Abstract
Cytochrome cm552 (cyt cm552) from the ammonia-oxidizing Nitrosomonas europaea is encoded by the cycB gene, which is preceded in a gene cluster by three genes encoding proteins involved in the oxidation of hydroxylamine: hao, hydroxylamine oxidoreductase; orf2, a putative membrane protein; cycA, cyt c554. By amino acid sequence alignment of the core tetraheme domain, cyt cm552 belongs to the NapC/TorC family of tetra- or pentaheme cytochrome c species involved in electron transport from membrane quinols to a variety of periplasmic electron shuttles leading to terminal reductases. However, cyt cm552 is thought to reduce quinone with electrons originating from HAO. In this work, the tetrahemic 27 kDa cyt cm552 from N. europaea was purified after extraction from membranes using Triton X-100 with subsequent exchange into n-dodecyl β-D-maltoside. The cytochrome had a propensity to form strong SDS-resistant dimers likely mediated by a conserved GXXXG motif present in the putative transmembrane segment. Optical spectra of the ferric protein contained a broad ligand–metal charge transfer band at ~625 nm indicative of a high-spin heme. Mössbauer spectroscopy of the reduced 57Fe-enriched protein revealed the presence of high-spin and low-spin hemes in a 1:3 ratio. Multimode EPR spectroscopy of the native state showed signals from an electronically interacting high-spin/low-spin pair of hemes. Upon partial reduction, a typical high-spin heme EPR signal was observed. No EPR signals were observed from the other two low-spin hemes, indicating an electronic interaction between these hemes as well. UV–vis absorption data indicate that CO (ferrous enzyme) or CN− (ferric or ferrous enzyme) bound to more than one and possibly all hemes. Other anionic ligands did not bind. The four ferrous hemes of the cytochrome were rapidly oxidized in the presence of oxygen. Comparative modeling, based on the crystal structure and conserved residues of the homologous NrfH protein from Desulfovibrio of cyt cm552, predicted some structural elements, including a Met-ligated high-spin heme in a quinone-binding pocket, and likely axial ligands to all four hemes.
Nitrifying bacteria, such as the chemolitho-autotrophic Nitrosomonas europaea, oxidize ammonium (NH+4) to nitrite (NO2−). This is the first and rate-limiting step in the twostep process of biological nitrification (NH3 → NO 2− → NO3−), the essential oxidative step in the global nitrogen cycle (1, 2). The initial step in ammonia oxidation is the generation of hydroxylamine (NH3 + 2e− + O2 + 2H+ → NH2OH + H2O) by the membrane-bound ammonia monooxygenase (AMO). Hydroxylamine in the periplasm is oxidized to nitrite (NH2OH + H2O → HNO2 + 4e− + 4H+) by hydroxylamine oxidoreducatse (HAO).1 Transfer of the four electrons from HAO to the membrane ubiquinone pool is thought to involve the periplasmic cyt c554 and the membrane-anchored cyt cm552, which may serve as a quinone reductase.
The X-ray analysis of the homotrimeric 24-heme HAO (3), and the monomeric tetraheme cyt c554 (4, 5), and the detailed spectroscopic (6–9) and redox analysis (10–12) of their hemes reveal a complex system of multiheme c cytochromes involved in the four-electron redox process. However, as a membrane protein, cyt cm552 has resisted purification and characterization. The gene encoding cyt cm552 was identified in N. europaea by Bergmann et al. (13), in a cluster containing hao (HAO), orf2 (encoding a putative membrane protein), cycA (c554), and cycB (cm552). The latter two genes share an operon. This cluster is highly conserved in the autotrophic ammonia-oxidizing bacteria (Nitrosococcus oceani, Nitrosospira multiformis, and Nitrosomonas multiformis) which would be expected if the products of the four genes are critical for hydroxylamine oxidation and electron transfer (14).
By sequence alignment of the core tetraheme domain, cyt cm552 has been shown to share common ancestry with the widespread bacterial NapC/NrfH/NirT/TorC family of tetra-and pentaheme quinol dehydrogenases (13, 14). Members of this family of relatively ubiquitous proteins in facultative anaerobes are involved in electron transfer from the membrane quinol pool to a range of periplasmic terminal reductases which employ electron acceptors other than O2: nitrite, nitrate, fumarate, dimethyl sulfoxide, or metal ions such as Fe(III) (15). In this work, we use the abbreviation NapC/NrfH for this family since they represent the two major evolutionary subfamilies (14). The C-terminal domains vary in sequence, possibly to correspond to the various periplasmic electron acceptor proteins. Strong sequence homology to the NapC/NrfH family suggests a quinol oxidoreductase function for cyt cm552, and the location of the cycB gene encoding cyt cm552 within the hao gene cluster suggests that the flux in vivo is in the direction of quinone reduction by electrons from HAO (16), possibly via cyt c554.
Proteins of the NapC/NrfH family that have been studied include the 19 kDa NrfH isolated from Desulfovibrio desulfuricans (17), the 22 kDa NrfH from Wolinella succinogenes (18), and the pentaheme NrfB from Escherichia coli (19), all of which transfer electrons to a specific periplasmic pentaheme cytochrome c nitrite reductase, NrfA; the 20 kDa CymA from Shewanella frigidmarina (20) that delivers electrons to insoluble Fe(III) terminal reductase; and the pentaheme 46 kDa TorC from E. coli (21) and 46 kDa DorC from Rhodobacter capsulatus (22) of the TMAO/DMSO reducing respiratory systems. Recently, the X-ray structure of the tetraheme NrfH from Desulfovibrio vulgaris was determined (23). The primarily α-helical protein is composed of a periplasmic domain that binds the four hemes and a single N-terminal transmembrane helix anchor. NrfH formed a dimer (which had not been predicted) in the crystal and was in complex with its NrfA, in an NrfA4H2 overall arrangement. A novel feature of NrfH, residing at the edge of the putative quinone reaction site, is the high-spin pentacoordinate heme 1 coordinated with Met from a CXXCHXM motif. Coordination of heme 4 of NrfH is also unusual: a Lys residue from NrfA provides the distal ligand. The pentaheme NrfB from E. coli represents another interesting variation within the NapC/NrfH family; it has, in effect, an added C-terminal heme thought to be involved in electron transfer to NrfA. Lacking a quinone-binding site, NrfB accepts electrons from a quinol dehydrogenase containing an iron–sulfur center (19).
The octaheme HAO and the pentaheme NrfA are hypothesized to have common ancestry on the basis of the similarity of the amino acid sequence within their pentaheme protein domains, secondary and tertiary structures, and the spatial arrangement of their hemes (14). Cyt cm552, which accepts electrons from HAO, and NrfH, the tetraheme electron donor to NrfA, are homologous on the basis of sequence and are found in a gene cluster with HAO and NrfA, respectively. Thus, a NrfA/NrfH system that facilitated the reduction of nitrite to ammonia by quinol is thought to have evolved, in a later oxygen-containing environment, to an HAO/cyt cm552 system which reduced quinone coupled to the oxidation of hydroxylamine to nitrite (14).
In N. europaea, the terminal oxidase of the cytochrome aa3 family (24) and ubiquinone-8 (25) have been purified, and a cytochrome bc1 complex has been shown to mediate electrons from ubiquinone to cytochrome aa3 (26). Two membrane-associated c-type cytochromes were previously isolated (27) but not characterized in detail. A preliminary enrichment and description of the optical spectrum of cyt cm552 has been reported (26, 28). With the purification and characterization of it described herein, membrane cyt cm552 is the first NapC/NrfH protein to be characterized whose role is hypothesized to be the reduction of quinone. Cyt cm552 contains the unusual heme-binding CXXCHXM motif seen in NrfH protein from D. vulgaris. We report the first optical, EPR, and Mössbauer spectroscopic evidence of the resulting high-spin heme. The similarity in sequence and predicted secondary structure between cyt cm552 and NrfH from D. vulgaris led to contruction of a homology model of cyt cm552, allowing the tentative identification of ligands to the hexacoordinate hemes and residues in and near a putative quinone binding site. The latter appear to be suited to the presumed quinol reductase function of cyt cm552.
MATERIALS AND METHODS
Growth of Cells and Preparation of Cell-Free Extracts
N. europaea cells were continuously cultured in ammonia minimal salt medium in a 14 L bioreactor maintained at 29 °C (29). For 57Fe-enriched cells, trace levels of 56Fe were removed from the medium using Chelex-100 dialysis and supplemented with 0.17 mg/L 57Fe dissolved in 6 N HCl (6). Cells were harvested utilizing a Millipore Pelicon tangential flow filtration system and centrifuged at 20000g for 1 h at 4 °C. The resulting cell paste was resuspended to a concentration of 20% wet cell weight per volume in 50 mM potassium phosphate (KPi, pH 7.8) and stored at −20 °C. For lysis, cells in 50 mM KPi (pH 7.8), containing 5 mM benzamidine, were subjected to three freeze–thaw cycles. After the first cycle, the cell suspension was treated with deoxyribonuclease II to reduce viscosity. Membranes were sedimented by centrifugation at 20000g and 4 °C for 1 h and washed with 50 mM KPi (pH 7.8), and a series of salt (1 M KCl in 50 mM KPi) and deionized water washes were performed to remove peripherally bound proteins (e.g., cytochrome c554 and HAO) (30). The resulting salt-washed membranes were suspended in 4 volumes (v/w) of 50 mM KPi and stored at −20 °C.
Isolation and Chromatographic Purification
Salt- and water-washed membranes (approximately 25 g) were suspended in 250 mL of solubilization buffer [50 mM KPi (pH 7.8), 10% Triton X-100, 20% ethylene glycol, 400 mM KCl, and 5 mM benzamidine] and stirred for 1 h (until the suspension became translucent) at room temperature or overnight at 4 °C. After centrifugation at 38000g for 30 min, a deep red supernate was collected. The solubilization step was repeated until the absorbency of the hemoprotein at 400 nm was no longer found in the supernate. The supernates were combined, concentrated to one-fifth of the original volume using an Amicon stir cell (YM-10 kDa), and dialyzed against 50 mM KPi.
