Abstract
Coliphage phi1, which was isolated for phage therapy in the Republic of Georgia, is closely related to the T-like myovirus RB49. The ∼275 open reading frames encoded by each phage have an average level of amino acid identity of 95.8%. RB49 lacks 7 phi1 genes while 10 phi1 genes are missing from RB49. Most of these unique genes encode functions without known homologs. Many of the insertion, deletion, and replacement events that distinguish the two phages are in the hyperplastic regions (HPRs) of their genomes. The HPRs are rich in both nonessential genes and small regulatory cassettes (promoterearly stem-loops [PeSLs]) composed of strong σ70-like promoters and stem-loop structures, which are effective transcription terminators. Modular shuffling mediated by recombination between PeSLs has caused much of the sequence divergence between RB49 and phi1. We show that exchanges between nearby PeSLs can also create small circular DNAs that are apparently encapsidated by the virus. Such PeSL “mini-circles” may be important vectors for horizontal gene transfer.
Keywords: T4-like phage, genome evolution, modular shuffling, regulatory cassette
Introduction
Several generations of molecular biologists, biochemists, and geneticists have used the large and complex bacteriophage T4 as a model system. The 169-kb genome of T4 has been sequenced, and the functions of many of its ∼300 genes are known (Miller et al. 2003). Although T4 is by far the best characterized T4-like phage, >200 related viruses have been described. All these share the basic T4 virion morphology—an elongated head, a contractile tail, a complex base-plate, and six radiating tail fibers. Most of the known, cultivated T4-like phages grow on Escherichia coli or other enterobacteria, but others grow on phylogenetically more distant bacteria (Aeromonas, Vibrio, cyanobacteria, etc.) and these can vary significantly in their virion morphology (Ackermann and Krisch 1997).
Despite the enormous diversity of the T4 superfamily, little genomic sequence data were available for these phages until recently. A high throughput, National Science Foundation (NSF)–funded sequencing project (Comeau et al. 2007) has obtained the complete genome sequences of a series of T4-like phages, including RB49 which is a divergent “Pseudo T-even” member (Monod et al. 1997) of the T4 superfamily. This coliphage was isolated in 1964 from a sewage treatment plant on Long Island, NY, by Rosina Berry (Russell and Huskey 1974). A similar coliphage, phi1, was isolated at the George Eliava Institute in Tbilisi in 1971 against pathogenic strains of enterobacteria, which caused hemolytic diarrhea that could be lethal for young children. phi1 was successfully adopted for use in phage therapy—a practice that continues to this day in the Republic of Georgia. Preliminary sequencing of essential structural and replication genes, along with random fragments, indicated a considerable level of nucleotide identity between these two phages, in spite of their different geographic origins.
A draft sequence of the phi1 genome was obtained in the NSF project; here, we present the analysis of the final version of the sequence and its comparison to the RB49 genome. The close similarity between these two phages, sharing 95% nucleotide identity over 94% of their genomes, gives us the opportunity to define the initial steps in the divergence of these genomes. The genome architecture of the T4 superfamily involves a conserved core of modules of essential genes that are punctuated by several regions that are extremely variable in their gene content (Comeau et al. 2007). The phi1/RB49 genome comparison supports the hypothesis that the generation of genomic diversity primarily occurs in the hyperplastic regions (HPRs) by the shuffling of database orphan open reading frames (ORFans) that frequently encode adaptive functions (Comeau et al. 2008). A novel mechanism of modular shuffling within the HPRs, mediated by flanking sequence motifs that contain both an early promoter and a stem-loop structure (PeSLs) associated preferentially with ORFans, seems to be responsible for much of this localized genomic variability.
Materials and Methods
Viral and Bacterial Strains
RB49 and phi1, as well as the E. coli BE and DH5α strains, were part of the collections of HMK at the LMGM-UMR5100 in Toulouse, France. phi1 was originally provided by M.K. and R.A. from the Tbilisi collection. All bacterial cultures were incubated at 37 °C in LB broth (Ausubel et al. 1992) under agitation (150 rpm).
Preparation of Phage Stocks
Phage stocks of phi1 and RB49 were prepared according to the method of Carlson and Miller (1994) by infecting an E. coli BE culture (optical density [OD]660 = 0.2–0.3) with a dilution of a concentrated phage stock. Incubation was continued until total bacterial lysis was observed. A few drops of chloroform were added to the lysate and, after 15 min at room temperature, it was vigorously agitated for 5 s before being centrifuged for 5 min at 2,300 × g. The supernatant containing the phage particles was enumerated by plaque assay (Carlson and Miller 1994) using BU buffer (7 g/L Na2HPO4, 4 g/L NaCl, 3 g/L KH2PO4) as the diluent.
Genome Sequencing/Correction and Bioinformatic Analysis
The sequencing/correction of phi1 was done from genomic DNA prepared by “hot-cold” lysis, which involves liberating the phage DNA from the capsid with a series of thermal shocks (Comeau et al. 2004), on 50 μL of phage stock (109 PFU/mL) diluted 10-fold in deionized sterile water. The 50 μL polymerase chain reactions (PCRs) contained the following components: 3 μL of template, 1× Taq polymerase buffer (NEB), 0.8 mM deoxynucleotide triphosphates (Sigma), 0.2 μM of each specific primer (Sigma), and 0.75 U of Taq polymerase (NEB). PCR cycling conditions were as follows: initial denaturation for 1 min at 95 °C; followed by 30 cycles of denaturation at 95 °C for 30 s, an annealing step for 30 s at the Tm of the specific primer set, and elongation for 2 min at 72 °C; with a final extension for 5 min at 72 °C. PCR products were migrated in 1.5% agarose gels in 0.5× TBE, with visualization by ethidium bromide or SYBR Safe (Invitrogen). Products were purified using the QIAquick PCR Purification Kit (QIAGEN) before being sequenced on a Beckman Ceq2000 sequencer (IFR109, Toulouse) with the CEQ DTCS Quick Start Kit (Beckman Coulter) according to the manufacturer's instructions.
The correction and analysis of the genome were done with the following programs: 1) Ceq2000XL (Beckman Coulter) for the correction of the raw sequencing results; 2) SeqMan (DNASTAR) for comparing the new genomic sequences against the “draft” phi1 genome available on the T4-type phage server at Tulane University (http://phage.bioc.tulane.edu); 3) GLIMMER (Delcher et al. 1999) and GeneMark (Besemer and Borodovsky 2005) for open reading frame (ORF) determination; and 4) Java Word Frequencies and Java Dotter (http://athena.bioc.uvic.ca/tools/) for the exploration of DNA “words”/patterns; and 5) the Blast tools at National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov) for the characterization of novel genes within phi1. The RB49 genome was also reanalyzed using these tools. The comparisons of the corrected phi1 and RB49 genomes were realized using the LAGAN program (Brudno et al. 2003), which permits comparative alignments of entire genomes on a nucleotide level, and the circular genome visualizations were generated using CGView (Stothard and Wishart 2005).
