Abstract
Whole-genome duplications (WGDs) have occurred repeatedly in the vertebrate lineage, but their evolutionary significance for phenotypic evolution remains elusive. Here, we have investigated the impact of the fish-specific genome duplication (FSGD) on the evolution of pigmentation pathways in teleost fishes. Pigmentation and color patterning are among the most diverse traits in teleosts, and their pigmentary system is the most complex of all vertebrate groups.
Using a comparative genomic approach including phylogenetic and synteny analyses, the evolution of 128 vertebrate pigmentation genes in five teleost genomes following the FSGD has been reconstructed. We show that pigmentation genes have been preferentially retained in duplicate after the FSGD, so that teleosts have 30% more pigmentation genes compared with tetrapods. This is significantly higher than genome-wide estimates of FSGD gene duplicate retention in teleosts. Large parts of the melanocyte regulatory network have been retained in two copies after the FSGD. Duplicated pigmentation genes follow general evolutionary patterns such as the preservation of protein complex stoichiometries and the overrepresentation of developmental genes among retained duplicates. These results suggest that the FSGD has made an important contribution to the evolution of teleost-specific features of pigmentation, which include novel pigment cell types or the division of existing pigment cell types into distinct subtypes. Furthermore, we have observed species-specific differences in duplicate retention and evolution that might contribute to pigmentary diversity among teleosts.
Our study therefore strongly supports the hypothesis that WGDs have promoted the increase of complexity and diversity during vertebrate phenotypic evolution.
Keywords: genome duplication, fish, conserved synteny, pigment cell, melanocyte, functional module
Introduction
It is now generally accepted that several rounds of polyploidization and rediploidization have occurred in vertebrates, including two rounds of whole-genome duplication (WGD) at the base of the vertebrate lineage (1R/2R duplications; Ohno 1970; Furlong and Holland 2002; Dehal and Boore 2005; Putnam et al. 2008). An additional fish-specific genome duplication (FSGD) has taken place in ray-finned fishes before the radiation of teleost fishes (Amores et al. 1998; Taylor et al. 2003; Jaillon et al. 2004; Meyer and Van de Peer 2005; Kasahara et al. 2007). In addition, more recent, lineage-specific WGDs have been observed in various vertebrate lineages (Otto 2007).
Although mutational inactivation and loss of redundant copies (nonfunctionalization) is the most common fate of one copy of duplicated genes, many duplicate genes are retained after rediploidization. Hence, WGDs add substantial amounts of genetic material to a genome (Lynch and Conery 2000; Blomme et al. 2006). An important question is, whether specific classes of genes are preferentially retained in duplicate after a WGD event. Recent studies on WGDs in eukaryotes as divergent as yeasts (Scannell et al. 2007; Wapinski et al. 2007), ciliates (Aury et al. 2006), plants (Maere et al. 2005), early vertebrates (Putnam et al. 2008), teleosts (Brunet et al. 2006), and the frog Xenopus leavis (Semon and Wolfe 2008) revealed that slowly evolving as well as highly expressed genes are preferentially retained, and gene dosage within protein complexes or metabolic pathways is generally kept. Furthermore, in multicellular organisms, genes involved in developmental processes, regulation of transcription and signal transduction are maintained at a high rate after WGDs. The retention of such genes is rather uncommon for paralogs generated through small-scale, more local duplications (Maere et al. 2005; Blomme et al. 2006; Brunet et al. 2006; Putnam et al. 2008).
The evolutionary consequences of WGDs and their potential contribution to the evolutionary success of anciently polyploid (paleopolyploid) species in the long term are not well understood (Otto 2007). Theoretical models predict that differential loss of gene duplicates in isolated populations may lead to genomic incompatibilities and ultimately speciation (Werth and Windham 1991; Lynch and Force 2000). Correlations of such reciprocal gene loss (RGL) and increased speciation rates after WGDs have been observed in yeasts (Scannell et al. 2006), ciliates (Aury et al. 2006), and teleost fish (Semon and Wolfe 2007). Furthermore, many authors have suggested that WGDs have provided the genetic raw material for important morphological transitions, key innovations, and increased phenotypic diversity and complexity (e.g., Ohno 1970; Holland et al. 1994; Aburomia et al. 2003; De Bodt et al. 2005), whereas this effect of WGDs was regarded as less important by others (Donoghue and Purnell 2005; Carroll 2008).
Here, we have analyzed the impact of the FSGD on the evolution of pigmentation pathways in teleost fish. Pigmentation and color patterning are among the most variable traits in vertebrates (Braasch et al. 2008; Protas and Patel 2008). Importantly, the pigmentary system of teleost fishes is the most diverse and complex of all vertebrates and therefore offers an excellent opportunity to study the effect of WGD on phenotypic evolution (Braasch et al. 2007, 2008). Teleosts have more neural crest-derived pigment cells types than all other vertebrate groups. Although black melanophores, reflecting iridophores and yellow-red xanthophores/erythrophores must have already been present in the common ancestor of ray-finned fish and tetrapods, teleosts have evolved an additional pigment cell type, the whitish leucophores, as well as distinct subtypes of the aforementioned pigment cell types (Mellgren and Johnson 2002; Bagnara and Matsumoto 2006; Braasch et al. 2008). In addition, blue cyanophores are present in some teleost lineages (Bagnara et al. 2007), and there is recent evidence for red fluorescent pigmentation in several reef fish species (Michiels et al. 2008).
It has been suggested that the FSGD was of major importance for the evolution of teleost pigmentation (Mellgren and Johnson 2002; Braasch et al. 2008). In a previous study, we could show that teleost fishes have more genes encoding pigment synthesis enzymes than any other vertebrate group as result of the FSGD (Braasch et al. 2007). Here, we have expanded our analysis to a list of all 128 known “vertebrate pigmentation genes.” We define vertebrate pigmentation genes as those genes that were shown in at least one vertebrate species to be involved in the development and/or differentiation of neural crest-derived pigment cells. The fact that almost all pigmentation genes found in mammals are also found to be involved in pigmentation in teleosts and vice versa (reviewed, e.g., in Rawls et al. 2001; Braasch et al. 2008) points to a general evolutionary conservation of vertebrate pigment cell development and justifies our assumption that a pigmentation gene of one vertebrate group is highly likely to have pigmentation functions in other lineages.
Importantly, many of the genes involved in pigment cell development have other functions not related to pigmentation. According to the duplication-degeneration-complementation model (Force et al. 1999), ancestral genes functions might be distributed among duplicated genes (subfunctionalization). Thus, in some cases, pigmentation functions could become separated from other functions after duplication. In this case, the duplication would not increase the number of pigmentation genes per se because only one paralog retains the pigmentation function. However, because the paralog that keeps the pigmentation function will be released from functional constraints imposed by other essential functions present in the ancestral gene, the duplication still might facilitate evolution of pigmentation by specializing one of the paralogs for the pigmentation function (Braasch et al. 2008). Thus, for the sake of the present study, the term “pigmentation gene” corresponds to the presumed ancestral gene's pigmentation function at the time of its duplication.
By reconstructing the evolutionary history of pigmentation gene families in the vertebrate lineage and by carefully distinguishing gene duplicates generated in the FSGD from those originating in the earlier rounds of WGD (1R, 2R) and in small-scale duplication events, we show that extant teleost genomes (zebrafish, medaka, stickleback, Tetraodon, fugu) contain ≈30% more pigmentation genes compared with tetrapods as result of FSGD. This is significantly higher than available estimates for genome-wide retention rates (≈12–24%). We found important differences in FSGD duplicate retention rates among functional categories of pigmentation genes, and most of the known major regulators of pigment cell development have been retained in duplicate. Our results therefore point to a major role of WGDs in the evolution of vertebrate phenotypes.
Materials and Methods
Sequence Database Surveys
Nucleotide sequences of pigmentation genes were identified using Blast searches against GenBank (http://www.ncbi.nlm.nih.gov/) and Ensembl genome assemblies (http://www.ensembl.org/index.html; version 50, July 2008) of human (Homo sapiens; Hsa), mouse (Mus musculus; Mmu), chicken (Gallus gallus: Gga), frog (Xenopus tropicalis; Xtr), zebrafish (Danio rerio; Dre), medaka (Oryzias latipes; Ola), spotted green pufferfish (Tetraodon nigroviridis; Tni), torafugu (Takifugu rubripes; Tru), and stickleback (Gasterosteus aculeatus; Gac). Usually, human sequences were used as initial queries. The results of the Blast searches were double-checked against the Ensembl gene families (if available) to collect missing sequences and to get an initial overview of the gene family tree topology. Pigmentation genes and their accession numbers are provided in supplementary tables S1–S2 and supplementary figures S1–S24 (Supplementary Material online).
