive loop by regulating each other's expression (11, 12). Although the enforced expression of C/EBPα in fibroblasts can trigger adipocytic differentiation, C/EBPα is incapable of inducing adipogenesis in the absence of PPARγ (13). In contrast, PPARγ can induce adipogenic differentiation in C/EBPα-null cells indicating that PPARγ is proximal in effecting adipogenesis (14). Apart from C/EBPβ, -δ, and -α, relatively few transcription factors have been described that bind to the Pparγ promoter and positively regulate Pparγ transcription during adipogenesis (15).
The nuclear factor erythroid-derived 2-related factor 2 (Nrf2) is a member of the cap-n-collar basic leucine zipper (CNC-bZIP) family of transcription factors and has been shown to play an important role in mediating cytoprotective responses to oxidative stress and electrophilic xenobiotics (16). Nrf2 forms heterodimers with small-maf proteins to up-regulate expression of genes encoding antioxidant enzymes and detoxifying proteins through the cis-active sequences known as antioxidant response elements. Although Nrf2 is dispensable for growth and development in mouse, Nrf2-deficient mice are sensitized to oxidative stress-related pathologies in various organs (16). Consistent with its role in xenobiotic metabolism, Nrf2 is highly expressed in tissues such as the liver, lung, kidney, and intestine. High levels of Nrf2 are also found in fat tissues (17, 18). In addition, Nrf2 expression is induced by a prolonged high fat diet (19). However, a potential role for Nrf2 in adipocyte development or lipid metabolism has not been fully explored. Here we investigated the role of Nrf2 in adipocyte differentiation. Mice deficient in Nrf2 display decreased fat mass in association with small adipocytes and are resistant to diet-induced obesity. Mouse embryonic fibroblasts (MEFs) deficient in Nrf2 show impaired adipogenesis, and in 3T3-L1 and human subcutaneous preadipocytes, shRNA-mediated knockdown of Nrf2 expression inhibits adipocyte differentiation. In addition, we further show that 1) PPARγ expression is Nrf2-dependent, 2) Nrf2 physically associates with the Pparγ promoter in vivo and stimulates Pparγ promoter activity, and 3) the impaired adipogenesis induced by knock-out or inhibition of Nrf2 is related at least in part to down-regulation of PPARγ expression. These studies provide evidence for a heretofore-unrecognized role of Nrf2 in adipogenesis and identify Nrf2 as a potentially important transcriptional regulator of PPARγ during the process of adipocyte differentiation.
EXPERIMENTAL PROCEDURES
Reagents
Insulin, dexamethasone, 3-isobutylmethylxanthine, indomethacin, and Oil Red O (ORO) were purchased from Sigma. Rosiglitazone maleate was from GlaxoSmithKline. Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), and calf serum were purchased from Invitrogen. Antibodies for Nrf2 (sc-13032; 1:500), lipoprotein lipase (sc-32382; 1:500), adipsin (ADPSN, sc-12402; 1:500), and C/EBPα (sc-61; 1:500) were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies for PPARγ (#2435; 1:1000) and fatty acid-binding protein 4 (aP2; #2120; 1:500) were from Cell Signaling Technology (Danvers, MA). Antibodies for Keap1 (MAB3024; 1:500) and fatty acid translocase (Cd36; ab36977; 1:1000) were from R&D Systems (Minneapolis, MN) and Abcam (Cambridge, MA), respectively. Antibodies for β-actin (A1978; 1:2000) and lamin A (L1293; 1:5000) were from Sigma. Pparγ2 reporter plasmid was a generous gift from Dr. Wen Xie (University of Pittsburgh) (20).
Animals and Feeding Studies
The Nrf2 knock-out (Nrf2 KO) mice, which are on C57BL6;129SV mix background, have been described previously (17). Wild type and Nrf2 KO littermates were maintained on a 12-h/12-h light/dark cycle. Food and water were provided ad libitum. Age-matched mice were housed together and provided either a regular diet (11% fat by calories, 4.0 kcal/g; Purina) or high fat diet (HFD; 41% fat by calories, 4.7 kcal/g; Research Diets). Body weights were monitored weekly. Food intake measurements were done on mice that were individually housed.
Gene Expression Analysis and Western Blotting
For quantitation of mRNA levels, total RNA was isolated and subjected to reverse transcription-PCR or real-time reverse transcription-PCR analyses using protocols described previously (21, 22). Gene-specific primers used are listed in supplemental Tables 1 and 2. Protein lysates were made using Nonidet P-40 lysis buffer and electrophoresed on SDS-polyacrylamide gels. Proteins were subsequently transferred onto nitrocellulose membranes and blocked in 5% milk at room temperature for 1 h. Primary antibody incubation was done overnight at 4 °C followed by incubation with a horseradish peroxidase-conjugated secondary antibody. Proteins were visualized using a chemiluminescent detection system (Pierce).
Locomotor Activity Measurements
Mice were housed individually in a beam-break locomotor activity chamber (San Diego Instruments, San Diego, CA). Ambulations were scored as interruption of beams in 30-min intervals and converted to meters based on the distance between the beams.
