Abstract
Basic helix-loop-helix (bHLH) proteins are a class of transcription factors found throughout eukaryotic organisms. Classification of the complete sets of bHLH proteins in the sequenced genomes of Arabidopsis thaliana and Oryza sativa (rice) has defined the diversity of these proteins among flowering plants. However, the evolutionary relationships of different plant bHLH groups and the diversity of bHLH proteins in more ancestral groups of plants are currently unknown. In this study, we use whole-genome sequences from nine species of land plants and algae to define the relationships between these proteins in plants. We show that few (less than 5) bHLH proteins are encoded in the genomes of chlorophytes and red algae. In contrast, many bHLH proteins (100–170) are encoded in the genomes of land plants (embryophytes). Phylogenetic analyses suggest that plant bHLH proteins are monophyletic and constitute 26 subfamilies. Twenty of these subfamilies existed in the common ancestors of extant mosses and vascular plants, whereas six further subfamilies evolved among the vascular plants. In addition to the conserved bHLH domains, most subfamilies are characterized by the presence of highly conserved short amino acid motifs. We conclude that much of the diversity of plant bHLH proteins was established in early land plants, over 440 million years ago.
Keywords: bHLH, transcription factor, plants, algae, evolution, phylogeny
Introduction
The basic helix-loop-helix (bHLH) domain is a highly conserved amino acid motif that defines a group of transcription factors. It was originally described in animals (Murre et al. 1989) and soon discovered in all the major eukaryotic lineages. Proteins that contain a bHLH domain (referred to as bHLH proteins) are involved in a myriad of regulatory processes. Their functions include regulating neurogenesis, myogenesis, and heart development in animals (Massari and Murre 2000; Jones 2004); controlling phosphate uptake and glycolysis in yeast (Robinson and Lopes 2000); or modulating secondary metabolism pathways, epidermal differentiation, and responses to environmental factors in plants (Ramsay and Glover 2005; Castillon et al. 2007).
The bHLH domain consists of 50–60 amino acids that form two distinct segments: a stretch of 10–15 predominantly basic amino acids (the basic region) and a section of roughly 40 amino acids predicted to form two amphipathic α-helices separated by a loop of variable length (the helix-loop-helix region). Structural analyses of mammalian and yeast bHLH proteins showed that the basic region forms the main interface where contact with DNA occurs, whereas the two helices promote the formation of homo- or heterodimers between bHLH proteins, a prerequisite for DNA binding to occur (Jones 2004).
Phylogenetic analyses have classified the diversity of bHLH proteins into a number of distinct groups. Over 50 bHLH proteins are encoded in the genomes of most animals (metazoans) and are typically classified into six major groups (A–F), based on their ability to bind DNA (Atchley and Fitch 1997; Ledent and Vervoort 2001; Jones 2004). Detailed analyses using whole-genome sequences showed that animal bHLH could be further classified in several smaller subfamilies that are highly conserved across major metazoan lineages (Ledent and Vervoort 2001; Simionato et al. 2007). Phylogenetic analyses indicate that 44 of these subfamilies were present in the common ancestor of all bilaterians, which is thought to have existed sometime before 600 million years ago (Ma) (Simionato et al. 2007). The genomes of Arabidopsis thaliana and Oryza sativa (rice) encode even more bHLH sequences than animals. Different phylogenetic studies proposed the classification of plant bHLH into 15–25 subgroups (Buck and Atchley 2003; Heim et al. 2003; Toledo-Ortiz et al. 2003; Li et al. 2006b). However, the origin and evolutionary history of these groups cannot be understood using A. thaliana and O. sativa sequences alone. The characterization of the evolution of plant bHLH diversity requires the phylogenetic analysis of bHLH proteins from a more diverse selection of plants, including algae, bryophytes, and different lineages of vascular plants.
In this study, we characterized the evolution of bHLH proteins in plants, defined here as the organisms that are likely to have been derived from the primary endosymbiotic event that gave rise to the red algae, chlorophytes, and land plants (Rodríguez-Ezpeleta et al. 2005). We show that the plant bHLH family is monophyletic and underwent a major radiation before the evolution of the mosses. The bHLH groups established in the early land plants over 400 Ma were conserved during subsequent plant evolution, although there were many gene duplications and losses within these groups. Our analysis defines 26 subfamilies that represent deep evolutionary relationships between plant bHLH proteins.
Materials and Methods
Sequence Retrieval
The A. thaliana bHLH reported by Bailey et al. (2003), Heim et al. (2003) and Toledo-Ortiz et al. (2003) were retrieved from The Arabidopsis Information Resource (http://www.arabidopsis.org/). A clear bHLH domain was not found in At1g31050 (AtbHLH111) and At1g22380 (AtbHLH152), so they were not further used in this study; we could not find At2g20095 (AtbHLH133) and At4g38071 (AtbHLH131) in any database. A data set of predicted O. sativa L. ssp. japonica bHLH proteins was retrieved from the Plant TFDB (Guo et al. 2008) and combined with the bHLH protein sequences reported by Li et al. (2006b), retrieved from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). Eleven new proteins were numbered following the nomenclature style of Li et al. (2006b), whereas a clear bHLH was not found in Os01g65080 (OsbHLH033), Os04g35000 (OsbHLH145), Os11g02054 (OsbHLH160), and Os12g02020 (OsbHLH161). A data set of predicted Physcomitrella patens bHLH was retrieved from the Plant TFDB (Guo et al. 2008). A direct search of genes annotated as bHLH was performed on the genome assembly of Selaginella moellendorffii v1.0 (http://www.jgi.doe.gov/). HMMsearch (Eddy 1998) was used to screen the genome assemblies of Cyanidioschyzon merolae (Matsuzaki et al. 2004), Chlamydomonas reinhardtii v3.0 (Merchant et al. 2007), Ostreoscoccus tauri v2.0 (Palenik et al. 2007), Thalassiosira pseudonana v3.0 (Armbrust et al. 2004), and the draft assemblies of Chlorella vulgaris C-169 and Volvox carteri (http://www.jgi.doe.gov/) with the PFAM profile hidden Markov model (pHMM) HLH_ls.hmm (http://pfam.sanger.ac.uk/).
Five Homo sapiens and four Amphimedon queenslandica (demosponge) representative sequences of the major metazoan groups of bHLH proteins (based on Jones 2004; Simionato et al. 2007) were retrieved from GenBank; group F proteins are not clearly alignable to other bHLH (Ledent et al. 2002) and so they were not used in this study. The Saccharomyces cerevisiae bHLH proteins reported by Robinson and Lopes (2000) were retrieved from http://www.yeastgenome.org/.
For simplicity, all sequences were renamed according to the supplementary table S1 (Supplementary Material online). The complete amino acid sequence of all proteins can be found in supplementary data 1 (Supplementary Material online).
Alignment and Phylogenetic Analysis
Protein sequences were prealigned using HMMalign (Eddy 1998) and the pHMM HLH_ls.hmm from PFAM (http://pfam.sanger.ac.uk/). The bHLH region was then extensively manually aligned in BioEdit (http://www.mbio.ncsu.edu/BioEdit/BioEdit.html). Unambiguous aligned positions were used for the subsequent phylogenetic analyses (supplementary fig. S1, Supplementary Material online). The Jones, Taylor, and Thorton (JTT) model was selected as the best-fitting amino acid substitution model with the Akaike information criterion implemented in ProtTest (Abascal et al. 2005). The maximum likelihood (ML) analyses were done with the program PhyML version 3.0.1 (Guindon and Gascuel 2003) using the JTT model of amino acid substitution, an estimated gamma distribution parameter and an Shimodaira-Hasegawa-like approximate likelihood ratio test. The PHYLIP package version 3.67 (Felsenstein 1989) was used to perform 100 bootstrap replicas of a neighbor joining (NJ) tree based on a JTT distance matrix. PAUP* version 4.0b10 (Swofford 2003) was used to perform 100 bootstrap replicas of a maximum parsimony (MP) tree. The Bayesian analysis was performed with MrBayes version 3.1.2 (http://mrbayes.csit.fsu.edu/): two independent runs were computed for 10 million generations, at which point the standard deviation of split frequencies was less than 0.01; one tree was saved every 100 generations, and 75,000 trees from each run were summarized to give rise to the final cladogram. All trees were visualized using the program Figtree (http://tree.bio.ed.ac.uk/software/figtree/).