Sequence Alignments and Phylogenetic Reconstructions
Nucleotide sequences obtained from Blast searches were loaded into BioEdit (Hall 1999), translated into proteins, and aligned using ClustalW (Thompson et al. 1994). Alignments were carefully checked and ambiguously aligned regions were removed before phylogenetic analyses. Maximum likelihood phylogenies based on protein data were computed with PHYML (Guindon et al. 2005) with 100 bootstrap replicates. Models of protein evolution (mainly JTT + I + G) and parameter values were determined with ProtTest (Abascal et al. 2005). MEGA4 (Tamura et al. 2007) was used to obtain Neighbor-Joining bootstrap values of 10,000 replicates and to draw phylogenies. Trees were rooted either with the closest vertebrate paralog in case of larger gene families or with an invertebrate ortholog. For some phylogenies, no suitable outgroup sequence was available (Dtnbp1, Atp6v0c, Nsf, Itgb1). Although Blast hits for potential outgroup sequences were available, these were too divergent for a reliable alignment and were thus excluded from the phylogenetic analyses.
Synteny Analyses
For microsynteny analyses, genes surrounding the human ortholog were used as initial queries for Blast searches against the teleost genome assemblies at Ensembl, followed by reciprocal Blast searches of the best hits against human and other teleost genomes.
For macrosynteny analysis, the last release of Tetraodon genome assembly (v8) was used to generate a rose-window representation as in Jaillon et al. (2004) and Brunet et al. (2006). In this meta-analysis, we took all Tetraodon proteins (available from the ftp site of Ensembl v50), did a Blast all against all of these proteins under default parameters, and selected the first hits for which genes are on different chromosomes. A cutoff P value ≤1 × 10−3 was applied, and the background was reduced by removing intrachromosomal links and links between chromosomes for which the total number is below eight. Of note, with this last release and this method, we increased the number of strong links between the chromosomes, corroborating the evidence for the FSGD obtained previously (Jaillon et al. 2004; Brunet et al. 2006). This whole-genome analysis of gene duplication was superimposed with the links collected for the duplicated pigmentation genes, which confirmed the FSGD origin for most of the duplicated pigmentation genes.
Functional Annotation Analysis
The Functional Annotation Tool of the DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/) was used to test for enrichment in gene ontology GO) functional terms (Huang da et al. 2009). From the lists of pigmentation and liver genes (supplementary tables S2–S3, Supplementary Material online), the Ensembl gene IDs of human orthologs were uploaded and compared with the human reference genome as background. Enrichments were considered significant if the modified Fisher Exact P value was <0.05.
Results
Vertebrate Pigmentation Genes
As a first step, we undertook database and literature surveys and generated a list of vertebrate pigmentation genes. From mammals, the list included over 100 known genes involved in coat color formation in mice (Bennett and Lamoreux 2003) from the Color Gene Database (http://www.espcr.org/micemut/) extended by genes important for skin and hair color differences between human populations, which were recently identified through genome-wide association studies (Bastian and Pinkel 2008). Many of these genes have also been identified as coat color genes in other mammals (e.g., rat, cat, dog, cow, sheep, pig, horse). Additional genes in the list affect plumage coloration in birds (e.g., Miwa et al. 2007). Pigmentation genes identified in teleosts were obtained from pigmentation mutant collections in zebrafish (Haffter et al. 1996; Kelsh et al. 1996; Odenthal et al. 1996) available from the Zebrafish Model Organism Database (ZFIN; http://zfin.org) and in medaka (Kelsh et al. 2004). Finally, we included information given by the Gene Ontology Database (GO term “pigmentation” GO:0043473 and children terms; http://www.geneontology.org/). From the initial pigmentation gene list, we omitted genes affecting pigmentation solely by their function in the development of hair or the retinal pigment epithelium (RPE). Genes required for the production of pheomelanin, a lighter type of melanin pigment not found in teleosts and other poikilotherm vertebrates (Fujii 1993b), were also not included. These genes are generally present in vertebrates including teleosts but have acquired their role in pheomelanin synthesis in the lineages of mammals and birds.
The final list of vertebrate pigmentation genes included in this study consists of 128 genes and is presented in supplementary table S1 (Supplementary Material online). Genes were grouped into ten functional categories adopting the classification for mouse coat color mutants (http://www.espcr.org/micemut/) and using additional information given by ZFIN (http://zfin.org) and the Gene Ontology Database (http://www.geneontology.org/): 1) melanophore development, 2) components of melanosomes, 3) melanosome biogenesis, 4) melanosome transport, 5) regulation of melanogenesis, 6) systemic effects, 7) xanthophore development, 8) pteridine synthesis, 9) iridophore development, and 10) uncategorized function (supplementary table S1, Supplementary Material online). Other categories may exist but are not represented by a known gene so far (e.g., pterinosome biogenesis, transport, etc.). Some genes were included into multiple categories because they are involved in the formation of different pigment cell types (e.g., Sox10, Dutton et al. 2001).
Furthermore, we chose “liver genes” to contrast our data on pigmentation genes with another category of functionally related genes that is not expected to be under high selection for duplicate gene retention. To this end, 187 liver genes obtained from the Gene Ontology Database (GO term “liver development” GO:0001889; http://www.geneontology.org/) and the collection of zebrafish liver mutants from ZFIN (http://zfin.org) were analyzed similarly as the pigmentation genes.
Identification of 46 Fish-Specific Pairs of Pigmentation Gene Duplicates
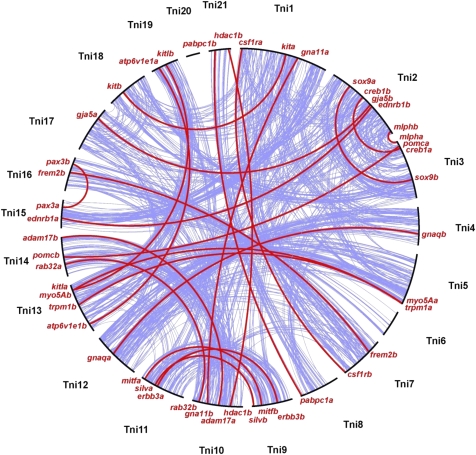
Fish-specific gene duplicates were identified by complementary approaches combining phylogenetic reconstructions and synteny analyses. Sequence information from the genome assemblies of five teleosts (zebrafish, medaka, stickleback, Tetraodon, fugu) and four tetrapods (frog, chicken, mouse, human) was included. This comparative genomic approach is particularly powerful to infer the duplication history of gene families in case of ambiguous tree topologies as well as species-specific gene order rearrangements or genome assembly problems (Braasch et al. 2008). Distinguishing FSGD duplicates from other types of WGD (1R/2R) or local gene duplications is essential because duplicates of varying age and type show different patterns of duplicate retention and functional evolution (Maere et al. 2005; Aury et al. 2006; Wapinski et al. 2007). Criteria that were used to conclude that pairs of gene duplicates were derived from the FSGD are illustrated by the example of Sox10 in figure 1. According to the tree topology, the duplication of the teleosts paralogs should date back to one common ancestor of the five teleost species but within the fish lineage after split from tetrapods (Fig. 1A). Furthermore, the gene duplicates should be part of paralogons (i.e., large chromosomal blocks of duplicated gene pairs) that show conserved synteny with each other and with an outgroup species, for example, human (double conserved synteny). On a fine scale, genes in the vicinity of the paralogs are connected in a pattern of “microsynteny” (fig. 1B). On the genome-wide scale, FSGD-duplicated pigmentation genes take part in a higher level pattern of “macrosynteny” connecting entire paralogous chromosomes (fig. 2). Macrosynteny patterns have been previously used to reconstruct the protochromosomes of the pre-FSGD karyotype of the ancestral euteleost (Jaillon et al. 2004; Woods et al. 2005; Kasahara et al. 2007). Here, we used the most recent analysis of Kasahara et al. (2007) to assign pigmentation gene duplicates to the 13 protochromosomes (a–m) from which they are derived.
FIG. 1.—
Evolution of the Sox10 transcription factor gene. (A) Maximum likelihood (ML) phylogeny of vertebrate Sox10 proteins based on 563 AA positions. The tree is rooted with human SOX9. Bootstrap values (ML/Neighbor-Joining [NJ]) above 50% are shown. The FSGD generated two Sox10 in teleosts, Sox10a (blue), and Sox10b (green). Sox10a is missing in zebrafish. (B) The microsynteny pattern of Sox10 regions in vertebrate genome shows double conserved synteny between the two teleost sox10 paralogons and human chr22q13. A sox10a paralogon could not be identified in zebrafish, and the zebrafish sox10b (dashed red bar) is not included in the Zv7 genome assembly but has been mapped to chr3 (Dutton et al. 2001). Numbered bars represent genes contributing to conserved synteny, and genes that do not contribute to conserved synteny are not shown. Dotted lines connect orthologous genes.
FIG. 2.—
Macrosynteny of FSGD-duplicated pigmentation genes. Blue lines connect paralogous genes on the 21 chromosomes in the Tetraodon genome (Tni1–Tni21). Red lines connect paralogous pigmentation gene duplicates, showing that they mostly follow the major routes of FSGD duplication. A highly similar pattern is also observed for stickleback, medaka, and zebrafish (not shown).