Cell Culture and Differentiation
MEFs were isolated as previously described (21) and were cultured in high glucose DMEM with 50 units/ml penicillin, 50 μg/ml streptomycin, and 10% calf serum until they became confluent. Induction of adipocyte differentiation in MEFs was achieved by culturing 2-day post-confluent cells in DMEM supplemented with 10% FBS, 0.5 mm 3-isobutylmethylxanthine, 10 μg/ml insulin, and 1 μm dexamethasone for 2 days. Cells were subsequently cultured in DMEM containing 10% FBS and insulin alone. 3T3-L1 preadipocytes were obtained from American Type Culture Collection (Manassas, VA). 3T3-L1 cells were differentiated 1 day after they became confluent (designated as day 0) by replacing calf serum with FBS and by adding 1 μm dexamethasone, 0.5 mm 3-isobutylmethylxanthine, 5 μg/ml insulin, 125 μm indomethacin, and 1 μm rosiglitazone maleate to the medium. After 3 days, cells were maintained for an additional 2–4 days in the same medium without additives, and fresh medium was replenished every 2 days. Cells were harvested on the indicated days in the figure legend after the induction for further analysis. Subcutaneous human preadipocytes were obtained from Zen-Bio, Inc. (Research Triangle Park, NC) and cultured in preadipocyte medium (Zen-Bio, catalog #PM-1) according to the manufacture's recommendation. To induce differentiation, confluent cells were kept in adipocyte differentiation medium (Zen-Bio #DM-2) for 7 days followed by 5 days of culture in adipocyte medium (Zen-Bio #DM-1). Detection of lipid droplets was achieved by staining with ORO. Briefly, cells were rinsed in phosphate-buffered saline and then fixed in 10% neutral-buffered formalin for 5 min. Fixed cells were rinsed 3 times in deionized water, stained with a solution of ORO for 30 min, and rinsed with deionized water. Quantification of the results was performed on a Typhoon scanner using Bio-Rad Gel Doc 2000TM Systems and Bio-Rad TDS Quantity One software.
Lentivirus-based shRNA Transduction
MISSION shRNA lentiviral particles were obtained from Sigma. Lentiviral transduction of 3T3-L1 cells with particles for shRNAs targeting Nrf2 (SHVRS-NM_010902), Keap1 (SHVRS-NM_016679), or scrambled non-target negative control (Scr, SHC002V) was performed based on manufacturer's protocol. Briefly, 24 h before transduction, 3T3-L1 cells were plated in 6-well plates at ∼40–50% confluency in complete medium described above. The following day hexadimethrine bromide (Sigma), a transduction enhancer, was added to each well at a concentration of 8 μg/ml, and viral particles were added to each well at a concentration of 2 × 105 transducing units/ml. After overnight incubation, medium containing viral particles was removed and replaced with fresh medium containing 2 μg/ml puromycin. Cells were grown to ∼90% confluency and subcultured in medium containing puromycin. Before lentiviral transduction, a puromycin titration was performed to identify the minimum concentration of puromycin that caused complete cell death of 3T3-L1 cells after 3–5 days. Knockdown of Nrf2 in human preadipocytes was performed using MISSION shRNA lentiviral particles targeting human Nrf2 (SHVRS-NM_006164).
Chromatin Immunoprecipitation Assay
3T3-L1 cells were used for chromatin immunoprecipitations using a kit from Upstate Biotechnology as described previously (23). A portion of the lysate was saved for subsequent purification of input genomic DNA. The remaining supernatant was precleared with protein A-Sepharose beads and sheared herring sperm DNA and followed by incubation with anti-Nrf2 rabbit polyclonal antibody or preimmune antibody control at 4 °C overnight. Purified DNA was PCR-amplified for 35 cycles (30 s at 95 °C, 30 s at 60 °C, and 45 s at 72 °C) with the primers indicated in supplemental Table 3 that span 3 kb of the Pparγ2 gene promoter. Non-precipitated (input) genomic DNA was amplified as input control, and NQO1 promoter was used as a positive control for Nrf2.
Statistical Analyses
All statistical analyses were performed using Graphpad Prism 4 (GraphPad Software, San Diego, CA) or Excel (Microsoft, Redmond, WA) with p < 0.05 taken as significant. Data are expressed as the mean ± S.E. For comparisons between groups, a Student's t test was performed. Two-way analysis of variance with Bonferroni post hoc testing was done to evaluate statistical significance of gene expression in 3T3-L1 cells.
RESULTS
Nrf2 Mutants Have Lower Amounts of Adipose Tissue
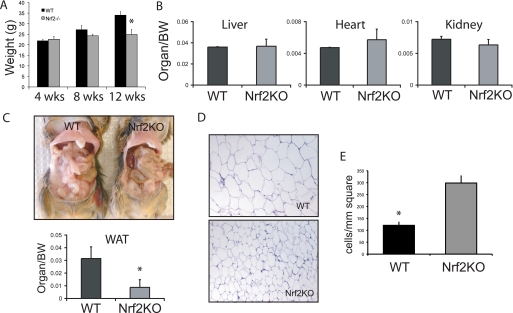
The body weights of wild type and Nrf2 KO littermates were indistinguishable at weaning (3–4 weeks). By 12 weeks, however, the mean body weight of adult Nrf2 KO mice under an ad libitum diet consisting of regular chow was 20% lower than that of wild type littermates (Fig. 1A). A comparison of organ weights normalized for body mass did not reveal a significant difference between Nrf2 KO and wild type littermate controls (Fig. 1B). However, the fat pads in Nrf2 KO mice appeared smaller in comparison to wild type littermates (Fig. 1C). In Nrf2 KO mice, white adipose tissue weight (epididymal, perirenal, and mesenteric weight normalized to body weight) was 60% lower than in wild type littermates (Fig. 1C). To investigate this further, we examined morphology of white adipose tissues. Fat cells in Nrf2 KO mice appeared smaller than cells in wild type mice (Fig. 1D). Quantitative analysis indicated that the adipocytes of Nrf2 KO mice were significantly smaller that those of wild type mice as reflected by a 3-fold greater number of adipocytes in cross-sections of fat tissue from knock-out mice versus from wild type mice (Fig. 1E). In addition, the amount of DNA per gram of adipose tissue was significantly greater in Nrf2 KO mice than in wild type animals (data not shown).
FIGURE 1.