Triton-solubilized membrane proteins were chromatographed on a Macro-Prep DEAE (Bio-Rad) anion-exchange column (2.5 cm × 20 cm) equilibrated with buffer A1 [20 mM Tris, 50 mM KPi (pH 7.8), 0.1% Triton, and 20% ethylene glycol]. Inclusion of 50 mM KPi in all running buffers provided dianions and improved elution of the acidic cyt cm552. Fractions were eluted using a linear salt gradient (from 0 to 1 M KCl) with buffer B1 (1 M KCl in buffer A) over 5 column volumes. Fractions containing cyt cm552, as determined by UV–vis spectra, were pooled and concentrated by ultrafiltration (YM-10) and dialyzed overnight against 20 mM Bis-Tris (pH 6.2). The dialyzed sample was further purified and detergent-exchanged on a SOURCE 15Q column (1.0 cm × 100 cm) equilibrated with buffer A2 [20 mM Bis-Tris (pH 6.2) and 0.01% DDM]. Fractions were eluted using a linear salt gradient (from 200 to 600 mM KCl) with buffer B2 [buffer A2 (pH 6.2) with 1 M KCl] over 10 column volumes. For cleanup and desalting, fractions containing cyt cm552 were pooled and concentrated to ~20 mg/mL protein using ultrafiltration and loaded onto a Toyopearl HW-55S column (1.6 cm × 70 cm) equilibrated with 20 mM NaHEPES (pH 7.2), 0.1% DDM, and 300 mM NaCl. Purity was confirmed using SDS–PAGE in conjunction with MALDI-TOF mass spectrometry; the identity was confirmed by N-terminal sequencing and the optical spectrum.
Biochemical Analyses
Protein concentrations were determined using the bicinchoninic acid (BCA) assay kit (Pierce) with bovine serum albumin as the standard. SDS–PAGE analyses were performed using NuPAGE 10% Bis-Tris gels with 2-morpholinoethanesulfonic acid (MES) running buffer. Gels were stained using Coomasie G-250 and examined for the presence of heme using peroxidase activity staining (31). N-Terminal sequencing was performed at the Mayo Clinic Protein Core Facility (Rochester, MN). Purified samples were submitted as solutions and applied to a PVDF membrane. Heme a, b, and c content was determined by the method described by Berry and Trumpower (32).
Preparation of Membrane Samples for EPR
N. europaea cells (23 g) were harvested and resuspended to 40% (w/v) in 50 mM KPi (pH 7.0), treated for 30 min at 30 °C with lysozyme, DNase II, and RNase A, and then subjected to three cycles of French pressure treatment at 15000 psi. Highmolecular mass components were removed by centrifugation at 35000g for 2 h at 4 °C, and then the supernate was ultracentrifuged at 100000g and 4 °C for 1.75 h. The pellet was washed in 50 mM KPi (pH 7.0), and twice with 1 M KCl, followed by a water-only wash. The resulting membranes were resuspended in 100 mM KPi (pH 7.0) to 75% (w/v) and homogenized on an orbital shaker at 100 rpm at 4 °C for 45 min. The EPR sample of oxidized membranes was prepared by adding 1 μL of 10 mM potassium ferricyanide in double-distilled H2O to 1 mL of membranes in a 2.5 mL syringe (net ferricyanide concentration of 10 mM). The sample was injected into the EPR tube at 4 °C using a Sage Instruments 341B Syringe Pump over approximately 1.5 h and then frozen rapidly in liquid N2.
EPR and Mössbauer Spectroscopy
EPR spectra of cyt cm552 samples were recorded on a Bruker 300 spectrometer equipped with an Oxford ESR-910 liquid helium cryostat and a Bruker bimodal cavity for generation of the microwave fields parallel and perpendicular to the static field. The quantification of all signals was relative to a CuEDTA spin standard. The microwave frequency was calibrated with a frequency counter and the magnetic field with a NMR gaussmeter. The sample temperature of the cryostat was calibrated using a calibrated carbon-glass resistor (LakeShore CGR-1-1000) placed in an EPR tube to mimic a sample. A modulation frequency of 100 kHz was used for all EPR spectra. All experimental data were collected under nonsaturating conditions. Specific experimental conditions are listed in the figure captions. Mössbauer spectra were obtained on a constant-acceleration instrument, and isomer shifts are reported with respect to an iron metal standard. All displayed spectra were recorded on 57Fe-enriched protein. The reduced cyt cm552 sample was prepared by the addition of an excess of an anaerobic sodium dithionite (Sigma) solution to a thoroughly degassed protein sample.
Optical Spectroscopy
UV–visible specta were collected using a Cary-14 spectrophotometer with an OLIS 4300S Spectroscopy Operating System (Online Instrument Systems, Inc., Bogart, GA) or on a Varian Cary 50 spectrophotometer thermostated with a Varian Peltier. CO was bound to a thoroughly degassed sample of 1.9 μM (protein concentration) cyt cm552 in a septum-sealed cuvette by either bubbling CO for a few seconds or by adding 100 μL via a Hamilton syringe and then adding an anaerobic sample of sodium dithionite. The CN− complex was obtained by adding a 100-fold molar excess of CN− to a 0.37 μM solution of cyt cm552 and following the spectral change for 60 min. Complete spectral shift occurred within 30 min. The reduced cyt cm552–CN− complex was obtained by adding sodium dithionite to the ferric complex.
Construction of the Homology Model
The computational three-dimensional (3D) coordinates for the raw backbone structure of cyt cm552 were constructed with the crystal structure of NrfH (PDB entry 2J7A) as a template using Swiss-Model server (swissmodel.expasy.org). Incorporation of the four hemes into the protein backbone was then performed using the Maestro modeling suite (Schrödinger) operating under UNIX at the University of Minnesota Supercomputing Institute. After heme incorporation, potential axial ligands to the heme irons were located by searching for atoms within a 3.5 Å sphere. This step was critical for identifying potential ligands for heme 1 (proximal) and heme 4 (distal); for all four hemes, atoms of no other likely residues besides the identified ligands were within the 3.5 Å sphere. After ligand identification, bonds were manually coordinated between the heme irons and the coordinating atoms of the ligands. Energy minimizations of the raw model were performed after heme incorporation and ligand coordination to provide the lowest-energy conformation. The final model was rendered for presentation using either Maestro or PyMOL (DeLano Scientific).
RESULTS
Extraction and Purification
Isolation of the membrane–envelope complex from Nitrosomonas cells by multiple freeze–thaw cycles and low-spin centrifugation has been shown to provide greater enrichment of membrane electron transport components compared to sonication, French pressure, or osmotic shock treatments (25, 30). Typically, 25 g of wet cell paste yielded approximately 9–10 g of washed membranes, of which 7 g could be solubilized with 10% Triton X-100. In general, both Triton X-100 and n-dodecyl β-D-maltoside were effective in solubilizing cyt cm552; however, using comparable detergent:protein ratios (approximately 0.5–1.5), Triton more selectively extracted cyt cm552 relative to other membrane proteins.
The first anion-exchange chromatography of the detergent-solubilized supernatant was carried out on a support of moderate bead size (50 μm) using Tris running buffers (pH 7.8). During the development of the step, it was noted that the majority of cyt cm552, in which 27 of the 44 C-terminal residues are aspartic or glutamic acids (see below), remained bound to the DEAE support even when the gradient of KCl in the elution buffer exceeded 1 M. Although the negatively charged HPO42− is typically unsuitable for anion-exchange chromatography, when KPi (100 mM) was included in the elution buffer, it apparently acted as counter-dianion to the DEAE and facilitated the elution of a narrow band of cyt cm552 relatively late in the salt gradient (>0.6 M KCl). This step separated most of the contaminating proteins. It also served to remove the majority of solubilized lipids and excess Triton X-100, which in turn improved separation achieved in the subsequent chromatographic step.
Pooled fractions were applied 1 pH unit above the protein's calculated pI of 4.2 to a higher-resolution anion-exchange column with a smaller bead size (15 μm) and eluted with a much narrower gradient. This step also served to exchange Triton with DDM as the detergent. After the two chromatographic steps, the protein was determined to be essentially pure on the basis of SDS–PAGE, gel filtration, and N-terminal sequencing analyses (see below). The best 410 nm (Soret) to 280 nm absorbency ratio was 5.6.
Heme Types
The presence of c-type but not a- or b-type heme was indicated by the presence of an absorbance maximum at only 550 nm in the α-band region of the pyridine ferrohemochrome spectrum.
Primary Structure, Electrophoretic Profile, and Oligomerization State
The predicted molecular mass of cyt cm552 is 27134 Da after cleavage of the signal peptide [MTR-LQKG7 (−814 Da) deduced from N-terminal sequencing analysis] and the attachment of the four c-type hemes (616 Da × 4); thus, it is one of the larger proteins in the NapC/NrfH family. Its theoretical pI is 4.2 due to the high abundance of Glu and Asp residues (12.4 and 9.7%, respectively, of total residue composition). Remarkably, these residues make up the two most abundant amino acids, whereas Glu and Asp occur on average in proteins at 6.2 and 5.5%, respectively (33). These two residues account for 27 of the last 44 residues of the C-terminus. To the best of our knowledge, no other protein is known to have such a localized high percentage of acidic residues.
