SUMMARY
Endogenous RNA-directed RNA polymerases (RdRPs) are cellular components capable of synthesizing new complementary RNAs from existing RNA templates. We present evidence for successive engagement of two different RdRPs in an endogenous siRNA-based mechanism targeting specific mRNAs in C. elegans soma. In the initiation stage of this process, a group of mRNA species are chosen as targets for downregulation, leading to accumulation of rare 26-nt 5′-phosphorylated antisense RNAs that depend on the RdRP homolog RRF-3, the argonaute ERGO-1, DICER, and a series of associated (“ERI”) factors. This primary process leads to production of a much more abundant class of 22-nt antisense RNAs, dependent on a secondary RdRP (RRF-1) and associating with at least one distinct Argonaute (NRDE-3). The requirement for two RdRP/Argonaute combinations and initiation by a rare class of uniquely-structured siRNAs in this pathway illustrate the caution and flexibility used as biological systems exploit the physiological copying of RNA.
INTRODUCTION
Gene silencing induced by small RNAs of approximately 20 to 30 nt in length is a widespread and diverse feature of eukaryotes (reviewed in Hutvagner and Simard, 2008). While small RNA pathways share certain characteristics such as reliance on Argonaute proteins and duplexed RNA structures, the modes of small RNA biogenesis and function in gene regulation show considerable diversity both within a single cell and between species. Small RNA machinery can be exploited for experimental purposes by delivery of foreign double-stranded RNA, which leads to silencing of matching endogenous RNAs by a process known as RNA interference (RNAi) (Fire et al., 1998). Single-stranded fragments of the input double-stranded RNA called short interfering RNAs guide the silencing machinery to their target mRNAs (Elbashir et al., 2001). In the nematode C. elegans, RNAi triggered by long double-stranded RNA involves two distinct phases: an initiating or primary response, and a secondary response. Each phase involves different sets of Argonautes, siRNAs, and other factors. Production of primary siRNAs involves dicing of long double-stranded RNA into siRNA duplexes by the RNase III-like enzyme DCR-1 (Bernstein et al., 2001; Grishok et al., 2001; Ketting et al., 2001; Knight and Bass, 2001). Production of secondary siRNAs involves RNA-directed RNA polymerase (RdRP) activity on target mRNAs, serving to amplify the silencing response (Sijen et al., 2001). The secondary response can also occur independently of DCR-1 when triggered by aberrant single-stranded RNAs (Aoki et al., 2007). One way of distinguishing primary and secondary siRNAs formed during RNAi is by 5′ chemical structure: primary siRNAs have a 5′ monophosphate characteristic of DCR-1 activity, while secondary siRNAs have a 5′ triphosphate characteristic of RdRP activity (Pak and Fire, 2007; Sijen et al., 2007).
Adding to the wealth of mechanistic data on exogenously induced RNAi, numerous studies have identified naturally occurring small RNAs with perfect complementarity to host transcripts from such diverse sources as fission yeast, nematodes, mouse oocytes, and immature maize ears (Ambros et al., 2003; Buhler et al., 2008; Nobuta et al., 2008; Reinhart and Bartel, 2002; Tam et al., 2008; Watanabe et al., 2008). These siRNAs are called endogenous siRNAs (endo siRNAs) to distinguish them from the siRNAs resulting from exogenously triggered RNAi (exo siRNAs). In C. elegans, both triphosphorylated and monophosphorylated endo siRNAs (hereafter referred to simply as siRNAs) have been identified. Known examples of these distinct types of siRNAs have characteristic length distributions, where the most abundant triphosphorylated species (secondary-type) are 22 nt, and the most abundant monophosphorylated species (primary-type) are 26-nt in length (Pak and Fire, 2007; Ruby et al., 2006). Both mono- and triphosphorylated siRNAs are predominantly antisense to corresponding mRNAs (Ambros et al., 2003; Pak and Fire, 2007; Ruby et al., 2006). Examples of siRNAs that are antisense to spliced exon-exon junctions indicate synthesis by a mechanism that can use a spliced mRNA transcript as a template (Pak and Fire, 2007; Ruby et al., 2006).
In many organisms, the RNAi machinery includes RdRPs, which use single-stranded RNA as templates for second strand synthesis (Cogoni and Macino, 1999; Dalmay et al., 2000; Makeyev and Bamford, 2002; Mourrain et al., 2000; Smardon et al., 2000). Three of the four putative C. elegans RdRPs have been implicated in siRNA-related activities: EGO-1 by a germline RNAi defective phenotype in loss-of-function mutants (Smardon et al., 2000), RRF-1 by a somatic RNAi defective phenotype in loss-of-function mutants as well as by direct assays for RRF-1-driven siRNA production in cell-free C. elegans extracts (Aoki et al., 2007; Sijen et al., 2001), and RRF-3 by the identification of certain genes whose endo siRNA accumulation is greatly reduced in loss-of-function mutants (Duchaine et al., 2006; Gent et al., 2009; Lee et al., 2006; Pavelec et al., 2009). Unlike EGO-1 and RRF-1, RRF-3 is dispensable for exogenously triggered RNAi; indeed, RNAi directed against many genes is more effective in rrf-3 mutants than in wildtype (Sijen et al., 2001; Simmer et al., 2002). These roles for RRF-3 in endogenous gene regulation and in exo RNAi efficacy are shared with a group of factors in the ERI pathway (so named because of Enhanced RNAi phenotypes). These include the widely conserved exoribonuclease and DCR-1 complex member ERI-1, the uncharacterized Dicer complex member ERI-9, DCR-1 itself, and the Argonaute ERGO-1 (Asikainen et al., 2007; Duchaine et al., 2006; Gabel and Ruvkun, 2008; Gent et al., 2009; Kennedy et al., 2004; Pavelec et al., 2009). All of these factors appear to have somatic roles, as their activity in siRNA production modulates the subcellular localization of a somatically expressed Argonaute NRDE-3, and their mutations increase efficacy of exo RNAi against somatic genes. Additional support for a somatic role comes from the results of Duchaine et al (2006), who found siRNAs from an ERI-regulated gene (K02E2.6) expressed in adult soma.
In this work, we characterize genetic requirements and small RNA products involved in a two-step RdRP pathway that modulates expression for a set of endogenous genes in C. elegans. RRF-3 initiates this pathway in concert with the Dicer/ERI complex and the Argonaute ERGO-1, leading to accumulation of a limited population of mRNA-complementary antisense siRNAs and engagement of a second RdRP, RRF-1. As a consequence of RRF-1 engagement, a much more abundant siRNA class is generated, with a distinct set of Argonaute interactions including NRDE-3. In addition to the differences in genetic requirements, we found that the siRNAs from each stage formed distinct structural populations, with the rare initial pool consisting of 26-nt 5′-monophosphorylated siRNAs while the more abundant RRF-1 pool consists of shorter (22 nt) siRNAs that evidently retain a 5′ triphosphate structure.