Of the 128 pigmentation genes analyzed, 46 genes (35.9%) were retained in two copies after the FSGD in at least 1 of the 5 teleost genomes and 65.2% (30/46) of the duplicated genes were present in two copies in all five teleost genomes (table 1; supplementary table S2, supplementary figs. S1–S24, Supplementary Material online). For 82 genes (64.1%), one of the paralogs has been lost in all five teleosts (supplementary table S2, Supplementary Material online). Twenty-two pairs of duplicates were previously shown to be derived from the FSGD, and 24 new pairs were identified in the present study (table 1). The 46 pigmentation gene paralogs are derived from the pre-FSGD karyotype with at least two pairs of duplicates descending from each protochromosome except protochromosome d (table 1). This shows that the duplication and retention of pigmentation genes is genome wide and not restricted to a particular chromosomal subset. Furthermore, we can exclude that some of the 46 paralog pairs were generated in an earlier round of WGD in vertebrates (1R or 2R) because such duplications would give different macrosynteny patterns.
Table 1.
FSGD-Duplicated Teleost Pigmentation Genes
| Genes | Function | Homo | Coorthologs | Danio | Oryzias | Gasterosteus | Tetraodon | Protochromosomes | Figures/ref. |
| Adam17 | a | Hsa2 | adam17a | Dre17 | Ola22 | GacXV | Tni10 | a | Figure S1 |
| adam17b | Dre20 | Ola24 | + | Tni14 | |||||
| Pomc | e | Hsa2 | pomca | Dre17 | Ola2 | GacX | + | a | Ref. 1 |
| pomcb | Dre20 | Ola24 | GacXVIII | Tni14 | |||||
| Rab32 | b | Hsa6 | rab32a | Dre20 | Ola24 | GacXVIII | Tni14 | a | Figure S2 |
| rab32b | Dre8 | − | GacXV | Tni10 | |||||
| Slc24a4 | b | Hsa14 | slc24a4a | Dre17 | − | − | − | a | Figure S3 |
| slc24ab | + | Ola24 | GacXVIII | + | |||||
| Dtnbp1 | c | Hsa6 | dtnbp1a | Dre19 | Ola16 | GacXX | + | b | Figure S4 |
| dtnbp1b | Dre16 | Ola11 | GacX | + | |||||
| Hdac1 | a | Hsa1 | hdac1a | Dre19 | Ola11 | GacX | Tni21 | b | Figure S5 |
| hdac1b | − | Ola22 | GacXV | Tni10 | |||||
| Pabpc1 | j | Hsa8 | pabpc1a | Dre16 | Ola16 | GacXX | Tni8 | b | Figure S6 |
| pabpc1b | Dre19 | Ola11 | GacX | Tni21 | |||||
| Creb1 | a, e | Hsa2 | creb1a | Dre1 | Ola2 | GacI | Tni3 | c | Figure S7 |
| creb1b | Dre20 | Ola21 | GacXVI | Tni2 | |||||
| Ednrb1 | a, g, i | Hsa13 | ednrb1a | Dre1 | Ola17 | GacIII | Tni15 | c | Ref. 2 |
| ednrb1b | Dre9 | Ola21 | GacXVI | Tni3 | |||||
| En1 | a | Hsa2 | eng1a | Dre9 | + | GacXVI | + | c | Ref. 3 |
| eng1b | Dre1 | Ola2 | GacI | + | |||||
| Gja5 | j | Hsa1 | gja5a | Dre1 | + | GacVI | Tni17 | c? | Figure S8 |
| gja5b | Dre9 | Ola21 | GacXVI | Tni2 | |||||
| Mlph | d | Hsa2 | mlpha | Dre6 | + | + | Tni3 | c | Figure S9 |
| mlphb | Dre9 | Ola21 | GacXVI | Tni2 | |||||
| Zic2 | a | Hsa13 | zic2a | Dre9 | Ola21 | GacXVI | Tni2 | c | Figure S10 |
| zic2b | Dre1 | − | − | − | |||||
| Atp6v0c | f | Hsa16 | atp6v0ca | Dre3 | Ola8 | GacXI | + | e | Figure S11 |
| atp6v0cb | Dre24 | Ola19 | GacV | Tni2 | |||||
| Mgrn1 | e | Hsa16 | mgrn1a | Dre12 | − | − | − | e | Figure S12 |
| mgrn1b | Dre3 | Ola8 | GacXI | Tni3 | |||||
| Nsf | c | Hsa17 | nsfa | Dre3 | + | GacXI | + | e | Figure S13 |
| nsfb | Dre12 | Ola19 | GacV | Tni2 | |||||
| Sox9 | a, i | Hsa17 | sox9a | Dre12 | + | GacV | Tni2 | e | Ref. 4 |
| sox9b | Dre3 | Ola8 | GacXI | Tni3 | |||||
| Sox10 | a, g, i | Hsa22 | sox10a | − | Ola1 | GacIX | Tni18 | e | Figure1 |
| sox10b | Dre3 | Ola8 | GacXI | + | |||||
| Kit | a | Hsa4 | kita | Dre20 | Ola4 | GacVIII | Tni1 | f | Ref. 5 |
| kitb | Dre1 | Ola1 | GacIX | Tni18 | |||||
| Qdpr | h | Hsa4 | qdpra | Dre20 | + | GacIV | Tni20 | f | Ref. 6 |
| qdprb | Dre1 | − | − | − | |||||
| Tyrp1 | b | Hsa9 | tyrp1a | Dre7 | Ola18 | GacVII | + | f | Ref. 6 |
| tyrp1b | Dre1 | Ola1 | GacIX | − | |||||
| Csf1r | g | Hsa5 | csf1ra | + | Ola10 | GacIV | Tni1 | g | Ref. 7 |
| csf1rb | − | Ola14 | GacVII | Tni7 | |||||
| Smtl | g | — | smtla | Dre14 | Ola13 | GacI | + | g | Refs. 8 and 9 |
| smtlb | Dre10 | − | − | − | |||||
| Frem2 | a | Hsa13 | frem2a | Dre15 | Ola13 | GacI | Tni16 | h | Figure S14 |
| frem2b | Dre10 | Ola14 | GacVII | Tni7 | |||||
| Myo7A | d | Hsa11 | myo7Aa | Dre18 | Ola13 | GacI | + | h | Figure S15 |
| myo7Ab | Dre21 | Ola14 | GacVII | + | |||||
| Rab38 | b | Hsa11 | rab38a | Dre15 | Ola13 | GacI | + | h | Figure S16 |
| rab38b | Dre10 | Ola14 | GacVII | Tni7 | |||||
| Tyr | b | Hsa11 | tyra | Dre15 | Ola13 | GacI | + | h | Ref. 6 |
| tyrb | − | Ola14 | GacVII | Tni7 | |||||
| Ghr | g | Hsa5 | ghra (slr) | Dre8 | Ola9 | GacXIII | Tni12 | i | Ref. 9 |
| ghrb | Dre21 | Ola12 | GacXIV | + | |||||
| Gnaq | a | Hsa9 | gnaqa | Dre5 | Ola9 | GacXIII | Tni12 | i | Figure S17 |
| gnaqb | − | Ola12 | GacVII/XIV | Tni4 | |||||
| Spr | h | Hsa2 | spra | Dre5 | Ola10 | GacXIV | + | i | Ref. 6 |
| sprb | Dre8 | − | GacXIII | − | |||||
| Myo5A | d | Hsa15 | myo5Aa | Dre18 | Ola3 | GacII | Tni5 | j | Figure S18 |
| myo5Ab | Dre25 | Ola6 | GacXIX | Tni13 | |||||
| Trpm1 | a | Hsa15 | trpm1a | Dre7 | Ola3 | GacII | Tni5 | j | Figure S19 |
| trpm1b | Dre25 | Ola6 | GacXIX | Tni13 | |||||
| Atp6v1e1 | f | Hsa22 | atp6v1e1a | Dre4 | Ola23 | GacIV | Tni19 | k | Figure S20 |
| atp6v1e1b | Dre25 | Ola6 | GacXIX | Tni13 | |||||
| Kitl | a | Hsa12 | kitla | Dre25 | + | GacXIX | Tni13 | k | Ref. 10 |
| kitlb | Dre4 | Ola23 | GacIV | Tni19 | |||||
| Edn3 | a, g, i | Hsa20 | edn3a | Dre11 | Ola5 | GacXVII | − | l | Ref. 2 |
| edn3b | Dre23 | Ola7 | GacXII | Tni9 | |||||
| Erbb3 | a | Hsa12 | erbb3a | Dre6 | Ola5 | + | Tni11 | l | Refs. 11 and 12 |
| erbb3b | Dre23 | Ola7 | GacXII | Tni9 | |||||
| Mitf | a | Hsa3 | mitfa | Dre6 | Ola5 | GacXVII | Tni11 | l | Ref. 13 |
| mitfb | Dre23 | Ola7 | GacXII | Tni9 | |||||
| Silv | b | Hsa12 | silva | Dre11 | Ola5 | + | Tni11 | l | Ref. 6 |
| silvb | Dre23 | Ola7 | GacXII | Tni9 | |||||
| Pax7 | g | Hsa1 | pax7a | Dre11 | Ola5 | + | Tni11 | l | Ref. 14 |
| pax7b | Dre23 | + | GacXII | + | |||||
| Egfr | a | Hsa7 | egfra | Dre2 | Ola17 | GacIII | Tni15 | m | Refs. 11 and 12 |
| egfrb | Dre24 | Ola20 | + | Tni4 | |||||
| Gna11 | a | Hsa19 | gna11a | Dre22 | Ola4 | GacVIII | Tni1 | m | Figure S17 |
| gna11b | Dre2 | Ola22 | GacXV | Tni10 | |||||
| Irf4 | j | Hsa6 | irf4a | Dre2 | Ola17 | GacIII | + | m | Figure S21 |
| irf4b | Dre20 | Ola4 | GacVIII | Tni1 | |||||
| Itgb1 | a | Hsa10 | itgb1a | Dre24 | Ola20 | GacXXI | + | m | Figure S22 |
| Itgb1b | Dre2 | Ola17 | + | Tni15 | |||||
| Mcoln3 | a | Hsa1 | mcoln3a | − | Ola17 | GacIII | Tni15 | m | Figure S23 |
| mcoln3b | Dre23 | Ola4 | GacVIII | Tni1 | |||||
| Myc | a | Hsa8 | myca | Dre24 | Ola20 | GacXXI | Tni6 | m | Figure S24 |
| mycb | Dre2 | Ola17 | − | − | |||||
| Pax3 | a, g | Hsa2 | pax3a | Dre2 | Ola17 | GacIII | Tni15 | m | Ref. 14 |
| pax3b | Dre15 | Ola13 | + | Tni16 |
NOTE.—Genomic location of the human gene and teleost coorthologs is indicated. Functions: a, melanophore development; b, components of melanosomes; c, melanosome construction; d, melanosome transport; e, regulation of melanogenesis; f, systemic effects; g, xanthophore development; h, pteridine synthesis; i, iridophores development; j, uncategorized function. Assignment to the pre-FSGD protochromosomes is according to Kasahara et al. (2007). Supplementary figures S1–S24 (phylogenetic trees) are found as Supplementary Material online. Key references (ref.) show the FSGD origin of teleost duplicates: 1, de Souza et al. (2005); 2, Braasch, Volff, and Schartl (2009); 3, Taylor et al. (2003); 4, Yan et al. (2005); 5, Siegel et al. (2007); 6, Braasch et al. (2007); 7, Braasch et al. (2006); 8, Zhu et al. (2004); 9, Fukamachi and Meyer (2007); 10, Hultman et al. (2007); 11, Gomez et al. (2004); 12, Froschauer et al. (2006); 13, Altschmied et al. (2002); 14, Minchin and Hughes (2008).