Alignments of the bHLH domain of related sequences were used to build pHMMs with HMMbuild (Eddy 1998). The pHMMs were used to classify proteins not used in the phylogenetic analyses in the plant bHLH subfamilies. The pHMMs were visualized with HMM Logo (Schuster-Bockler et al. 2004).
Detection of Conserved Motifs
The MEME and FIMO software (Bailey and Elkan 1994) were used to discover patterns in the complete amino acid sequences of plant bHLH proteins. Each motif was individually checked so that incorrect or insignificant matches were discarded. The complete plant amino acid sequences were also screened against the PFAM 23.0 database (http://pfam.sanger.ac.uk/).
Results
All the Major Groups of Land Plants Have Large Numbers of bHLH Proteins
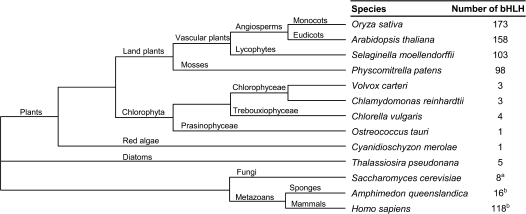
Previous phylogenetic analyses of plant bHLH proteins were based on the genome sequences of A. thaliana and O. sativa (Buck and Atchley 2003; Heim et al. 2003; Toledo-Ortiz et al. 2003; Li et al. 2006b). This provided a useful, but limited, phylogenetic framework for the classification of bHLH proteins in flowering plants (angiosperms). Nevertheless, it provided no insight into the diversity of this family in the earlier diverging groups of land plants. To determine if these subfamilies were angiosperm specific or if they arose earlier in plant evolution and to understand the deeper evolutionary history of this family in plants, we searched for bHLH protein coding sequences in the complete genome of the lycophyte S. moellendorffii, the moss P. patens, the chlorophytes V. carteri, C. reinhardtii, C. vulgaris, and O. tauri, and the red alga C. merolae. These sequences were combined with the previously reported A. thaliana and O. sativa sequences to generate a primary data set consisting of 544 bHLH sequences representing the major evolutionary lineages of plants (fig. 1). We then extended this data set to include proteins from selected eukaryotic groups: the full set of bHLH proteins encoded in the genomes of the diatom T. pseudonana and the fungi S. cerevisiae, plus representative bHLH sequences from the sponge A. queenslandica and H. sapiens (fig. 1).
FIG. 1.
Phylogenetic relationships of the species used in this study. The total number of bHLH proteins found in the genome of each species is indicated. The cladogram is based on the current view of plant and eukaryotic phylogeny (Baldauf 2003; Lewis and McCourt 2004; Rodríguez-Ezpeleta et al. 2005); aRobinson and Lopes (2000); bSimionato et al. (2007).
There are large numbers of bHLH proteins in all species of land plants (embryophytes) sequenced to date. A. thaliana and O. sativa have over 150 bHLH sequences in their genomes, making it the second largest family of transcription factors in angiosperms (Xiong et al. 2005). Approximately 100 bHLH proteins are encoded in the genomes of the lycophyte S. moellendorffii and the moss P. patens (fig. 1). In contrast, we found less than five bHLH-encoding sequences in the genome of each chlorophyte and red alga examined. Other unicellular eukaryotic organisms such as the diatom T. pseudonana and S. cerevisiae also have small numbers (less than 10) of bHLH proteins (fig. 1, Robinson and Lopes 2000). In animals, the sponge A. queenslandica has 16 bHLH-encoding genes, whereas most bilaterians have over 50 genes (Simionato et al. 2007).
Animals and land plants have considerably more bHLH sequences than other eukaryotic organisms. This suggests that the increase in the number of bHLH proteins occurred independently during the evolution of plants and animals.
Key Amino Acid Residues Are Highly Conserved Between Plant and Metazoan bHLH Proteins
To characterize the molecular evolution of plant bHLH proteins, we aligned the retrieved amino acid sequences in the conserved bHLH region (fig. 2, supplementary fig. S1, Supplementary Material online). The first 10–15 amino acids correspond to the basic region, where most interactions with the DNA are made (Ferré-D'Amaré et al. 1993). Most animal bHLH proteins bind to hexanucleotide sequences (5′-CANNTG-3′) known as E-boxes. All E-box–binding bHLH proteins have a glutamic acid (E) residue at position 9 that directly contacts the DNA at the CA nucleotides of the hexanucleotide sequence (Ferré-D'Amaré et al. 1993; Atchley et al. 1999). In plants, the critical E9 residue is present in 74% of the proteins analyzed (supplementary fig. S1, Supplementary Material online). Other positions of the basic region allow a better discrimination of the target DNA sequences and are easily distinguishable in the major animal bHLH groups (Atchley and Fitch 1997; Ledent and Vervoort 2001; Jones 2004; Atchley and Zhao 2007). Animal group A proteins bind the CAGCTG (or CACCTG) E-box configuration and have a diagnostic arginine (R) at position 8. Animal group B proteins have a lysine (K) or histidine (H) residue at position 5 and an R at position 13 and bind the CACGTG (or CATGTTG) E-box configuration. In plants, 53% of the bHLH proteins have the characteristic animal group B configuration H5-E9-R13 and only 8% have the typical R8-E9 found in animal group A. This suggests that most plant bHLH proteins also bind to E-boxes. Indeed, a number of plant bHLH proteins have been shown to bind the CACGTG sequence (e.g., Martínez-García et al. 2000; Toledo-Ortiz et al. 2003; Qian et al. 2007), which is classically known in plants as a G-box motif (Giuliano et al. 1988). Group E animal proteins, that bind N-boxes (CACGCG or CACGAG), have the same H5-E9-R13 configuration as group B and a proline (P) at position 6. This configuration is absent in all the 544 plant bHLH proteins analyzed. The remaining animal bHLH groups C and F proteins contain extra PAS and COE domains, not found in plant bHLH proteins, whereas group D proteins are atypical bHLH without a basic domain; 11% of the plant proteins have a conserved Q5-A9-R13 motif (supplementary fig. S1, Supplementary Material online), not present in animals. This raises the interesting possibility that these proteins bind to a novel target DNA sequence. Other frequent basic amino acids in animal bHLH, such as R in positions 10 and 12, are also highly conserved in plants (73% and 90%, respectively).
FIG. 2.
Alignment of the bHLH domain of representative plant proteins. A representative of each of the 26 subfamilies of plant bHLH is shown, together with the human protein Max, a well-characterized bHLH protein. The shaded boxes indicate the position of the DNA-binding basic region, the two α-helixes, and the variable loop region (Ferré-D'Amaré et al. 1993). The numbering of the amino acids follows (Atchley and Fitch 1997). This is a subset of the full alignment with all the proteins used in this study (supplementary fig. 1, Supplementary Material online).