RESULTS
Identification of somatic RRF-3 targets
The coexistence of two RdRPs in somatic cell populations in C. elegans (Sijen et al. 2001; Simmer et al. 2002) suggests the possibility of multistage regulation of small RNA responses. To investigate potential interplay of these two RdRPs in gene regulation, we first identified a representative set of RRF-3 targets. Our recent studies of RRF-3 roles in spermatogenesis have identified a group of target genes that are downregulated through RRF-3 function during spermatogenesis (Gent et al., 2009; Pavelec et al., 2009). To extend target characterization to somatic tissues, we first analyzed siRNA sequence datasets from three independent pairs of wildtype and rrf-3 mutant animals. For the initial analysis, we used an RNA library preparation protocol that can capture siRNAs with 5′ structures with or without a triphosphate. The three sequence datasets used for this analysis were (i) a paired set of larval (L4 stage) hermaphrodite animals (mutant and wildtype) raised at 16°C and (ii) two paired sets of adult males (mutant and wildtype) raised at 25°C. Comparisons within each pair allowed us to identify a set of genes with strong dependence on RRF-3 for siRNA accumulation in populations of different sex, stage, and temperature. Among these candidate RRF-3 targets, we compared each gene s siRNA counts from the rrf-3 mutant and control samples and looked for genes with at least a fivefold decrease in siRNA counts (normalized by total number of siRNAs) and a P-value of less than 0.001 for each of the three comparisons (Table 1; Figures 1A and S1A).
Table 1. An exemplary list of genes for which siRNA accumulation depends on RRF-3 (RRF-3 target genes).
The 23 genes in the table were chosen by having at least 5-fold reduction in siRNA count with a P-value lower then 0.001 in the rrf-3 mutant relative to control sample in the three different paired sample comparisons (two male and one hermaphrodite). The protein domain annotations were taken from Wormbase (www.wormbase.org) or from the GO annotation database (http://www.geneontology.org).
| Gene name | Position | Protein Domains |
|---|---|---|
| C18D4.4 | ChrV:17531478..17530471 | VMO-I domain |
| C36A4.11 | ChrIII:3840574..3841129 | |
| C44B11.6 | ChrIII:3152726..3153450 | |
| E01G4.5 | ChrII:13472340..13473832 | Protease inhibitor I4, serpin Lipocalin domains |
| F07G6.6 fbxa-52 | ChrX:1711208..1710289 | FTH domain |
| F14F7.5 | ChrIII:13030800..13036223 | WSN domain |
| F39E9.7 | ChrII:3307113..3306027 | Double-stranded RNA binding domain |
| F52D2.6 | ChrX:1974908..1980932 | |
| F55A4.4 | ChrX:1026174..1024644 | Double-stranded RNA binding domain |
| H09G03.1 | ChrIII:3226341..3230641 | |
| H16D19.4 | ChrI:12644416..12640727 | DNA helicase PIF1/RRM3 |
| K02E2.6 | ChrV:20375969..20377425 | Retroviral aspartyl protease |
| K06B9.6 | ChrIV:4196688..4196166 | |
| T08B6.2 | ChrIV:4896819..4894362 | 7-transmembrane receptor |
| W04B5.1 | ChrIII:2428212..2429351 | |
| W04B5.2 | ChrIII:2424343..2427460 | |
| W05H12.2 | ChrI:13412867..13411785 | 3′-5′ exonuclease activity |
| Y17D7B.4 | ChrV:18780182..18776168 | |
| Y17D7C.1 | ChrV:18707977..18711610 | |
| Y37E11B.2 | ChrIV:3578441..3579386 | |
| Y43F8B.9 | ChrV:19479168..19480372 | |
| Y46D2A.1 | ChrII:3323335..3326426 | DUF274 family |
| ZK380.5 | ChrX:1649233..1647987 |
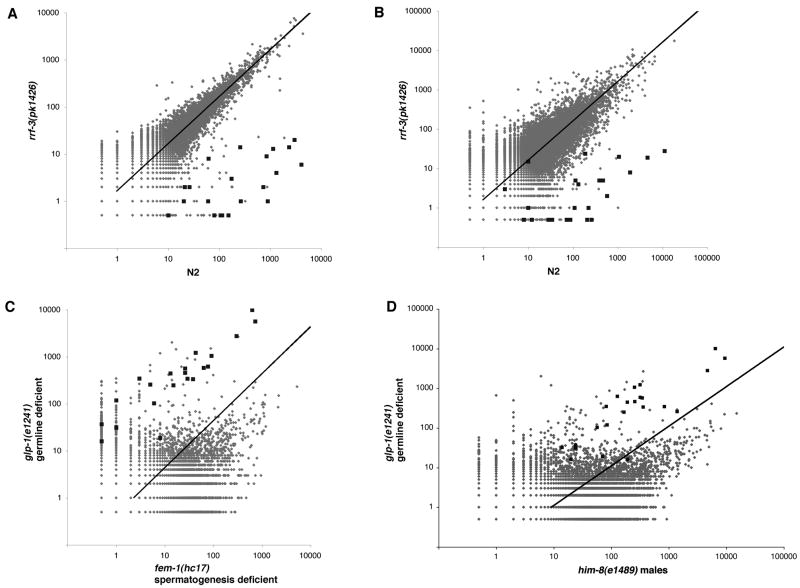
Figure 1. Identification of an exemplary set of somatic RRF-3 targets.
(A, B) Scatter plots depict comparisons of gene-by-gene siRNA counts from one of the three paired rrf-3 mutant and wildtype samples used to identify RRF-3 target genes (A) and from an independent pair of rrf-3 mutant and wildtype samples (B). The siRNA counts were obtained using a 5′-phosphate-independent small RNA capture and sequencing method. The resulting sequences were aligned to a WS190 cDNA dataset. Each dot s X- and Y-coordinates represents a gene s siRNA count from the indicated samples. The dark squares correspond to the 23 RRF-3 target genes identified by reduced siRNA count in rrf-3 mutant v. wildtype in all three comparisons (at least fivefold reduction with a P-value of less than 0.001). Lighter dots correspond to the rest of the 21133 genes in the cDNA reference dataset. Genes with siRNA counts of zero were assigned a value of 0.5 to avoid exclusion from the plot (axes are in logarithmic scale). The trendline denotes the expected siRNA counts per gene had the samples been derived from identical populations. The trendline varies from the diagonal due to difference in total sequence counts between samples. (C,D) Scatter plots depict gene-by-gene siRNA counts from germline mutants. Germline-deficient glp-1(e2141) animals were compared with female (fem-1(hc17): [C]) and male (him-8(e1489): [D]) animals. The RRF-3 targets (dark squares) and other genes above the trendline have higher relative siRNA abundance in glp-1(e2141) soma-only animals than in either fem-1(hc17) or him-8(e1489) animals with their combined somas and germlines. See also supplementary Figure S1.