Nrf2 knock-out mice have lower amounts of adipose tissue. A, shown is body weight in adult (age 12 weeks) male wild type (WT; n = 8) and Nrf2 KO (n = 10) mice fed a regular chow diet. B, shown is organ weight in wild type and Nrf2 KO mice. BW, body weight. C, the top panel shows the white adipose tissue (WAT) in wild type and Nrf2 KO mice. The bottom panel shows the total fat pad weight (epididymal, retro-peritoneal, and inguinal) in wild type and Nrf2 KO mice. All the measurements were normalized to body weights of individual mice, n = 8 per group. *, p < 0.05. D, representative histological sections of white adipose tissues stained with hematoxylin and eosin are shown. E, shown are cell numbers per mm2 in the sections of adipose tissues shown in panel D. *, p < 0.05.
Nrf2 Mutants Are Resistant to Diet-induced Obesity
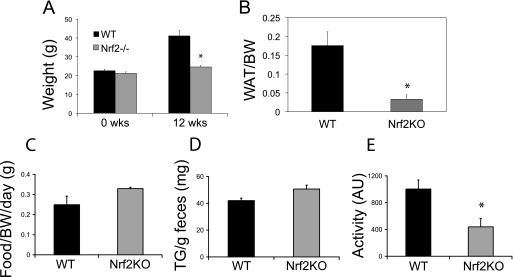
To further assess the potential role of Nrf2 in adipocyte function, we challenged adult mice with a HFD and examined whether deficiency of Nrf2 protects against diet-induced obesity. After 12 weeks on the HFD, wild type mice had gained 75% of their starting weight, whereas Nrf2 KO mice had gained only 15% of their starting weight (Fig. 2A). Similar to chow-fed animals, the group differences in body weight in HFD-fed animals were due to a decrease in fat mass in Nrf2 KO mice. Marked enlargement of fat pads was observed in wild type mice compared with Nrf2 KO mice (data not shown), and white adipose tissue weight was significantly greater in wild type mice compared with Nrf2 KO mice (Fig. 2B). To examine if the lower adipose tissue mass observed in Nrf2 KO mice was caused by a decrease in caloric intake or fat absorption, we measured food intake and fecal fat over a 1-week period. The amount of food consumed was similar between mutant and control mice (Fig. 2C), and there was no difference in the fecal lipid levels between the two groups (Fig. 2D). Thus, the lower adipose tissue mass of Nrf2 KO animals does not appear to be due to changes in food consumption or intestinal fat absorption. To examine if physical activity plays a role in the lean phenotype, locomotor activity was monitored using a beam break apparatus. Nrf2 KO mice did not demonstrate hyperactivity compared with wild type animals (Fig. 2E). These results suggest that the defect is intrinsic to adipose cells that are deficient in Nrf2.
FIGURE 2.
Nrf2 knock-out animals on HFD gain less weight over time. A, shown are body weights of 4-week-old wild type (WT) and Nrf2 KO mice before high fat feeding and weights after 12 weeks on a high fat diet. All mice in this experiment were males (n = 8 per group). *, p < 0.05. B, shown are total fat pad weights (epididymal, retro-peritoneal, and inguinal) of wild type and Nrf2 KO mice on a high fat diet for 12 weeks. n = 8; *, p < 0.05. WAT, white adipose tissue; BW, body weight. C, shown is the measurement of daily food intake of wild type and Nrf2 KO mice on HFD. D, fecal triglyceride (TG) levels are shown of wild type and Nrf2 KO mice. n = 8; *, p < 0.05. E, horizontal activity of wild type and Nrf2 KO mice is shown. AU, arbitrary units. n = 4; *, p < 0.05.
Nrf2 Deficiency Impairs Adipogenesis in Mouse Embryonic Fibroblasts
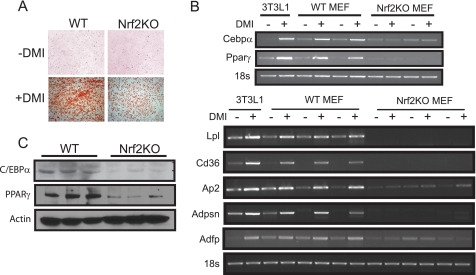
MEFs, isolated from embryos at gestation E12–14, can be induced to differentiate into adipocytes when exposed to a mixture consisting of dexamethasone, 3-isobutylmethylxanthine, and insulin (DMI) (24). Therefore, we compared adipocyte differentiation capacity of MEFs isolated from wild type and Nrf2-deficient embryos to determine whether Nrf2 is required for adipogenesis. Wild type MEFs cultured with DMI showed a dramatic accumulation of intracellular lipid as determined by ORO staining (Fig. 3A). In contrast, lipid accumulation was significantly lower in Nrf2-deficient MEFs treated with DMI, suggesting that loss of Nrf2 impairs differentiation of MEFs into adipocytes (Fig. 3A). To investigate the underlying mechanism of impaired fat accumulation, we analyzed the expression of various genes related to adipocyte differentiation. Although Cebpβ and Cebp δ expression patterns were similar in wild type and Nrf2 KO fibroblasts (data not shown), Pparγ and Cebpα expression were markedly lower in Nrf2 KO fibroblasts compared with wild type cells (Fig. 3B). Western blotting also demonstrated that PPARγ and C/EBPα protein levels were lower in white adipose tissues of Nrf2 knockouts (Fig. 3C). Consistent with the decreased expression of Pparγ, both basal and DMI-induced expressions of downstream target genes involved in adipogenesis including lipoprotein lipase (Lpl), complement factor D (Adpsn), aP2, Cd36, and adipose differentiation-related protein (Adfp) were all significantly lower in Nrf2 KO MEFs compared with wild type (Fig. 3B). These results indicate that adipocyte differentiation was impaired in Nrf2 mutants.