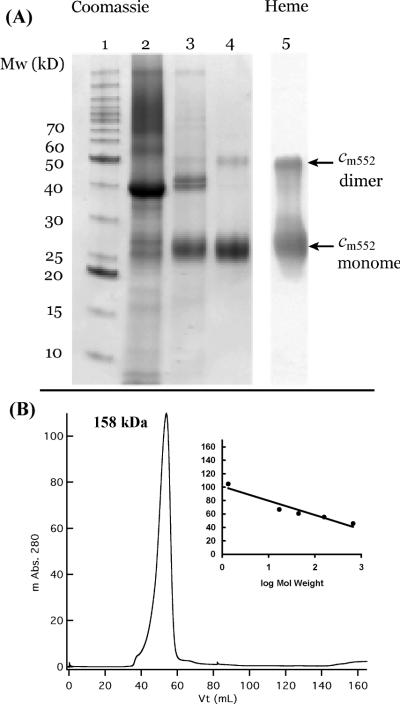
SDS–PAGE analysis of purified cyt cm552 showed a major band at 27 kDa and a relatively minor but strong band at 49 kDa (Figure 1A, lane 4). Both bands stained positively for c-type heme using heme-linked peroxidase staining. The band at 27 kDa is in agreement with the molecular mass of 27.1 kDa calculated from the amino acid sequence. The 49 kDa band was not a contaminant protein; N-terminal sequencing of the purified sample showed >90% purity when mole ratios of major to minor amino acids detected in each cycle were compared (from two independent samples). The detected sequence (SIGTLLTGAL) corresponded exactly to residues 8–18 of cyt cm552.
Figure 1.
Electrophoretic profile of cyt cm552 and oligomerization state of the purified product. (A) SDS–PAGE analysis of fractions during purification: lane 1, molecular mass standards; lane 2, Triton X-100-solubilized membrane fraction; lane 3, post-first anion-exchange chromatography (Macro-Prep DEAE) in the presence of Triton X-100; lane 4, post-second anion-exchange chromatography (Source Q)/detergent exchange with DDM and size exclusion (Toyopearl HW 55S) chromatography; and lane 5, heme stain of lane 4. (B) Gel filtration chromatography of purified cyt cm552 in the presence of the detergent DDM showing its oligomerization state. The inset shows column calibration with thyroglobulin (670 kDa), gamma globulin (158 kDa), ovalbumin (44 kDa), and myoglobin (17 kDa).
The 49 kDa band strongly suggests the presence of high-affinity dimers that are characterized by a migration rate faster than that of the monomeric species, and an equilibrium between the monomeric and dimeric species in the presence of SDS (34). This dimer behavior resembles that of glycophorin A in vitro, which is mediated by a tight transmembrane helix–helix interaction (35, 36). These transmembrane helices primarily associate by a GXXXG sequence motif that can be found in the transmembrane segments of cyt cm552 and NapC/NrfH proteins in general (Figure S1 of the Supporting Information).
Micellar gel filtration of purified cyt cm552 on Toyopearl HW-55S in the presence of DDM resulted in a single, relatively symmetric peak and further confirmed a multimeric fold. The peak, at 158 kDa (Figure 1B) based on column calibration, corresponds to a dimer with an expected DDM micelle size between 40 and 50 kDa (37), indicating a monomer plus micelle apparent molecular mass of 79 kDa. This interpretation is in keeping with the crystal structure of the cyt cm552 homologue NrfH (23) and the electrophoretic profile observed here. However, we cannot completely exclude the existence of a tetramer containing a classic four-helix bundle assuming a single micelle per tetramer (27 × 4 + 56 ≈ 166 kDa).
Homology-Based Structure of Cyt cm552 by Comparison with the NrfH X-ray Structure
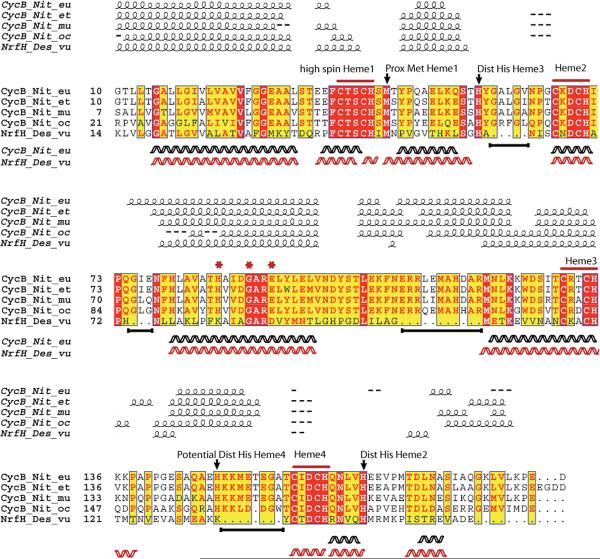
Cyt cm552 is a close homologue of NrfH (13–15). An alignment of the relevant regions of the primary structure of NrfH of D.vulgarus and cyt cm552 of N. europaea showed 37.6% identical sequence (Figure 2). The inclusion of CycB proteins (cyt cm552) from other ammonia-oxidizing bacteria helped support the identification of key residues in both NrfH and CycB proteins. This analysis was also extended to other NapC/NrfH family members to provide additional confidence (Figure S1). Figure 2 also shows the locations of α-helices in the crystal structure of NrfH, elements of the predicted secondary structure of NrfH and cyt cm552, and amino acid residues of interest.
Figure 2.
Clustal-W multiple-sequence alignment of NrfH from D.vulgaris and CycB proteins from ammonia oxidizers. Predicted secondary structure (helical) elements are shown above the alignment, while helices below represent elements based on the crystal structure (NrfH) and homology modeling (cyt cm552). Alignment analyzed using ESPript (Easy Sequencing in PostScript) visualization. A white letter with a red background designates strict identity; a red letter designates similarity in a group, and a yellow background designates similarity across groups. Arrows indicate probable axial ligands in CycB proteins based on alignment to NrfH (see the text). Asterisks indicate aligned ligands proposed to be involved in Q binding in NrfH (see the text). Black lines below the alignment represent homologous regions in CycB not present in NrfH.
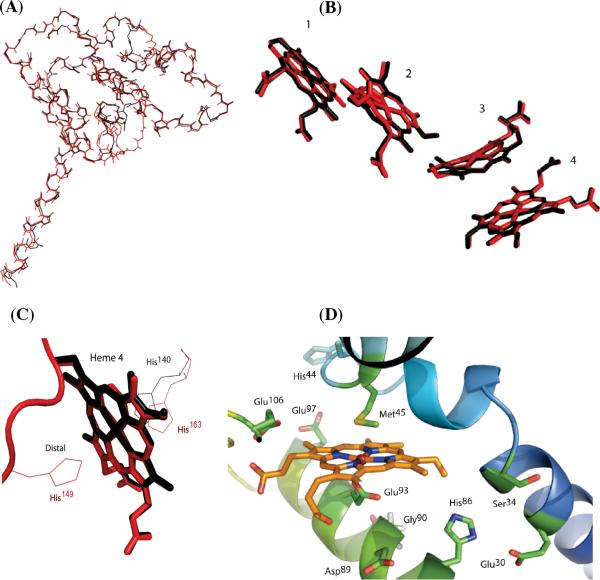
Comparison of the primary sequence of NrfH to that of CycB revealed regions of 5, 3, 11, and 8 residues present in cyt cm552 but not in NrfH (Figure 2). These segments were not modeled into the backbone structure of cyt cm552. Interestingly, these four regions are found in all nitrifiers and contain similar amino acid sequences. Secondary structure prediction for the CycB proteins in the nitrifiers indicates helical regions that correspond reasonably well to the predicted and experimental (based on X-ray crystallography) helical regions of NrfH. This supported the possibility that the proteins have a sufficiently similar general fold to enable modeling. The resulting 3D model suggests that cyt cm552 is similar to NrfH in overall tertiary structure (Figure 3A) and approximate location of the four hemes (Figure 3B).
Figure 3.
Superimposed structures of NrfH (black) from D.vulgaris (PDB entry 2J7A) and cyt cm552 model (red) coordinates: (A) main chain peptide backbone, (B) hemes 1–4, (C) heme 4 showing the potential distal His ligand to cyt cm552, and (D) putative Q-binding pocket in cyt cm552 according to molecular modeling.
The distal His axial ligands to hemes 2 and 3 in NrfH correspond to His57and His168 in cyt cm552. The axial ligand to heme 4 of NrfH of D.vulgarus is a Lys of the electron-accepting partner protein, the nitrite reductase (NrfA), which crystallized in a very tight complex with NrfH. In contrast, cyt cm552 was readily purified in pure form as a dimer and, as shown in subsequent sections, contains three low-spin hexacoordinate hemes. On the basis of the hypothesized structure of cyt cm552 shown in Figure 3, a His-containing strand of residues between helix 4 and heme 4 appears to lie in a resonable spatial position for ligation of His149 to heme 4 (Figure 3C). That His is conserved in all the nitrifiers shown.
Q-Binding Site and Tentative Assignment of Heme Ligands
Figure 3D represents the putative Q-binding site for cyt cm552 in the homology model based on the NrfH structure. The unusual proximal Met ligand to pentacoordinate heme 1 is predicted to be structurally retained. The negatively charged Asp residue that occupies the heme 1 distal position (not coordinated to iron) of NrfH is replaced with a Glu residue in nitrifiers and is structurally conserved. Of particular interest is the presence of several acidic residues in and near the Q-binding pocket that are not present in NrfH (namely, Glu106, Glu97, Asp89, and Glu30). Their presence effectively doubles the number of acidic residues in or near the site as compared to NrfH.
UV–Vis Spectra
(1) Ferric and Ferrous Cytochrome
The UV–visible spectrum of the as-isolated ferricytochrome cm552 at pH 7.8 was characterized by a maximum at 408 nm for the Soret (γ) band and, in the Q-band region, a broad peak at 532 nm with a weak shoulder at 550 nm (Figure 4). These features are found with other cytochromes belonging to the NapC/NrfH family. The ferricytochrome spectrum was also characterized by a weak band at ~625 nm, which is not seen with other NapC/NrfH cytochromes, and indicating a ligand–metal charge transfer (LMCT) typically observed for high-spin heme iron (38). Although homology modeling predicts a Met-ligated high-spin heme for cyt cm552, it must be emphasized that His, or other ligands to the high-spin heme iron, could also have resulted in a 625 nm band. The intensity of this band varied slightly between preparations but was consistently present at neutral pH. Prolonged (>48 h) exposure to room temperature did not alter the intensity of this band. A band at 695 nm indicative of His–Fe–Met coordination was not seen in the spectrum. Addition of ferricyanide did not alter the as-isolated spectrum.