Among the 23 genes that met these criteria were K02E2.6 and C44B11.6, two genes whose effective siRNA accumulation has been shown in other studies to require RRF-3 (Duchaine et al., 2006; Lee et al., 2006). For brevity, we refer to the 23 genes as exemplary RRF-3 targets; we emphasize that these represent a sample of RRF-3 regulatory targets, not an exhaustive list. Other genes behaved similarly but did not quite reach our threshold values or expressed siRNAs at too low a level in one or more samples to detect definitive differences between rrf-3 mutant and control. To guard against bias due to overrepresentation of individual sequences in our siRNA dataset, we carried out an independent analysis with “collapsed” sets of sequences (allowing each sequence to occur only once in each sample). After collapsing the dataset, the 23 RRF-3 targets still had greatly reduced siRNA counts in the rrf-3 mutant as compared to the N2 sample (Figure S1A). To validate the RRF-3 targets, we sequenced small RNA libraries from a fourth biological replicate (i.e., an independently prepared pair of samples from rrf-3(pk1426) and N2 L4 hermaphrodites). All but 2 of the 23 RRF-3 targets had greater than fivefold reduction in siRNA counts; 16 had greater than 100-fold reduction (Figure 1A,B). F52D2.6, the more significant of the two exceptions, was validated as a target in further experiments (Figures 3 and S3). The other, C18D4.4, produced too few siRNAs in the fourth replicate to make a confident call.
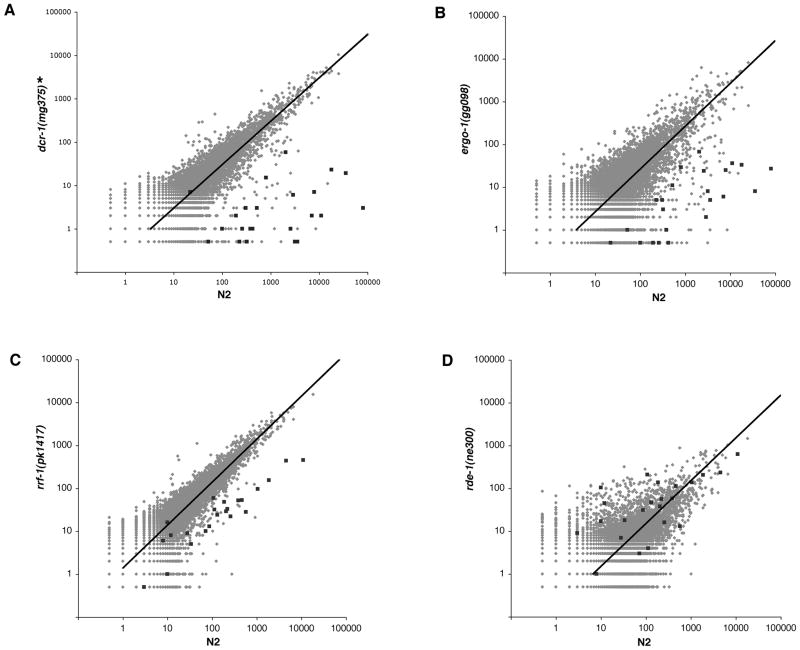
Figure 3. Identification of additional RNAi factors in the RRF-3 pathway.
(A–D) Scatter plots depict gene-by gene siRNA counts from RNAi mutants and N2 control. Strains carrying mutations in dcr-1 (A), ergo-1 (B), and rrf-1 (C) showed decreased siRNA counts for the 23 RRF-3 targets (dark squares), while the rde-1 mutant (D) did not. (*) As described in Experimental Procedures, the characterized dcr-1(mg375) strain YY011 has recently been shown to carry a background mutation upstream of the mut-16 gene; while this mutation does not account for the ~100 fold effect on numerous RRF-3 target RNAs, we cannot rule out a contribution of background to the observed effects (see Experimental Procedures). See also Figure S3.
To further investigate the source of the putative non-spermatogenesis-associated siRNAs, we compared RNA profiles from two germline deficient mutants, glp-1(e2141) and glp-4(bn2) with patterns from males (derived from him-8(e1489)) and females (derived from fem-1(hc17)). Three of these strains (glp-1(e2141), glp-4(bn2), and fem-1(hc17)) are temperature-sensitive such that they produce self-fertile hermaphrodites at 16°C. For each sample, we thus prepared libraries from adult animals of each of the four strains raised at the nonpermissive temperature of 25°C. We found that all of the 23 RRF-3 targets had high relative siRNA counts in the germline deficient mutants, consistent with these siRNAs being substantially enriched in somatic tissue (Figure 1C,D, S1B, Table S1). Interestingly, these data also indicate that a preponderance of the total siRNA population was present in the germline rather than soma.
Analysis of genome-wide, RRF-3 signatures on siRNA length distributions, orientations, and nucleotide frequencies
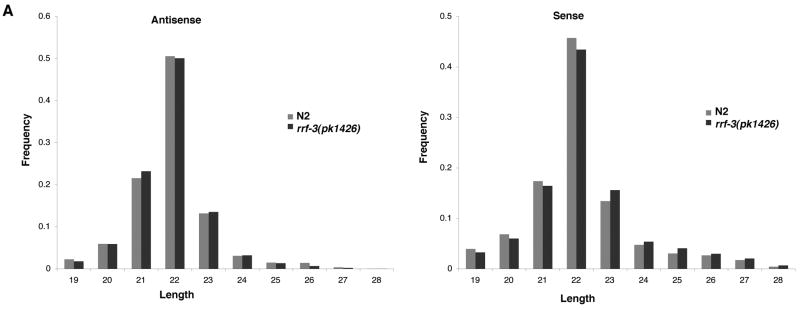
Given that detectable changes in siRNA counts between rrf-3 mutant and wildtype samples were limited to a small number of genes, we asked whether effects of RRF-3 might be evident genome-wide if we extended the analysis beyond simply siRNA counts to include lengths, nucleotide frequencies, and abundance of sense verses antisense siRNAs. In both rrf-3 mutant and wildtype samples, the antisense-oriented sequences accounted for greater than 95% of the total sequences (Table S1). Sense and antisense oriented siRNAs from both samples showed a predominant length of 22 nt (Figure 2) and a strong enrichment for a 5′ guanosine (~75%, regardless of genotype, Figure S2A). The siRNAs from the 23 exemplary RRF-3 targets showed the same biases towards a length of 22 nt and preferred starting base of guanosine, even among the residual siRNAs present in the rrf-3 mutant (Figures S2A,B). We conclude from these results that RRF-3 has subtle effects on total siRNAs at a genome-wide level in terms of abundance, lengths, and nucleotide preferences; but specific genes do depend upon RRF-3 for siRNA accumulation.
Figure 2. Analysis of genome-wide RRF-3 effects on siRNA lengths.
Histograms of siRNA length distributions indicate similar enrichment for 22 nt siRNAs from the 5′-phosphate-independent sequence libraries for rrf-3 mutant and wildtype. The reads from the samples represented in Figure 1B were separated based on alignment orientation, and the frequency of each length between 19 and 28 nt tabulated for both antisense (left) and sense (right). See also Figure S2.