For ten pigmentation genes, we also identified lineage-specific single-gene duplications (supplementary tables S1–S2, supplementary figs. S1–S24, Supplementary Material online) such as the presence of two gnaqb genes in stickleback (supplementary fig. S17, Supplementary Material online). In tetrapods, there were only five lineage-specific gene duplications (e.g., duplication of Atp6v1e1 in mammals; supplementary fig. S20, Supplementary Material online), suggesting that vertebrate pigmentation genes have a rather low rate of paralogy outside teleosts.
Of the liver genes, 19.3% (36/187 genes) were retained in two copies after the FSGD with 50.0% (18/36) being present in duplicate in all five teleost genomes. For 81.7% (151/187 genes) of liver genes, one of the duplicates has been lost in all five teleosts (supplementary table S3, Supplementary Material online). The FSGD duplicate retention rate for liver genes is significantly lower than for pigmentation genes (36/187 vs. 46/128 genes, respectively; χ2 test, P = 0.00083).
Lineage-Specific Patterns of Duplicate Loss and Retention
Because in 46 cases a FSGD-duplicated pigmentation gene is present in at least one of the analyzed teleost genomes, both paralogs must have been present in their euteleost common ancestor. However, this retention rate of 35.9% is just a parsimonious lower estimate, as only five of the ≈22,000 euteleost species (Nelson 2006) have been analyzed, and independent FSGD duplicate loss might have occurred in different lineages. A fraction of independent duplicate losses can be inferred from synteny data if RGL has occurred. In this case, the remaining singleton genes are located on paralogous (and not orthologous) chromosomes when two lineages are compared.
Using the paralogy/orthology assignment of teleost chromosomes established by Kasahara et al. (2007), we identified four such cases (ebna1bp2, eda, drd2, snai2) through comparison of the location of singletons on zebrafish chromosomes to the chromosomal position in medaka and Tetraodon (supplementary table S2, Supplementary Material online). This gives a RGL rate of 3% (4/128 genes) between zebrafish and acanthomporphs for pigmentation genes. Adding the four RGL genes to the 46 retained duplicates at least 50 (39.1%) pigmentation gene duplicates had been retained in the last common euteleost ancestor of the five analyzed teleosts (fig. 3). This might still be an underestimation, as possible independent loss of orthologous gene copies in divergent lineages is not taken into account. Convergent duplicate loss becomes progressively more likely compared with RGL with advancing time since the duplication event and increasing sequence divergence of paralogs (Scannell et al. 2007). A similar analysis of liver genes revealed 4/187 instances (2%) of RGL (supplementary table S3, Supplementary Material online). Thus, the estimate for FSGD duplicate retention in the euteleost ancestor for liver genes is 21.4% (40/187 genes; fig. 3; supplementary fig. S25, Supplementary Material online). Within the ∼50 Myr between the FSGD and the divergence of zebrafish from the acanthomorphs (medaka, stickleback, pufferfishes; Kasahara et al. 2007), both, pigmentation and liver genes, have lost the majority of FSGD duplicates. However, the loss rate has been significantly lower (χ2 test, P = 0.00065) for pigmentation genes (60.9%) compared with liver genes (78.6%).
FIG. 3.—
Evolution of teleost pigmentation gene repertoires following the FSGD. (A) In the five extant teleost genomes, around 30% of FSGD-duplicated pigmentation genes (P; n = 128) have been retained. Duplicated liver genes (L; n = 187) have been retained by only 14% in extant teleosts. In the hypothetical euteleost ancestor, almost twice as many duplicates had been retained for pigmentation genes compared with liver genes (39.1% vs. 21.4%). For all five teleost genomes as well as for the reconstructed euteleost ancestor, the retention rates of pigmentation versus liver genes are significantly different (χ2 tests; **P < 0.01, ***P < 0.001). (B) Losses of pigmentation gene duplicates mapped onto the teleost phylogeny (Setiamarga et al. 2008). Numbers of pigmentation genes retained in duplicate are given in boxes. Gene loss rates in percent are based on the number of retained duplicates at the previous node in the phylogeny. The majority of duplicate losses have occurred before the divergence of the five euteleost species (60.9%). Within acanthomorphs (medaka, stickleback, pufferfishes), further losses have occurred shortly after the split from the zebrafish lineage (18.0%) and lineage-specific gene losses are relatively rare. Gene loss rates of pigmentation genes are generally lower than of liver genes (L; see also supplementary fig. S25, Supplementary Material online).
During the divergence of euteleosts, further lineage-specific duplicate losses have occurred that can be mapped onto the phylogeny (fig. 3). Similarly, independent loss of several pigmentation gene duplicates has occurred in the lineage leading to zebrafish (ten losses) as well as in the lineage leading to acanthomorphs (nine losses). Within acanthomorphs, most gene losses have occurred shortly after the split from the zebrafish lineage and later lineage-specific loss of duplicates is rare compared with the liver genes (fig. 3).
In the extant five teleost genomes, finally, a similar FSGD-duplicated retention rate of ≈30% is observed for pigmentation genes across the euteleost phylogeny. Pufferfishes do not deviate from this trend despite the massive genome compaction that has occurred in their lineage (Brenner et al. 1993). Liver genes, in contrast, have retained only around 12–16% of FSGD duplicates in the extant teleosts due to their higher loss rates before and after the euteleost divergence (fig. 3).
Duplicate Retention Rates Differ among Functional Categories
Next, we asked about the functional distribution of duplicated pigmentation genes. A comparison of the different pigment cell types shows (table 2) that the retention rate for melanophores (37.0%; 37/100 genes; categories a–f) and iridophores (36.4%; 4/11 genes; category i) are similar to the rate for all pigmentation genes (35.9%; 46/128 genes). For xanthophores, the retention rate seems to be elevated (50%; 11/22 genes; categories g and h) but this is not statistically significant (χ2 test, P = 0.20937). Noticeably, genes that have been assigned to more than one pigment cell type are retained at a rate of 63.6% (7/11 genes; supplementary table S1, Supplementary Material online).
Table 2.