The α-helices promote the formation of homo- or heterodimeric complexes between bHLH proteins. The structure of a dimer is stabilized by the hydrophobic amino acids isoleucine (I), leucine (L), and valine (V) in conserved positions in the bHLH domain (Ferré-D'Amaré et al. 1993). These positions are highly conserved in animals (Atchley et al. 1999) and in plants (fig. 2). An L residue is present in sites 23 and 64 in 99% and 96% of the plant proteins and in 98% and 80% of the animal proteins, respectively. Sites 54 and 61 have an I, L, or V in 99% and 93% of the plant proteins and in 98% and 93% of the animal proteins, respectively. A conserved P breaks the first helix and starts a loop of variable length (usually six to nine residues in plants). Some loop residues are also conserved: site 47 is K or R in 88% of the plant proteins (supplementary fig. S1, Supplementary Material online) and 82% of the animal proteins (Atchley et al. 1999).
The high degree of sequence similarity between the bHLH domain of plant and animal proteins, particularly in key DNA-interacting basic amino acids and in helix-stabilizing hydrophobic amino acids, indicates that the molecular structure and transcription factor activity of bHLH proteins are conserved between animals and plants.
Twenty bHLH Subfamilies Found in Flowering Plants Were also Present in Early Land Plants
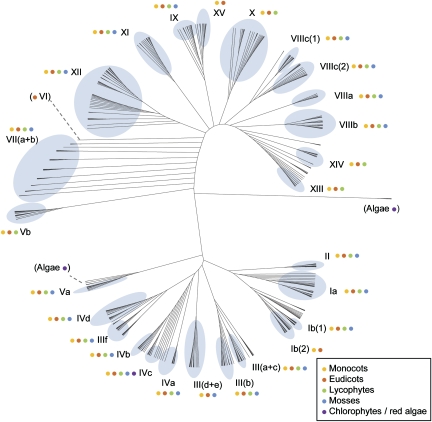
To understand the evolutionary relationships between plant bHLH proteins, we used conserved regions of the alignment shown in supplementary figure S1 (Supplementary Material online) to compute phylogenetic trees. An ML analysis shows that proteins from different species cluster together in compact clades with high support values (fig. 3, supplementary fig. S2, Supplementary Material online). MP and NJ analyses support the existence of most of these clades (supplementary fig. S2, Supplementary Material online). Based on the topology of the trees, clade support values, branch lengths, and visual inspection of the bHLH amino acid sequences, we defined 26 subfamilies of bHLH proteins (fig. 3, supplementary fig. S2, Supplementary Material online). These subfamilies are mostly consistent with the groups proposed by previous phylogenetic analyses of plant bHLH using A. thaliana and O. sativa sequences alone (Buck and Atchley 2003; Heim et al. 2003; Toledo-Ortiz et al. 2003; Li et al. 2006b). We adopted the A. thaliana bHLH group nomenclature proposed by Heim et al. (2003) to label these subfamilies, with some modifications, for example, Ib was divided in Ib(1) and Ib(2), and IIIa and IIIc were combined into III(a + c). We also defined three new groups (XIII, XIV, and XV) that include 28 A. thaliana sequences not present in the analysis by Heim et al. Of the 544 proteins analyzed, 10% do not clearly fall in any of the 26 subfamilies and were classified as “orphans” (supplementary fig. S2, Supplementary Material online). These proteins often have a high degree of sequence divergence from other bHLH: This may be due to lineage-specific specializations or, alternatively, they may correspond to pseudogene sequences. One of the A. thaliana groups proposed by Heim and colleagues (group VI, consisting of only two proteins) falls in this “orphan” category.
FIG. 3.
Twenty subfamilies of bHLH were already established in the common ancestral of vascular plants and mosses. Maximum likelihood analysis of 544 plant bHLH, shown as an unrooted cladogram. The blue balloons delineate the 26 subfamilies of plant bHLH proteins. Colored dots symbolize the species to which the proteins in each group belong (yellow: Oryza sativa [monocot]; red: Arabidopsis thaliana [eudicot]; green: Selaginella moellendorffii [lycophyte]; blue: Physcomitrella patens [moss]; purple: Volvox carteri, Chlamydomonas reinhardtii, Chlorella vulgaris, Ostreococcus tauri, and Cyanidioschyzon merolae (chlorophytes and red algae). A full tree with protein names, proportional branch lengths, and clade support values is given in supplementary fig. S2 (Supplementary Material online).
Of the 26 plant bHLH subfamilies, 3 include only angiosperm proteins and 23 include angiosperm and lycophyte proteins (fig. 3). Because the last common ancestor of angiosperms and lycophytes lived sometime in the Upper Silurian period before 415 Ma (Kenrick and Crane 1997), this implies that these 23 bHLH subfamilies are at least 415 million years (My) old. Interestingly, 20 of these subfamilies include not only vascular plants but also moss proteins. Given that the oldest evidence for the existence of vascular plants is trilete spores in Upper Ordovician sediments (Steemans et al. 2009), it suggests that these subfamilies are more than 443 My old. A bHLH protein from the chlorophyte algae O. tauri is a member of subfamily IVc (fig. 3). This suggests that this subfamily may be over 1 billion years old (Heckman et al. 2001).
A clade composed of V. carteri, C. reinhardtii, and C. vulgaris bHLH proteins is sister to the proteins in subfamily Va. However, we did not include these chlorophyte proteins into the Va subfamily as this relationship is not strongly supported. Nevertheless, this relationship suggests that subfamily Va is phylogenetically closer to chlorophyte proteins than to any other land plant proteins. Another group of V. carteri, C. reinhardtii, and C. vulgaris proteins forms a clade that is clearly distinct from other plant proteins (fig. 3); this probably represents a group that evolved among the chlorophytes or, alternatively, was present in the common ancestors of the chlorophytes and land plants but maintained among the chlorophytes and lost in the ancestors of land plants. The only bHLH-encoding gene found in the genome of the red algae C. merolae could not be allocated to any chlorophyte or land plant bHLH clade.
In summary, the phylogenetic analysis shows that plant bHLH proteins form 26 distinct subfamilies or evolutionary lineages; 20 of these subfamilies were already present in early land plants 443 Ma, by which time the mosses had diverged from the vascular plants. Despite several rounds of gene duplications and losses in different plant lineages, these subfamilies have been highly conserved throughout plant evolution.