Requirements for the Dicer complex, the ERGO-1 Argonaute, and the RdRP RRF-1 in the somatic RRF-3 pathway
To identify other factors required with RRF-3 in somatic gene regulation, we measured siRNA abundance for the RRF-3 targets in various mutants of the somatic exogenous RNAi pathway and ERI pathway by deep sequencing. Loss-of-function mutations of dcr-1 itself were not useful tools for such comparisons due to early and severe effects of complete loss of function on development (Grishok et al., 2001; Ketting et al., 2001; Knight and Bass, 2001). Instead, we made use of a specific helicase domain allele, dcr-1(mg375), which loses DCR-1 contribution to ERI function but which retains miRNA, piRNA, and exo RNAi function and allows viability (Pavelec et al., 2009). In dcr-1(mg375) mutant populations, siRNA counts for the 23 RRF-3 targets dropped dramatically (15 had greater than 100-fold reduction; Figure 3A). A complicating feature of this analysis is a genetic background difference in the characterized dcr-1(mg375) strain YY011 [see Experimental Procedures]. Although numerous strains that share the background show much less dramatic decreases in RRF-3 target siRNA levels, it remains possible that the dcr-1(mg375) effect is potentiated in this genetic background. In exo RNAi, DCR-1 functions in concert with the double-stranded RNA binding protein RDE-4 (Parrish and Fire, 2001; Tabara et al., 2002). Northern blots for siRNAs corresponding to one of the 23 RRF-3 targets, C44B11.6, have shown a requirement for RDE-4 in C44B11.6 siRNA accumulation (Lee et al., 2006). RDE-4 was also required for RRF-3 target siRNA accumulation: in rde-4(ne299) animals, 18 of the 23 RRF-3 targets had greater than 5-fold reduction and 14 had greater than 100-fold reduction (Figure S3C). Similarly, both ERI-1 and ERI-9, members of the DCR-1 complex, were required: in eri-1(mg366) animals, 22 had greater than 5-fold reduction, and 15 had greater than 100-fold reduction (Figure S3A). In eri-9(gg106) animals, 23 had greater than 5-fold reduction, and 16 had greater than 100-fold reduction (Figure S3B).
Two Argonautes were strong candidates for the primary Argonaute in the somatic ERI pathway—RDE-1, because of its known function in exo RNA pathways following DCR-1 activity (Parish and Fire), and ERGO-1, because of its ERI phenotype and its interaction with downstream ERI Argonaute NRDE-3 (Pavelec et al., 2009; Yigit et al., 2006). We found that ERGO-1 but not RDE-1 was required for siRNA accumulation. In ergo-1(gg098) animals, 23 of the 23 RRF-3 targets had greater than 5-fold reduction, and 11 had greater than 100-fold reduction (Figure 3B); while in rde-1(ne300) only 1 had greater than 5-fold reduction, and none had greater than 100-fold (Figure 3D).
While the exo RNAi component RDE-1 was dispensable for RRF-3 target siRNA accumulation, another exo RNAi component, the RdRP RRF-1, was evidently required for efficient accumulation. In siRNAs sequenced from NL2098 [rrf-1(pk1417)] animals, 18 of 23 RRF-3 targets had greater than 5-fold reduction, and 13 had greater than 100-fold reduction (Figure 3C). This result was notable in the indication that efficient accumulation of these siRNAs required the activity of two RdRPs. Combined with the fact that RRF-1 produces secondary siRNAs during exo RNAi, this suggested a process involving multiple stages of siRNA synthesis and potentially two distinct subsets of siRNAs, one dependent upon RRF-3 and the DCR-1 complex, and the other upon RRF-1.
ERI pathway siRNAs reduce target mRNA levels
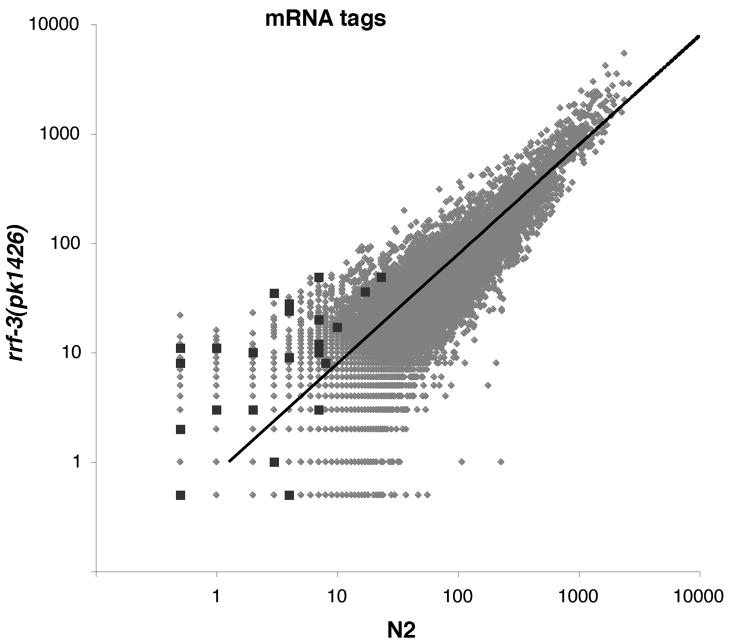
A possible consequence of the DCR-1 complex in the RRF-3 pathway would be cleavage of the transcripts being used for double-stranded RNA synthesis. In addition, downstream silencing steps could involve downregulation of mRNA levels. In particular, siRNAs synthesized by RRF-1 have been shown to induce target degradation (Aoki et al., 2007). In mutants of the ERI pathway, in which siRNAs levels are reduced, mRNA levels would be predicted to be elevated. This is indeed the case for ERI targets during spermatogenesis (Asikainan et al., 2007; Gent et al., 2009; Pavelec et al., 2009). To measure RRF-3 effects on mRNA levels, we prepared sequencing libraries from mRNA fragments (tags) from rrf-3 mutant and N2 L4 hermaphrodites raised at 16°C. Although the ability to measure mRNA abundance changes can be limited for genes with relatively small mRNA tag counts, we observed a strong trend toward increased RRF-3 target mRNA levels in the rrf-3 mutant (on average 3.8-fold increase; Figure 4).
Figure 4. Inverse association between siRNA and mRNA levels for RRF-3 target genes.
The scatter plot depicts gene-by-gene mRNA tag counts from RNA-seq libraries derived from N2 and rrf-3(pk1426). Genes with mRNA tag counts of zero were assigned a value of 0.5 to avoid exclusion from the plot (axes are in logarithmic scale). Dark squares correspond to the 23 exemplary RRF-3 target genes. Lighter dots correspond to the rest of the 21133 genes in the cDNA reference dataset. The trendline denotes the expected mRNA fragment alignment counts had the samples been derived from identical populations. The trendline varies from the diagonal due to difference in total read counts between samples. See also Table S2.