FIGURE 3.
Adipogenesis and induction of adipogenic genes are impaired in Nrf2-deficient MEFs. A, MEFs derived from E13.5 wild type (WT) and Nrf2 KO embryos were cultured with DMI to induce differentiation into adipocytes. Cells were then stained after 7 days with ORO to visualize lipid accumulation. A representative of three independent experiments is shown. B, shown is reverse transcription-PCR analysis of key lipogenic genes in wild type and Nrf2 KO MEFs differentiated for 5 days. Differentiated 3T3-L1 cells were used as positive control for adipogenic induction. C, protein levels of PPARγ and C/EBPγ in wild type and Nrf2 KO white adipose tissue (epididymal, retro-peritoneal, and inguinal) extracts measured by Western blotting. β-Actin levels are shown as loading controls.
Nrf2 Is Required for Adipogenesis in 3T3-L1 and Human Subcutaneous Preadipocytes
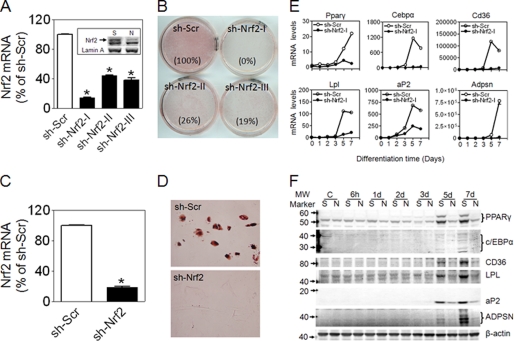
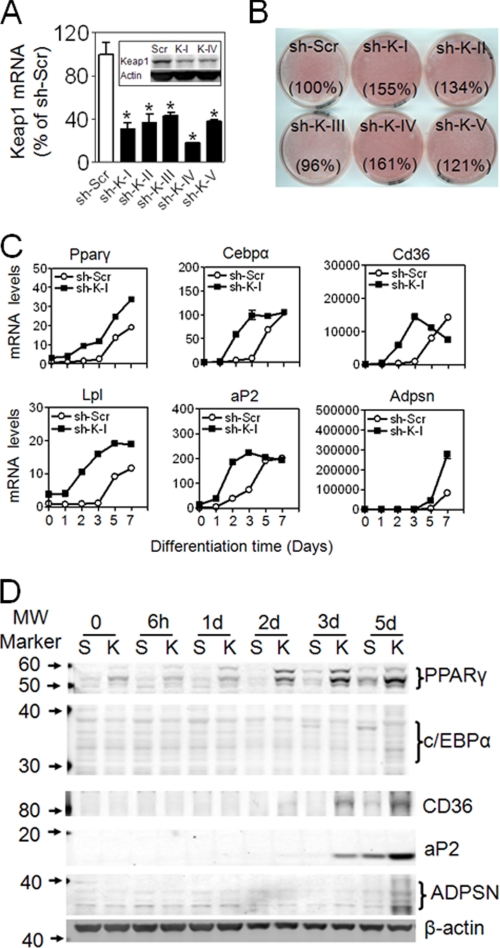
To confirm that Nrf2 is involved in adipogenesis and exclude nonspecific effects associated with development in MEF cells, we performed shRNA-mediated knockdown of endogenous Nrf2 in 3T3-L1 cells. Several shRNAs against Nrf2 were used for transduction. As shown in Fig. 4A, one of the constructs (sh-Nrf2-I) markedly silenced Nrf2 expression, whereas two other constructs (sh-Nrf2-II, sh-Nrf2-III) had a moderate silencing effect. Oxidative stress induction of Nrf2 protein accumulation and NAD(P)H:quinone oxidoreductase 1 (Nqo1), a well established Nrf2 target gene, was attenuated, indicating that Nrf2 activity is suppressed in the knockdown cells (supplemental Fig. S1). To determine whether reduction in Nrf2 influences adipogenesis, knockdown cells were cultured in differentiation medium and monitored for lipid accumulation by ORO staining. After 6 days of exposure to differentiation medium, lipid accumulation was significantly decreased in Nrf2 knockdown cells compared with scramble (Fig. 4B). Consistent with these results, adipogenic differentiation in human subcutaneous preadipocytes was also suppressed by knockdown of Nrf2 (Fig. 4, C and D). In 3T3-L1 Nrf2 knockdown cells, gene (Fig. 4E) and protein (Fig. 4F) expression of PPARγ, C/EBPα, aP2, CD36, LPL, and ADPSN induced by hormonal mixture was significantly lower than that of scramble cells. To further substantiate these findings, we examined the effects of increased Nrf2 function on adipogenesis by knocking down Keap1, which is a negative regulator of Nrf2. 3T3-L1 cells transduced with Keap1-shRNAs showed efficient knockdown of Keap1 expression (Fig. 5A). Nqo1 expression was induced by knockdown of Keap1, confirming that Nrf2 pathway is activated in these cells (supplemental Fig. S2). Under differentiation protocol, ORO staining demonstrated increased lipid accumulation in Keap1-knockdown cells compared with scramble cells (Fig. 5B). In addition, induction of PPARγ, C/EBPα, and PPARγ target genes were accelerated and enhanced, suggesting that differentiation is enhanced by Nrf2 activation (Fig. 5, C and D).
FIGURE 4.