Figure 4.
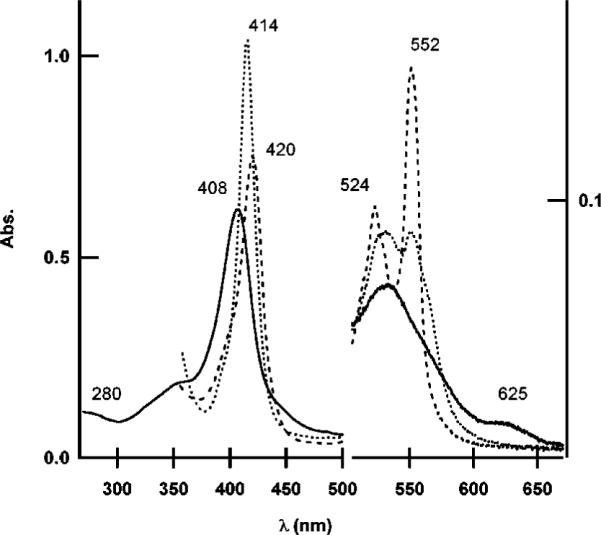
UV–vis spectra of cyt cm552: as-isolated ferric (—), dithionite-reduced ferrous (---), and equilibrium ferrous–CO complex after saturation with CO gas (• • •). All spectra were recorded at pH 7.8 in 20 mM Tris-HCl buffer and 0.02% DDM.
Dithionite reduction of cyt cm552 resulted in Soret (γ), β, and α bands at 420, 524, and 552 nm, respectively. Typically, the ferrous γ absorbency increased by ~20% relative to the ferric γ absorbency. No significant reduction was observed in the presence of ascorbate and TMPD even under anaerobic conditions.
(2) Apparent Displacement of the Distal Ligand by Small Molecules
The following small molecules were tested for binding to oxidized and/or reduced cyt cm552 at neutral pH: CO, CN−, F−, NO2−, and N3−. The anions F−, NO2−, and N3− did not bind. As shown below, CN− and CO appeared to displace the distal heme ligand and bind with all or most hemes of the enzyme. All ferrous hemes were very rapidly oxidized in the presence of O2.
Ferrous cyt cm552 formed a CO complex. The transitions upon binding of CO to dithionite-reduced cyt cm552 (Figure 4) included a Soret shift to 414 nm with an increase in peak sharpness and intensity, and a decrease in β and α bands with new maxima at 534 and 550 nm. The sharp, symmetric shape of the Soret band suggests a single heme environment; hence, more than one and possibly all hemes were accessible to solvent and bound CO, indicating a weak iron–distal His bond that can be displaced by CO. Introduction of O2 into the sealed cuvette containing the anaerobic CO–ferrocytochrome cm552 complex resulted in full return to its original ferricytochrome spectrum. Incubation of ferric cyt cm552 (pH 7.8) in the presence of a 100-fold excess of KCN for 30 min resulted in a red shift of the Soret band to 413 nm and a decrease in intensity (Figure 5). The broad peak at ~532 nm in the Q-band region was slightly red-shifted to ~538 nm. The CT band at ~625 nm was lost, suggesting that the high-spin heme formed a complex with CN−. Ferrous cyt cm552 also binds cyanide, resulting a red shift of the Soret band to 414 nm and an increase in intensity. In the Q-band region, the α and β peaks were centered at 526 and 555 nm, respectively, with increased sharpness relative to those of ferrous cyt cm552. Cyanide appears to have bound to more than one and possibly all hemes of ferrous cyt cm552.
Figure 5.
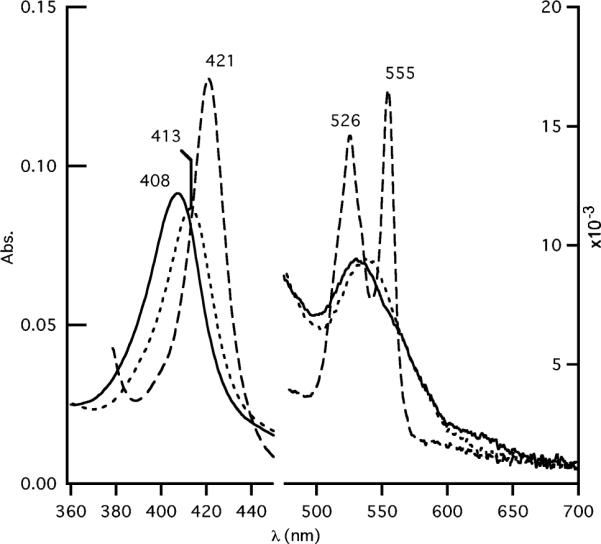
CN− complexes of ferrous and ferric cyt cm552 at pH 7.8: (—) oxidized without CN−, (• • •) ferricytochrome after incubation for 30 min with a 100-fold molar excess of KCN, and (---) dithionite reduction of the CN− complex. All spectra were recorded at pH 7.8 in 20 mM Tris-HCl buffer and 0.02% DDM.
Mössbauer Spectroscopy
Mössbauer spectra at 4.2 K of 57Fe-enriched as-isolated and reduced cyt cm552 are shown in Figure 6. The as-isolated spectrum (Figure 6A) in the absence of a magnetic field shows a set of broad unresolved quadrupole doublets. Spectra recorded at higher temperatures (>100 K, not shown) display a similar broad unresolved doublet pattern. In the presence of a small magnetic field (0.45 mT, not shown), the pattern broadens further and produces no features that can be assigned to typical low-spin ferric hemes. The positions of these broad doublets are consistent with several overlapping low-spin or high-spin Fe(III) heme species. Such heme species are paramagnetic and would normally display a six-line pattern in a magnetic field. However, this pattern is not observed, indicating that the Fe atoms of all hemes in the protein experience a relatively low hyperfine field due to a small electronic spin expectation. The low-spin expectation can occur as a result of electronic exchange interactions between hemes. Thus, all hemes in cyt cm552 are part of a spin-exchange interaction with the other hemes.
Figure 6.
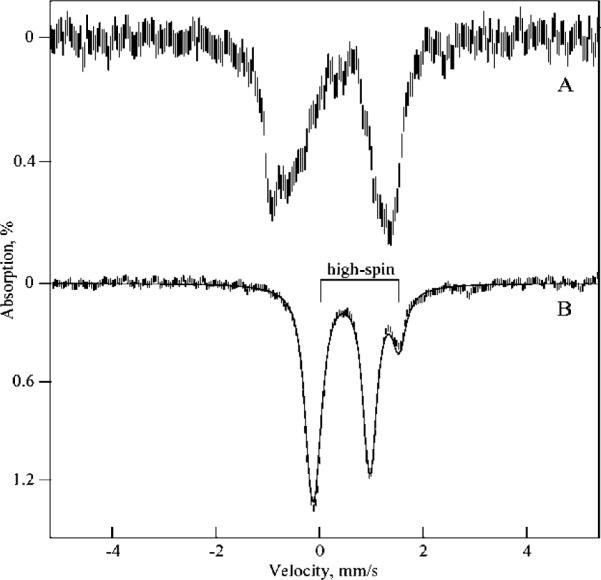
Low-temperature (4.2 K) Mössbauer spectra of (A) as-isolated and (B) dithionite-reduced cyt cm552, recorded in the absence of an applied magnetic field. The bracket marks the doublet arising from a ferrous high-spin heme. The solid line is a fit composed of four species of equal amounts with parameters typical of low- and high-spin ferrous hemes.
Upon addition of excess sodium dithionite to an anaerobic sample of cyt cm552, a sharper quadrupole doublet pattern is observed (Figure 6B). The simulation shown (solid line) is composed of one Fe site (25% of the area, doublet marked) where δ = 0.76 mm/s, ΔEQ = 1.55 mm/s, and three Fe sites (75% of the area) with 0.40 mm/s < δ < 0.41 mm/s and 1.00 mm/s < ΔEQ < 1.25 mm/s. These parameters are typical of high-spin (S = 2) and low-spin (S = 0) reduced heme centers, respectively. Thus, the Mössbauer data of the reduced protein clearly indicate the presence of high-spin and low-spin hemes in a ratio of 1:3. No effects of an exchange interaction between the hemes would be expected for this state of the protein since three of the hemes are now in S = 0 diamagnetic states.