To test whether RRF-1, ERI-1, and RDE-4 are required for mRNA downregulation in soma, we compared the 23 RRF-3 targets with previously published microarray analyses of mRNA levels in mutants of rrf-1, eri-1, and rde-4 (Lee et al., 2006; Welker et al., 2007). All three mutant datasets show a strong trend toward increased RRF-3 target mRNA levels (Table S2). In contrast, an rde-1 mutant shows no change. An alternative explanation for the effect of RRF-1, ERI-1, and RDE-4 on RRF-3 target mRNA levels could have been that they might regulate rrf-3 transcription. Such regulation is unlikely to occur at the level of steady state RRF-3 levels as the microarray analyses show no significant change in rrf-3 mRNA expression.
RRF-3 and ERI factors function in the production and stabilization of a class of 26-nt, 5′G, 5′-monophosphorylated siRNAs
Our finding that strains lacking rrf-3 or rrf-1 are each defective in both siRNA production and mRNA downregulation for the 23 exemplary RRF-3 targets led us to speculate that two populations of siRNAs might be produced by the ERI pathway. To test this, we made use of the fact that two chemical classes of siRNA molecules, based on 5′ structures, are known (Pak and Fire, 2007; Ruby et al., 2006). One class, which we will call “type A,” carry a 5′ monophosphate and can be captured by direct ligation to an adapter on each end. The second class, which we will call “type B”, have a 5′ end that cannot be captured by simple ligation but can be captured following treatment of RNAs with phosphatase and kinase. Type B siRNAs include a category of siRNAs with 5′-triphosphate ends, consistent with retention of an unprocessed 5′ nucleotide from synthesis by an RNA polymerase. Type A siRNAs are presumably processed to yield the monophosphate at the 5′ ends; enzymes capable of such processing include (but are not limited to) Dicer and Argonaute class nucleases (Bernstein et al., 2001; Elbashir et al., 2001; Liu et al., 2004). For C. elegans, type B siRNAs are the much more abundant class of both endo and exo siRNAs and these therefore dominate the populations that are obtained using 5′-phosphate-independent capture strategies; 5′ phosphate-dependent capture strategies, by contrast, enrich strongly for type A siRNAs, although some type B siRNA sequence may be present in such libraries due to possible non-enzymatic or post-extraction loss of 5′ phosphates (Pak and Fire, 2007).
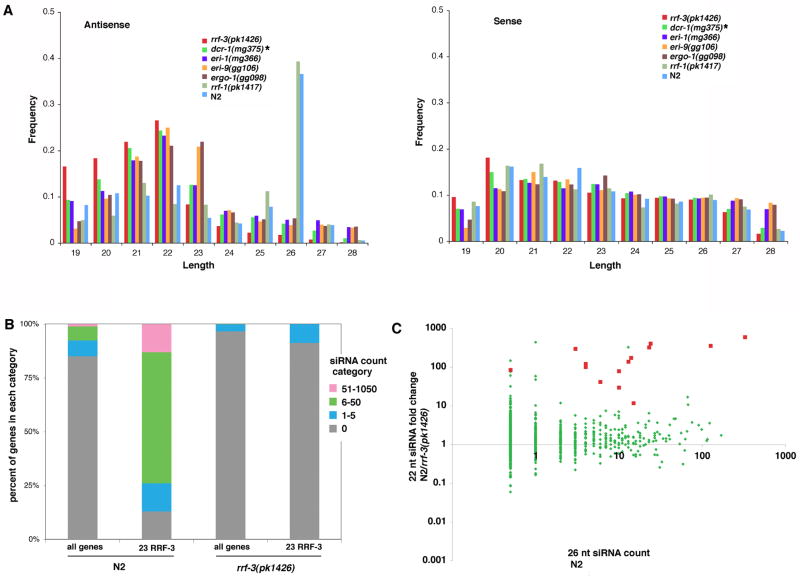
To search for RRF-3 effects on type A siRNAs, we prepared small RNA sequencing libraries using a 5′-phosphate-dependent method. The fact that monophosphorylated siRNAs (type A) are present at very low levels relative to other monophosphorylated RNAs such as microRNAs, piRNAs, and ribosomal RNA fragments would necessitate prohibitively large sequence datasets to obtain significant siRNA counts for each of the individual 23 RRF-3 targets. However, at the sequencing depth we obtained, the behavior of this class of RNA was evident through aggregation across the whole set of genes. Consistent with previous work by Ruby et al. (2006), in wildtype animals the type A siRNAs had a predominant length of 26 nt (Figure 5A) and a strong enrichment for a 5 -guanosine (Figure S4A). Strikingly, in the rrf-3 mutant, the peak at 26 nt was absent genome-wide. Consistent with this, factors predicted to be required for either production or stabilization of RRF-3 products—DCR-1, ERI-1, ERGO-1, and ERI-9—were also required for the 26-nt species. In contrast, RRF-1, which was evidently required for efficient production of the type B siRNAs (Figure 3C), was dispensable for the 26-nt, type A species (Figure 5 and S4B). These results are consistent with each RdRP producing a different population of siRNAs, with RRF-3 functioning upstream of RRF-1.
Figure 5. RRF-3 is required genome-wide for production of 26-nt, type A siRNAs.
Type A RNAs were sequenced using a method that selects for small RNAs with 5′ monophosphates. (A) Histograms of type A siRNA length distributions showing that the antisense-oriented, 26-nt peak is absent in rrf-3, dcr-1, eri-1, eri-9, and ergo-1 mutant strains but not rrf-1. After aligning to the reference cDNA set, reads were separated based on alignment orientation, and the frequency of each length between 19 and 28 nt tabulated for both antisense (left) and sense (right). Sense-oriented alignments served as an internal control to show that all lengths were represented in each sample. (B) Stacked bar charts indicate a decreased number of genes produced 26-nt, antisense, type A siRNAs in the rrf-3 mutant sample relative to N2. Genes were categorized according to siRNA counts, and the percent of genes in each category is shown on the Y-axis. For both the rrf-3(pk1426) and N2 samples, the set of 23 RRF-3 target genes are shown separately from the set of 21133 genes in the WS190 reference set. For categorization, the siRNA counts were normalized by the number of microRNAs in each library to account for differences in sample sizes. (C) Many genes with large numbers of 26-nt type A siRNAs (dependent upon RRF-3) can produce type B siRNAs independently of RRF-3. In the scatter plot, each gene is represented by a dot where the X-coordinate indicates the antisense, 26-nt, type A siRNA count, and the Y-coordinate indicates the fold change in type B siRNA counts between N2 and rrf-3(pk1426). Because the type A siRNA analysis included only siRNAs of the predominant length (26-nt) and orientation (antisense), the type B siRNAs were processed in a parallel fashion: only siRNAs of 22-nt length and antisense orientation were included. Also for the type B siRNAs, each gene s alignment count was increased by 1 to prevent division by zero. To avoid outliers arising from ratios of small numbers, genes with type B siRNA counts of less than 50 from the N2 sample were excluded from the analysis. The 14 RRF-3 targets that met this criterion are represented by the red squares, and the rest of the 1164 genes are represented by green dots. For the type A siRNAs, genes with alignment counts of zero were assigned a value of 0.5 to avoid exclusion from the plot s logarithmic-scaled axis. See also Figure S4.