FSGD Duplicate Retention Rates According to Pigmentation Gene Functions
| Category | Number of genes | FSGD duplicates | Retention rate (%) |
| All pigmentation genes | 128 | 46 | 35.9 |
| Melanophore genes | 100 | 37 | 37.0 |
| Melanophore development | 45 | 22 | 48.9 |
| Melanosome components | 10 | 6 | 60.0 |
| Melanosome biogenesis* | 22 | 2 | 9.1 |
| Melanosome transport | 4 | 3 | 75.0 |
| Regulation of melanogenesis | 7 | 3 | 42.0 |
| Systemic effects | 13 | 2 | 15.4 |
| xanthophore genes | 22 | 11 | 50.0 |
| Xanthophore development** | 11 | 9 | 81.8 |
| Pteridine synthesis | 11 | 2 | 18.2 |
| Iridophore genes | 11 | 4 | 36.4 |
| Uncategorized | 10 | 3 | 30.0 |
| Genes in multiple categories | 11 | 7 | 63.6 |
NOTE.—*P < 0.05, **P < 0.01 in χ2 tests compared with the category “all pigmentation genes.”
There are important differences among the different functional classes of pigmentation genes (table 2). Within the six melanophore categories, four of them have a higher retention rate than the overall average for pigmentation genes: melanophore development (48.9%; 22/45 genes), melanosome components (60.0%; 6/10 genes), melanosome transport (75.0%; 3/4 genes), and regulation of melanogenesis (42.9%; 3/7 genes). However, none of them is significantly different from the pigmentation gene average. In contrast, genes involved in melanosome biogenesis have a significantly reduced FSGD duplicate retention rate of 9.1% compared with the pigmentation gene average (2/22 genes; χ2 test, P = 0.01264). The category of systemic effect genes also has a comparatively low retention rate (15.4%; 2/13 genes).
The two xanthophore categories deviate in their FSGD duplicate retention rate. The rate for pteridine synthesis genes, which are involved in the formation of the yellow pigment, is comparatively low (18.2%; 2/11 genes). Interestingly, sepiapterin reductase, an enzyme that is involved in multiple steps of this metabolic pathway (Ziegler 2003), is encoded by two FSGD duplicates in teleosts (Braasch et al. 2007).
Xanthophore development genes, finally, have the highest retention rate of all categories (81.8%), which is significantly higher than the pigment cell average rate (χ2 test, P = 0.00282). Due to this high retention rate in xanthophore development, xanthophore genes have a higher FSGD duplicate retention rate than melanophores and iridophores.
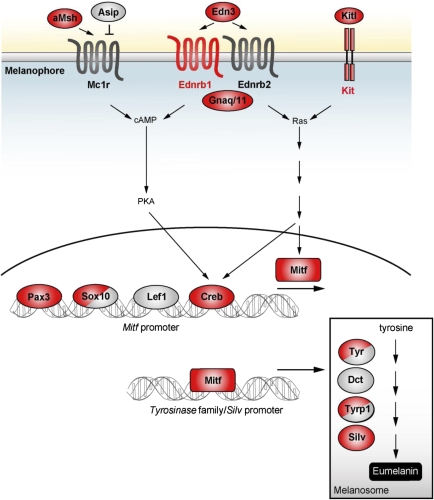
Duplications in the Pigment Cell Regulatory Networks
Crucial questions are how the duplicated teleost pigmentation genes are connected to each other and at which positions FSGD duplicates have been retained in the regulatory network of pigment cell development. Figure 4 illustrates our current understanding of the signaling network in vertebrate melanocyte/melanophore development with the Mitf transcription factor as master regulator in the central position (Béjar et al. 2003; Tachibana et al. 2003; Levy et al. 2006; Lin and Fisher 2007). Nearly all known key players of this network, that is, Sox10, Pax3, Kit, Kitl, Ednrb1, Edn3, and Mitf, have been retained in duplicate after FSGD. Thus, it appears that large parts of this pathway are present in two copies in teleosts. However, there are also subtle differences between teleost species in the retention of pathway component duplicates (fig. 4).
FIG. 4.—
Impact of the FSGD on the melanocyte/-phore signaling network. Many components have been identified by pigmentation mutants in both, mammals and teleosts. Diverse external signals are integrated on the promoter of the Mitf transcription factor gene, the master regulator of melanophore development (Béjar et al. 2003; Levy et al. 2006). Mitf regulates the expression of melanogenic enzymes, which catalyze the biosynthesis of melanin from tyrosine in melanosomes. The αMSH peptide is encoded by the Pomc gene. Lef1 is a downstream target of the Wnt signaling pathway. Red indicates duplications as result of the FSGD present in all five teleost genomes analyzed and gray indicates singleton genes. Molecules with divided red/gray shading indicate genes retained in duplicate in some teleost lineages but singleton in others (see table 1). Most of the key regulators of the melanophore signaling network have been retained in two copies in teleosts. The signaling network is adapted from Tachibana et al. (2003), Levy et al. (2006), and Lin and Fisher (2007).
Almost nothing is known about the composition of the regulatory networks in other pigment cell types. The current knowledge of the xanthophore network is summarized in supplementary figure S26 (Supplementary Material online). Also here, large parts have been retained in two copies after the FSGD and differences between teleost lineages are apparent.
Discussion
Expansion of the Pigmentation Gene Repertoire in Teleosts as Result of the FSGD
The listing of vertebrate pigmentation genes assembled in the present study (supplementary table S1, Supplementary Material online) shows that in many cases orthologous genes are affected in pigmentation mutants in mammals, in birds, and also in teleosts. This points, once again, to the evolutionary conservation of the pigmentary system, at least with respect to the development of melanophores/melanocytes, the only pigment cell type present in all vertebrate groups.
However, an important difference between tetrapods and teleosts is that the latter, as result of the FSGD, have an increased repertoire of genes potentially involved in pigmentation. Extant teleost genomes have retained around 30% of duplicated pigmentation genes from the FSGD. This FSGD duplicate retention rate for teleost pigmentation genes is higher than several genome-wide estimates for teleosts that range between 12% and 24% based on different approaches and data sets (Postlethwait et al. 2000; Jaillon et al. 2004; Woods et al. 2005; Brunet et al. 2006; Kassahn et al. 2009). Importantly, genome-wide FSGD duplicate retention does not differ significantly between the five teleost genomes (Kassahn et al. 2009).
We compared our data on pigmentation and liver genes to the genome-wide FSGD-duplicated retention rates from Brunet et al. (2006) and Kassahn et al. (2009) that have used synteny and phylogenetic methods as well. This revealed that the FSGD duplicate retention rate is significantly higher than for the genome-wide estimates (supplementary table S4, Supplementary Material online). For example, 29.7% (38/128) of FSGD pigmentation gene duplicates have been retained in Tetraodon (fig. 2). In contrast, Brunet et al. (2006), which used the most comparable approach to ours to detect FSGD duplicates, reported a significantly lower (χ2 test, P = 1.7 × 10−5) genome-wide FSGD duplicate retention rate in Tetraodon of 15.4% (364/2,371 analyzed genes). The genome-wide estimates for duplicate retention from Kassahn et al. (2009) are generally also significantly lower than for pigmentation genes (supplementary table S4, Supplementary Material online). Liver gene duplicates, in contrast, only have been retained in 12–16% of cases, which is similar or even lower than the genome-wide retention estimates (supplementary table S4, Supplementary Material online). Hence, pigmentation genes seem to have been preferentially retained in two copies following the FSGD. The FSGD therefore has generated a significant proportion of additional genes potentially involved in pigmentation.
Sato et al. (2009) recently reported FSGD duplicate retention rates for signal transduction and metabolic pathways in zebrafish, medaka, stickleback, and Tetraodon. Particularly, the retention rates for the signal transductions pathways involved in long-term potentiation of synaptic transmission, olfactory transduction, and taste transduction (25–35%) were higher than genome-wide estimates. These retention rates for signal transduction are in the range of pigmentation genes and do not significantly differ from them (supplementary table S4, Supplementary Material online). Thus, besides pigmentation, other pathways also show high rates of FSGD duplicate retention.
An important question is whether the high retention rate of duplicated pigmentation genes is due to the overrepresentation of certain functional categories among pigmentation genes. For example, enrichment for transcription factors, developmental genes, and cell communication proteins has been found for retained duplicates after the FSGD (Brunet et al. 2006; Kassahn et al. 2009) and the earlier vertebrate genome duplications (Putnam et al. 2008). Pigmentation genes are not enriched for transcription factor activity or cell communication. More general, developmental terms are enriched in pigmentation genes but likewise this also applies, for example, for liver genes (supplementary table S5, Supplementary Material online). This shows that although the enrichment of developmental terms among pigmentation genes may contribute to their retention, it is unlikely that this functional category alone is responsible for the expansion of the pigmentation gene repertoire after the FSGD. In fact, some specific groups of pigmentation genes, which are not enriched for developmental terms, have an exceptionally high FSGD duplicate retention rate (see below). It remains to be determined, if other properties like the number of gene functions or network connections could explain the differences in retention rate between pigmentation genes and other functional groups such as the liver genes.