Plant bHLH Proteins Are Monophyletic
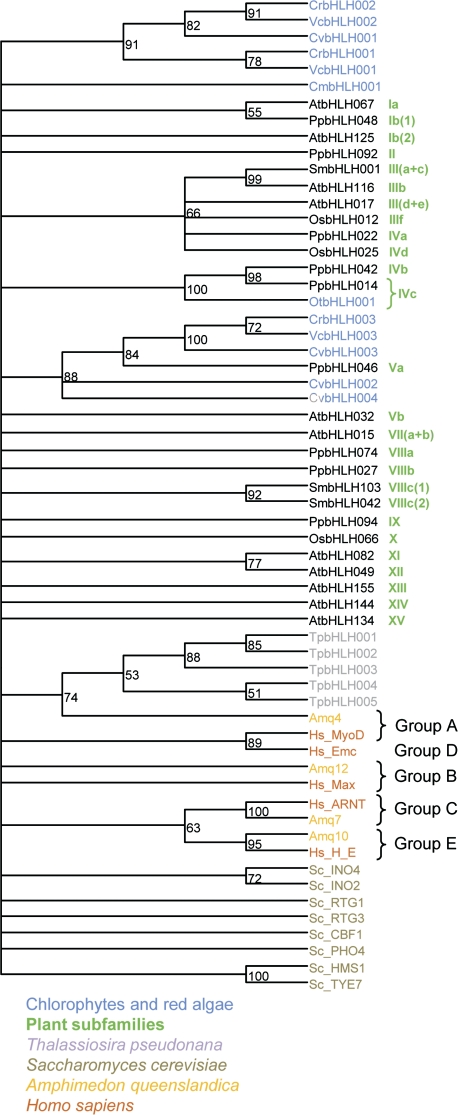
The phylogenetic information contained in the 50–60 amino acids of the bHLH allows delimitation of major evolutionary lineages of proteins in plants but does not allow good resolution of deeper nodes that represent the phylogenetic relationships between different bHLH subfamilies; these basal nodes often have low support values (supplementary fig. S2, Supplementary Material online) and vary when using NJ or MP analyses (data not shown). Similar poor resolution was observed in previous classifications of bHLH proteins in other groups of organisms (Atchley and Fitch 1997; Ledent and Vervoort 2001; Buck and Atchley 2003; Toledo-Ortiz et al. 2003; Li et al. 2006b). Thus, the inter-subfamily relationships shown in figure 3 should be interpreted cautiously. We initially tried to incorporate non-plant bHLH sequences in the ML analysis. However, the large number of proteins and the great evolutionary distances (and consequent high degree of sequence divergence) caused the non-plant proteins to form very long branches, nested within plant clades with no obvious sequence similarity (data not shown). To circumvent this problem, we opted to perform a phylogenetic analysis on a simplified alignment (supplementary fig. S1, Supplementary Material online) that includes chlorophytes, red algae, diatom, and yeast proteins plus representatives of the 26 plant subfamilies and of the higher order metazoan groups (Atchley and Fitch 1997; Simionato et al. 2007).
The deep evolutionary relationships between many proteins were still not resolved: most branches in the Bayesian phylogenetic tree had low support values (fig. 4). However, some close relationships between different plant bHLH subfamilies (fig. 3) were supported by this analysis. For example, subfamilies IVc and Va were probably established in the common ancestors of chlorophyte algae and land plants; subfamily IVb possibly evolved later among land plants from subfamily IVc proteins. Pairs of subfamilies such as VIIIc(1)/VIIIc(2), XI/XII, and III(a + c)/IIIb seem to form monophyletic lineages. Interestingly, the five diatom sequences and a sponge group A protein form a well-supported clade. Closer examination of the amino acid sequence of the five diatom bHLH proteins reveals that each of these proteins have an arginine in position 8 of the bHLH domain, a defining characteristic of group A proteins (Atchley and Fitch 1997). Although beyond the scope of this study, this suggests that group A might predate the origin of opisthokontes, the eukaryotic lineage that includes fungi and animals.
FIG. 4.
Plant bHLH do not group with other eukaryote bHLH. A Bayesian analysis was performed on an alignment of the bHLH sequence of one representative of each of the 26 subfamilies of plant bHLH, all the chlorophyte and red algae proteins, 5 proteins found in diatom Thalassiosira pseudonana, 8 Saccharomyces cerevisiae proteins, and representatives of 5 major groups of metazoan bHLH in the sponge Amphimedon queenslandica and Homo sapiens. The tree is unrooted. The numbers in the clades are posterior probability values; clades with less than 50% support were collapsed.
No clustering of plant proteins with proteins from other eukaryotic organisms is found on the Bayesian tree (fig. 4). The small number of bHLH proteins found in the genomes of different chlorophytes and red algae (fig. 1) suggests that the first plants had one or a few bHLH proteins, from which all modern plant bHLH descended and radiated. This view is consistent with previous analyses that highlighted the distant relationship of angiosperm and animal bHLH proteins (Ledent and Vervoort 2001; Buck and Atchley 2003; Toledo-Ortiz et al. 2003). The lack of discernible phylogenetic relationships between bHLH subfamilies in plants and other eukaryotic organisms supports the hypothesis that plant bHLH proteins are monophyletic.
Conserved Non-bHLH Motifs Are Present in Most Plant bHLH Subfamilies
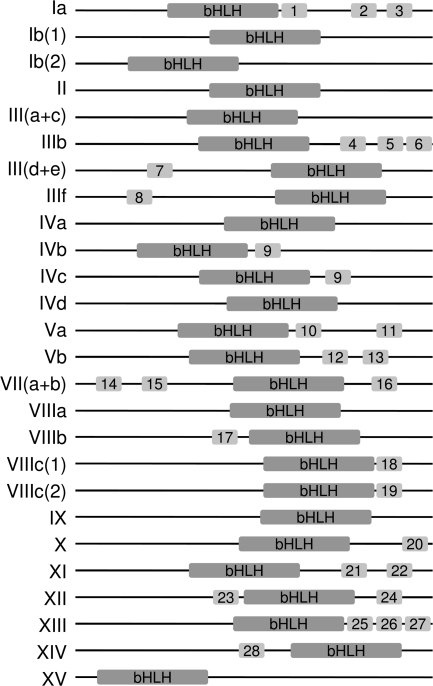
The amino acids sequences outside the bHLH region are generally divergent, even in closely related proteins from the same species. Nevertheless, it has been reported that short conserved amino acid motifs are often present in related plant bHLH proteins (Heim et al. 2003; Li et al. 2006b). If our plant bHLH classification were correct, then we expected that such motifs should be conserved within subfamilies. To determine if non-bHLH motifs were conserved throughout plant evolution, we searched for amino acid patterns in our data set of plant bHLH proteins. We found 28 motifs that are represented in both angiosperm and non-angiosperm proteins (supplementary table S2, Supplementary Material online). The relative position of each of these motifs is conserved (fig. 5): most are located C-terminal to the bHLH domain, which itself is generally located toward the C-terminal half of plant proteins. Each of these motifs is only found in members of the same subfamily, apart from motif 9, which is found in both IVb and IVc proteins (fig. 5). None of the 28 conserved motifs corresponds to known domains in the PFAM database. Motifs 14 and 15, present in several proteins of subfamily VII(a + b), overlap with the active phytochrome binding (APB) motif, shown to mediate the binding of several A. thaliana bHLH proteins to phytochrome B (Khanna et al. 2004). Motif 9 (present in IVb and IVc proteins) has a typical leucine zipper (LZ) conformation. The LZ is a dimerization domain that occurs in several regulatory proteins and consists of a periodic repetition of leucine followed by six other residues (Bornberg-Bauer et al. 1998). Several animal bHLH proteins also have an LZ immediately C-terminal to the second helix (Atchley and Fitch 1997). However, its presence in unrelated bHLH proteins suggested a multiple origin of the LZ domain in animal bHLH proteins (Atchley and Fitch 1997; Morgenstern and Atchley 1999). We could not find similarities between the bHLH sequences of IVb/IVc proteins and animal bHLH–LZ proteins. Therefore, it is likely that the acquisition of an LZ motif in bHLH proteins occurred independently in plant and animals. The occurrence of conserved domains outside the bHLH domain strongly supports the classification made on the basis of alignments of the bHLH sequence.
FIG. 5.
Non-bHLH amino acid motifs are highly conserved in each bHLH subfamily. An idealized representation of a typical member of each bHLH subfamily is shown, with the bHLH domain and other conserved motifs drawn as shaded boxes. The diagrams are not drawn to scale. The sequences of each motif in individual proteins are given in supplementary table S2 (Supplementary Material online).