EPR Spectroscopy
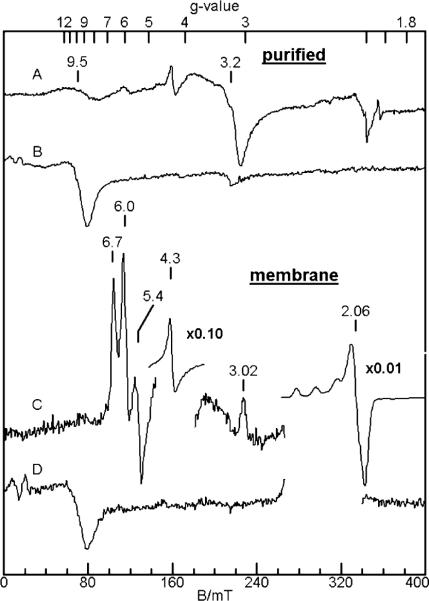
EPR spectra of purified cyt cm552 are shown in parts A and B of Figure 7 for orientations of the microwave field perpendicular (B1 ⊥ B) and parallel (B1 ∥ B) to the static magnetic field, respectively. The parallel mode spectra are shown at 2 K due to a better signal-to-noise ratio. The perpendicular mode spectrum shows signals at g = 9.5 and 3.2; no change in the spectrum is observed at 12 K in comparison to 2 K, except that the intensity was greater at the lower temperature. The spectrum is shown at 12 K to allow comparison to the spectrum of the membrane sample (see below), as some of the species in the membrane sample saturate at the lower temperature. The signals at g = 4.3 and 2.06 are from adventitious Fe(III) and Cu(II) minority species, respectively; both species have concentrations of <1% of the protein concentration. None of the signals from cyt cm552 can be associated with that expected for isolated low-spin or high-spin heme species. Indeed, the g = 9.5 signal intensifies in parallel mode (Figure 7B), indicating the presence of an electronic interaction between hemes of cyt cm552. Since no standard heme signals are observed, and to be consistent with the Mössbauer results, all hemes of cyt cm552 must be involved in spin-exchange interactions. The temperature dependence of the g = 9.5 and 3.2 signals between 2 and 20 K is the same, with signal intensities that are proportional to 1/T, with some diminution of intensity (relative to 1/T) near 20 K (data not shown). This dependence indicates that these signals are, or are very near, the ground doublet. These signals are consistent with an assignment to a high-spin/low-spin pair of hemes, similar to those observed for cyt c554 (a). It is unexpected that EPR signals are not observed from the other heme pair, since interacting low-spin heme pairs show signals in other multi-heme proteins, such as cyt c554 (9) and HAO (7). Presumably, one or both of these low-spin hemes exhibit a highly anisotropic type of symmetry; such hemes are referred to as large gmax hemes (39). Such anisotropic low-spin hemes display broad, low-intensity features in isolation and, when spin-coupled, may be too broad for observation.
Figure 7.
Perpendicular (A and C) and parallel (B and D) mode X-band EPR spectra of purified cyt cm552 (A and B), and N. europaea membrane (C and D) and purified cyt cm552 (C and D). Experimental conditions: microwave frequency, 9.6 GHz (B1 ⊥ B) or 9.3 GHz (B1 ∥ B); temperature, 12 K (B1 ⊥ B) or 2 K (B1 ∥ B); microwave power (A) 2 mW, (B and C) 0.2 mW, or (D) 0.02 mW.
Parts C and D of Figure 7 are EPR spectra of the cell membranes of N. europaea. The perpendicular mode spectrum (Figure 7C) shows intense signals from rhombic high-spin heme (g = 6.7 and 5.4; E/D = 0.026), axial high-spin heme (g = 6.0; E/D = 0), low-spin heme (g = 3.0), rhombic Fe(III) species (g = 4.3), and Cu(II) (g = 2.06). Spin quantification of these signals indicates that the rhombic high-spin heme, low-spin heme, and rhombic Fe(III) species are comparable in concentration, 0.02 mM. The concentration of axial high-spin heme species (0.002 mM) is 1 order of magnitude lower, and the concentraton of Cu(II) species (1 mM) significantly higher. These signals are from metal centers with half-integer spin and thus dominate the perpendicular mode spectrum. In parallel mode (Figure 7D), however, all signals from isolated half-integer spin species are forbidden and therefore vanish. The parallel mode spectrum shows the same g = 9.5 signal observed in the spectrum of purified cyt cm552. The intensity of the signal is comparable to that obtained from a 0.2 mM sample of purified protein, indicating the high abundance of cyt cm552 in the bacterial membrane. This also indicates that the unique integer-spin signal was not an artifact of purification of the enzyme, which started with washes of the membrane with 1 M KCl in 50 mM KPi and then deionized water and then detergent solublilization. The Cu(II) signal (g ~ 2) in parallel mode is attenuated by more than 100-fold but still present due to imperfect alignment of B1 and B.
Figure 8A shows the perpendicular (solid lines) and parallel mode (dashed lines) EPR spectra of an anaerobic sample of cyt cm552. The results of titration with sodium dithionite follow. The first addition (Figure 8B) of reductant results in a decrease in the g = 9.5 and 3.2 signals and the appearance of a new isolated high-spin heme species with g = 6.09 and 5.73 signals (E/D = 0.007). The next addition (Figure 8C) results in loss of the g = 9.5 and 3.2 signals and a further increase in the level of the isolated high-spin species, yet further addition of reductant results in a loss of signal intensity from the high-spin heme species (Figure 8D). The reduction of cyt cm552 also results in the appearance of a minority heme species (<5%) at g = 2.01. Closer examination of the latter signal reveals a triplet hyperfine pattern with splitting of 2 mT (not shown). This is typical of a five-coordinate ferrous heme–nitrosyl species. At present, the origin of the nitric oxide, and this resulting heme–nitrosyl species, is unknown. The exposure of the completely reduced sample to air resulted in the immediate recovery of the as-isolated protein signals, again indicating that the hemes are highly autoxidizable.
Figure 8.
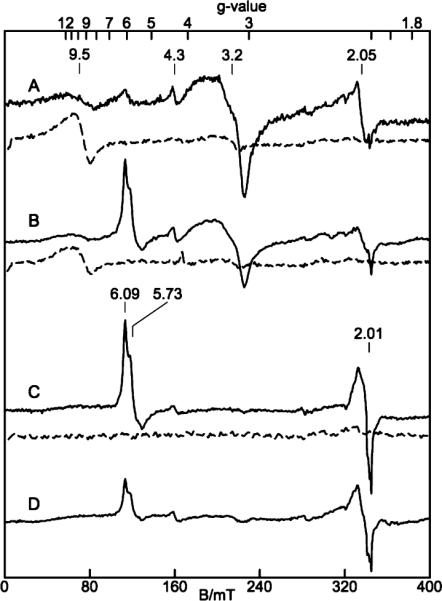
EPR spectra of a reductive titration of an anaerobic sample of cyt cm552 (~0.2 mM). Solid lines are for B1 ⊥ B (9.65 GHz) and dashed lines for B1 ∥ B (9.30 GHz): (A) purified protein and (B–D) successive additions of sodium dithionite. Experimental conditions: temperature, 14 K (B1 ⊥ B)or2K(B1 ∥ B); microwave power for parallel mode spectra, 0.2 mW; microwave power for perpendicular mode spectra, (A) 0.2 or (B–D) 2 mW.
DISCUSSION
Putative Q Reductase versus Q Dehydrogenase
The large NapC/NrfH family of tetraheme c cytochromes transfer electrons from quinol to a variety of periplasmic terminal oxidoreductases which reduce compounds such as nitrate, nitrite, DMSO, etc., for anaerobic respiration. Cyt cm552, encoded by the cycB gene from aerobic ammonia-oxidizing N. europaea, is the first member of the NapC/NrfH family to be characterized whose role is hypothesized to be the reduction of quinone. Several observations support the role of cyt cm552 as a quinone reductase rather than a dehydrogenase. First, the inclusion of its gene in the HAO gene cluster with cyt c554 suggests that cyt cm552 accepts electrons from either HAO or cyt c554. This genetic evidence is significant because the structural genes of cyt cm552 homologues have been found to lie in clusters with other genes that are redox partners (e.g., NirT lies in the NirSTBM cluster where NirS is the cyt cd1 nitrite reductase; NrfH lies in the NrfHAIJ cluster where NrfA is the redox partner cyt c nitrite reductase). Second, the genomes of nitrifiers N. europaea and Nitrosomonas oceanus lack the periplasmic terminal oxidoreductases usually associated with anaerobic respiration; only one type of a terminal oxidase (cytochrome aa3 oxidase) is present (24, 40), suggesting an inability to “respire anaerobically” by those pathways. At low oxygen concentrations, N. europaea has been shown to possess dissimilatory nitrite reductase activity (denitrification) possibly catalyzed by the nirK protein, a copper-type nitrite reductase (41). However, there is no precedent for a NapC/NrfH family member having a redox relationship with a NirK protein.
In this study, cyt cm552 was isolated and purified from N. europaea cultured under aerobic conditions. A small-scale preparation using cultures grown in a bioreactor with regulated dissolved oxygen levels confirmed that cyt cm552 is expressed in cells grown at an 80% saturation dissolved oxygen concentration (~0.2 mM O2 at 28 °C and 1 atm). In contrast, the members of the NapC/NrfH family involved in anaerobic respiration can be isolated only in cultures grown under anaerobic conditions. This observation argues against a NapC/NrfH-like role of cyt cm552 in anaerobic metabolism.
High-Spin Heme Site and Heme–Heme Interactions
The tetraheme NrfH of D. vulgarus contains a high-spin heme and a CXXCHXM heme c binding motif in which Met rather than His is coordinated to the heme iron (23). The quinone-binding site has been suggested to be near this heme. Amino acid sequence analysis indicates that this crucial heme binding motif is conserved in the vast majority of NapC/NrfH members. The rare exceptions are notable in that the replacement has a carboxamide side chain; the NapC proteins from Haemophilus ducreyi, Pasteurella multocida, and Photobacterium profundum and the CymA of Shewanella putrefaciens (Figure S1) all contain an Asn in place of the Met residue. A Gln for Met replacement is seen in the NapC from Haemophilus influenzae. The structural similarity of the Met versus Asn/Gln side chains and putative “conservative replacement” of the Met with Asn or Gln suggest that iron ligation by the nitrogen of the carboxamide side chain of Asn or Gln deserves consideration. Thus, it could be argued, on the basis of sequence information alone, that the residue position (CXXCHXM/N/Q) appears significant relative to iron binding. There is precedent for a similar displacement of the His in the CXXCH motif of a high-spin active site c heme; a lysine residue serves as the axial ligand to the active site of the octaheme tetrathionate reductase from Shewanella oneidensis (42). In the crystal structure of both tetrathionate reductase and NrfH, the His points away from the heme iron.