Previous studies showed that there is broad variation in the enrichment/depletion of GO terms among FSGD duplicates (Brunet et al. 2006; Kassahn et al. 2009). Our data on functional categories of pigmentation genes (table 2) further suggest that FSGD duplicate retention rates can deviate considerably from the genome-wide estimates. In future studies, it will be thus necessary to further study in more detail FSGD gene duplicate retention and loss to identify functional groups, pathways, and networks with particularly high or low retention rates. Those pathways will then be interesting candidates to further study the impact of the FSGD for teleost evolution on the functional level.
Patterns of Pigmentation Gene Duplicate Loss and Retention
The reconstruction of pigmentation gene repertoire evolution after the FSGD revealed that, taking RGL into account, at least around 40% of FSGD-duplicated pigmentation genes have been retained in the last common euteleost ancestor of zebrafish, medaka, stickleback, and pufferfishes. Unfortunately, comparable genome-wide estimates are not available at present. Sato et al. (2009) estimated the FSGD duplicate retention rate for signal transduction pathway genes in the last euteleost ancestor to be 42.7%. For liver genes (fig. 3) and the metabolic tricarboxylic acid cycle (Sato et al. 2009), in contrast, the FSGD retention rate in the euteleost ancestor had been around 20% and 26%, respectively. Taken together, these data point to a comparatively low rate of duplicate pigmentation gene loss before the euteleost divergence.
But also euteleosts appear to have lost comparatively few pigmentation duplicates. According to previous estimates, only around 50% of genes retained in duplicate in zebrafish have also been retained in two copies in pufferfishes (Taylor et al. 2003; Woods et al. 2005). Similarly, for liver genes, this value is estimated around 57% (17/30). For pigmentation genes, in contrast, 80% (32/40 genes) of retained duplicates in zebrafish are also retained in pufferfishes. Thus, lineage-specific loss of gene duplicates seems to have been comparatively low for pigmentation genes. This pattern would be expected if the duplicated genes have kept only low redundancy levels and both have become essential before the divergence of euteleosts, precluding later gene loss. As the pigmentary innovations of the teleosts lineage are thought to predate the split of zebrafish and acanthomorphs (Braasch et al. 2008), this observation points to a possible phylogenetic correlation of the refinement of the teleost pigmentary system and the acquisition of important functions by duplicated pigmentation genes.
Pigmentation Genes Follow General Patterns of WGD Duplicate Retention
Our analyses suggest that there are important differences in the rate of FSGD duplicate retention among pigment cell types as well as among the different functional categories for a given cell type. Although many of the comparisons are statistically not significant due to the relative small set of genes analyzed for most categories, several interesting trends become apparent.
For example, almost all genes involved in melanosome biogenesis have been reverted to the single-gene status after the FSGD. Genes of this category encode proteins with general functions in the formation melanosomes and other lysosome-related organelles. These proteins build large complexes that route other proteins to organelles including melanosomes (Bennett and Lamoreux 2003; Raposo and Marks 2007). Genes of this category seem to have been lost concertedly after the FSGD to avoid detrimental imbalances and to keep stoichiometry of complex units, following the predictions of the “gene balance hypothesis” (Papp et al. 2003). Furthermore, this group of genes is usually prone to the loss of duplicated genes because they are generally singleton genes in vertebrates, that is, reduction to the single-gene status has also occurred after the 1R/2R WGDs (data not shown). Theoretically, stoichiometry would also be maintained if all subunit duplicates had been retained after the FSGD. However, it has been previously shown for Paramecium, which has also undergone multiple rounds of WGD, that this is only a transient phase and complex constituents tend to return to the single-gene status after old WGDs (Aury et al. 2006). Here, this also is the case for 1R/2R/FSGD duplicates of melanosome biogenesis protein complex genes.
Sixty percent of genes encoding the components of melanosomes, that is, proteins present in mature melanosomes (Bennett and Lamoreux 2003), have retained both copies after the duplication in the FSGD. To this category belong mainly the enzymes involved in melanin synthesis. The components of melanosomes are specialized for pigmentary functions and are not enriched for developmental genes. The high retention rate of this group has been also observed in our previous analysis of teleost pigment synthesis pathways (Braasch et al. 2007). It might be related to subfunctionalization of paralogs for melanogenesis within the melanosomes of melanophores versus those of the RPE, for which there is some evidence from teleost pigmentation mutants (reviewed in Braasch et al. 2008). Furthermore, our functional analysis of tyrp1 gene duplicates revealed that FSGD paralogs at least from this functional group diverge in a lineage-specific manner (Braasch, Liedtke, et al. 2009).
Genes involved in melanosome transport within the pigment cell also might be preferentially retained after the FSGD. Although this observation is based on four genes only, it might reveal important innovations in physiological color change. Physiological color change by moving melanosomes from the cell center to the periphery (darkening) or vice versa (lightening) is very important for many behavioral aspects and is highly sophisticated in teleosts compared with other vertebrates (Fujii 1993a; Fujii 2000).
Genes involved in the development of pigment cells are retained in duplicate at a high rate. This is the case for genes required for the development of melanophores as well as for xanthophore development. These two categories contain many transcription factors and signaling pathway components (supplementary table S1, Supplementary Material online). Retention and functional divergence of duplicated developmental regulatory genes would be expected if the FSGD has made an important contribution to teleost-specific features of pigment cell evolution such as the partitioning of melanophores and xanthophores into distinct subpopulations that are under different genetic controls (Odenthal et al. 1996; Mellgren and Johnson 2002).
Most striking is the high retention rate for genes involved in xanthophore development with around 82%. This category consists mainly of genes for signaling and transcriptional control, reflecting the general high retention rate of development genes after the FSGD (Brunet et al. 2006). Furthermore, most of them (Edn3, Ednrb1, Sox10, Pax3, etc.) are involved in the development of more than one pigment cell type. Thus, it remains open whether the high FSGD retention rate for xanthophore developmental genes is indeed founded in xanthophore-specific characteristics. If so, one would expect also an exceptional high FSGD retention rate for xanthophore-specific genes, of which, unfortunately, only few have been identified so far. Interestingly, xanthophores are the evolutionary youngest pigment cell type that seems to have emerged in the gnathostome lineage after the divergence of agnathans. Melanophores and iridophores, in contrast, were already present in the vertebrate ancestor (Mellgren and Johnson 2002; Braasch et al. 2008).
In conclusion, the comparison of functional categories of pigmentation genes reveals that the retention of FSGD duplicates, being noticeably high, follows more general WGD duplicate retention patterns, that is, preservation of protein complex stoichiometries and overrepresentation of developmental genes among retained duplicates. Genes with multiple functions for pigment cell development also seem to be preferentially retained. Other studies also found multifunctionality to be an important determinant for duplicate retention after the FSGD (Sato et al. 2009) and the early vertebrate WGDs (Hufton et al. 2009).
Evolution of Teleost Pigmentation Pathways by Genome Duplication
Looking at duplicated pigmentation genes in the context of regulatory networks revealed that large parts of the melanophore and xanthophore pathways (fig. 4; supplementary fig. S26, Supplementary Material online) are present in two copies in teleosts. As all pigment cell types develop from a common precursor cell (Bagnara et al. 1979), a similar situation can be expected for the developmental pathways of other pigment cells.
According to a model recently developed by Freeling and Thomas (2006), increasing morphological complexity in the evolution of plants and animals is passively driven by recurrent WGDs. As result of WGDs, duplicated functional modules would accumulate in the gene pool and provide the genetic fundament for morphological innovations. For example, this may be the case when duplicated pathway components become specialized for sister cell types derived from a common evolutionary precursor (Arendt 2008). Despite these and other hypotheses based on theoretical models, the understanding of duplicated functional modules, pathways, and networks in multicellular organisms remains rudimentary. There are so far only few examples for WGD-derived functional module or pathway duplications: the starch biosynthesis pathway in rice (Wu et al. 2008), the C4 photosynthetic pathway in grasses (Wang et al. 2009), the egesterol pathway in the fungus Rhizopus oryzae (Ma et al. 2009), and presumably the core network of cartilage development in vertebrates (Hecht et al. 2008). The present study suggests that the teleost pigmentation pathways represent further examples of WGD-duplicated functional modules. In contrast to the aforementioned studies, the pigmentation pathways analyzed here include in addition to the enzymes implicated in the biosynthesis of defined products such diverse gene categories as developmental genes for lineage specification and differentiation, subcellular organelle formation, intracellular transport, and homeostasis.
Duplications at different levels of organismal organization (e.g., gene network, cell type, tissue, body segment, behavioral program) are considered major sources of evolutionary novelties (West-Eberhard 2003). Oakley et al. (2007) proposed that “coduplication”, that is, the phylogenetic coincidence of gene duplications with the appearance of new structures should be considered as a null model for the origin of evolutionary novelties. In the present case, there seems to be a phylogenetic correlation between the pigmentary innovations such as leucophores that appear to predate the euteleost lineage (Braasch et al. 2008) and the generation of additional pigmentation genes by the FSGD, which is in line with the coduplication null model.