In all backgrounds tested, the sense-oriented type A population lacked a predominant length (in contrast to type B, which showed a strong peak at 22 nt; compare Figures 2 and 5A). Likewise, the-sense oriented type A species lacked an enrichment for a 5′ guanosine, suggesting that they could be produced by mRNA metabolism independently of the ERI pathway (Figure S4A).
Dependency of type B siRNAs on type A siRNAs
While certain genes produced large numbers of 26-nt, type A siRNAs in wildtype L4 stage animals, the majority of genes (85% of the genes in the WS190 cDNA reference set) yielded none (Figure 5B). The set of 23 RRF-3 targets were enriched for their expression, as only three had a 26-nt, type A siRNA count of zero, and 17 of the genes had at least six. Intriguingly, high 26-nt type A siRNA levels alone do not indicate type B siRNA dependence upon RRF-3. Many genes produced similar numbers of type B siRNAs in rrf-3 mutant and wildtype despite their substantial production of RRF-3-dependent, 26-nt, type A siRNAs (Figure 5C). This suggests that RRF-3-driven production of 26-nt, type A siRNAs is not the only means of initiating production of type B siRNAs. The exemplary 23 somatic RRF-3 targets and other genes, however, showed a strong dependence upon RRF-3 for accumulation of the type B siRNAs.
Since the ERI pathway is required for accumulation of type B siRNAs for certain genes during spermatogenesis (Gent et al., 2009; Pavelec et al., 2009), we asked whether there could be a connection between type B siRNAs and type A siRNAs during spermatogenesis as well. We found that 26-nt type A siRNAs in isolated spermatogenic cells can be abundant and are dependent upon RRF-3 (Figure S4C, Table S1).
To further characterize the relationship between type A and type B siRNAs, we compared their nucleotide composition and position preferences. In addition to the enrichment for a 5′ guanosine, both type A and type B species were enriched for adenosine and guanosine throughout their lengths, marking a pyrimidine-rich siRNA footprint in the corresponding mRNAs (Figure S2A, S4A). Similarly, both type A and type B shared position preferences in that siRNAs mapping to the same loci were highly abundant from both sets (Figure S4D,E). While certain positions were highly enriched, there was also flexibility in that multiple positions were present within single transcripts. Analysis of siRNA start-to-start distances revealed several intriguing features of siRNA positioning (where the siRNA position is represented by its starting, 5′ nucleotide). First, type A start positions were strongly correlated with type B start positions (Figure S4E). In other words, the same cytosine that templated the monophosphorylated guanosine at the 5′ end of a 26-nt siRNA synthesized by RRF-3 also appears to have templated the triphosphorylated guanosine at the 5′ end of the 22-nt siRNA synthesized by RRF-1. Second, residual type B siRNAs retained the same start site preferences in rrf-1 and rrf-3 mutants (Figure S4E). These data indicate that both stages of siRNA production in the ERI pathway result in similar siRNA nucleotide and position preferences despite the differences in the machinery required for synthesis of each siRNA population.
DISCUSSION
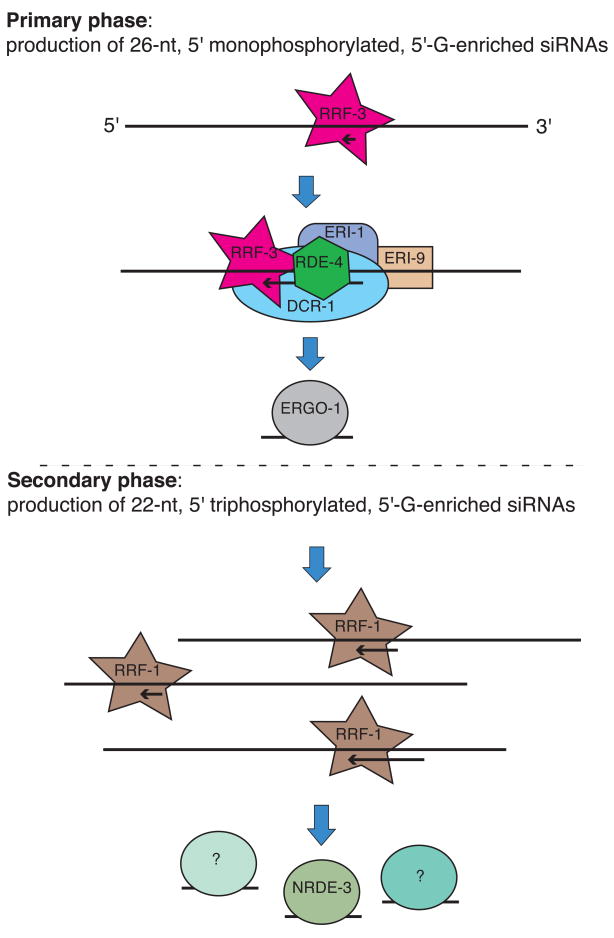
Taken together with other studies of RNAi mechanisms, our data suggest a working model, illustrated in Figure 6, in which RRF-3 interacts with a complex of proteins including Dicer (DCR-1), its double-stranded RNA-binding partner RDE-4, the riboexonuclease ERI-1, and the uncharacterized Dicer interactor ERI-9 to produce and stabilize primary siRNAs (type A siRNAs). Our data do not reveal whether RRF-3 produces long antisense RNA to be diced to into siRNAs with 5′ monophosphates, or whether RRF-3 produces shorter antisense RNA to be diced only at the 3′ end (or neither end should the DCR-1 complex provide a structural rather than enzymatic role in this context). If the 5′ end is not diced, the initial 5′ triphosphate could be converted to a monophosphate by another enzyme such as the pyrophosphatase homolog PIR-1, which is also known to physically interact with DCR-1 (Duchaine et al., 2006). Alternatively, the triphosphorylated 5′-most nucleotide could be removed by a 5′ exonuclease. Following engagement of the DCR-1 complex in the exo RNAi pathway, the primary siRNAs are loaded into a complex containing the Argonaute RDE-1. Since RDE-1 is dispensable for the RRF-3 pathway, and ERGO-1 was required for accumulation of both primary and secondary (type B) siRNAs, ERGO-1 takes the central role as the primary Argonaute, validating previous speculations (Yigit et al., 2006).
Figure 6. A model of the somatic ERI pathway.
All factors implicated by our data in the somatic ERI pathway are included in the model. The dashed line through the center represents a conceptual division between the primary and secondary phases of the pathway; our data do not exclude more complicated feedback mechanisms in either direction. The Argonautes labeled with a “?” are included to indicate that NRDE-3 may be one of multiple Argonautes with a substantial role in the secondary phase. The NIHMS has received the file ‘mmc1.pdf’ as supplementary data. The file will not appear in this PDF Receipt, but it will be linked to the web version of your manuscript.