PeSL Mini-circle Characterization.
A culture of E. coli BE (OD660 = 0.2) was infected with the phage phi1 or RB49 at a multiplicity of infection of 5. The infection was followed for 45 min with samples taken every 5 min. A few drops of chloroform were mixed into each of these samples (0.5 mL), and the aqueous phase was immediately frozen at -20 °C.
PCR amplification of the PeSL “mini-circles” was done using specific primers chosen with inverted orientations to each other (inverse-PCR) that were located in the central region of the ORF in question (supplementary table S1, Supplementary Material online), with the spacing between the primers being 25–150 bp. These primers were also selected to avoid homodimerization in order to prevent the formation of PCR artifacts. PCRs and cycling conditions were as previously mentioned above, except for the following modifications: 3 μL of DNA template (obtained either from infected cells or mature phage particles); an extension time of 5 min; and a final extension of 9 min. PCR products were visualized as previously mentioned, and the only digital manipulations done on the gel images (Adobe Photoshop) were to adjust the brightness and contrast equally throughout all regions of the gels. In parallel, we performed negative controls using the same conditions used to obtain the PeSL mini-circles. These consisted of inverse-PCRs on essential genes (g17 and g46) chosen in the conserved genome modules and not bordered by PeSL sequences. To demonstrate that PeSL mini-circles were within the capsids of mature particles, small aliquots of phage stock were treated with DNase I (NEB) that was then heat inactivated according to the manufacturer's instructions. These “cleaned” aliquots were then subjected to hot-cold lysis to release the packaged DNA, and inverse-PCR amplification was performed as described as above.
The inverse-PCR products were purified by gel extraction using the QIAquick Gel Extraction Kit (QIAGEN) before being sequenced as above. Three types of sequencing protocols were used: the standard and betaine (technical note CEQ-AI-2006) protocols recommended by the manufacturer, and a modified betaine protocol (initial hot start at 96 °C for 2.5 min; follow by 40 cycles of 96 °C denaturation for 60 s, primer Tm + 5 °C annealing for 45 s, and 60 °C extension for 4 min). In some cases, the extremities of the PeSL PCR products could not be precisely determined by directly sequencing the products; therefore, it was first necessary to ligate these products into a cloning vector (pGEM-T Vector System I TA Cloning Kit; Promega). The ligation products were then subjected to a specific PCR using the M13 primers, followed by migration and gel extraction before sequencing.
Ultrafiltration experiments were performed using 30/100K Nanosep and 1000K Microsep centrifugation devices (Pall Life Sciences). Fifty-microliter aliquots of RB49-extracted DNA were filtered through the Nanosep devices (∼50 μL flow-through), and the retentates were resuspended in 50 μL of sterile water. For the larger Microsep devices, the 50-μL DNA aliquot was diluted to 500 μL in sterile water and filtered through the membrane (∼500 μL flow-through). The membrane (and retained DNA) was then washed five times with 3 mL of sterile water per wash and spun until a final hold-up volume (retentate) of 50 μL was remaining. The various fractions were then used as templates in normal PCR to detect genomic DNA (a “normal” genome sequence; locus 6.1 in supplementary table S1, Supplementary Material online) and in inverse-PCR to detect the presence of PeSL192 and PeSL210 mini-circles as described above.
PeSL Promoter and Terminator Functions.
To measure the strength/function of the PeSL motifs as promoters and terminators, a selection of representative PeSLs were cloned into the pRS551 transcriptional fusion vector system (Simons et al. 1987) containing lacZ as the reported gene. In order to test promoter function, the entire PeSL sequence (from the upstream stop codon to the nucleotide just before the downstream ATG) of PeSL193/192 and PeSL206/205, as well as a characterized strong promoter (lacUV5; Higashitani et al. 1997), were synthesized as single-stranded oligonucleotides (Sigma) with EcoRI and BamHI adaptors. Corresponding forward and reverse oligonucleotides were annealed (95 °C for 5 min, 60 °C for 15 min) and directionally cloned into pRS551. Competent E. coli DH5α was transformed with these different ligation reactions, and the cells were selected for resistance to ampicillin (100 μg/mL). To test terminator function, the majority of the kanR gene and the strong terminators (T1 rrnB terminators) following it were excised upstream from the lacZ reporter gene and replaced by the stem-loop of PeSL211/210 or a control non–stem-loop-forming sequence of similar size (supplementary fig. S1, Supplementary Material online). The terminator sequences were synthesized with HindIII and BamHI adaptors, then annealed and cloned as above. All clones were verified by sequencing isolated plasmid DNA. β-Galactosidase assays were carried out on independent duplicate cultures, without induction, as described by Miller (1992).
Data Deposition
The GenBank (http://www.ncbi.nlm.nih.gov/Genbank) accession number for the phi1 complete genome sequence presented in this article is EF437941.