In light of the prevalent CXXCHXM motif in the NapC/NrfH family, and the crystallographic evidence for a high-spin heme in NrfH from D. vulgaris, it is surprising that spectroscopic characterization of isolated NapC/NrfH members has thus far indicated four low-spin, bis-His-ligated hemes (16–21), and spectroscopic indication of a possible Met-ligated heme in NrfH from D. desulfuricans has not been reported (17). It must be stated, however, that there is only one example of a methionine-coordinated high-spin heme described in the literature; however, the corresponding EPR spectrum consists of two species, and an assignment of E/D to the methionine-ligated heme species is not available (43). In our work, the EPR spectra of partially reduced cyt cm552 (Figure 8B–D) revealed signals typical for an isolated high-spin heme at g = 6.09 and 5.73 and E/D = 0.007. This E/D value is within the range observed for histidine coordination. This value increases to 0.04 or 0.09 for tyrosine or cysteine axial ligation, respectively (44). Thus, while the E/D value observed for cyt cm552 is similar to that of histidine-ligated high-spin hemes, a definite conclusion about the nature of the proximal ligand cannot be made on the basis of EPR spectra. Considering the interesting disconnect between spectroscopic data (present and previous) and crystallographic/sequence data, the possibility of ligand switching between His and Met residues of the CXXCHXM motif ought to be considered.
From this work, it can be concluded that Mössbauer, EPR, and optical spectroscopies here all indicate the presence of high-spin and low-spin heme centers in cyt cm552. The Mössbauer spectrum of fully reduced cyt cm552 shows doublets with parameters typical of high- and low-spin ferrous heme species, in a ratio of 1:3. The Mössbauer and EPR data both indicate that all hemes in cyt cm552 are involved in spin interactions. The spin-coupled EPR signals of as-isolated cyt cm552 are similar to those observed from the high-spin/low-spin heme pairs of cyt c554 (9) and NiR (45, 46), suggesting that cyt cm552 also contains diheme motifs similar to those observed in other multi-heme proteins (47). The addition of an excess of ferricyanide to an as-isolated cyt cm552 did not change the EPR spectrum, indicating that all four hemes are oxidized in the as-isolated state. Partial reduction of cyt cm552 results in the loss of the spin-coupled EPR signals and the appearance of an isolated Fe(III) heme species with nearly axial symmetry. This is attributed to reduction of the low-spin heme of the high-spin/low-spin heme pair. Since no other low-spin heme signals are observed, this implies that the hemes are arranged in two separated pairs, the other pair consisting of two spin-interacting low-spin hemes. NrfH (23) shows a physical arrangement of four hemes forming two pairs (hemes 1 and 2 and hemes 3 and 4). On the basis of the sequence homology with NrfH and the EPR data presented here, the hemes of cyt cm552 may form similar pairings.
As shown here, membrane samples of Nitrosomonas show the same parallel mode EPR signal as that observed from purified cyt cm552. Hence, the signal was not an artifact of solubilization and purification. This signal is distinct from other proteins in Nitrosomonas known to have heme–heme interactions: HAO (7) and cyt c554 (9). Similar spininteracting EPR signals have not been observed for other proteins of the NapC/NrfH family (17, 18).
Ligand Binding
The binding of small exogenous ligands to cytochromes in the NapC/NrfH family has not been previously demonstrated, but a variety of c cytochromes containing at least one high-spin heme bind small ligands like CO, N 3−, and CN−. Cyt cm552 gave no evidence of binding the anionic ligands (F−, NO2−, and N3−), which may be due to charge repulsion from the proposed acidic residue(s) in the heme pocket. Since the high-spin heme environment of cyt cm552 has more negatively charged residues than the corresponding site of NrfH, it will be interesting to see whether NrfH binds anionic ligands.
CO and CN− appear to bind with multiple hemes of cyt cm552. This is reminiscent of ligand binding to the tetraheme cyt c554, where a high-pH-induced form containing three low-spin hemes apparently reacts with CO and CN− (6). A similar result is observed for the tetraheme cyt c3 that contains four low-spin hemes and each heme has a different Kd for CO (48). These data suggest a relatively weak distal His–Fe bond in the low-spin hemes and that the hemes are accessible to solvent.
Dimerization Motif
We report here the first isolation of a protein from the NapC/NrfH family as a homodimer which is stable in SDS–PAGE. The multimeric state of cyt cm552 was evidenced by its migration profile on an SDS–PAGE gel and from its elution volume on gel filtration. No redox partner protein was co-isolated with cyt cm552, although HAO was present in solution throughout the purification prior to the final chromatographic step. We also note, in the transmembrane helix predicted from the amino acid sequence of cyt cm552, the GXXXG motif which has been shown to mediate a dimeric fold (34, 35). Although previously unrecognized, the G(X)3–4G motif is well represented in an amino acid sequence alignment of various NapC/NrfH members (Figure S1), suggesting that these other members may adopt a similar dimeric or other multimeric state. In fact, a close examination of the dimeric NrfH crystal structure (23) shows the two glycines of the GXXXG motif as the interfacial residues lining each transmembrane helix at the packing contact.
Axial Ligands of the Three Hexacoordinate Hemes
Homology modeling strongly suggested the identity of the distal His axial ligands of hemes 2 and 3 in cyt cm552 of Nitrosomonas to be His57 and His168, respectively, which are conserved in all the nitrifiers shown. The axial ligand to heme 4 of NrfH of D. vulgarus is a Lys of the electron-receiving partner protein, NrfA. A corresponding Lys is absent in cyt cm552, but His142 is modeled here in a reasonable position for ligation to heme 4. His142 is conserved in all ammonia-oxidizing nitrifiers. An alignment of cyt cm552 from Nitrosomonas with representative members of the NapC/NrfH family of cytochromes (Figure S1) shows His142 to align with either His (in NapC, CymA, and NirT) or Met (in TorC and DorC) residues. Mutation of this His residue in Paracoccus pantotrophus NapC and/or W. succinogenes NrfH (49, 50) has provided evidence that they are essential for electron transfer function and are likely to be distal ligands to a heme. Interestingly, on the basis of alignments of the NapC/NrfH family, NrfH proteins appear to be the only subgroup of the family to have a Lys from a partner protein as a heme ligand.
Quinone-Binding Site of Heme 1
The finding of an apparent Q-binding site in the homology model is the strongest evidence to date that cyt cm552 is a Q oxidoreductase. The Q-binding site near the Met-ligated heme 1 cavity in NrfH is lined with Asp89, Gly86, and Lys82 residues that are proposed to be involved in quinol binding and an Asp38 residue proposed to be involved in shuttling of protons to the periplasm upon dehydrogenation of quinol (23). Analysis of the corresponding region in cyt cm552 using both sequence alignment and the homology model shows a cavity lined with His86, Gly90, and Glu93 residues (possibly involved in Q binding) and a Ser34 residue (possibly involved in proton shuttling). Thus, the quinone-binding residues appear to be conserved, with a His (a weaker base) replacing a Lys. It is not obvious how this arrangement in cyt cm552 might better promote reduction and protonation of quinone in contrast to oxidation and deprotonation of quinol in NrfH. However, we note that there are twice as many (two additional) acidic residues in the putative Q-binding pocket in cyt cm552 (Glu97 replacing Asn93 and Asp89 replacing Ala85). Acidic residues within the putative Q-binding site could facilitate reduction by H-bonding with the quinol product and/or via movement of protons toward the site of protonation and reduction of quinone. The involvement of acid-rich protein regions in the movement of protons relative to a proton-active enzyme site has been reviewed (51).
A similar role is possible for the C-terminal, acid-rich domain (24 of 60 residues are Asp or Glu) of cyt cm552 if it is ultimately determined to be on the surface of the membrane in a position to concentrate protons near a point of entry to the Q-binding site. The acid-rich domain is also found in cycB protein (analogous to cyt cm552) from Nitrosomonas eutropha and Nitrosospira multiformis but not in other members of the NapC/NrfH family, suggesting that it may be relevant to quinone reduction on the P-face of the membrane. The absence of this feature in the marine ammonia oxidizer Nitrosococcus oceanus counters that hypothesis, although pH values may be different within the very unique stacks of membrane found in Nitrosococcus (52).
Identity of the Cyt cm552 Reductase
The identity of the immediate electron donor to cyt cm552 is unclear. Some observations implicate cyt c554, which shares an operon with cyt cm552 in the HAO gene cluster. A major fraction (50–100%) of cytochrome cyt c554 is found in fresh bufferwashed membranes, requires a 1 M KCl wash for removal (27), and could, in theory, have been associated with cyt cm552. An excellent docking site has been modeled from X-ray structures which places a heme of cyt c554 close enough to heme 1 of HAO for rapid electron exchange (4, 5). Furthermore, it has been demonstrated that cyt c554 can be reduced by HAO in the presence of hydroxylamine (53).
Some observations suggest possible roles for cyt c554 other than the reduction of cyt cm552. The highest observed in vitro rate of reduction of cyt c554 by hydroxylamine and HAO does not account for the aerobic in vivo turnover of hydroxylamine (53); however, the in vitro rate of reduction of the soluble monoheme cyt c552 by hydroxylamine and catalytic amounts of HAO and cyt c554 (54) would suffice. Electron flow from HAO to cyt c554 to cyt c552 would channel electrons into periplasmic reductive reactions, including cytochrome c peroxidase (55) or nitrite reduction. Reduction of the soluble cyt c552 would bypass the cyt cm552 Q reductase and the bc1 complex, possibly accounting for the observed in vivo reduction of oxygen by hydroxylamine in the presence of bc1 inhibitors (26). Cytochrome c554 can reduce NO (56), suggesting a role in the detoxification of NO.