We also queried the PFAM database of protein domains with the 544 plant bHLH proteins and found significant matches to an ACT domain in several unrelated proteins (OsbHLH036, VcbHLH001, CrbHLH002, OsbHLH170, VcbHLH002, and PpbHLH097). The ACT is a regulatory ligand-binding domain found in a diverse group of proteins, mostly metabolic enzymes (Chipman and Shaanan 2001). The occurrence of the ACT domain in plant bHLH proteins was previously reported (Anantharaman et al. 2001), and an ACT-like domain was found to mediate homodimerization of the maize R protein (Feller et al. 2006). Feller et al. also found ACT-like domains in over 30 A. thaliana proteins using low-stringency structure-based searches, but we could not confirm this using our stringent motif-based search methods. The occurrence of the ACT domain in a few proteins from different bHLH subfamilies suggests that the ACT–bHLH association occurred multiple times, possibly through domain-shuffling processes. Such mechanisms have been proposed to play an important role in the evolution of several metazoan bHLH proteins (Morgenstern and Atchley 1999; Ledent and Vervoort 2001).
The presence of highly conserved motifs among proteins of the same subfamily supports the phylogenetic relationships inferred from the bHLH domain sequence alone. The conservation of these extra domains during plant evolution suggests that they are essential for the function of the bHLH proteins in the respective subfamilies. Nevertheless, the presence of the ACT domain in a few unrelated proteins also indicates that domain-shuffling processes may have played a small role in plant bHLH evolution.
Discussion
Our analysis shows that most of the major subfamilies of plant bHLH transcription factors were already present in early land plants, before the divergence of mosses and vascular plants. The recent advent of large-scale sequencing projects has shown that many of the gene families that control angiosperm development were present in early land plants (Floyd and Bowman 2007). However, unlike the bHLH family, many of these families (such as the MIKCC MADS-box and TCP transcription factors) diversified after the divergence of lycophytes from the other vascular plants (Floyd and Bowman 2007). We envisage two major alternative hypotheses that would explain the early radiation of the bHLH proteins in plants. The first is that the radiation occurred in parallel with the evolution of multicellularity, before the transition of plants to terrestrial environments. The increase in the number of cell types and morphological complexity brought about by multicellularity would have been programmed by increasingly elaborate gene regulatory networks. bHLH proteins, with their ability to heterodimerize and differentially control gene expression, might have become an ideal tool to assemble such complex regulatory pathways. Consistent with this view is the observation that the first large radiation of the bHLH family in metazoans may have accompanied the evolution of multicellularity (Simionato et al. 2007). A second hypothesis is that the diversification of plant bHLH proteins accompanied the colonization of the land. The challenges faced by plants in a dry terrestrial environment led to the evolution of many novel structures and physiological mechanisms, orchestrated by versatile gene regulatory networks. Distinguishing between these alternatives will require knowledge of the number of bHLH proteins encoded in the genomes of multicellular algae. The sequence of a charophycean (multicellular aquatic algae, sister group to land plants) genome would allow the testing of these hypotheses, but unfortunately only a handful of expressed sequence tags are currently available.
All sequenced genomes of chlorophytes and red algae encode few bHLH proteins (fig. 1). We detected three distinct evolutionary lineages in chlorophytes (fig. 3). One lineage includes both chlorophytes and land plants (subfamilies IVc and IVb), implying that it predates the divergence of chlorophytes from the ancestors of land plants, over 1 billion years ago (Heckman et al. 2001). Interestingly, a characteristic of these two subfamilies is the presence of an LZ motif associated with the bHLH domain. This association has also occurred, independently, in animals. A second lineage of chlorophyte proteins is more similar to subfamily Va than to any other bHLH subfamily, although support for monophyly is poor. A third lineage is distinct from all other plant bHLH proteins and possibly evolved only in chlorophytes. The only bHLH protein found in red algae could not be clearly allocated to any clade. This suggests that none of the 26 subfamilies of plant bHLH proteins was established at the time of divergence of red algae from other plants, 1.5 billion years ago (Yoon et al. 2004). Alternatively, these protein lineages were lost in a C. merolae ancestor but are still present in other red algae; the availability of additional whole-genome sequences from red algae will help to clarify this. However, the small number of bHLH found in all the chlorophytes and red algae examined (fig. 1) and the lack of clear phylogenetic relationships with other eukaryotic bHLH proteins (fig. 4) allows us to confidently deduce that all bHLH proteins found in plants evolved after the primary endosymbiotic event that led to the evolution of plastids and are not represented in other eukaryotic groups.
Plant transcription factor families usually have high expansion rates compared with metazoan families, caused by elevated rates of retention of duplicated genes (Shiu et al. 2005). Accordingly, there are usually many (1–12) proteins per species in each of the 26 plant bHLH subfamilies (supplementary fig. S2, Supplementary Material online), in contrast with the small number (1–4) of genes found in each of the 44 metazoan subfamilies (Ledent and Vervoort 2001; Simionato et al. 2007). Members of the same plant bHLH subfamily are frequently involved in the same biological process (table 1). Usually the functions of these proteins overlap, causing them to be partially or totally redundant (e.g., HEC or BEE proteins). A striking exception comes from three A. thaliana subfamily Ia proteins, MUTE, SPEECHLESS, and FAMA: they play nonoverlapping roles in controlling sequential cell fate specification during stomatal differentiation, in a pathway surprisingly similar to metazoan bHLH proteins controlling muscle and neural development (Nadeau 2009; Serna 2009). Interestingly, the function of these proteins seems to be mostly conserved in rice and maize homologs, despite these species having considerably different stomata morphology and differentiation patterns (Liu et al. 2009). Other examples of members of the same bHLH subfamily regulating similar processes in different species are currently known (table 1). A new challenge will be to understand how the function of bHLH proteins has changed during plant evolution. An interesting glimpse comes from subfamily VIIIc(1), where the P. patens proteins PpRSL1 and PpRSL2—the only moss bHLH proteins that have been characterized so far—were shown to be required for the development of rhizoids (Menand et al. 2007). Rhizoids were lost during vascular plant evolution but the two representatives of subfamily VIIIc(1) in A. thaliana (AtRHD6 and AtRSL1) are required for the formation of root hairs, analogous structures to rhizoids with a similar rooting function (Menand et al. 2007). This suggests that these proteins were independently recruited to fulfil similar functions during land plant evolution.
Table 1.
Functionally Characterized bHLH Proteins from Different Plant Species.