Stable knockdown of Nrf2 by shRNA lentivirus suppresses adipogenesis in 3T3-L1 cells and primary human preadipocytes. A, shown is expression of Nrf2 mRNA and protein (inset: S, sh-Scr; N, sh-Nrf2-I) in 3T3-L1 cells transduced with shRNA lentivirus targeted against mouse Nrf2. n = 3. B, ORO staining of scramble (Scr) and knockdown cells treated with a hormonal mixture (detailed under “Experimental Procedures”) for 3 days followed by 3 days of culture in DMEM supplemented with 10% FBS. The number in parentheses after each plate name is the relative volume (density × area) of ORO staining. A representative of 15 independent experiments is shown. C, expression is shown of Nrf2 in subcutaneous human preadipocytes infected with shRNA lentivirus targeting human Nrf2. n = 3. D, shown is ORO staining of differentiated human adipocytes (ORO staining). Confluent cells were kept in adipocyte differentiation medium (Zen-Bio #DM-2) for 7 days followed by 5 days of culture in adipocyte medium (Zen-Bio #AM-1). A representative of three independent experiments is shown. E, expression of adipogenic genes is shown. Levels of mRNA are expressed as -fold of sh-Scr at day 0, n = 3. F, protein levels of adipogenic markers are shown. A representative experiment of 3–5 independent experiments is shown. C, control.
FIGURE 5.
Stable knockdown of Keap1 by shRNA lentivirus enhances hormonal mixture-induced adipogenesis in 3T3-L1 cells. A, expression is shown of Keap1 mRNA and protein (inset) in 3T3-L1 cells transduced with shRNA lentivirus targeting mouse Keap1. Sh-K-I∼V represent five different shRNAs targeting Keap1. n = 3. B, ORO staining of differentiated cells is shown. Cells were treated with differentiation medium for 3 days followed by 3 days of culture in DMEM supplemented with 10% FBS. The number in parentheses after each plate name is the relative volume of ORO staining. A representative of five independent experiments is shown. C, expression of adipogenic genes is shown. Levels of mRNA are expressed as -fold sh-Scr at day 0. n = 3. D, shown are protein levels of adipogenic markers. A representative of 3–5 independent experiments is shown.
Nrf2 Activates Pparγ Promoter Activity
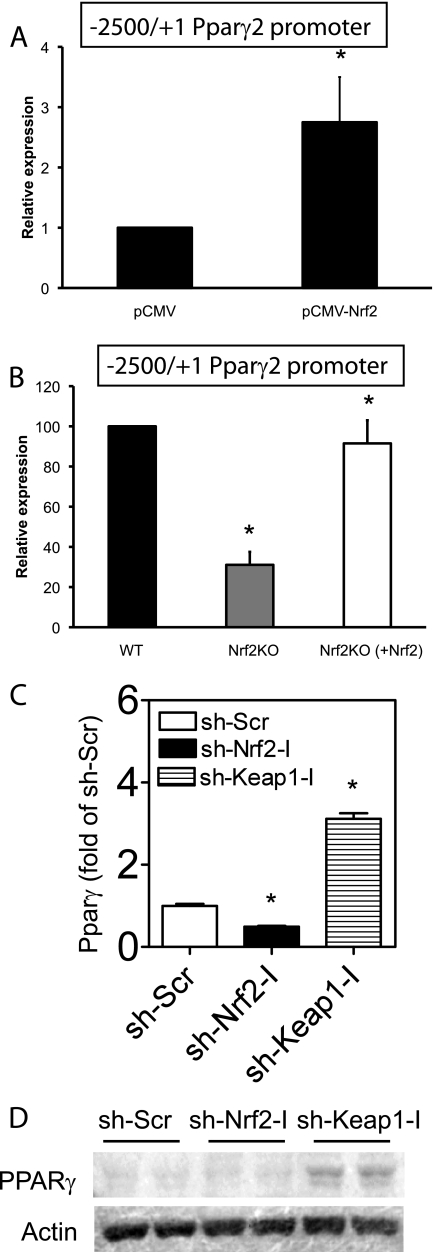
The Pparγ gene gives rise to two isoforms, γ1 and γ2, through the use of different start sites and alternate splicing (25). The γ2 isoform is highly enriched in adipose tissue (3). To determine whether Nrf2 regulates Pparγ2 promoter activity, a luciferase reporter gene driven by the Pparγ2 promoter was transfected into wild type MEF cells along with Nrf2 expression plasmid. Coexpression of Nrf2 increased luciferase activity driven by the Pparγ2 promoter ∼3-fold relative to cells transfected with vector alone (Fig. 6A). To determine whether Pparγ2 promoter activity is Nrf2-dependent, we compared expression of the luciferase reporter plasmid transfected into wild type and Nrf2 KO MEFs. Luciferase expression in Nrf2-deficient MEFs was 3-fold lower compared with wild type MEFs (Fig. 6B), and low expression in Nrf2 KO MEFs was rescued by cotransfection of Nrf2 expression plasmid (Fig. 6B). Consistent with promoter assays, up-regulation of Nrf2 by knockdown of Keap1 in 3T3-L1 cells was accompanied by up-regulation of Pparγ2 expression (Fig. 6, C and D). These data suggest that the decrease in Pparγ2 expression in Nrf2 KO mice was due to loss of Nrf2 activation of the Pparγ2 promoter.
FIGURE 6.
Nrf2 regulates PPARγ expression. A, shown is transactivation of the mouse Pparγ promoter by Nrf2. MEF cells were transfected with the luciferase gene under the mouse Pparγ promoter along with either vector or Nrf2 cDNA. Cells were harvested 48 h later, and luciferase activity was determined. Transfection efficiency was normalized to Renilla luciferase under control of the cytomegalovirus promoter. Bars represent the mean of three independent experiments ± S.E. (*, p < 0.05). B, the luciferase gene under the mouse Ppar γ promoter was transfected into wild type (WT), Nrf2 KO, or Nrf2 KO fibroblasts along with Nrf2 cDNA. Luciferase activity was measured 48 h after. Bars represent the mean ± S.E., n = 3 independent experiments; *, p < 0.05. C, expression of Pparγ in 3T3-L1 cells with Nrf2- or Keap1 knockdown is shown. *, p < 0.05 versus sh-Scr. D, Western blotting of PPARγ in 3T3-L1 cells with Nrf2- or Keap1 knockdown is shown.