Reduction of cyt cm552 directly by HAO would provide an electron path to quinone and then the bc1 complex. Evolutionary evidence suggests a binding site for cyt cm552 on HAO. The putative hydroxylamine-oxidizing protein pair of HAO and cyt cm552 (eight and four hemes, respectively) shares common ancestry with the nitrite-reducing protein pair of NrfA and NrfH (five and four hemes, respectively) (14). The catalytic heme 1 and the four electron transfer hemes of NrfA can be superimposed on the catalytic heme 6, and four of the seven electron transfer hemes of HAO (57, 58). Furthermore, homology in amino acid sequence is seen between the corresponding pentaheme domains of NrfA and HAO and also between the protein domains containing hemes 1–4 of NrfH and cyt cm552 (14). Many lines of evidence suggest that intermolecular electron transfer takes place at heme 2 of NrfA and heme 4 of NrfH, which are 12.1 Å apart, edge to edge (22, 23). By analogy, heme 5 of HAO [which corresponds to heme 2 of NrfA and has a proprionate exposed to solvent (3)] and heme 4 of cyt cm552 deserve consideration as a possible intermolecular electron transfer pair.
Supplementary Material
ACKNOWLEDGMENT
We thank Yuk Sham and the University of Minnesota Supercomputing Institute for assistance and resources, Tyler Yin for help with heme incorporation and energy minimizations in the cyt cm552 model, and Brad Elmore for review of the manuscript.
This work was supported by grants from the U.S. Department of Energy (DE-FG02-95ER20191) and the National Science Foundation (9723608) to A.B.H. and from the National Institutes of Health (GM-077387) to M.P.H.
Footnotes
Abbreviations: AMO, ammonia monooxygenase; cyt, cytochrome; HAO, hydroxylamine oxidoreductase; DDM, n-dodecyl β-D-maltoside; EPR, electron paramagnetic resonance; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate; Orf, open reading frame; TMPD, N,N,N,N-tetramethyl-p-phenylenediamine; LMCT, ligand–metal charge transfer.
SUPPORTING INFORMATION AVAILABLE An extended amino acid sequence alignment of NapC/NrfH/TorC family members that have been isolated and characterized. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Blackburn TH. The microbial nitrogen cycle. In: Krumbein WW, editor. Microbial Geochemistry. Blackwell Scientific Publications; Oxford, England: 1983. pp. 63–89. [Google Scholar]
- 2.Kowalchuk GA, Stephen JR. Ammonia-oxidizing bacteria: A model for molecular microbial ecology. Annu. Rev. Microbiol. 2001;55:485–529. doi: 10.1146/annurev.micro.55.1.485. [DOI] [PubMed] [Google Scholar]
- 3.Igarashi N, Moriyama H, Fujiwara T, Fukumori Y, Tanaka N. The 2.8 Å structure of hydroxylamine oxidoreductase from a nitrifying chemoautotrophic bacterium, Nitrosomonas europaea. Nat. Struct. Biol. 1997;4:276–284. doi: 10.1038/nsb0497-276. [DOI] [PubMed] [Google Scholar]
- 4.Iverson TM, Arciero DM, Hsu BT, Logan MS, Hooper AB, Rees DC. Heme packing motifs revealed by the crystal structure of the tetra-heme cytochrome c554 from Nitrosomonas europaea. Nat. Struct. Biol. 1998;11:1005–1012. doi: 10.1038/2975. [DOI] [PubMed] [Google Scholar]
- 5.Iverson TM, Arciero DM, Hooper AB, Rees DC. High-resolution structures of the oxidized and reduced states of cytochrome c554 from Nitrosomonas europaea. J. Biol. Inorg. Chem. 2001;4:390–397. doi: 10.1007/s007750100213. [DOI] [PubMed] [Google Scholar]
- 6.Andersson K, Lipscomb J, Valentine M, Munck E, Hooper A. Tetraheme cytochrome c-554 from Nitrosomonas europaea. Heme-heme interactions and ligand binding. J. Biol. Chem. 1986;261:1126–1138. [PubMed] [Google Scholar]
- 7.Hendrich M, Petasis D, Arciero D, Hooper A. Correlations of structure and electronic properties from EPR spectroscopy of hydroxylamine oxidoreductase. J. Am. Chem. Soc. 2001;123:2997–3005. doi: 10.1021/ja002982d. [DOI] [PubMed] [Google Scholar]
- 8.Hendrich M, Upadhyay A, Riga J, Arciero D, Hooper A. Spectroscopic characterization of the NO adduct of hydroxylamine oxidoreductase. Biochemistry. 2002;41:4603–4611. doi: 10.1021/bi011332z. [DOI] [PubMed] [Google Scholar]
- 9.Upadhyay A, Petasis D, Arciero D, Hooper A, Hendrich M. Spectroscopic characterization and assignment of reduction potentials in the tetraheme cytochrome c554 from Nitrosomonas europaea. J. Am. Chem. Soc. 2003;125:1738–1747. doi: 10.1021/ja020922x. [DOI] [PubMed] [Google Scholar]
- 10.Arciero D, Collins M, Haladjian J, Bianco P, Hooper A. Resolution of the four hemes of cytochrome c554 from Nitrosomonas europaea by redox potentiometry and optical spectroscopy. Biochemistry. 1991;30:11459–11465. doi: 10.1021/bi00112a013. [DOI] [PubMed] [Google Scholar]
- 11.Collins M, Arciero D, Hooper A. Optical spectro-potentiometric resolution of the hemes of hydroxylamine oxidoreductase. Heme quantitation and pH dependence of Em. J. Biol. Chem. 1993;268:14655–14662. [PubMed] [Google Scholar]
- 12.Kurnikov I, Ratner M, Pacheco A. Redox equilibria in hydroxylamine oxidoreductase. Electrostatic control of electron redistribution in multielectron oxidative processes. Biochemistry. 2005;44:1856–1863. doi: 10.1021/bi048060v. [DOI] [PubMed] [Google Scholar]
- 13.Bergmann DJ, Arciero DM, Hooper AB. Organization of the hao gene cluster of Nitrosomonas europaea: Genes for two tetraheme c cytochromes. J. Bacteriol. 1994;176:3148–3153. doi: 10.1128/jb.176.11.3148-3153.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergmann DJ, Hooper AB, Klotz MG. Structure and Sequence Conservation of hao Cluster Genes of Autotrophic Ammonia-Oxidizing Bacteria: Evidence for Their Evolutionary History. Appl. Environ. Microbiol. 2005;71:5371–5382. doi: 10.1128/AEM.71.9.5371-5382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson DJ. Bacterial respiration: A flexible process for a changing environment. Microbiology. 2000;146:551–571. doi: 10.1099/00221287-146-3-551. [DOI] [PubMed] [Google Scholar]
- 16.Roldan MD, Sears HJ, Cheesman MR, Ferguson SJ, Thomson AJ, Berks BC, Richardson DJ. Spectroscopic Characterization of a Novel Multiheme c-Type Cytochrome Widely Implicated in Bacterial Electron Transport. J. Biol. Chem. 1998;273:28785–28790. doi: 10.1074/jbc.273.44.28785. [DOI] [PubMed] [Google Scholar]
- 17.Almeida MG, Macieira S, Goncalves LL, Huber R, Cunha CA, Romao MJ, Costa C, Lampreia J, Moura JJG, Moura I. The isolation and characterization of cytochrome c nitrite reductase subunits (NrfA and NrfH) from Desulfovibrio desulfuricans ATCC 27774: Re-evaluation of the spectroscopic data and redox properties. Eur. J. Biochem. 2003;270:3904–3915. doi: 10.1046/j.1432-1033.2003.03772.x. [DOI] [PubMed] [Google Scholar]
- 18.Simon J, Gross R, Einsle O, Kroneck PMH, Kroger A, Klimmek O. A NapC/NirT-type cytochrome c (NrfH) is the mediator between the quinone pool and the cytochrome c nitrite reductase of Wolinella succinogenes. Mol. Microbiol. 2000;35:686–696. doi: 10.1046/j.1365-2958.2000.01742.x. [DOI] [PubMed] [Google Scholar]
- 19.Clarke T, Cole J, Richardson D, Hemmings A. The crystal structure of the pentahaem c-type cytochrome NrfB and characterisation of its solution-state interaction with the pentahaem nitrite reductase NrfA. Biochem. J. 2007;406:19–30. doi: 10.1042/BJ20070321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Field SJ, Dobbin PS, Cheesman MR, Watmough NJ, Thomson AJ, Richardson DJ. Purification and Magneto-optical Spectroscopic Characterization of Cytoplasmic Membrane and Outer Membrane Multiheme c-Type Cytochromes from Shewanella frigidimarina NCIMB40. J. Biol. Chem. 2000;275:8515–8522. doi: 10.1074/jbc.275.12.8515. [DOI] [PubMed] [Google Scholar]
- 21.Gon S, Giudici-Orticoni MT, Mejean V, Iobbi-Nivol C. Electron Transfer and Binding of the c-Type Cytochrome TorC to the Trimethylamine N-Oxide Reductase in Escherichia coli. J. Biol. Chem. 2001;276:11545–11551. doi: 10.1074/jbc.M008875200. [DOI] [PubMed] [Google Scholar]
- 22.Shaw AL, Hochkoeppler A, Bonora P, Zannoni D, Hanson GR, McEwan AG. Characterization of DorC from Rhodobacter capsulatus, a c-type Cytochrome Involved in Electron Transfer to Dimethyl Sulfoxide Reductase. J. Biol. Chem. 1999;274:9911–9914. doi: 10.1074/jbc.274.15.9911. [DOI] [PubMed] [Google Scholar]
- 23.Rodrigues M, Oliveira T, Pereira I, Archer M. X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel haem coordination. EMBO J. 2006;25:5951–5960. doi: 10.1038/sj.emboj.7601439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dispirito A, Lipscomb J, Hooper AB. Cytochrome aa3 from Nitrosomonas europaea. J. Biol. Chem. 1986;261:17048–17056. [PubMed] [Google Scholar]