| Name | bHLH Number | Function | Reference |
| Subfamily Ia | |||
| AtMUTE | AtbHLH045 | Control sequential cell fate specification during stomatal differentiation | Nadeau (2009); Serna (2009) |
| AtFAMA | AtbHLH097 | ||
| AtSPCH | AtbHLH098 | ||
| OsMUTE | OsbHLH055 | Control stomata development | Liu et al. (2009) |
| OsFAMA | OsbHLH051 | ||
| OsSPCH2 | OsbHLH053 | ||
| Subfamily Ib(1) | |||
| RGE1/ZHOUPI | AtbHLH095 | Regulates embryonic development and endosperm breakdown | Kondou et al. (2008); Yang et al. (2008) |
| Subfamily Ib(2) | |||
| OsIRO2 | OsbHLH056 | Regulates genes involved in Fe uptake under Fe-deficiency conditions | Yuko et al. (2007) |
| Subfamily III(a + c) | |||
| FIT | AtbHLH029 | Required for the up-regulation of responses to iron deficiency in Arabidopsis roots | Bauer et al. (2007) |
| RERJ1 | OsbHLH006 | Involved in the rice shoot growth inhibition caused by jasmonic acid | Kiribuchi et al. (2004) |
| Subfamily IIIb | |||
| ICE/SCRM | AtbHLH116 | Control stomatal development; implicated in the cold acclimation response and freezing tolerance | Chinnusamy et al. (2003); Kanaoka et al. (2008); Fursova et al. (2009) |
| ICE2/SCRM2 | AtbHLH033 | ||
| TaICE41 | Wheata | Potential activators of the cold-responsive genes | Badawi et al. (2008) |
| TaICE87 | |||
| Subfamily III(d + e) | |||
| MYC2/JAI1/JIN1 | AtbHLH006 | Involved in abscisic acid, jasmonic acid and light signalling pathways | Abe et al. (2003); Lorenzo et al. (2004); Yadav et al. (2005) |
| AIB | AtbHLH017 | Involved in abscisic acid signalling | Li et al. (2007) |
| PsGBF | Peaa | Regulates phenylpropanoid biosynthetic pathway | Qian et al. (2007) |
| Subfamily IIIf | |||
| TT8 | AtbHLH042 | Partially redundantly regulate anthocyanin biosynthesis, trichome and root hair development | Nesi et al. (2000); Payne et al. (2000); Bernhardt et al. (2003); Zhang et al. (2003) |
| GL3 | AtbHLH001 | ||
| EGL3 | AtbHLH002 | ||
| Ra/OSB1 | OsbHLH013 | Regulate the anthocyanin biosynthetic pathway | Ludwig et al. (1989); Burr et al. (1996); Hu et al. (2000); Spelt et al. (2000); Sakamoto et al. (2001); Sweeney et al. (2006) |
| Rb | OsbHLH165 | ||
| Rc | OsbHLH017 | ||
| OSB2 | OsbHLH016 | ||
| Lc | Maizea | ||
| IN1 | Maizea | ||
| An1 | Petuniaa | ||
| Subfamily IVa | |||
| NAI1 | AtbHLH020 | Required for the formation of an ER-derived structure, the ER body | Matsushima et al. (2004) |
| Subfamily IVc | |||
| ILR3 | AtbHLH105 | Modulate metal homeostasis and auxin-conjugate metabolism | Rampey et al. (2006) |
| Subfamily Va | |||
| BIM1 | AtbHLH046 | Implicated in brassinosteroid signaling | Yin et al. (2005) |
| BIM2 | AtbHLH102 | ||
| BIM3 | AtbHLH141 | ||
| Subfamily VII(a + b) | |||
| PIF1/PIL5 | AtbHLH015 | Bind to activated phytochromes and mediate light and gibberellin signaling responses; PIF4 was recently shown to also mediate plant architecture responses to high temperatures | Castillon et al. (2007); de Lucas et al. (2008); Leivar et al. (2008); Koini et al. (2009) |
| PIF3 | AtbHLH008 | ||
| PIF4 | AtbHLH009 | ||
| PIF5/PIL6 | AtbHLH065 | ||
| PIF7 | AtbHLH072 | ||
| HFR1 | AtbHLH026 | Mediate both phytochrome and cryptochrome signaling | Duek and Fankhauser (2003) |
| SPATULA | AtbHLH024 | Regulator of carpel margin development; mediator of germination responses to light and temperature | Heisler et al. (2001); Penfield et al. (2005) |
| ALCATRAZ | AtbHLH073 | Required for the formation of a cell layer necessary for fruit dehiscence | Rajani and Sundaresan (2001) |
| UNE10 | AtbHLH016 | Involved in the fertilization process | Pagnussat et al. (2005) |
| BP-5 | OsbHLH102 | Involved in the regulation of amylose synthesis in the rice endosperm | Zhu et al. (2003) |
| Subfamily VIIIb | |||
| HEC1 | AtbHLH088 | Redundantly control the development of the transmitting tract and stigma; each of these proteins can form heterodimers with SPATULA | Gremski et al. (2007) |
| HEC2 | AtbHLH037 | ||
| HEC3 | AtbHLH043 | ||
| LAX | OsbHLH123 | Regulator of axillary meristem generation in rice | Komatsu et al. (2003) |
| INDEHISCENT | AtbHLH040 | Required for the differentiation, in the Arabidopsis fruit, of three cell types involved in seed dispersal | Liljegren et al. (2004) |
| Subfamily VIIIc(1) | |||
| AtRHD6 | AtbHLH083 | Required for the formation of root hairs | Menand et al. (2007) |
| AtRSL1 | AtbHLH086 | ||
| PpRSL1 | PpbHLH043 | Redundantly required for the development of rhizoids and caulonemata | Menand et al. (2007) |
| PpRSL2 | PpbHLH033 | ||
| Subfamily VIIIc(2) | |||
| RSL2 | AtbHLH085 | Partially redundant and involved in root hair development | Yi (2008) |
| RSL3 | AtbHLH084 | ||
| RSL4 | AtbHLH054 | ||
| RSL5 | AtbHLH139 | ||
| Subfamily XI | |||
| UNE12 | AtbHLH059 | Involved in the fertilization process | Pagnussat et al. (2005) |
| PTF1 | OsbHLH096 | Involved in the responses to phosphate deficiency stress | Yi et al. (2005) |
| Subfamily XII | |||
| ZCW32/BPE | AtbHLH031 | Controls petal size | Szecsi et al. (2006) |
| BEE1 | AtbHLH044 | Redundant positive regulators of brassinosteroid signalling | Friedrichsen et al. (2002) |
| BEE2 | AtbHLH058 | ||
| BEE3 | AtbHLH050 | ||
| CIB1 | AtbHLH063 | Shown to interact with the blue-light receptor CRY2 and promote floral initiation | Liu et al. (2008) |
| CIB5 | AtbHLH076 | ||
| Subfamily XIII | |||
| LHW | AtbHLH156 | Regulates the size of the vascular initial population in the root meristem | Ohashi-Ito and Bergmann (2007) |
| Subfamily XIV | |||
| SAC51 | AtbHLH142 | Involved in a spermidine synthase mediated stem elongation process | Imai et al. (2006) |
| Subfamily XV | |||
| PRE1 | AtbHLH136 | Proposed to act as positive regulators of gibberellin signalling | Lee et al. (2006) |
| PRE2 | AtbHLH134 | ||
| PRE3 | AtbHLH135 | ||
| PRE4 | AtbHLH161 | ||
| PRE5 | At3g28857a | ||
| PRE6 | At1g26945a | ||
| KIDARI | At1g26945a | Represses light signal transduction; interacts and negatively regulates HFR1 | Hyun and Lee (2006) |
| Orphans | |||
| AMS | AtbHLH021 | Required for correct anther development, particularly tapetum development | Sorensen et al. (2003) |
| DYT1 | AtbHLH022 | Zhang et al. (2006) | |
| TDR | OsbHLH005 | Li et al. (2006a) | |
| Udt1 | OsbHLH164 | Jung et al. (2005) | |
| MEE8 | AtbHLH108 | Required for early embryo development | Pagnussat et al. (2005) |
| Fer | Tomatoa | Controls iron-uptake responses in roots | Ling et al. (2002) |
| Gmyc1 | Gerberaa | Regulates the expression of an anthocyanin pathway enzyme | Elomaa et al. (1998) |
| delila | Antirrhinuma | Regulates the pattern of anthocyanin pigmentation | Goodrich et al. (1992) |
| JAF13 | Petuniaa | Regulates the anthocyanin biosynthetic pathway | Quatrochio et al. (1998) |
| PAR1 | At2g42870a | Negatively control growth and metabolic shade avoidance responses | Roig-Villanova et al. (2007) |
| PAR2 | At3g58850a | ||
NOTE.—ER, endoplasmic reticulum.
These proteins were not included in our phylogenetic analysis; their classification was based on pHMM scores to subfamily-specific pHMMs.