Nrf2 Binds to the Pparγ Promoter
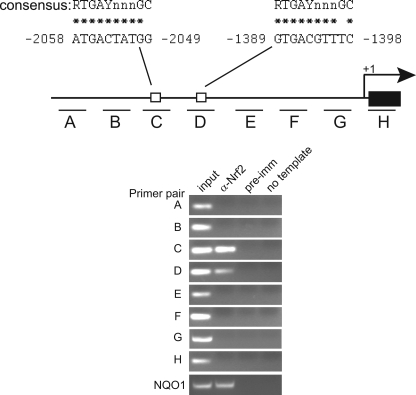
To determine whether Nrf2 binds the endogenous Pparγ promoter, chromatin immunoprecipitation assay were carried out. Chromatin isolated from 3T3-L1 cells was immunoprecipitated with an anti-Nrf2 antibody or preimmune sera as control and subjected to PCR amplification using primers spanning 3 kb of the Pparγ promoter region (Fig. 7A). Occupancy by Nrf2 was detected with primer pairs C and D (Fig. 7B). No signal was observed when preimmune rabbit IgG was used for precipitation. Sequence inspection in the region spanned by primer pairs C and D revealed two consensus binding sites for Nrf2. One is located in the sense orientation at nucleotide position −1389 to −1398 and another at nucleotide position −2049 to −2058 in the negative strand. Together with the results in Nrf2 KO mice and the luciferase reporter studies, these findings suggest that Nrf2 binds the Pparγ promoter and contributes to its activation in vivo.
FIGURE 7.
Chromatin immunoprecipitation assays indicate a physical association between Nrf2 and the Pparγ promoter region. Chromatin immunoprecipitation assays were performed on 3T3-L1 chromatin using either Nrf2 or preimmune antisera as the negative control. A primer walk was performed across a 3-kb region encompassing the Pparγ2 promoter. The diagram indicates the location of the primer sets. Open boxes indicate the locations of the Nrf2 consensus binding site. PCR amplification of the Nqo1 promoter was used as a positive control. Non-immunoprecipitated chromatin (1%) was used as an input control.
DISCUSSION
Nrf2 is a basic leucine zipper transcription factor that is activated by oxidative stress and regulates the expression of cytoprotective and antioxidant genes (16). Consistent with this function, Nrf2 is expressed in various organs including liver, lung, kidney, and digestive tract, which are involved in detoxification. In addition, Nrf2 is expressed in fat tissue, but its role in adipocyte differentiation has not been directly investigated (17). In this study we have discovered that Nrf2 plays an important role in adipogenesis. Nrf2 knock-out mice have lower adipose tissue mass, smaller adipocytes, and are resistant to HFD-induced obesity. Hormonal mixture-induced adipogenesis is depressed in embryonic fibroblasts derived from Nrf2 knock-out mice and in 3T3-L1 and primary human preadipocyte cells in which shRNA has been used to knock down Nrf2 expression. Conversely, adipocyte differentiation of 3T3-L1 cells is enhanced by activation of Nrf2 through knockdown of its negative regulator Keap1. Data from these cell-based assays showing effects of Nrf2 on adipocyte differentiation suggest that the defect of adipogenesis in knock-out mice is intrinsic to fat cells and/or fat cell precursors and is not simply secondary to functional disturbances in other tissues.
PPARγ is considered to be a master regulator of adipogenesis, and we have found that PPARγ gene expression and protein levels as well as expression levels of downstream target genes of Pparγ are greatly attenuated in adipose tissue of Nrf2 KO mice. In 3T3-L1 cells and human preadipocytes, we have also found that shRNA knockdown of Nrf2 impairs PPARγ expression and adipogenesis in a parallel fashion. Previous studies have hinted at a role for Nrf2 in Pparγ expression. Decreased PPARγ levels have been reported in liver and lung tissues of Nrf2 KO mice, and hepatic induction of Pparγ expression by a short term (4 weeks) high fat feeding is also decreased in Nrf2 mutant mice (26, 27). In addition to discovering that adipose tissue Pparγ gene expression and protein levels are significantly attenuated by deficiency of Nrf2, we have found that Nrf2 positively regulates Pparγ promoter activity and binds directly in the region of an Nrf2 consensus binding sequence in the transcriptional start site of the Pparγ promoter.
Although the current studies indicate that Nrf2 may influence adipogenesis via direct interaction with the Pparγ promoter, it is also possible that Nrf2 affects fat cell differentiation through its known ability to modulate intracellular reactive oxygen species. Reactive oxygen species have been reported to inhibit adipocyte differentiation in part by activating CHOP-10, which in turn reduces C/EBP DNA binding activity (28). Reactive oxygen species may also promote increased activity of hypoxia-inducible factor 1, which can repress Pparγ promoter function (28). Thus, deficiency of Nrf2 could affect adipocyte differentiation through both direct and indirect effects on PPARγ activity and perhaps other pathways as well.