Results
Sequencing and Annotation of the phi1 Genome
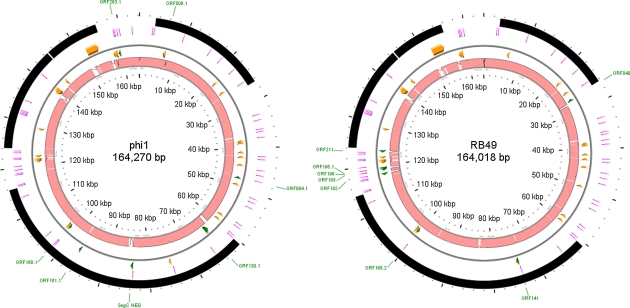
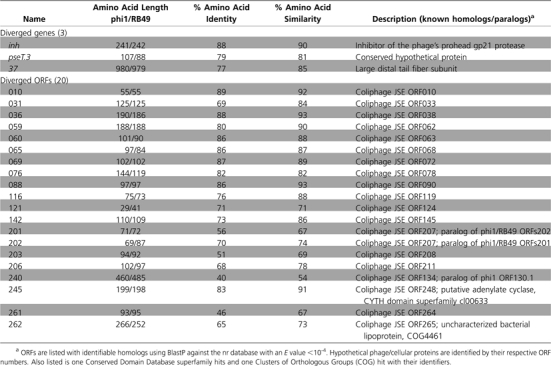
Preliminary sequencing in several regions of the phi1 and RB49 genomes indicated an extremely close phylogenetic relation between them. However, these phages had been isolated from different hosts, at different times, and in different countries. The correction of the phi1 genome sequence (NC_009821), and its comparison with RB49, revealed a limited number of sequence differences between the two phages and also brought to light a few errors in the original annotation of RB49 (NC_005066). We have added five new RB49 ORFs (006.1, 168.2, 189.1, 196.1, and 256.1) and removed ORF237 (finding no convincing bioinformatic support for it). The comparison of the two genomes is illustrated in fig. 1, which represents an alignment of the entire nucleotide sequences of the two phages, showing an identity of ≥95% over 94% of their genomes. The phi1 genome has 276 genes, composed of 154 ORFs and 122 T4-type genes. Among the latter, 23 are of unknown function. Our reanalysis of the RB49 genome shows 277 genes, with 157 ORFs and 120 T4-type genes, 22 of which have an unknown function. phi1 possesses seven unique genes, whereas RB49 has eight (table 1). The majority of these unique genes are ORFans (or related to other phage ORFans), but some have homology (E value <10-4) to genes of known functions, notably the phi1 segC gene, which is a homing endonuclease present in T4 (Sharma et al. 1992), and RB49 ORF040, which is a novel HNH-type homing endonuclease. RB49 ORF211 is a homolog of ORF1 (of unknown function) from a Vibrio fischeri superintegron. In RB49, the majority of the unique genes are located in the HPRs, either in one cluster (ORFs 192, 195, 196, 196.1, and 211) or as isolated genes (e.g., ORF040). However, in phi1, surprisingly only two (ORFans 84.1 and 263.1) of the seven unique ORFs are in the HPRs; the others are located in more conserved genome regions. However, ORFan009.1 is within a small group of ORFans present in both phages and ORFan168.1 is close to the hoc gene, an auxiliary component of the capsid that is known to be variable and poorly represented in the T4 superfamily (Comeau and Krisch 2008). In this cluster is also found the inh gene, which is divergent between phi1 and RB49 (table 2), and other genes of unknown function present in T4. There are 23 divergent genes shared by phi1 and RB49 that have <90% amino acid identity at the protein level (table 2). These genes code mostly for unknown functions, but of the few known genes that are divergent, it is interesting to note the presence of gene 37, which codes for the large distal subunit of the long tail fibers that are involved in the determination of phage host range (Tétart et al. 1998). Fourteen of the divergent genes are localized in the HPRs, often clustered together, whereas the nine others are located in the conserved core regions of the genome, again often near each other, suggesting the existence of zones (or “hotspots”) where genomic rearrangements preferentially occur.
FIG. 1.—
Genome comparison of coliphages phi1 and RB49. The inner pink circles represent reciprocal whole-genome alignments of RB49 and phi1 generated by LAGAN (Brudno et al. 2003), the areas indicated in pink shading have a nucleotide homology of >95%. The second and third circles (separated by a thin gray line) are the reverse and forward strands with the divergent (<90% protein identity; orange arrows) and unique (green arrows) genes/ORFs indicated. The fourth circle shows the locations of the PeSLs (radiating magenta lines), and the outermost circle indicates the conserved structural and replication modules (black arcs). The names of the unique genes/ORFs are indicated in green labels outside the circles.
Table 1.
Differential Loci between phi1 and RB49, along with Their Known or Putative Functions
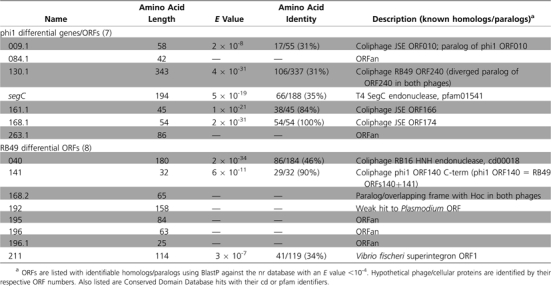 |
Table 2.
Known Genes and ORFs Showing Significant Sequence Divergence between Phi1 and RB49 (<90% Protein Identity), along with Their Known or Putative Functions
 |
Identification and Functional Analysis of Conserved Sequence Motifs in the HPRs—PeSLs
PeSL Characteristics and Functions.
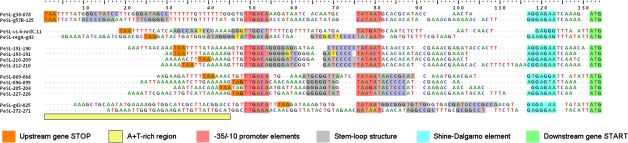
Bioinformatic analyses of the phi1 and RB49 genomes revealed the presence of a series of repeated motifs in both genomes that are preferentially located in the HPRs. These motifs contained sequences that are identical to the E. coli σ70 promoter consensus (Sinoquet et al. 2008), indicating they would act as phage early promoters (Pearly). This sequence similarity was not surprising because immediately after infection, the T4-like phages completely co-opt the host transcriptional machinery, which is largely σ70 dependent. Furthermore, these repeated phage σ70-like recognition sequences also have an AT-rich region upstream of the -35 element (fig. 2). Finally, the motifs have another striking component—a sequence capable of forming a stable stem-loop structure (SLS) that frequently contains tracts of polyG followed by polyC. The intergenic location of the PeSL's SLS suggests that it plays the role of a terminator/attenuator for any transcription initiated upstream of the PeSL element. In the HPRs, which are generally densely populated by PeSL motifs, their constituent SLSs could attenuate or block the accumulation of transcription from distal PeSLs and hence prevent overexpression of downstream genes. For example, such PeSL-mediated termination would also prevent inappropriate spill-over of early ORF(an) gene expression into the downstream late-expressed virion genes. Five subclasses of PeSL motifs can be distinguished on the basis of the position of their SLSs relative to their promoter sequence. These SLSs can occur between the -35 and -10 boxes, or they can overlap with either the -35 or -10 boxes. Additionally, they can be placed in the AT-rich region upstream of the -35 box or in the region downstream of the -10, just before the site of transcription initiation (fig. 2).
FIG. 2.—
A selection of PeSLs from the phi1 genome. The phi1 (and RB49) PeSLs are composed of multiple motifs, starting with the stop codons of the upstream genes (orange) which are typically found in AT-rich regions (pale yellow) located upstream of the -35 and -10 boxes (rose) of the σ70-like promoters. The SLSs (gray stems, with the G-T pairs in bright yellow) are placed in various positions, with the most common positioning being either between the -35 and the -10 boxes or overlapping the -10 box. The PeSLs have at their 3′ extremity a Shine-Dalgarno sequence (blue) and the start codon (green) of the downstream genes. The PeSL sequences were primarily aligned on the basis of the promoter and Shine-Dalgarno sequences.