The presence of highly conserved motifs (such as the APB motif in PIF proteins) in the different plant bHLH subfamilies (fig. 5) indicates that the partners of molecular interactions are also conserved. This is particularly exciting because it suggests that protein interactions that are at the base of gene regulatory networks are highly conserved across plants. Several plant bHLH proteins are known to form transcription complexes with MYB proteins (Ramsay and Glover 2005). Although the early evolution of MYB proteins in plants has not been characterized, we found over 30 MYB sequences in the genome of C. reinhardtii and more than 150 sequences in P. patens (data not shown). Given the large number of both bHLH and MYB proteins in mosses, it is appealing to hypothesize that the bHLH–MYB complex had evolved early in land plant evolution. The picture that emerges from this and other studies is that much of the complex regulatory machinery that we are currently dissecting in “higher” plants was actually invented by very “simple” ones, early in land plant evolution. The recent reappraisal of algae, bryophytes, and lycophytes as experimental organisms will be an excellent tool to clarify the molecular and biological foundations of many of these processes.
Supplementary Material
Supplementary tables S1 and S2, figures S1 and S2, and data 1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We would like to thank Lars Østergaard, Keke Yi, and Rita Galhano for critical comments on the manuscript and Julie Hawkins and Alastair Culham for teaching us phylogenetics. This work was supported by a grant (SFRH/BD/28100/2006) to N.P. from the Portuguese Fundação para a Ciência e a Tecnologia; a grant in aid to L.D. and the John Innes Centre from The Biotechnology and Biological Research Council of the United Kingdom; and a grant from the Human Frontiers in Science Program to L.D.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J Mol Biol. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, et al. (45 co-authors) The genome of the diatom Thalassiosira Pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci USA. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Terhalle W, Dress A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol. 1999;48:501–516. doi: 10.1007/pl00006494. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Zhao J. Molecular architecture of the DNA-binding region and its relationship to classification of basic helix-loop-helix proteins. Mol Biol Evol. 2007;24:192–202. doi: 10.1093/molbev/msl143. [DOI] [PubMed] [Google Scholar]
- Badawi M, Reddy YV, Agharbaoui Z, Tominaga Y, Danyluk J, Sarhan F, Houde M. Structure and functional analysis of wheat ICE (inducer of CBF expression) genes. Plant Cell Physiol. 2008;49:1237–1249. doi: 10.1093/pcp/pcn100. [DOI] [PubMed] [Google Scholar]
- Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, Jakoby M, Werber M, Weisshaar B. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell. 2003;15:2497–2502. doi: 10.1105/tpc.151140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- Bauer P, Ling H-Q, Guerinot ML. FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol Biochem. 2007;45:260–261. doi: 10.1016/j.plaphy.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development. 2003;130:6431–6439. doi: 10.1242/dev.00880. [DOI] [PubMed] [Google Scholar]
- Bornberg-Bauer E, Rivals E, Vingron M. Computational approaches to identify leucine zippers. Nucleic Acids Res. 1998;26:2740–2746. doi: 10.1093/nar/26.11.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M, Atchley W. Phylogenetic analysis of plant basic helix-loop-helix proteins. J Mol Evol. 2003;56:742–750. doi: 10.1007/s00239-002-2449-3. [DOI] [PubMed] [Google Scholar]
- Burr FA, Burr B, Scheffler BE, Blewitt M, Wienand U, Matz EC. The maize repressor-like gene intensifier1 shares homology with the r1/b1 multigene family of transcription factors and exhibits missplicing. Plant Cell. 1996;8:1249–1259. doi: 10.1105/tpc.8.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007;12:514–521. doi: 10.1016/j.tplants.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee B-h, Hong X, Agarwal M, Zhu J-K. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman DM, Shaanan B. The ACT domain family. Curr Opin Struct Biol. 2001;11:694–700. doi: 10.1016/s0959-440x(01)00272-x. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Daviere J-M, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C. HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J. 2003;34:827–836. doi: 10.1046/j.1365-313x.2003.01770.x. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Elomaa P, Mehto M, Kotilainen M, Helariutta Y, Nevalainen L, Teeri TH. A bHLH transcription factor mediates organ, region and flower type specific signals on dihydroflavonol-4-reductase (dfr) gene expression in the inflorescence of Gerbera hybrida (Asteraceae) Plant J. 1998;16:93–99. doi: 10.1046/j.1365-313x.1998.00273.x. [DOI] [PubMed] [Google Scholar]
- Feller A, Hernandez JM, Grotewold E. An ACT-like domain participates in the dimerization of several plant basic-helix-loop-helix transcription factors. J Biol Chem. 2006;281:28964–28974. doi: 10.1074/jbc.M603262200. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Ferré-D’Amaré A, Prendergast G, Ziff E, Burley S. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL. The ancestral developmental tool kit of land plants. Int J Plant Sci. 2007;168:1–35. [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162:1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fursova OV, Pogorelko GV, Tarasov VA. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene. 2009;429:98–103. doi: 10.1016/j.gene.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA. 1988;85:7089–7093. doi: 10.1073/pnas.85.19.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J, Carpenter R, Coen ES. A common gene regulates pigmentation pattern in diverse plant species. Cell. 1992;68:955–964. doi: 10.1016/0092-8674(92)90038-e. [DOI] [PubMed] [Google Scholar]
- Gremski K, Ditta G, Yanofsky MF. The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development. 2007;134:3593–3601. doi: 10.1242/dev.011510. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guo A-Y, Chen X, Gao G, Zhang H, Zhu Q-H, Liu X-C, Zhong Y-F, Gu X, He K, Luo J. PlantTFDB: a comprehensive plant transcription factor database. Nucleic Acids Res. 2008;36:D966–D969. doi: 10.1093/nar/gkm841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB. Molecular evidence for the early colonization of land by fungi and plants. Science. 2001;293:1129–1133. doi: 10.1126/science.1061457. [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20:735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- Heisler MG, Atkinson A, Bylstra YH, Walsh R, Smyth DR. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development. 2001;128:1089–1098. doi: 10.1242/dev.128.7.1089. [DOI] [PubMed] [Google Scholar]
- Hu J, Reddy VS, Wessler SR. The rice R gene family: two distinct subfamilies containing several miniature inverted-repeat transposable elements. Plant Mol Biol. 2000;42:667–678. doi: 10.1023/a:1006355510883. [DOI] [PubMed] [Google Scholar]
- Hyun Y, Lee I. KIDARI, encoding a non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol Biol. 2006;61:283–296. doi: 10.1007/s11103-006-0010-2. [DOI] [PubMed] [Google Scholar]
- Imai A, Hanzawa Y, Komura M, Yamamoto KT, Komeda Y, Takahashi T. The dwarf phenotype of the Arabidopsis acl5 mutant is suppressed by a mutation in an upstream ORF of a bHLH gene. Development. 2006;133:3575–3585. doi: 10.1242/dev.02535. [DOI] [PubMed] [Google Scholar]
- Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004;5:226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K-H, Han M-J, Lee Y-S, Kim Y-W, Hwang I, Kim M-J, Kim Y-K, Nahm BH, An G. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell. 2005;17:2705–2722. doi: 10.1105/tpc.105.034090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu J-K, Torii KU. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell. 2008;20:1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P, Crane P. The origin and early evolution of plants on land. Nature. 1997;389:33–39. [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiribuchi K, Sugimori M, Takeda M, et al. (16 co-authors) RERJ1, a jasmonic acid-responsive gene from rice, encodes a basic helix-loop-helix protein. Biochem Biophys Res Commun. 2004;325:857–863. doi: 10.1016/j.bbrc.2004.10.126. [DOI] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J. LAX and SPA: major regulators of shoot branching in rice. Proc Natl Acad Sci USA. 2003;100:11765–11770. doi: 10.1073/pnas.1932414100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondou Y, Nakazawa M, Kawashima M, et al. (11 co-authors) RETARDED GROWTH OF EMBRYO1, a new basic helix-loop-helix protein, expresses in endosperm to control embryo growth. Plant Physiol. 2008;147:1924–1935. doi: 10.1104/pp.108.118364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledent V, Paquet O, Vervoort M. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 2002;3:research0030.0031–0030.0018. doi: 10.1186/gb-2002-3-6-research0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee S, Yang K-Y, Kim Y-M, Park S-Y, Kim SY, Soh M-S. Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:591–600. doi: 10.1093/pcp/pcj026. [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell. 2008;20:337–352. doi: 10.1105/tpc.107.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LA, McCourt RM. Green algae and the origin of land plants. Am J Bot. 2004;91:1535–1556. doi: 10.3732/ajb.91.10.1535. [DOI] [PubMed] [Google Scholar]