Relatively little is known about how the components of the pigment cell pathways have evolved after duplication with respect to functional divergence. For example, they could have evolved new functions (neofunctionalization) involved in teleost pigmentary novelties. Another evolutionary fate of functional module components could have been subfunctionalization of ancestral functions, as observed for the mitf paralogs in zebrafish (Lister et al. 2001; Altschmied et al. 2002). The ancestral functions in RPE and in melanophore development have been distributed with mitfa being expressed in melanophores and mitfb in the RPE (Lister et al. 2001). This functional specialization is also seen for the silv paralogs in zebrafish (Schonthaler et al. 2005) and the tyrp1 duplicates in medaka (Braasch, Liedtke, et al. 2009). The expression of silv and tyrp1 genes is under the transcriptional control of Mitf (fig. 4). Thus, it appears that at least parts of the duplicated functional module shown in figure 4 have become specialized for the melanophore or the RPE (for a review, see also Braasch et al. 2008).
More generally, however, the function of both paralogs as pigmentation genes remains to be demonstrated. In some cases, one of the two duplicates might have become specialized for the pigmentation function. This is seen for the Kit ligand–receptor pair. After FSGD, only kitla and kita are still involved in melanophore development, whereas kitlb and kitb have different, yet unknown functions not related to pigmentation (Mellgren and Johnson 2005; Hultman et al. 2007). Nevertheless, such cases can increase the evolvability of the pigmentary system because it releases it from functional constraints imposed by other essential functions of the ancestral gene.
Finally, our analysis also gives some insights into the potential contribution of the FSGD to the diversity of pigmentation among teleosts. There are subtle differences between teleost species in the retention of pathway component duplicates (table 1 and fig. 4). Such modifications (e.g., loss of sox10a in zebrafish) might be the genomic basis of species-specific differences in pigment cell repertoires like the absence of leucophores from larval pigment patterns in the zebrafish. On top of that, lineage-specific divergence of retained pigmentation paralogs provides potential for further pigmentary divergence among teleosts.
Conclusions
Our study shows that in teleosts the increase of complexity in the pigmentary system is correlated with an increase in the repertoire of pigmentation genes. Modern teleosts have around 30% more pigmentation genes than tetrapods as result of FSGD. Large parts of the functional modules of pigment cell development and differentiation have been retained after the FSGD, presumably providing the genetic raw material for teleost-specific pigment cell innovations. Importantly, WGDs enable at the same time the duplication of all genes found in a given, fine-tuned pathway, generating a complete copy of the ancestral network upon which selection can act. Such duplication of entire pathways would hardly be possible by succeeding simple gene duplications. It has been proposed that genetic novelties arising from 1R/2R in early vertebrates might have enabled the evolution of major vertebrate innovations such as the endoskeleton or the neural crest and its derivatives (Ohno 1970; Shimeld and Holland 2000; Wada and Makabe 2006; Zhang and Cohn 2008). However, empirical data supporting these hypotheses are mainly restricted to few genes. Despite the fact that protein interaction is an important parameter for paralog evolution (Bridgham et al. 2008) and that network duplication might substantially contribute to cell type diversification (Arendt 2008), information on how developmental signaling cascades and regulatory networks have been influenced by WGDs is sparse. By the example of pigmentation pathways, the present study provides evidence for existence of duplicates of regulatory networks as result of the FSGD in teleosts, supporting an important role of WGDs for the phenotypic evolution in vertebrates.
The present study and the recent study by Sato et al. (2009) have presented a framework for future evolutionary investigation of other networks following WGD. For example, visual perception networks might also have been particularly prone for duplicate retention (Bowmaker 2008; Gojobori and Innan 2009) and could have evolved concomitantly to pigmentary innovations. Finally, our survey for pigmentation genes will be a helpful resource for ongoing studies using teleosts as models for pigmentary diseases and melanoma formation and investigating the evolution and development of pigmentation in colorful teleosts such as cichlids and guppies.
Supplementary Material
Supplementary tables S1–S5 and figures S1–S26 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Supplementary Material
Acknowledgments
We would like to thank the members of our laboratories for helpful discussions. This work was supported by grants from the German Science Foundation (DFG, to M.S. and J.N.V.) and by the Biofuture program of the Bundesministerium für Bildung und Forschung (BMBF) (to J.N.V.), the Association pour la Recherche contre le Cancer (ARC) (to J.N.V.), the French Institute for Agronomy Research (Institut National de Recherche Agronomique PHASE) (to J.N.V.), the French Research Agency (ANR) (to J.N.V.), and the Foundation pour la Recherche Médicale (FRM) (to J.N.V.).
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Aburomia R, Khaner O, Sidow A. Functional evolution in the ancestral lineage of vertebrates or when genomic complexity was wagging its morphological tail. J Struct Funct Genomics. 2003;3:45–52. [PubMed] [Google Scholar]
- Altschmied J, et al. Subfunctionalization of duplicate mitf genes associated with differential degeneration of alternative exons in fish. Genetics. 2002;161:259–267. doi: 10.1093/genetics/161.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet. 2008;9:868–882. doi: 10.1038/nrg2416. [DOI] [PubMed] [Google Scholar]
- Aury JM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Bagnara JT, Fernandez PJ, Fujii R. On the blue coloration of vertebrates. Pigment Cell Res. 2007;20:14–26. doi: 10.1111/j.1600-0749.2006.00360.x. [DOI] [PubMed] [Google Scholar]
- Bagnara JT, Matsumoto J. Comparative anatomy and physiology of pigment cells in nonmammalian tissues. In: Nordlund JJ, Boissy RE, Hearing VJ, King RA, Ortonne J-P, editors. The pigmentary system: physiology and pathophysiology. New York: Oxford University Press; 2006. pp. 9–40. [Google Scholar]
- Bagnara JT, et al. Common origin of pigment cells. Science. 1979;203:410–415. doi: 10.1126/science.760198. [DOI] [PubMed] [Google Scholar]
- Bastian BC, Pinkel D. Expanding the genetic spectrum of pigmentation. Pigment Cell Melanoma Res. 2008;21:507–508. doi: 10.1111/j.1755-148X.2008.00490.x. [DOI] [PubMed] [Google Scholar]
- Béjar J, Hong Y, Schartl M. Mitf expression is sufficient to direct differentiation of medaka blastula derived stem cells to melanocytes. Development. 2003;130:6545–6553. doi: 10.1242/dev.00872. [DOI] [PubMed] [Google Scholar]
- Bennett DC, Lamoreux ML. The color loci of mice—a genetic century. Pigment Cell Res. 2003;16:333–344. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- Blomme T, et al. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowmaker JK. Evolution of vertebrate visual pigments. Vision Res. 2008;48:2022–2041. doi: 10.1016/j.visres.2008.03.025. [DOI] [PubMed] [Google Scholar]
- Braasch I, Liedtke D, Volff JN, Schartl M. Pigmentary function and evolution of tyrp1 gene duplicate in fish. Pigment Cell Melanoma Res. 2009;22:839–850. doi: 10.1111/j.1755-148X.2009.00614.x. [DOI] [PubMed] [Google Scholar]
- Braasch I, Salzburger W, Meyer A. Asymmetric evolution in two fish-specifically duplicated receptor tyrosine kinase paralogons involved in teleost coloration. Mol Biol Evol. 2006;23:1192–1202. doi: 10.1093/molbev/msk003. [DOI] [PubMed] [Google Scholar]
- Braasch I, Schartl M, Volff JN. Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol Biol. 2007;7:74. doi: 10.1186/1471-2148-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Volff JN, Schartl M. The evolution of teleost pigmentation and the fish-specific genome duplication. J Fish Biol. 2008;73:1891–1918. [Google Scholar]
- Braasch I, Volff JN, Schartl M. The endothelin system: evolution of vertebrate-specific ligand-receptor interactions by three rounds of genome duplication. Mol Biol Evol. 2009;26:783–799. doi: 10.1093/molbev/msp015. [DOI] [PubMed] [Google Scholar]
- Brenner S, et al. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature. 1993;366:265–268. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- Bridgham JT, Brown JE, Rodriguez-Mari A, Catchen JM, Thornton JW. Evolution of a new function by degenerative mutation in cephalochordate steroid receptors. PLoS Genet. 2008;4:e1000191. doi: 10.1371/journal.pgen.1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet FG, et al. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol Biol Evol. 2006;23:1808–1816. doi: 10.1093/molbev/msl049. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol. 2005;20:591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza FS, Bumaschny VF, Low MJ, Rubinstein M. Subfunctionalization of expression and peptide domains following the ancient duplication of the proopiomelanocortin gene in teleost fishes. Mol Biol Evol. 2005;22:2417–2427. doi: 10.1093/molbev/msi236. [DOI] [PubMed] [Google Scholar]
- Donoghue PCJ, Purnell MA. Genome duplication, extinction and vertebrate evolution. Trends Ecol Evol. 2005;20:312–319. doi: 10.1016/j.tree.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Dutton KA, et al. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- Froschauer A, Braasch I, Volff JN. Fish genomes, comparative genomics and vertebrate evolution. Curr Genomics. 2006;7:43–57. [Google Scholar]