We have demonstrated the activities of the constituent promoter and attenuator/terminators of the PeSLs using a combination of in vivo expression and in vitro sequencing experiments. For the PeSL terminator/attenuator function, we confirmed the existence of the SLSs by demonstrating that they form during DNA sequencing reactions. Without betaine, a substance that relaxes secondary structures (Rees et al. 1993), sequencing reactions (which use a single strand of the template) were blocked by the SLSs of the PeSLs, whereas the sequencing reactions could be extended downstream by the addition of betaine (data not shown). In vivo, the insertion of the SLS of PeSL211/210 (intergenic of ORFs 210–211) between a strong promoter and a lacZ reporter gene resulted in reduction in lacZ expression comparable with that of a very strong terminator (T1 rrnB; Simons et al. 1987) (supplementary fig. S1, Supplementary Material online). Similarly, PeSL promoter function was confirmed by insertion of the entire PeSL sequences of PeSL193/192, PeSL206/205, or a control lacUV5 promoter, into a promoterless lacZ reporter cassette (Simons et al. 1987). Nearly all the isolated clones showed single point mutations specifically in the -35 or -10 regions, indicating that the native PeSL expression level was probably so strong as to be deleterious. However, one wild-type clone of PeSL206/205 was obtained and it showed high lacZ expression levels (∼13,000 Miller units; supplementary fig. S1, Supplementary Material online).
PeSL Genome Distribution.
Among the ∼100 putative PeSL motifs we have identified (52 in phi1 and 51 in RB49), the vast majority of these are located in the HPRs and these are most frequently positioned immediately upstream of an ORFan (fig. 1). Nevertheless, there are a few PeSLs located within the replication and virion modules that constitute the conserved cores of the phage genomes. Such PeSLs are almost always associated with genes of unknown function. In the few cases where the PeSLs are associated with genes of known function, these genes are associated with unusual genetic plasticity, such as the gene 43 DNA polymerase necessary for phage genome replication (Karam and Konigsberg 2000) that is bordered by PeSLs on either side in both RB49 and phi1. There are several variants of the structural organizations of the g43 locus in the T4 superfamily—although it is often encoded, as in T4, by a single large polypeptide (monocistronic) with a variably sized linker between two major functional domains. In the Aeromonas and Acinetobacter phages, however, the enzyme is encoded by two distinct subunits (bicistronic; Petrov et al. 2006). For two of the Aeromonas phages (44RR2.8t and 25), we have found sequences similar to PeSLs between these two cistrons (g43A and g43B; data not shown). For three other phages (Aeromonas phage Aeh1 and coliphages RB43 and JS98), various other PeSL-like sequences are also found in the plastic g43 locus region. The other known genes associated with PeSLs are genes such as nrdD and hoc, which are auxiliary metabolic and structural phage components that have an uncharacteristically high plasticity. The PeSLs seem, therefore, preferentially associated with the most variable regions/genes of the genome.
PeSLs: A Driving Force of Genomic Variability
The HPRs are characterized by the presence of numerous ORFans (database orphan ORFs), some of which are unique to either the phi1 or RB49 genomes (table 1). In these two phages, such differential ORFans can, for the most part (10 of 14), be associated with the presence of a contiguous PeSL. When this is not the case, there is invariably one or more PeSLs located in the close vicinity (1–4 kb away). Such observations suggest that the PeSLs may play an important role in generating genomic plasticity in these phages. Because their sequence conservation is distributed over nearly a ∼100-bp interval, PeSL sequences could be preferred sites for recombination in the HPRs and thus mediate efficient shuffling of the genes that they flank, while simultaneously bringing their own transcription promoters and terminators.
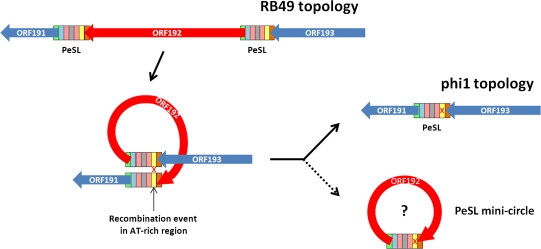
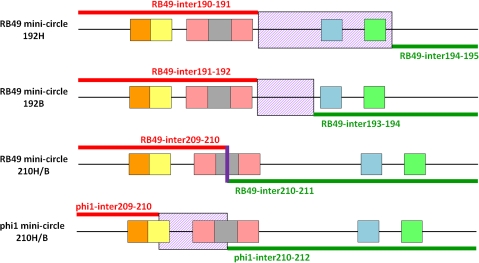
Figure 3 illustrates the mechanism that PeSLs may employ to create genomic variability, using as an example ORF192 of RB49. This unique ORFan is flanked in RB49 by PeSL motifs, whereas in phi1 this cassette is replaced by a single PeSL element between ORFans 191 and 193. Starting from the RB49 genome topology, an excision event involving recombination between the PeSLs flanking ORFan192 could generate the phi1 topology. This recombination event would generate a “PeSL mini-circle” containing the deleted ORFan192 sequence and a single, chimeric PeSL derived from the left- and right-flanking PeSLs. It is also possible that the reverse exchange could occur—involving the ORFan192 PeSL mini-circle and a PeSL resident in the genome that would insert this ORFan into the same, or a different, genomic context. Such a generic shuffling mechanism could provide a simple and plausible explanation for a significant part of the genomic plasticity observed in the T4 superfamily of phages. Such a process has a substantially higher rate of generating viable recombinants compared with random illegitimate recombination and hence, on an evolutionary time scale, it could be responsible for a significant part of the genetic plasticity of phage genomes. These insertion or excision events would be targeted to occur in generally “acceptable” locations (intergenic sites within HPRs) and thus would avoid most of the problems associated with the creation of unviable chimeras.
FIG. 3.—
Proposed mechanism for PeSL-mediated modular shuffling. ORF192 in RB49 is flanked by two PeSLs (multicolored rectangles whose constituent elements are color coded as in fig. 2), and a recombination between these two elements, probably in the AT-rich region (marked with an X), has generated a deletion of this ORF from phi1. Such an excision event could also generate a PeSL mini-circle containing ORF192. Note that this process could be reversible and work in the opposite direction to insert ORF192 into phi1 to generate the RB49 topology.
Formation of PeSL Mini-circles by phi1 and RB49
To determine if PeSL mini-circles were actually formed by PeSL-mediated recombination, we performed inverse-PCRs on E. coli cultures infected by either phi1 or RB49. Such samples were analyzed at various time points during infection to follow the kinetics of PeSL mini-circle production. These inverse-PCRs were done with primers targeted near the middle of the ORFan coding sequence, but oriented outward from each other. Such an “inverse” PCR can only generate an amplification product if the target sequence is located in a circular DNA molecule formed, for example, by recombination between two flanking PeSLs. As negative controls, inverse-PCRs were done with primers targeted to essential conserved genes (g17 in the virion module and g46 in the replication module) that lack flanking PeSL motifs or Pearly consensus sequences. No amplification products were obtained from such controls under any conditions (data not shown).
A series of PeSLs motifs in the vicinity of a variety of ORFans have now been analyzed, but we will limit our presentation to the results obtained on only a few PeSL mini-circles. These examples were chosen to be representative of the various types of PeSLs motifs we found and also because the results cannot be easily dismissed as PCR artifacts. All the PCRs and sequencing reactions presented were repeated multiple times in order to verify their reproducibility.
ORFan192 of RB49.