Recently, it was reported that Nrf2 interferes with adipogenic differentiation of MEF cells (29). Nrf2-deficient MEFs were found to exhibit accelerated adipogenesis as compared with wild type MEFs. This finding differs from our results demonstrating that deficiency of Nrf2 inhibits adipogenesis. The basis for these apparently discrepant results is not known but could be related to the different types of MEF cells used in the adipocyte differentiation experiments. Specifically, Shin et al. (29) used immortalized MEFs, whereas we used primary MEFs derived from E13.5 embryos in our differentiation assays. Primary and immortalized MEFs differ in their capacity to differentiate into adipocytes. Although primary MEFs readily differentiate in the presence of hormonal inducers, most immortalized cells are incapable of differentiating unless additional pro-adipogenic factors are incorporated (24). Another possible explanation for the potentially discrepant findings between their study and ours could involve differences in the genetic backgrounds of the mouse strains from which the MEFs were originally derived. However, Shin et al. (30) have recently published a study in Nrf2 knock-out mice derived on a C57BL/6 background that showed effects on body weight and adipose tissue development similar to our current findings in Nrf2 knockouts derived on a mixed genetic background. Specifically, in the setting of a HFD, the Nrf2 KO mice in the study of Shin et al. (30) also had lower amounts of fat and gained much less weight than the wild type controls (30). Based on these and other observations, Shin et al. (30) suggested that constitutive levels of Nrf2 may contribute to accumulation of fat during administration of a HFD. In addition, Shin et al. (30) noted that initial body weights were smaller in their Nrf2 knock-out mice than in controls even before administration of a high fat diet. In the current studies, we also observed that body weights of Nrf2 knock-out mice were lower than those of controls during feeding with regular chow. Thus, notwithstanding the contrasting findings between our group and those of Shin et al. (29) with respect to adipogenesis in primary versus immortalized MEFs from Nrf2 KO mice), our current results and those of Shin et al. (30) on body weight and adipose tissue development in separate strains of Nrf2 KO mice appear reasonably consistent. Finally, in addition to studies in primary MEFs, we have demonstrated that knockdown of Nrf2 impairs adipogenesis in human preadipocytes and in 3T3-L1 cells, both well established and widely used models of adipocyte differentiation. These results together with our current findings and those of Shin et al. (30) in Nrf2 knock-out mice are compatible with a positive role for Nrf2 in promoting fat cell development.
In the current studies it should be noted that we did not detect any differences between Nrf2 KO animals and their wild type controls with respect to feeding behavior, intestinal fat absorption, or locomotor activity. Given that the Nrf2 KO animals gained less weight than the wild type mice despite relatively similar food intakes, it appears that deficiency of Nrf2 may affect energy expenditure. Deficiency of Nrf2 is known to promote oxidative stress that could serve as a stimulant for mitochondrial biogenesis. This raises the possibility that the lean phenotype and resistance to diet-induced obesity in the Nrf2 KO animals could be related to increases in mitochondrial activity secondary to increases in oxidative stress that stem from deficiency of Nrf2. For example, superoxide has been reported to activate mitochondrial uncoupling proteins, which can dissipate caloric energy as heat by uncoupling mitochondrial respiration from ATP production (31). Thus, future studies on mitochondrial oxygen consumption and on uncoupling protein activity in fat and muscle could give further insight into the mechanisms whereby Nrf2 deficiency influences body weight, particularly in the setting of high fat diets that are known to up-regulate uncoupling protein gene expression (32). It is also possible that variability in the genetic backgrounds of the Nrf2 KO mice and their wild type controls could contribute to the observed differences in body weight and adiposity between the KO mice and the controls. As previously noted, however, Shin et al. (30) have also observed attenuated weight gain and adipose tissue formation in studies comparing other strains of Nrf2 KO mice and controls. Nevertheless, future studies to determine the extent to which variation in genetic background might influence the effects of Nrf2 knock-out on energy metabolism and fat cell development could still be of interest. Finally, because of the important role of adipose tissue in regulating glucose and lipid metabolism, the current findings should serve to motivate follow-up studies of the effects of Nrf2 activity on risk for insulin resistance, dyslipidemia, and diabetes.
In summary, the results presented here suggest an important biologic role for Nrf2 beyond its effects on cellular detoxification and the oxidative stress response. We show that the loss of Nrf2 results in decreased adipose tissue mass and resistance to diet-induced obesity in the absence of any obvious changes in food intake or intestinal fat absorption. In both animal studies and in cell-based assays, our findings indicate that loss of Nrf2 leads to impaired adipogenesis. We also show that Nrf2 regulates Pparγ promoter activity and Pparγ gene and protein expression levels in a manner that could mediate its effects on adipocyte differentiation. Thus, Nrf2 appears to belong to the relatively small group of transcription factors known to directly regulate transcription of Pparγ during adipogenesis. Together these findings suggest that variations in Nrf2 activity can drive functionally important steps in the process of adipocyte differentiation and may also induce significant changes in energy metabolism. Finally, the results raise the possibility that Nrf2 may be an interesting new target for exploring pharmacologic interventions directed at prevention and treatment of obesity and related clinical disorders.
Supplementary Material
Acknowledgments
We are grateful to Drs. Jingqi Fu, Courtney G. Woods, Rui Zhao, Sheila Collins, and Melvin E. Andersen for their contributions to this study. We also thank Zen-Bio for providing subcutaneous human preadipocytes and culture media for this study.
This work was supported, in whole or in part, by National Institutes of Health Grants DK76788 and ES016005 (to J. P.) and CA091907 (to J. Y. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Tables 1–3.
- PPAR
- peroxisome proliferator-activated receptors
- C/EBP
- CCAAT-enhancer-binding protein
- Nrf2
- nuclear factor E2-related factor 2
- ORO
- Oil Red O
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- HFD
- high fat diet
- DMI
- dexamethasone, 3-isobutylmethylxanthine, and insulin
- LPL
- lipoprotein lipase
- ADPSN
- adipsin
- Adfp
- adipose differentiation-related protein
- Cd36
- fatty acid translocase
- aP2
- fatty acid-binding protein 4
- FBS
- fetal bovine serum
- MEF
- mouse embryonic fibroblast
- KO
- knock-out
- shRNA
- short hairpin RNA.