- Li H, Sun J, Xu Y, Jiang H, Wu X, Li C. The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol Biol. 2007;65:655–665. doi: 10.1007/s11103-007-9230-3. [DOI] [PubMed] [Google Scholar]
- Li N, Zhang D-S, Liu H-S, et al. (15 co-authors) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006a;18:2999–3014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, et al. (13 co-authors) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006b;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Roeder AHK, Kempin SA, Gremski K, Østergaard L, Guimil S, Reyes DK, Yanofsky MF. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell. 2004;116:843–853. doi: 10.1016/s0092-8674(04)00217-x. [DOI] [PubMed] [Google Scholar]
- Ling H-Q, Bauer P, Bereczky Z, Keller B, Ganal M. The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA. 2002;99:13938–13943. doi: 10.1073/pnas.212448699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- Liu T, Ohashi-Ito K, Bergmann DC. Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development. 2009;136:2265–2276. doi: 10.1242/dev.032938. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci USA. 1989;86:7092–7096. doi: 10.1073/pnas.86.18.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Fukao Y, Nishimura M, Hara-Nishimura I. NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body. Plant Cell. 2004;16:1536–1549. doi: 10.1105/tpc.021154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin-i T, et al. (42 co-authors) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- Menand B, Yi K, Jouannic S, Hoffmann L, Ryan E, Linstead P, Schaefer DG, Dolan L. An ancient mechanism controls the development of cells with a rooting function in land plants. Science. 2007;316:1477–1480. doi: 10.1126/science.1142618. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, et al. (117 co-authors) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B, Atchley WR. Evolution of bHLH transcription factors: modular evolution by domain shuffling? Mol Biol Evol. 1999;16:1654–1663. doi: 10.1093/oxfordjournals.molbev.a026079. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nadeau JA. Stomatal development: new signals and fate determinants. Curr Opin Plant Biol. 2009;12:29–35. doi: 10.1016/j.pbi.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development. 2007;134:2959–2968. doi: 10.1242/dev.006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu H-J, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie L-F, Ye D, Sundaresan V. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development. 2005;132:603–614. doi: 10.1242/dev.01595. [DOI] [PubMed] [Google Scholar]
- Palenik B, Grimwood J, Aerts A, et al. (38 co-authors) The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci USA. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Josse E-M, Kannangara R, Gilday AD, Halliday KJ, Graham IA. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr Biol. 2005;15:1998–2006. doi: 10.1016/j.cub.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Qian W, Tan G, Liu H, He S, Gao Y, An C. Identification of a bHLH-type G-box binding factor and its regulation activity with G-box and Box I elements of the PsCHS1 promoter. Plant Cell Rep. 2007;26:85–93. doi: 10.1007/s00299-006-0202-x. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, van der Woude K, Mol JN, Koes R. Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 1998;13:475–488. doi: 10.1046/j.1365-313x.1998.00046.x. [DOI] [PubMed] [Google Scholar]
- Rajani S, Sundaresan V. The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr Biol. 2001;11:1914–1922. doi: 10.1016/s0960-9822(01)00593-0. [DOI] [PubMed] [Google Scholar]
- Rampey RA, Woodward AW, Hobbs BN, Tierney MP, Lahner B, Salt DE, Bartel B. An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics. 2006;174:1841–1857. doi: 10.1534/genetics.106.061044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Robinson KA, Lopes JM. Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic Acids Res. 2000;28:1499–1505. doi: 10.1093/nar/28.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Löffelhardt W, Bohnert HJ, Philippe H, Lang BF. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portoles S, Rodriguez-Concepcion M, Martinez-Garcia JF. Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 2007;26:4756–4767. doi: 10.1038/sj.emboj.7601890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W, Ohmori T, Kageyama K, Miyazaki C, Saito A, Murata M, Noda K, Maekawa M. The Purple leaf (Pl) locus of rice: the Plw allele has a complex organization and includes two genes encoding basic helix-loop-helix proteins involved in anthocyanin biosynthesis. Plant Cell Physiol. 2001;42:982–991. doi: 10.1093/pcp/pce128. [DOI] [PubMed] [Google Scholar]
- Schuster-Bockler B, Schultz J, Rahmann S. HMM logos for visualization of protein families. BMC Bioinformatics. 2004;5:7. doi: 10.1186/1471-2105-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna L. Emerging parallels between stomatal and muscle cell lineages. Plant Physiol. 2009;149:1625–1631. doi: 10.1104/pp.108.133090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S-H, Shih M-C, Li W-H. Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol. 2005;139:18–26. doi: 10.1104/pp.105.065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan B, Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen A-M, Kröber S, Unte US, Huijser P, Dekker K, Saedler H. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 2003;33:413–423. doi: 10.1046/j.1365-313x.2003.01644.x. [DOI] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol JNM, Koes R. Anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell. 2000;12:1619–1632. doi: 10.1105/tpc.12.9.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steemans P, Herisse AL, Melvin J, Miller MA, Paris F, Verniers J, Wellman CH. Origin and radiation of the earliest vascular land plants. Science. 2009;324:353. doi: 10.1126/science.1169659. [DOI] [PubMed] [Google Scholar]
- Sweeney MT, Thomson MJ, Pfeil BE, McCouch S. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell. 2006;18:283–294. doi: 10.1105/tpc.105.038430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods) Sunderland (MA): Sinauer Associates; 2003. [Google Scholar]
- Szecsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 2006;25:3912–3920. doi: 10.1038/sj.emboj.7601270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Liu T, Tian C, Sun S, Li J, Chen M. Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol Biol. 2005;59:191–203. doi: 10.1007/s11103-005-6503-6. [DOI] [PubMed] [Google Scholar]
- Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S. A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell. 2005;17:1953–1966. doi: 10.1105/tpc.105.032060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Johnston N, Talideh E, Mitchell S, Jeffree C, Goodrich J, Ingram G. The endosperm-specific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal development. Development. 2008;135:3501–3509. doi: 10.1242/dev.026708. [DOI] [PubMed] [Google Scholar]
- Yi K. Temporal regulation of root hair development by RHD6 family genes [PhD thesis] [Norwich (UK)]: University of East Anglia; 2008. [Google Scholar]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P. OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol. 2005;138:2087–2096. doi: 10.1104/pp.105.063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- Yuko O, Reiko Nakanishi I, Hiromi N, Takanori K, Michiko T, Satoshi M, Naoko KN. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 2007;51:366–377. doi: 10.1111/j.1365-313X.2007.03149.x. [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development. 2006;133:3085–3095. doi: 10.1242/dev.02463. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Cai X-L, Wang Z-Y, Hong M-M. An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. J Biol Chem. 2003;278:47803–47811. doi: 10.1074/jbc.M302806200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.