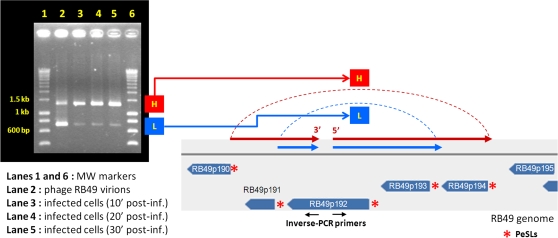
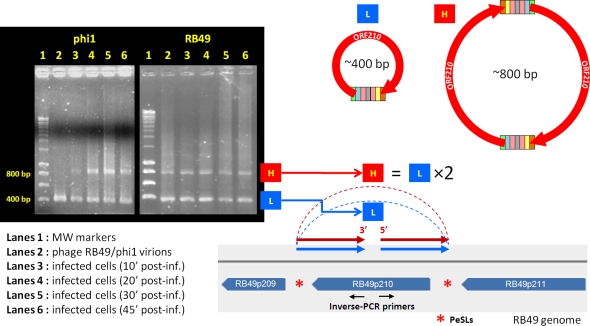
ORFan192 is located within a cluster of ORFans in HPR3 of the RB49 genome (fig. 1); this ORFan is absent from phage phi1. Our analysis of the RB49 PeSL motifs in this region revealed that ORFan192 was actually directly bordered on both sides by PeSLs and that several additional PeSLs are in close proximity (eight PeSLs in an ∼5-kb area). Due to the high density of PeSLs in this small region of the genome, we anticipated the possibility of detecting multiple species of differently sized PeSL mini-circles by the inverse-PCR. PCR amplification of the DNA extracted from an E. coli–infected culture after 10, 20, and 30 min of infection revealed two prominent products (fig. 4): an H band (high molecular weight [MW]) with a size between 1,400 and 1,500 bp and an L band (low MW) of 700–800 bp. These results suggest that recombination involving different PeSL motifs were generating mini-circles containing the same ORFan192 sequence. At the different time points after infection, although the size of these bands remained constant, the quantity of the material in them changed as the infection progressed. The second lane of fig. 4 corresponds to the same inverse-PCR, but in this case it was performed on an RB49 phage stock. The identical bands were present, but their relative quantities differed, the smaller band being less prevalent than that during infection. Such results raise two interesting questions. The fact that the relative band intensities change reproducibly depending on the source of template DNA (virion or infected cells) strongly argues against some sort of PCR artifact. The larger band is preferentially present during infection, whereas the smaller band is present in lower quantities. However, the smaller band is present in relatively higher quantities in the phage stock. The clear presence of the mini-circle bands in the phage stock poses the problem of how the small circular DNA molecules get into the phage capsids. It seems very unlikely the PeSL mini-circles are merely bound to the exterior surface of the virion or are leftover recombinational intermediates from the host cytoplasm, which contaminate the stock because phage stocks treated with DNase I prior to nucleic acid extraction still produced the same inverse-PCR bands (supplementary fig. S2, Supplementary Material online). Although the experiments presented here are most simply interpreted as indicating the presence of some the PeSL mini-circles within the phage head, we cannot exclude the possibility (however remote) that the mini-circles are agglomerated to the exterior surface of the protein lattice of the virion in such a way that they are resistant to DNase activity.
FIG. 4.—
PeSL mini-circles created by recombination between neighboring PeSLs in the region of ORF192. Using two primers (the oppositely oriented black arrows) located in the middle of ORF192 in inverse-PCR, two major bands (H: high MW, L: low MW) were detected inside the phage particles and in infected cultures of Escherichia coli after 10 min. Sequencing of these two bands showed that they were circular and contained either two ORFs (192 and 193 for band L) or four ORFs (191–194 for band H), each resulting from an exchange between two pairs of PeSLs (red asterisks) in the vicinity.
Linear double-stranded DNA (dsDNA) concatemers of the phage genomes are produced by the phage recombination and replication apparatus, and these are packaged by a phage-encoded nanomachine that is transiently associated with preformed empty heads. Once the virion is completely filled with DNA, the genome concatemers are cleaved and the packaging apparatus dissociates (Leiman et al. 2003). The inclusion of PeSL mini-circles within the phage head would require 1) that the packaging apparatus, or other proteins such as the encapsidated internal proteins (IPs; Comeau et al. 2007), recognizes them; 2) that they somehow become entangled in the large genomic DNA during packaging; or 3) that they be accidentally enclosed in the prohead during assembly before regular DNA packaging. Regardless of which mechanism is responsible, this appears to be an inefficient process. Inverse-PCR on dilutions of an extracted DNA template allowed us, for example, to estimate the copy number of encapsidated PeSL ORF210 mini-circles. According to these calculations, the smallest ORF210 mini-circle would be present in one out of ∼105 virions. Although this frequency is low, on an evolutionary time scale it is probably sufficient to have an impact on the genome's evolution. Furthermore, this frequency is estimated for a single mini-circle species and there could be numerous different mini-circles generated by the diverse PeSLs in the genome. Hence the chances that any one particle has at least one PeSL mini-circle may be significantly greater.
Sequencing of bands H and L revealed that these PCR products had a sequence that was compatible with their being produced by a circular template. Sequencing of band H with the reverse primer, for example, produced a sequence starting in ORFan192 and continuing upstream into the intergenic space between ORFans 190–191 and then jumping downstream into the intergenic space between ORFs 194–195 (fig. 4). The sequence then continued reading into ORFs 194 and 193, and then terminated at the forward-primer location within ORFan192. This sequence is compatible with a circular DNA template containing four entire ORFans (191–194). The crossover that formed this sequence occurred between the PeSLs in the intergenic spaces of ORFans 190–191 and ORFs 194–195, and more precisely at a site between the ATG and the -10 element of these PeSLs (fig. 5). The size of the sequenced band was in agreement with that estimated for band H on the agarose gel. In a similar fashion, band L was a PeSL mini-circle containing only ORFans 192 and 193 (figs. 4 and 5) that formed by a crossover between the sequences of the Shine-Dalgarno and the -10 boxes of the flanking PeSLs. Here again, the length of the sequence obtained corresponded to the size of the band observed in the gel. Such results offer unambiguous support for the formation of PeSL mini-circles by recombination between PeSLs.
FIG. 5.—
Schematic diagram of the observed PeSL mini-circles indicating the positions/zones where recombination between the pairs of PeSLs occurred. Alignments of the sequenced PeSL mini-circle sequences compared with the phi1 and RB49 genomes showing the regions of 100% homology on either side of the crossover (red and green lines), as well as the zones (violet hashed box) or exact point (violet line) of exchange events. The different PeSL elements are color coded as in fig. 3.
ORFan210 in phi1 and RB49.