REFERENCES
- 1.Rosen E. D., Spiegelman B. M. (2006) Nature 444, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feige J. N., Gelman L., Michalik L., Desvergne B., Wahli W. (2006) Prog. Lipid Res. 45, 120–159 [DOI] [PubMed] [Google Scholar]
- 3.Tontonoz P., Spiegelman B. M. (2008) Annu. Rev. Biochem. 77, 289–312 [DOI] [PubMed] [Google Scholar]
- 4.Darlington G. J., Ross S. E., MacDougald O. A. (1998) J. Biol. Chem. 273, 30057–30060 [DOI] [PubMed] [Google Scholar]
- 5.Otto T. C., Lane M. D. (2005) Crit. Rev. Biochem. Mol. Biol. 40, 229–242 [DOI] [PubMed] [Google Scholar]
- 6.Cao Z., Umek R. M., McKnight S. L. (1991) Genes Dev. 5, 1538–1552 [DOI] [PubMed] [Google Scholar]
- 7.Yeh W. C., Cao Z., Classon M., McKnight S. L. (1995) Genes Dev. 9, 168–181 [DOI] [PubMed] [Google Scholar]
- 8.Christy R. J., Kaestner K. H., Geiman D. E., Lane M. D. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 2593–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke S. L., Robinson C. E., Gimble J. M. (1997) Biochem. Biophys. Res. Commun. 240, 99–103 [DOI] [PubMed] [Google Scholar]
- 10.Wu Z., Xie Y., Bucher N. L., Farmer S. R. (1995) Genes Dev. 9, 2350–2363 [DOI] [PubMed] [Google Scholar]
- 11.Shao D., Lazar M. A. (1997) J. Biol. Chem. 272, 21473–21478 [DOI] [PubMed] [Google Scholar]
- 12.Wu Z., Rosen E. D., Brun R., Hauser S., Adelmant G., Troy A. E., McKeon C., Darlington G. J., Spiegelman B. M. (1999) Mol. Cell 3, 151–158 [DOI] [PubMed] [Google Scholar]
- 13.Freytag S. O., Paielli D. L., Gilbert J. D. (1994) Genes Dev. 8, 1654–1663 [DOI] [PubMed] [Google Scholar]
- 14.Rosen E. D., Hsu C. H., Wang X., Sakai S., Freeman M. W., Gonzalez F. J., Spiegelman B. M. (2002) Genes Dev. 16, 22–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefterova M. I., Lazar M. A. (2009) Trends Endocrinol. Metab. 20, 107–114 [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi M., Yamamoto M. (2006) Adv. Enzyme Regul. 46, 113–140 [DOI] [PubMed] [Google Scholar]
- 17.Chan K., Lu R., Chang J. C., Kan Y. W. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13943–13948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshinari K., Okino N., Sato T., Sugatani J., Miwa M. (2006) Drug Metab. Dispos. 34, 1081–1089 [DOI] [PubMed] [Google Scholar]
- 19.Kim S., Sohn I., Ahn J. I., Lee K. H., Lee Y. S., Lee Y. S. (2004) Gene 340, 99–109 [DOI] [PubMed] [Google Scholar]
- 20.Zhou J., Febbraio M., Wada T., Zhai Y., Kuruba R., He J., Lee J. H., Khadem S., Ren S., Li S., Silverstein R. L., Xie W. (2008) Gastroenterology 134, 556–567 [DOI] [PubMed] [Google Scholar]
- 21.Leung L., Kwong M., Hou S., Lee C., Chan J. Y. (2003) J. Biol. Chem. 278, 48021–48029 [DOI] [PubMed] [Google Scholar]
- 22.Pi J., Bai Y., Daniel K. W., Liu D., Lyght O., Edelstein D., Brownlee M., Corkey B. E., Collins S. (2009) Endocrinology 150, 3040–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W., Kwok A. M., Chan J. Y. (2007) J. Biol. Chem. 282, 24670–24678 [DOI] [PubMed] [Google Scholar]
- 24.Rosen E. D., MacDougald O. A. (2006) Nat. Rev. Mol. Cell Biol. 7, 885–896 [DOI] [PubMed] [Google Scholar]
- 25.Zhu Y., Qi C., Korenberg J. R., Chen X. N., Noya D., Rao M. S., Reddy J. K. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7921–7925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho H. Y., Reddy S. P., Debiase A., Yamamoto M., Kleeberger S. R. (2005) Free Radic. Biol. Med. 38, 325–343 [DOI] [PubMed] [Google Scholar]
- 27.Tanaka Y., Aleksunes L. M., Yeager R. L., Gyamfi M. A., Esterly N., Guo G. L., Klaassen C. D. (2008) J. Pharmacol. Exp. Ther. 325, 655–664 [DOI] [PubMed] [Google Scholar]
- 28.Gummersbach C., Hemmrich K., Kröncke K. D., Suschek C. V., Fehsel K., Pallua N. (2009) Differentiation 77, 115–120 [DOI] [PubMed] [Google Scholar]
- 29.Shin S., Wakabayashi N., Misra V., Biswal S., Lee G. H., Agoston E. S., Yamamoto M., Kensler T. W. (2007) Mol. Cell. Biol. 27, 7188–7197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin S., Wakabayashi J., Yates M. S., Wakabayashi N., Dolan P. M., Aja S., Liby K. T., Sporn M. B., Yamamoto M., Kensler T. W. (2009) Eur. J. Pharmacol. 620, 138–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Echtay K. S., Roussel D., St.-Pierre J., Jekabsons M. B., Cadenas S., Stuart J. A., Harper J. A., Roebuck S. J., Morrison A., Pickering S., Clapham J. C., Brand M. D. (2002) Nature 415, 96–99 [DOI] [PubMed] [Google Scholar]
- 32.Surwit R. S., Wang S., Petro A. E., Sanchis D., Raimbault S., Ricquier D., Collins S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4061–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.