The secondary response, in which the majority of siRNAs are produced and which is essential for effective gene silencing, is poorly understood even in the exogenous RNAi pathway. In somatic tissues, RRF-1 is known to function in secondary siRNA production in response to exogenously triggered RNAi. Our data indicate that RRF-1 is also involved in the somatic endogenous ERI pathway, specifically in production of secondary siRNAs downstream of RRF-3 activity. However, we note that RRF-1 is not the only RdRP that can function in this capacity, since large numbers of secondary siRNAs were still present in the rrf-1 mutant. C. elegans has two additional RdRPs, EGO-1 and RRF-2, which could function redundantly with RRF-1 (it is also conceivable that RRF-3 could function at this stage in addition to functioning in the initiation phase).
While multiple secondary Argonautes may function in the ERI pathway, our data indicates the identity of one in particular: NRDE-3. Its regulatory targets include E01G4.5 and others of our 23 exemplary RRF-3 targets, and its subcellular location in the soma is determined by RRF-3, DCR-1, ERI-1, ERI-9, RDE-4, and ERGO-1 (Guang et al., 2008; Pavelec et al., 2009).
While the involvement of DCR-1 and ERGO-1 with their predicted RNase capabilities suggests that transcript downregulation is at least partly due to cleavage, the pathway could also involve other forms of gene silencing in the secondary response. One intriguing possibility is that RdRP activity may be associated with chromatin modifications and transcriptional gene silencing, as shown previously in fission yeast, plants, and nematodes (Chan et al., 2004; Hall et al., 2002; Maine et al., 2005; Volpe et al., 2002).
An area of great interest not addressed by our results is the choice of RRF-3 targets. Our analysis does not reveal the full complement of genes associated with primary siRNA production; however it does suggest that the number is greater than 1500 (Figure 5B). This number could increase drastically with deeper sequencing. We fortuitously identified a handful of these genes because of their dependence on RRF-3 for production of secondary siRNAs. This set of 23 genes may be somewhat unique in that they engage RRF-3 in the soma rather than germline where diversity of RNAi-related pathways active in the germline may allow for compensation such that loss of one initiation mechanism is masked by increased activity of another.
The presence of multiple stages of small RNA production characterized by distinct small RNA populations appears to be a widespread feature of small RNA-mediated silencing. Among other examples are the piwi-interacting RNA pathway in insects (Brennecke et al., 2007; Gunawardane et al., 2007); the nat-siRNA, ta-siRNA, and RNA-directed DNA methylation pathways in plants (Axtell et al., 2006; Borsani et al., 2005; Daxinger et al., 2009); and hairpin-induced RNAi in amoeba (Martens et al., 2002). In addition to the potential advantage in amplifying an initial silencing trigger into a secondary response with distinct silencing capabilities, such systems could also provide potential for multiple steps of regulation of the gene silencing mechanism. From the organism s perspective, multistage processes could provide a safe and flexible regulatory response in which the amplified secondary effectors are able to enforce potent gene regulation while only the initial primer effectors can trigger amplification.
EXPERIMENTAL PROCEDURES
C. elegans strains
The following strains were used in this study: Bristol N2 (Brenner, 1974), BA17 fem-1(hc17) IV (Nelson et al., 1978), CB4037 glp-1(e2141) III (Priess et al., 1987), JK816 fem-3(q20) IV [see below] (Barton et al., 1987), NL2098 rrf-1(pk1417) I (Sijen et al., 2001), NL2099 rrf-3(pk1426) II (Sijen et al., 2001), PD3307 rrf-3(pk1426) II; fem-3(q20) IV, PD3330 rrf-3(pk1426) II; him-8(e1489) IV, PD3331 him-8(e1489) IV, SS104 glp-4(bn2) I (Beanan and Strome, 1992), WM48 rde-4(ne299) III (Tabara et al., 1999), rde-1(ne300) V (Tabara et al., 1999). YY009 eri-1(mg366) IV, YY011 dcr-1(mg375) III [see below], YY166 ergo-1(gg098) V, and YY216 eri-9(gg106) III (Pavelec et al., 2009).
Recent work from other groups (H. Gabel, C. Zhang, S. Fischer, T. Montgomery, and G. Ruvkun; L. Frater and P. Anderson personal communications) has identified a dimorphism between laboratory strains of C. elegans affecting a 451 nt segment upstream of the mut-16 gene. A majority of laboratory strains have an ‘ancestral’ structure at this locus, while a subset of strains, likely derived from a single animal, carry a 451 bp deletion. Of the stocks used in this study, two strains have the shorter upstream fragment [JK816 fem-3(q20) and YY011 dcr-1(mg375)], while the remainder of strains carry the cannonical/nondeleted mut-16 allele. The ability of both mut-16 types to produce RRF-3-dependent siRNAs has been confirmed by examining sequence datasets from these strains and from a set of independently analyzed strains that were part of other small RNA sequencing projects to be described elsewhere (L. Frater, J. Maniar, D. Pavelec, S. Kennedy, A. Fire and P. Anderson, personal communication). Although some quantitative variation in RRF-3-dependent siRNA levels was evident in strains with the mut-16 upstream deletion background, this variation was modest (within an order of magnitude for class B siRNAs and subtle if any for class A siRNAs).
C. elegans population synchronization and harvesting
All strains used in this study were harvested as synchronized populations, where synchronization was obtained by treatment with sodium hypochlorite solution to dissolve animals of all stages but embryos. Growth temperatures and stage at harvesting were as follows: rrf-3 mutant and control strains: The two sets of rrf-3(pk1426); him-8(e1489) and him-8(e1489) males were raised at 25°C and harvested as adults after separation from hermaphrodites by filtration through 35 μm filter cloth. These were the same samples analyzed previously (Gent et al., 2009). The hermaphrodites at this stage were gravid and had begun to lay eggs. The two sets of rrf-3(pk1426) and N2 hermaphrodites were raised at 16°C and harvested at the L4 larval stage. Germline mutant strains: glp-1(e2141), glp-4(bn2), fem-3(q20), fem-1(hc17), and rrf-3(pk1426); fem-3(q20) were raised at 25°C and harvested as young adults (equivalent stage as the him-8(e1489) males). Spermatogenic cells were isolated as previously described (Gent et al., 2009). Additional RNAi and ERI mutant strains: rrf-1(pk1417) was raised at 16°C. dcr-1(mg375), ergo-1(gg098), eri-1(mg366), eri-9(gg106), rde-1(ne300), and rde-4(ne299) were raised at 20°C. All seven RNAi and ERI mutant strains were harvested at L4 stage.