- 25.Hooper AB, Erickson RH, Terry KR. Electron transport systems of Nitrosomonas: Isolation of a membrane-envelope fraction. J. Bacteriol. 1972;110:430–438. doi: 10.1128/jb.110.1.430-438.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittaker M, Bergmann D, Arciero D, Hooper A. Electron transfer during the oxidation of ammonia by the chem-olithotrophic bacterium Nitrosomonas europaea. Biochim. Biophys. Acta. 2000;1459:346–355. doi: 10.1016/s0005-2728(00)00171-7. [DOI] [PubMed] [Google Scholar]
- 27.DiSpirito AA, Taaffe LR, Hooper AB. Localization and concentration of hydroxylamine oxidoreductase and cytochromes c552, c554, cm552, cm553, and a in Nitrosomonas europaea. Biochim. Biophys. Acta. 1985;806:320–330. [Google Scholar]
- 28.Vannelli T, Bergmann D, Arciero DM, Hooper AB. Mechanism of N-Oxidation and Electron Transfer in the Ammonia-Oxidizing Autotrophs. In: Lidstrom ME, Tabita FR, editors. Proceedings of the 8th International Symposium on Microbial Growth on C1 Compounds; Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 80–87. [Google Scholar]
- 29.Logan MSP, Hooper AB. Organo-hydrazines are Suicide Substrates of Hydroxylamine Oxidoreductase of Nitrosomonas europaea. Biochemistry. 1995;34:9257–9264. doi: 10.1021/bi00028a039. [DOI] [PubMed] [Google Scholar]
- 30.McTavish H, Arciero DM, Hooper AB. Interactions with Membranes of Cytochrome c554 from Nitrosomonas europaea. Arch. Biochem. Biophys. 1995;324:53–58. doi: 10.1006/abbi.1995.9930. [DOI] [PubMed] [Google Scholar]
- 31.Thomas P, Ryan D, Levin W. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 32.Berry E, Trumpower B. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal. Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 33.Lehmann WD, Bohne A, von der Lieth CW. The information encrypted in accurate peptide masses: Improved protein identification and assistance in glycopeptide identification and characterization. J. Mass. Spectrom. 2000;35:1335–1341. doi: 10.1002/1096-9888(200011)35:11<1335::AID-JMS70>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 34.Melnyk RA, Kim S, Curran AR, Engelman DM, Bowie JU, Deber CM. The Affinity of GXXXG Motifs in Transmembrane Helix-Helix Interactions Is Modulated by Long-range Communication. J. Biol. Chem. 2004;279:16591–16597. doi: 10.1074/jbc.M313936200. [DOI] [PubMed] [Google Scholar]
- 35.MacKenzie KR, Prestegard JH, Engelman DM. A Transmembrane Helix Dimer: Structure and Implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 36.Smith S, Song D, Shekar S, Groesbeek M, Ziliox M, Aimoto S. Structure of the transmembrane dimer interface of glycophorin A in membrane bilayers. Biochemistry. 2001;40:6553–6558. doi: 10.1021/bi010357v. [DOI] [PubMed] [Google Scholar]
- 37.Warr GG, Drummond CJ, Grieser F, Ninham BW, Evans DF. Aqueous Solution Properties of Nonionic n-Dodecyl-β-D-Maltoside Micelles. J. Phys. Chem. 1986;90:4581–4586. [Google Scholar]
- 38.Makinen MW, Churg AK. Structural and Analytical Aspects of the Electronic Spectra of Hemeproteins. In: Lever ABP, Gray HB, editors. Iron Porphyrins, Part 1. Addison-Wesley; London: 1983. pp. 141–235. [Google Scholar]
- 39.Walker FA. Magnetic spectroscopic (EPR, ESEEM, Mössbauer, MCD and NMR) studies of low-spin ferriheme centers and their corresponding heme proteins. Coord. Chem. Rev. 1999;186:471–534. [Google Scholar]
- 40.Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, Hauser L, Hooper A, Klotz M, Norton J, Sayavedra-Soto L, Arciero D, Hommes N, Whittaker M, Arp D. Complete Genome Sequence of the Ammonia-Oxidizing Bacterium and Obligate Chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 2003;185:2759–2773. doi: 10.1128/JB.185.9.2759-2773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beaumont HJE, Hommes NG, Sayavedra-Soto LA, Arp DJ, Arciero DM, Hooper AB, Westerhoff HV, van Spanning RJM. Nitrite Reductase of Nitrosomonas europaea Is Not Essential for Production of Gaseous Nitrogen Oxides and Confers Tolerance to Nitrite. J. Bacteriol. 2002;184:2557–2560. doi: 10.1128/JB.184.9.2557-2560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mowat CG, Rothery E, Miles CS, McIver L, Doherty MK, Drewette K, Taylor P, Walkinshaw MD, Chapman SK, Reid GA. Octaheme tetrathionate reductase is a respiratory enzyme with novel heme ligation. Nat. Struct. Mol. Biol. 2004;11:1023–1024. doi: 10.1038/nsmb827. [DOI] [PubMed] [Google Scholar]
- 43.Barker PD, Nerou EP, Cheesman MR, Thomson AJ, de Oliveira P, Hill HAO. Bis-Methionine Ligation to Heme Iron in Mutants of Cytochrome b562. 1. Spectroscopic and Electrochemical Characterization of the Electronic Properties. Biochemistry. 1996;35:13618–13626. doi: 10.1021/bi961127x. [DOI] [PubMed] [Google Scholar]
- 44.Palmer G. In: The Porphirins. Dolphin D, editor. Vol. IV. Academic Press; San Diego: 1979. p. 328. [Google Scholar]
- 45.Bamford VA, Angove HC, Seward HE, Thomson AJ, Cole JA, Butt JN, Hemmings AM, Richardson DJ. Structure and spectroscopy of the periplasmic cytochrome c nitrite reductase from Escherichia coli. Biochemistry. 2002;41:2921–2931. doi: 10.1021/bi015765d. [DOI] [PubMed] [Google Scholar]
- 46.Costa C, Moura JJG, Moura I, Liu MY, Peck HD, LeGall J, Wang Y, Huynh Boi H. Hexaheme nitrite reductase from Desulfovibrio desulfuricans. Mossbauer and EPR characterization of the heme groups. J. Biol. Chem. 1990;265:14382–14388. [PubMed] [Google Scholar]
- 47.Brige A, Leys D, Meyer TE, Cusanovich MA, Van Beeumen JJ. The 1.25 Å Resolution Structure of the Diheme NapB Subunit of Soluble Nitrate Reductase Reveals a Novel Cytochrome c Fold with a Stacked Heme Arrangement. Biochemistry. 2002;41:4827–4836. doi: 10.1021/bi012144b. [DOI] [PubMed] [Google Scholar]
- 48.Takayama Y, Kobayashi Y, Yahata N, Saitoh T, Hori H, Ikegami T, Akutsu H. Specific binding of CO to tetraheme cytochrome c3. Biochemistry. 2006;45:3163–3169. doi: 10.1021/bi051867i. [DOI] [PubMed] [Google Scholar]
- 49.Cartron M, Roldan M, Ferguson S, Berks B, Richardson D. Identification of two domains and distal histidine ligands to the four haems in the bacterial c-type cytochrome NapC: The prototype connector between quinol/quinone and periplasmic oxidoreductases. Biochem. J. 2002;368:425–432. doi: 10.1042/BJ20020865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gross R, Eichler R, Simon J. Site-directed modifications indicate differences in axial haem c iron ligation between the related NrfH and NapC families of multihaem c-type cytochromes. Biochem. J. 2005;390:689–693. doi: 10.1042/BJ20050448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shutova T, Klimov VV, Andersson B, Samuelsson G. A cluster of carboxylic groups in PsbO protein is involved in proton transfer from the water oxidizing complex of Photosystem II. Biochim. Biophys. Acta. 2007;1767:434–440. doi: 10.1016/j.bbabio.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 52.Murray RGE, Watson SW. Structure of Nitrosocystis oceanus and Comparison with Nitrosomonas and Nitrobacter. J. Bacteriol. 1965;89:1594–1609. doi: 10.1128/jb.89.6.1594-1609.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arciero D, Balny C, Hooper AB. Spectroscopic and Rapid Kinetic Studies of Reduction of Cytochrome c554 by Hydroxylamine Oxidoreductase from Nitrosomonas europaea. Biochemistry. 1991;30:11466–11472. doi: 10.1021/bi00112a014. [DOI] [PubMed] [Google Scholar]
- 54.Yamanaka T, Shinra M. Cytochrome c552 and cytochrome c554 derived from Nitrosomonas europaea. Purification, properties and their function in hydroxylamine oxidation. J. Biochem. 1974;75:1265–1273. doi: 10.1093/oxfordjournals.jbchem.a130510. [DOI] [PubMed] [Google Scholar]
- 55.Arciero D, Hooper A. A di-heme cytochrome c peroxidase from Nitrosomonas europaea catalytically active in both the oxidized and half-reduced states. J. Biol. Chem. 1994;269:11878–11886. [PubMed] [Google Scholar]
- 56.Upadhyay A, Hooper A, Hendrich M. NO reductase activity of the tetraheme cytochrome c554 of Nitrosomonas europaea. J. Am. Chem. Soc. 2006;128:4330–4337. doi: 10.1021/ja055183+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Einsle O, Messerschmidt A, Stach P, Bourenkov GB, Bartunik HD, Huber R, Kroneck PMH. Structure of cytochrome c nitrite reductase. Nature. 1999;400:476–480. doi: 10.1038/22802. [DOI] [PubMed] [Google Scholar]
- 58.Einsle O, Stach P, Messerschmidt A, Simon J, Kroger A, Huber R, Kroneck PMH. Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 Å resolution, inhibitor binding, and heme-packing motifs. J. Biol. Chem. 2000;275:39608–3961. doi: 10.1074/jbc.M006188200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.