- Fujii R. Cytophysiology of fish chromatophores. Int Rev Cytol. 1993a;143:191–255. [Google Scholar]
- Fujii R. Coloration and chromatophores. In: Evans DH, editor. The physiology of fishes. Boca Raton (FL): CRC Press; 1993b. pp. 535–562. [Google Scholar]
- Fujii R. The regulation of motile activity in fish chromatophores. Pigment Cell Res. 2000;13:300–319. doi: 10.1034/j.1600-0749.2000.130502.x. [DOI] [PubMed] [Google Scholar]
- Fukamachi S, Meyer A. Evolution of receptors for growth hormone and somatolactin in fish and land vertebrates: lessons from the lungfish and sturgeon orthologues. J Mol Evol. 2007;65:359–372. doi: 10.1007/s00239-007-9035-7. [DOI] [PubMed] [Google Scholar]
- Furlong RF, Holland PW. Were vertebrates octoploid? Philos Trans R Soc Lond B Biol Sci. 2002;357:531–544. doi: 10.1098/rstb.2001.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojobori J, Innan H. Potential of fish opsin gene duplications to evolve new adaptive functions. Trends Genet. 2009;25:198–202. doi: 10.1016/j.tig.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Gomez A, Volff JN, Hornung U, Schartl M, Wellbrock C. Identification of a second egfr gene in Xiphophorus uncovers an expansion of the epidermal growth factor receptor family in fish. Mol Biol Evol. 2004;21:266–275. doi: 10.1093/molbev/msh017. [DOI] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, et al. Mutations affecting pigmentation and shape of the adult zebrafish. Dev Genes Evol. 1996;206:260–276. doi: 10.1007/s004270050051. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hecht J, et al. Evolution of a core gene network for skeletogenesis in chordates. PLoS Genet. 2008;4:e1000025. doi: 10.1371/journal.pgen.1000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994:125–133. [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hufton AL, et al. Deeply conserved chordate non-coding sequences preserve genome synteny but do not drive gene duplicate retention. Genome Res. 2009;19:2036–2051. doi: 10.1101/gr.093237.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman KA, Bahary N, Zon LI, Johnson SL. Gene duplication of the zebrafish kit ligand and partitioning of melanocyte development functions to kit ligand a. PLoS Genet. 2007;3:e17. doi: 10.1371/journal.pgen.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Kasahara M, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Kassahn KS, Dang VT, Wilkins SJ, Perkins AC, Ragan MA. Evolution of gene function and regulatory control after whole-genome duplication: comparative analyses in vertebrates. Genome Res. 2009;19:404–1418. doi: 10.1101/gr.086827.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsh RN, et al. Zebrafish pigmentation mutations and the processes of neural crest development. Development. 1996;123:369–389. doi: 10.1242/dev.123.1.369. [DOI] [PubMed] [Google Scholar]
- Kelsh RN, et al. The Tomita collection of medaka pigmentation mutants as a resource for understanding neural crest cell development. Mech Dev. 2004;121:841–859. doi: 10.1016/j.mod.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- Lister JA, Close J, Raible DW. Duplicate mitf genes in zebrafish: complementary expression and conservation of melanogenic potential. Dev Biol. 2001;237:333–344. doi: 10.1006/dbio.2001.0379. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A. The origin of interspecific genomic incompatibility via gene duplication. Am Nat. 2000;156:590–605. doi: 10.1086/316992. [DOI] [PubMed] [Google Scholar]
- Ma LJ, et al. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 2009;5:e1000549. doi: 10.1371/journal.pgen.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellgren EM, Johnson SL. The evolution of morphological complexity in zebrafish stripes. Trends Genet. 2002;18:128–134. doi: 10.1016/s0168-9525(01)02614-2. [DOI] [PubMed] [Google Scholar]
- Mellgren EM, Johnson SL. kitb, a second zebrafish ortholog of mouse Kit. Dev Genes Evol. 2005;215:470–477. doi: 10.1007/s00427-005-0001-3. [DOI] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Michiels NK, et al. Red fluorescence in reef fish: a novel signalling mechanism? BMC Ecol. 2008;8:16. doi: 10.1186/1472-6785-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin JE, Hughes SM. Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev Biol. 2008;317:508–522. doi: 10.1016/j.ydbio.2008.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa M, et al. Endothelin receptor B2 (EDNRB2) is associated with the panda plumage colour mutation in Japanese quail. Anim Genet. 2007;38:103–108. doi: 10.1111/j.1365-2052.2007.01568.x. [DOI] [PubMed] [Google Scholar]
- Nelson JS. Fishes of the world. New York: John Wiley & Sons; 2006. [Google Scholar]
- Oakley TH, Plachetzki DC, Rivera AS. Furcation, field-splitting, and the evolutionary origins of novelty in arthropod photoreceptors. Arthropod Struct Dev. 2007;36:386–400. doi: 10.1016/j.asd.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Odenthal J, et al. Mutations affecting xanthophore pigmentation in the zebrafish, Danio rerio. Development. 1996;123:391–398. doi: 10.1242/dev.123.1.391. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, et al. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- Protas ME, Patel NH. Evolution of coloration patterns. Annu Rev Cell Dev Biol. 2008;24:425–446. doi: 10.1146/annurev.cellbio.24.110707.175302. [DOI] [PubMed] [Google Scholar]
- Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Raposo G, Marks MS. Melanosomes—dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Mellgren EM, Johnson SL. How the zebrafish gets its stripes. Dev Biol. 2001;240:301–314. doi: 10.1006/dbio.2001.0418. [DOI] [PubMed] [Google Scholar]
- Sato Y, Hashiguchi Y, Nishida M. Temporal pattern of loss/persistence of duplicate genes involved in signal transduction and metabolic pathways after teleost-specific genome duplication. BMC Evol Biol. 2009;9:127. doi: 10.1186/1471-2148-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- Scannell DR, et al. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc Natl Acad Sci USA. 2007;104:8397–8402. doi: 10.1073/pnas.0608218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonthaler HB, et al. A mutation in the silver gene leads to defects in melanosome biogenesis and alterations in the visual system in the zebrafish mutant fading vision. Dev Biol. 2005;284:421–436. doi: 10.1016/j.ydbio.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Semon M, Wolfe KH. Reciprocal gene loss between Tetraodon and zebrafish after whole genome duplication in their ancestor. Trends Genet. 2007;23:108–112. doi: 10.1016/j.tig.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Semon M, Wolfe KH. Preferential subfunctionalization of slow-evolving genes after allopolyploidization in Xenopus laevis. Proc Natl Acad Sci USA. 2008;105:8333–8338. doi: 10.1073/pnas.0708705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiamarga DH, et al. Interrelationships of Atherinomorpha (medakas, flyingfishes, killifishes, silversides, and their relatives): the first evidence based on whole mitogenome sequences. Mol Phylogenet Evol. 2008;49:598–605. doi: 10.1016/j.ympev.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Shimeld SM, Holland PW. Vertebrate innovations. Proc Natl Acad Sci USA. 2000;97:4449–4452. doi: 10.1073/pnas.97.9.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel N, Hoegg S, Salzburger W, Braasch I, Meyer A. Comparative genomics of ParaHox clusters of teleost fishes: gene cluster breakup and the retention of gene sets following whole genome duplications. BMC Genomics. 2007;8:312. doi: 10.1186/1471-2164-8-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Kobayashi Y, Matsushima Y. Mouse models for four types of Waardenburg syndrome. Pigment Cell Res. 2003;16:448–454. doi: 10.1034/j.1600-0749.2003.00066.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Makabe K. Genome duplications of early vertebrates as a possible chronicle of the evolutionary history of the neural crest. Int J Biol Sci. 2006;2:133–141. doi: 10.7150/ijbs.2.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biol. 2009;10:R68. doi: 10.1186/gb-2009-10-6-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Werth CR, Windham MD. A model for divergent, allopatric speciation of polyploid pteridophytes resulting from silencing of duplicate-gene expression. Am Nat. 1991;137:515–526. [Google Scholar]
- West-Eberhard MJ. Developmental plasticity and evolution. New York: Oxford University Press; 2003. [Google Scholar]
- Woods IG, et al. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005;15:1307–1314. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhu Z, Ma L, Chen M. The preferential retention of starch synthesis genes reveals the impact of whole-genome duplication on grass evolution. Mol Biol Evol. 2008;25:1003–1006. doi: 10.1093/molbev/msn052. [DOI] [PubMed] [Google Scholar]
- Yan YL, et al. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- Zhang G, Cohn MJ. Genome duplication and the origin of the vertebrate skeleton. Curr Opin Genet Dev. 2008;18:387–393. doi: 10.1016/j.gde.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Zhu Y, et al. Cloning of somatolactin alpha and beta cDNAs in zebrafish and phylogenetic analysis of two distinct somatolactin subtypes in fish. J Endocrinol. 2004;182:509–518. doi: 10.1677/joe.0.1820509. [DOI] [PubMed] [Google Scholar]
- Ziegler I. The pteridine pathway in zebrafish: regulation and specification during the determination of neural crest cell-fate. Pigment Cell Res. 2003;16:172–182. doi: 10.1034/j.1600-0749.2003.00044.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.