ORFan210 is well conserved in both the RB49 and phi1 genomes (93% amino acid identity); however, the cluster of ORFs downstream of ORFan210 differs in phi1 and RB49. The first of these ORFs in RB49 is ORF211 (homolog of an integron ORF), and we wanted to determine what effect, if any, these differences in genome topology would have on the formation of PeSL mini-circles containing ORFan210. Unexpectedly, the number and size of the bands in the inverse-PCR on infected cultures were identical in both phages (fig. 6). The two bands present, band H (∼800 bp) and band L (∼400 bp), differed only slightly in their relative intensities during the course of infection. However, in the virions only band L was visible in RB49, whereas both bands were present in phi1. Sequencing of bands H and L revealed that the sequences of the comparably sized bands were nearly identical for both phages (there are only a few single-nucleotide polymorphisms between them) and each of them was the product of recombination between pairs of PeSLs. The crossover points were identical for both bands within one phage (i.e., H and L from phi1), but they were slightly different between the two phages (fig. 5). For RB49, the recombination could be localized within the loop of the stem-loop (fig. 5, violet line), whereas the recombination in phi1 was within a region between the stem-loop and the AT-rich region. The sequence of the 210L band indicated that it contained, as expected from its size, only ORFan210 and a chimeric PeSL derived from the flanking PeSL sequences (fig. 6). Surprisingly, the larger H band was a dimer of the 210L sequence. Such a dimer could be formed by frequent recombination between 210L monomers. More importantly, the sequence of this dimer convincingly argues that PeSL mini-circles cannot be explained away as inverse-PCR artifacts because 210H contained a copy of the sequence between the two inverse primers. This “interprimer” region would not be expected to be found if the products were generated by any known PCR artifact.
FIG. 6.—
PeSL mini-circles created by recombination between neighboring PeSLs in the region of ORF210. Using two primers (the oppositely oriented black arrows) located in the middle of ORF210 in inverse-PCR, two major bands (H: high MW, L: low MW) could be detected inside phage particles and in infected cultures of Escherichia coli after 10 min. Sequencing of these two bands showed that they were circular and were the products of recombination between the PeSLs flanking ORF210 (red asterisks). The L band contained a monomer of the ORF210 mini-circle sequence, whereas the H band was a dimer of the same sequence.
Physical Evidence of PeSL Mini-circles.
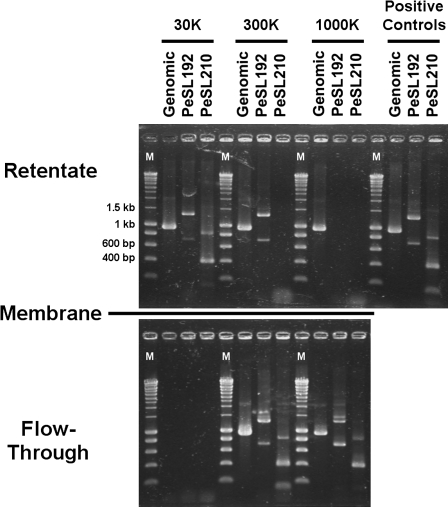
We attempted to directly observe the formation of PeSL mini-circles 192H/B via Southern hybridization (data not shown); however, these attempts were unsuccessful—presumably because of their low copy numbers (see calculations mentioned above) the mini-circles were below the technique's detection limit. The extremely small quantities of each specific PeSL mini-circle required their physical characterization by a separation technique where their presence could be assayed by inverse-PCR. To do this we employed filtration of extracted RB49 DNA through differing pore-size ultrafiltration membranes (30–1000K) to separate large molecules (remaining in the retentate) from small ones (flowing through the membrane). The various filtration fractions were then used as templates in normal PCR to detect genomic DNA (a “normal” genome sequence) and inverse-PCR to detect the presence of PeSL192 and PeSL210 mini-circles (as in figs. 4 and 6). These results (fig. 7) demonstrate that the PeSL mini-circles behave as small DNA molecules that are physically separate from the genomic DNA. As anticipated, none of the DNA molecules passed through the 30K membrane with a MW cutoff of ∼150–300 bp. The PeSL210 mini-circles (∼400/800 bp) were not retained by the 300K membrane (∼1.5- to 3-kb cutoff), whereas about half of the PeSL192 mini-circles (∼700/1400 bp) were. Finally, none of the PeSL mini-circles were retained by the 1000K membrane (∼5- to 10-kb cutoff), whereas the genomic DNA was retained. The presence of some genomic DNA in the 300/1000K filtrates is expected because of the shearing of the DNA during extraction and due to the nature of ultrafiltration. The membrane MW cutoffs are the molecule size where 90% of the material is retained by the membrane (i.e., 90% efficiency), the remaining 10% “bleed-through” is detected due to the sensitivity of the PCR assay. The more relevant criterion for this experiment is the molecule retained by the filter. Consequently, the larger pore-sized membranes above showed that the PeSL mini-circles were not efficiently retained, whereas the genomic DNA was. This result allows us to conclude two things: 1) that the mini-circles are not PCR artifacts as they were not produced in the 1000K retentate (containing substantial genomic DNA); and 2) that the mini-circles exist, at least transiently, and behave as extra-chromosomal elements.
FIG. 7.—
Direct evidence of PeSL mini-circle independent DNA molecules. Extracted RB49 DNA was filtered through three differing pore-size ultrafiltration membranes, which separate larger molecules (remain in retentate) from smaller ones (flow through the filter). The various fractions were used as templates in normal PCR to detect genomic DNA and inverse-PCR to detect the presence of PeSL192 and PeSL210 mini-circles (as in figs. 4 and 6). Lanes labeled with an “M” contain molecular weight markers, with pertinent sizes indicated. The approximate dsDNA cutoffs (defined as 90% retention) for the membranes are as follows: 30K ≈ 150–300 bp; 300K ≈ 1.5–3 kb; 1000K ≈ 5–10 kb.
Discussion
The comparative analysis of the final versions of the phage RB49 and phi1 genome sequences showed that these extremely closely related phages have diverged to a very limited extent. Most of their differences are localized in the HPRs of these genomes, and these are primarily the consequence of the insertion, deletion, or replacement of ORFans. This analysis resulted in the discovery of a new class of regulatory elements involved in genetic plasticity, the PeSLs, conserved composite elements which contain both consensus promoter sequences and adjacent stem-loops structures. We suggest that PeSLs motifs are key elements in the generation of T4-like phage genomic variability by providing the sites for homologous recombination that allow the excision or insertion of ORFans associated with PeSLs. Such PeSL + ORFan cassettes would permit modification of the genome in a more efficient manner than random illegitimate recombination because their genetic shuffling would be preferentially targeted to the resident PeSLs of the HPRs and thus would avoid the disruption of essential genes and transcription units of the genome's conserved core modules. The organization of the PeSL + ORFan cassettes could be compared with integrons—mobile elements which bring exogenous ORFs/genes and a promoter to ensure expression of the cassette into a new genomic context (Mazel 2006). The various experimental approaches employed in this work demonstrate that extra-chromosomal PeSL mini-circles can be generated and that these could be intermediates involved in the horizontal gene transfer of ORFans. Although the creation of circular intermediates/by-products during recombination is rare, but not unheard of (e.g., during V[D]J recombination; Gellert 2002), mechanisms to recombine circular molecules are common (e.g., integrative phage/plasmids, integrons). All these mechanisms tend to employ integrases, excisionases, and/or recombinases to accomplish the task, none of which, beyond the typical T4-like recombination machinery, have been identified in RB49/phi1. The PeSL mini-circles therefore either exploit the existing phage/host recombination machineries or interact with as-yet unidentified phage proteins. There are a few ORFs (004, 113, 182, 194, 211; table 1 and supplementary table S2, Supplementary Material online) in the RB49/phi1 genomes that are associated with pathogenicity islands, plasmids, or integrons, which could be suspected to be involved.