RNA sequencing library preparation
To characterize small RNA pools, 5′-monophosphate-dependent and independent libraries were prepared essentially as described previously (Lau et. al., 2001, Lui et al., 2007, Gent et al., 2009).
mRNA sequencing libraries were prepared using a direct linker scheme similar to that used for small RNA sequencing libraries. mRNA isolation (Ambion MicroPoly(A) purist kit) was followed by fragmentation (Ambion Fragmentation Reagents) and treatment with T4 polynucleotide kinase (New England Biolabs) and ATP to produce a population of fragments with 5′ monophosphorylated and 3′ hydroxylated ends. We then removed the ATP (Illustra microspin G-25 column), ligated a pre-adenylated 3′ adapter using T4 RNA ligase 1 (with no ATP), and a 5′ adapter using T4 RNA ligase 1 (with ATP). Linkers, primers, reverse transcription and PCR amplification of these pools were as described [Lui et al., 2007] with the addition of 3-nt barcodes (CCC or GGG) in the 5′ adapter to allow for pooling of samples and as a guard against contamination.
Sequence processing and alignment
36-nt reads were generated using the Illumina Genome Analyzer system for all samples. Processing of siRNA reads was done as previously described (Gent et al., 2009). For mRNA sample processing, multiple incidences of identical reads from the same sample were counted only once to avoid potential “jackpots” that can be introduced during amplification. All reads were aligned to a cDNA reference (WS190 annotation) set using MAQ (http://maq.sourceforge.net; Li et al., 2008) using the first 19 nt of each read as the alignment query for siRNAs and 28 nt for mRNA tags. To confirm the results, we also aligned the reads to both the WS190 cDNA set and the WS190 C. elegans genome using BLAT alignment software (default parameters except -tileSize=10 and -stepSize=5; http://www.soe.ucsc.edu/~kent; Kent, 2002). For the 5′-monophosphate-dependent reads we also used BLAT to align to a microRNA dataset downloaded from mirBASE (http://microrna.sanger.ac.uk/sequences/). The cDNA dataset was downloaded from http://www.wormbase.org:80/biomart/martview. The source reads are available at the NCBI short read archive (accession number GSE19414).
siRNA start-to-start distance analysis
This analysis measured the tendencies or lack thereof for siRNAs to start at particular distances from each other. Samples were analyzed in pairs so that siRNA start positions from one sample (reference) were compared with start positions from another (query). For example, a reference siRNA starting at position 10 would give a start-to-start distance of 30 relative to a query siRNA starting at position 40. To measure the prevalence of each start-to-start distance for a single gene, the number of reference siRNAs starting at each position along the length of the gene was counted to produce a start incidence associated with each position. Next, the reference start incidence from a single position was compared with the query start incidence for all positions within an arbitrary distance. For each distance, the reference and query start incidences were multiplied to yield a distance incidence. Since positions with large start incidences dominated the outcome, we utilized a cube root transformation inspired by SAM (Significance Analysis of Microarrays) (Tusher et al., 2001): instead of the product of the start incidences, we used the cube root of the product, a number we call the distance index. The calculations were then repeated for each query position, compared again to all possible reference positions. The resulting distance indexes were summed to produce a distance coincidence index or simply coincidence index for each possible start-to-start distance. To analyze start-to-start distances from siRNAs aligned to multiple genes, the coincidence indexes from each gene were calculated individually and summed.
Supplementary Material
Acknowledgments
We are grateful to Sam Gu, Julia Pak, Leah Frater, Phil Anderson, Chaya Krishna, Cheryl Smith, Ziming Weng, Phil Lacroute, Anton Valouev, and Arend Sidow for their help with experimental protocols and analysis, Virginia Walbot and Anne Villenueve for comments on the manuscript, the Caenorhabditis Genetics Center for strains, and for financial support from NIH (R01GM37706 [AF], T32HG00044 [JIG and JM], R01GM076619 [SK and DP]), Stanford Graduate Fellowship (PP), NSF Graduate Fellowship (JM), and Stanford Dean’s Fellowship and Machiah Foundation (AL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. Embo J. 2007;26:5007–5019. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asikainen S, Storvik M, Lakso M, Wong G. Whole genome microarray analysis of C. elegans rrf-3 and eri-1 mutants. FEBS Lett. 2007;581:5050–5054. doi: 10.1016/j.febslet.2007.09.043. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Barton MK, Schedl TB, Kimble J. Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics. 1987;115:107–119. doi: 10.1093/genetics/115.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992;116:755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol. 2008;15:1015–1023. doi: 10.1038/nsmb.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Kanno T, Bucher E, van der Winden J, Naumann U, Matzke AJ, Matzke M. A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. Embo J. 2009;28:48–57. doi: 10.1038/emboj.2008.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gabel HW, Ruvkun G. The exonuclease ERI-1 has a conserved dual role in 5.8S rRNA processing and RNAi. Nat Struct Mol Biol. 2008;15:531–533. doi: 10.1038/nsmb.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Schvarzstein M, Villeneuve AM, Gu SG, Jantsch V, Fire AZ, Baudrimont A. A Caenorhabditis elegans RNA-directed RNA Polymerase in Sperm Development and Endogenous RNAi. Genetics. 2009 doi: 10.1534/genetics.109.109686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. Rna. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- Maine EM, Hauth J, Ratliff T, Vought VE, She X, Kelly WG. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired dna during C. elegans meiosis. Curr Biol. 2005;15:1972–1978. doi: 10.1016/j.cub.2005.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Bamford DH. Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol Cell. 2002;10:1417–1427. doi: 10.1016/s1097-2765(02)00780-3. [DOI] [PubMed] [Google Scholar]
- Martens H, Novotny J, Oberstrass J, Steck TL, Postlethwait P, Nellen W. RNAi in Dictyostelium: the role of RNA-directed RNA polymerases and double-stranded RNase. Mol Biol Cell. 2002;13:445–453. doi: 10.1091/mbc.01-04-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Nelson GA, Lew KK, Ward S. Intersex, a temperature-sensitive mutant of the nematode Caenorhabditis elegans. Dev Biol. 1978;66:386–409. doi: 10.1016/0012-1606(78)90247-6. [DOI] [PubMed] [Google Scholar]
- Nobuta K, Lu C, Shrivastava R, Pillay M, De Paoli E, Accerbi M, Arteaga-Vazquez M, Sidorenko L, Jeong DH, Yen Y, et al. Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc Natl Acad Sci U S A. 2008;105:14958–14963. doi: 10.1073/pnas.0808066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Parrish S, Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. Rna. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- Pavelec DM, Lachowiec J, Duchaine TF, Smith HE, Kennedy S. Requirement for ERI/DICER Complex in Endogenous RNAi and Sperm Development in Caenorhabditis elegans. Genetics. 2009 doi: 10.1534/genetics.109.108134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR, Schnabel H, Schnabel R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Welker NC, Habig JW, Bass BL. Genes misregulated in C. elegans deficient in Dicer, RDE-4, or RDE-1 are enriched for innate immunity genes. Rna. 2007;13:1090–1102. doi: 10.1261/rna.542107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.