This proposed PeSL cassette-mediated gene shuffling furnishes a possible mechanism for phage modular evolution suggested by Botstein and Campbell nearly 30 years ago (Botstein 1980; Campbell and Botstein 1983). In the 1970s, the detailed study of phages λ and P22, lead to the conclusion that phage functions were maintained in the genome as modules, and that these homologous or analogous sequences could be interchanged easily between different phages (Susskind and Botstein 1978). In Botstein and Campbell's formulation, modules (composed of either a single gene or more often a group of functionally related ones) are exchanged via flanking conserved “linker” sequences that ensure both the proper placement and regulation of the module in its new phage genomic context (Botstein 1980). Modular shuffling was observed, using λ/P22 hybrids, to be very efficient due to the linker sequences and to result in a high frequency of viable recombinants due to the insertions/exchanges being preferentially targeted to “suitable” genomic contexts.
Twenty years later, Hendrix et al. (2000) postulated a variant of this idea, the “moron accretion” hypothesis, to explain the accumulation of new, solitary genes (generally ORFans) that were, in general, inserted opposite to the sense of transcription in the lambdoid phages. In their scheme to explain such events, genome evolution would result from an undefined (perhaps illegitimate recombination) mechanism, which would result in the accumulation of “morons” in new genomic contexts. Morons were composed of individual genes that brought with them both their own promoters and terminators. These regulatory sequences are not thought to be the vehicles of mobility, as it appears to be in the case of PeSLs. Moron accretion does not seem to depend on conserved flanking sequences and therefore was postulated to be relatively inefficient, with nonhomologous recombination generating numerous nonviable chimeras and only a few rare ones that were both viable and had a selective advantage because of the moron gene function.
Our PeSL-mediated modular shuffling obviously differs significantly from this latter hypothesis and is more congruent with the original theory of modular evolution. PeSL-mediated recombination allows for targeted genome exchanges between phages containing the conserved PeSL elements (acting as linker sequences) and allows for the insertion/deletion of endogenously expressed modules without disrupting neighboring gene regulation (due to their terminators). The transcriptional isolation of the PeSL cassettes should also make this type of shuffling relatively efficient because the formation of nonviable recombinants would be significantly reduced. It remains to be determined if PeSL-mediated recombination, and PeSL mini-circle formation, benefits from an active, targeted recombinase (or “helper” protein[s]), or whether it is simply the by-product of the highly efficient phage replication/recombination system (Mosig 1994) that only requires small (<50 bp) patches of homology (Singer et al. 1982). It is tempting to speculate that the relatively conserved PeSL stem-loop motif (generally involving GGGG…CCCC) may be a target for a specific DNA-binding protein involved in recombination. PeSL-like sequences could be responsible for two interesting observations previously made in phage T4. In 1998, Mosig et al. characterized a series of 13 T4 deletion mutants and about half of these events involved a GGGC motif, sometimes paired with the sequence GCCC as inverted repeats. These deletion events were responsible for removing several small, nonessential ORFs from a T4 HPR. The authors suggested that such exchanges could be the consequence of a sequence-specific DNA-binding protein initiating a novel, targeted pathway of recombination. Four years earlier, Repoila et al. (1994) observed what we would now call PeSL-like sequences associated with the two highly variable IP genome loci in the T4-even coliphages. The IPs, of which there are more than 30 known variants (Comeau et al. 2007), are encapsidated in mature particles and then injected into the bacterial cell during infection for phage defensive/adaptive functions. The T-even phage IP-associated elements are not homologs of the PeSLs, but are analogs—they are based on the T4 early promoters (i.e., not σ70-like) and contain weaker, more AT-rich SLSs of variable sizes that generally lack polyG/polyC stems. Nevertheless, they may be responsible for the substantial plasticity within the two small IP loci and may limit IP shuffling to these specific segments and thus explain the absence of IP genes elsewhere in the T-even genomes.
In summary, a consensus is emerging that the design of the larger phage genomes, such as the T4- and SPO1-likes (Stewart et al. 2009; and probably large viruses in general), is based upon a conserved core genome of essential genes and a large and variable set of facultative genes. The conserved core genes show either strong (e.g., myoviruses; Filée et al. 2006; Comeau et al. 2007) or moderate (e.g., siphoviruses; Brüssow and Desiere 2001) vertical evolution, depending upon the phage family in question. The plastic regions of phage genomes show rampant horizontal gene transfer, accumulating and shuffling cellular and phage genes/ORFs alike, seemingly only limited by the physical constraint of the size of the genome that can be encapsidated. We have previously discussed (Krisch and Comeau 2008) that the primitive T4-like phages probably had a much more fluid genome content/organization (more HPR-like) and that the current-day HPRs are all that remains of the original widespread genomic plasticity. This shift may have occurred once the phage core modules were sufficiently perfected to become an effectively fixed entity by evolution, as they lost the modularity of their constituent components. Regardless, there is a certain unanimity regarding the existence and evolutionary “utility/logic” of phage HPRs, yet the details of their formation and maintenance remain obscure. Here, we have demonstrated a mechanism that is potentially responsible for generating phage genomic plasticity. Our experimental results and discovery of PeSL-like sequences in other T4-like phages imply that modular shuffling mediated by PeSLs, or analogous conserved regulatory cassettes, may be an important driving force in phage genome evolution and adaptation. Finally, our results represent the clearest and most convincing evidence yet available that extra-chromosomal circular intermediates could play a significant role in modular shuffling. Although there is no evidence yet to suggest that this sort of genomic plasticity occurs in other viruses or in cellular organisms, this question obviously merits investigation.
Supplementary Material
Supplementary tables S1 and S2 and figures S1 and S2 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
We thank our colleagues M. Codeville, D. Lane, S. Ait-Bara, and K. Cam for help with and discussions relating to Southern hybridizations, vector constructions, and enzyme assays. We thank C. Monod for aiding M.K. in the initial discovery of the extreme sequence similarity between phi1 and RB49. Finally, we would like to particularly thank the directors of the IFR109, H. Richard-Foy and P. Cochard, for making this joint venture possible. This work was supported by intramural funding from the CNRS and by services from the CNRS's IFR109 Sequencing Platform. A.M.C. was supported by a scientific prize from the Les Treilles Foundation. H.M.K. was supported by the Kribu Foundation.
References
- Ackermann HW, Krisch HM. A catalogue of T4-type bacteriophages. Arch Virol. 1997;142:2329–2345. doi: 10.1007/s007050050246. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, et al. Short protocols in molecular biology. New York: J. Wiley and Sons; 1992. [Google Scholar]
- Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D. A theory of modular evolution for bacteriophages. Ann N Y Acad Sci. 1980;354:484–491. doi: 10.1111/j.1749-6632.1980.tb27987.x. [DOI] [PubMed] [Google Scholar]
- Brudno M, et al. LAGAN and MULTI-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüssow H, Desiere F. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol Microbiol. 2001;39:213–222. doi: 10.1046/j.1365-2958.2001.02228.x. [DOI] [PubMed] [Google Scholar]
- Campbell A, Botstein D. Evolution of the lambdoid phages. In: Hendrix R, Roberts J, Stahl F, Weisberg R, editors. Lambda II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1983. pp. 365–380. [Google Scholar]
- Carlson K, Miller ES. Experiments in T4 genetics. In: Karam JD, editor. Molecular biology of bacteriophage T4. Washington: ASM Press; 1994. pp. 421–483. [Google Scholar]
- Comeau AM, Bertrand C, Letarov A, Tétart F, Krisch HM. Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology. 2007;362:384–396. doi: 10.1016/j.virol.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Comeau AM, et al. Exploring the prokaryotic virosphere. Res Microbiol. 2008;159:306–313. doi: 10.1016/j.resmic.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Comeau AM, Krisch HM. The capsid of the T4 phage superfamily: the evolution, diversity and structure of some of the most prevalent proteins in the biosphere. Mol Biol Evol. 2008;25:1321–1332. doi: 10.1093/molbev/msn080. [DOI] [PubMed] [Google Scholar]
- Comeau AM, Short S, Suttle CA. The use of degenerate-primed random amplification of polymorphic DNA (DP-RAPD) for strain-typing and inferring the genetic similarity among closely related viruses. J Virol Methods. 2004;118:95–100. doi: 10.1016/j.jviromet.2004.01.020. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with Glimmer. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filée J, Bapteste E, Susko E, Krisch HM. A selective barrier to horizontal gene transfer in the T4-type bacteriophages that has preserved a core genome with the viral replication and structural genes. Mol Biol Evol. 2006;23:1688–1696. doi: 10.1093/molbev/msl036. [DOI] [PubMed] [Google Scholar]
- Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Lawrence JG, Hatfull GF, Casjens S. The origins and ongoing evolution of viruses. Trends Microbiol. 2000;8:504–508. doi: 10.1016/s0966-842x(00)01863-1. [DOI] [PubMed] [Google Scholar]
- Higashitani A, Higashitani N, Horiuchi K. Minus-strand origin of filamentous phage versus transcriptional promoters in recognition of RNA polymerase. Proc Natl Acad Sci USA. 1997;94:2909–2914. doi: 10.1073/pnas.94.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam JD, Konigsberg WH. DNA polymerase of the t4-related bacteriophages. Prog Nucleic Acid Res Mol Biol. 2000;64:65–96. doi: 10.1016/s0079-6603(00)64002-3. [DOI] [PubMed] [Google Scholar]
- Krisch HM, Comeau AM. The immense journey of bacteriophage T4—from d'Hérelle to Delbrück and then to Darwin and beyond. Res Microbiol. 2008;159:314–324. doi: 10.1016/j.resmic.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure and morphogenesis of bacteriophage T4. Cell Mol Life Sci. 2003;60:2356–2370. doi: 10.1007/s00018-003-3072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- Miller ES, et al. Bacteriophage T4 genome. Microbiol Mol Biol Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A short course in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Monod C, Repoila F, Kutateladze M, Tétart F, Krisch HM. The genome of the pseudo T-even bacteriophages, a diverse group that resembles T4. J Mol Biol. 1997;267:237–249. doi: 10.1006/jmbi.1996.0867. [DOI] [PubMed] [Google Scholar]
- Mosig G. Homologous recombination. In: Karam JD, editor. Molecular biology of bacteriophage T4. Washington: ASM Press; 1994. pp. 421–483. [Google Scholar]
- Mosig G, Colowick NE, Pietz BC. Several new bacteriophage T4 genes, mapped by sequencing deletion endpoints between genes 56 (dCTPase) and dda (a DNA-dependent ATPase-helicase) modulate transcription. Gene. 1998;223:143–155. doi: 10.1016/s0378-1119(98)00238-8. [DOI] [PubMed] [Google Scholar]
- Petrov VM, et al. Plasticity of the gene functions for DNA replication in the T4-like phages. J Mol Biol. 2006;361:46–68. doi: 10.1016/j.jmb.2006.05.071. [DOI] [PubMed] [Google Scholar]
- Rees WA, Yager TD, Korte J, Vonhippel PH. Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry. 1993;32:137–144. doi: 10.1021/bi00052a019. [DOI] [PubMed] [Google Scholar]
- Repoila F, Tétart F, Bouet JY, Krisch HM. Genomic polymorphism in the T-even bacteriophages. EMBO J. 1994;13:4181–4192. doi: 10.1002/j.1460-2075.1994.tb06736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RL, Huskey RJ. Partial exclusion between T-even bacteriophages—incipient genetic isolation mechanism. Genetics. 1974;78:989–1014. doi: 10.1093/genetics/78.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Ellis RL, Hinton DM. Identification of a family of bacteriophage-T4 genes encoding proteins similar to those present in group-I introns of fungi and phage. Proc Natl Acad Sci USA. 1992;89:6658–6662. doi: 10.1073/pnas.89.14.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Singer BS, Gold L, Gauss P, Doherty DH. Determination of the amount of homology required for recombination in bacteriophage T4. Cell. 1982;31:25–33. doi: 10.1016/0092-8674(82)90401-9. [DOI] [PubMed] [Google Scholar]
- Sinoquet C, Demey S, Braun F. Large-scale computational and statistical analyses of high transcription potentialities in 32 prokaryotic genomes. Nucleic Acids Res. 2008;36:3332–3340. doi: 10.1093/nar/gkn135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CR, et al. The genome of Bacillus subtilis bacteriophage SPO1. J Mol Biol. 2009;388:48–70. doi: 10.1016/j.jmb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- Susskind MM, Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978;42:385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tétart F, Desplats C, Krisch HM. Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: recombination between conserved motifs swaps adhesin specificity. J Mol Biol. 1998;282:543–556. doi: 10.1006/jmbi.1998.2047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.