Abstract
Correction of genetic diseases requires integration of the therapeutic gene copy into the genome of patient cells. Retroviruses are commonly used as delivery vehicles because of their precise integration mechanism, but their use has led to adverse events in which vector integration activated proto-oncogenes and contributed to leukemogenesis. Here, we show that integration by lentiviral vectors can be targeted away from genes using an artificial tethering factor. During normal lentivirus infection, the host cell–encoded transcriptional coactivator lens epithelium–derived growth factor/p75 (LEDGF/p75) binds lentiviral integrase (IN), thereby targeting integration to active transcription units and increasing the efficiency of infection. We replaced the LEDGF/p75 chromatin interaction–binding domain with CBX1. CBX1 binds histone H3 di- or trimethylated on K9, which is associated with pericentric heterochromatin and intergenic regions. The chimeric protein supported efficient transduction of lentiviral vectors and directed the integration outside of genes, near bound CBX1. Despite integration in regions rich in epigenetic marks associated with gene silencing, lentiviral vector expression remained efficient. Thus, engineered LEDGF/p75 chimeras provide technology for controlling integration site selection by lentiviral vectors.
Introduction
Lens epithelium–derived growth factor/p75 (LEDGF/p75) is a transcriptional coactivator1,2 that colocalizes with chromatin3 and interacts with the integrase (IN) of the human immunodeficiency virus type-1 (HIV-1) and other lentivirinae4,5,6,7 (Figure 1). RNA interference (RNAi)–mediated depletion of LEDGF/p75 results in the relocalization of IN to the cytoplasm and blocks HIV replication at the integration step of the viral life cycle.8,9,10,11 In addition, LEDGF/p75 depletion alters the genomic distribution of lentiviral integration sites.12,13,14 Lentiviruses preferentially integrate in active transcription units and disfavor promoter regions and locations within 1 kb of CpG islands.13,14,15,16,17 For both HIV and EIAV (equine infectious anemia virus), integration in LEDGF/p75-depleted cells is reduced in transcription units, but enriched in CpG islands and at the 5′-end of genes, together with an increased GC content of regions surrounding integration sites. A model has therefore been proposed in which LEDGF/p75 functions as a molecular tether, bridging IN in the viral preintegration complex and host chromatin.11,18
Figure 1.
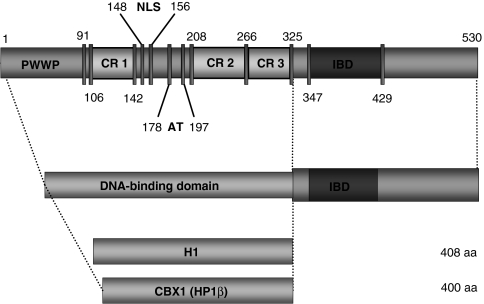
Domain structure of LEDGF/p75 and schematic representation of LEDGF325–530 fusions. LEDGF/p75 contains an integrase-binding domain (IBD) in the C-terminus and a combination of chromatin-interacting modules located in the N-terminal end, most notably the PWWP domain, the AT-hook domain, and three relatively charged regions (CR1–CR3) influence chromatin binding. In the lower panel the DNA-binding domain fusions with LEDGF325–530 are depicted, H1-LEDGF325–530, and CBX1-LEDGF325–530, respectively. Protein elements are drawn to scale. Numbers indicate amino acids of each domain. CBX1, heterochromatin protein 1β (formerly HP1β); H1, histone H1; LEDGF, lens epithelium–derived growth factor; NLS, nuclear localization signal.
Overexpression of a C-terminal fragment of LEDGF/p75 (amino acid 325–530; LEDGF325–530) or the IN-binding domain alone does not mediate chromatin binding, but relocates HIV-IN to the cytoplasm and blocks HIV replication.18,19 The mechanism of chromatin association is poorly understood, but elements in the N-terminal portion of LEDGF/p75 have been shown to be necessary. These include a PWWP domain, which contains a Pro-Trp-Trp-Pro signature related to the Tudor domain “Royal Family”,20 a nuclear localization signal, and two AT hooks11,21,22,23 (Figure 1).
Meehan et al. recently showed that LEDGF proteins bearing H1.1, H1.5, and LANA in place of LEDGF's first 199 amino acids are functional HIV-1 cofactors.24 Here, we used the LEDGF–IN interaction to retarget lentiviral integration to alternative regions of the genome. We engineered artificial chromatin tethers by fusing the C-terminal IN-binding fragment of LEDGF/p75 to alternative chromatin-binding proteins, expressed these in LEDGF/p75-depleted cells, and asked whether (i) infection was rescued and (ii) integration was retargeted to the regions bound by the chimeric protein. In a previous study, Ciuffi et al. created fusions of LEDGF/p75 IN-binding domain and the λ repressor DNA–binding domain and found increased in vitro strand transfer activity near λ repressor–binding sites.25 However, this approach has not yet been used to redirect viral integration in cells.
We compared integration targeting for many hybrids between chromatin-binding proteins and LEDGF/p75, with particular focus on domains with binding specificities that might be useful during human gene therapy. The heterochromatin protein 1β (CBX1, formerly HP1β) binds to sites enriched in histone H3K9 di- and trimethylation at centromeric heterochromatin and transcriptionally silent regions—this provides a chromosomal target present at high copy in gene-sparse regions. We found that a fusion in which CBX1 replaced the chromatin-interaction domain of LEDGF/p75 rescued the infection block in LEDGF/p75-depleted cells. We characterized proviral integration sites using 454 pyrosequencing and found integration to be retargeted in the presence of the fusion to genomic sites bound by CBX1. These regions are low in gene expression and normally disfavored for lentiviral integration, but transgene expression from the vector was nevertheless efficient. These findings open possibilities for targeting of gene therapy vectors by using the LEDGF/p75–IN interaction, potentially to gene-poor regions where their genotoxic potential may be reduced.
Results
Generation of cell lines depleted for LEDGF/p75
To study the role of LEDGF/p75 in tethering and targeting, we generated potent knockdown (KD) cell lines using murine leukemia virus (MLV)– based retroviral vectors encoding two miRNA-based short-hairpin RNAs26 and a zeocin resistance cassette (Supplementary Figure S1a). Transduction of HeLaP4-CCR5 cells and subsequent selection resulted in a polyclonal cell line suppressing LEDGF/p75 mRNA levels to 7% of parental HeLaP4-CCR5 cells (Supplementary Figure S1b). Monoclonal lines were established, selecting the most potent LEDGF KD cells. Three monoclonal lines were isolated that contain <4% of the wild-type (WT) LEDGF/p75 mRNA, referred to as A3, B5, and D11 (96.7, 97, and 97.6% KD, respectively) (Supplementary Figure S1b). LEDGF/p75 protein was undetectable by western blot analysis in either the polyclonal or the monoclonal cell lines (Supplementary Figure S1c). Whereas immunocytochemistry showed that LEDGF/p75 was not depleted from all nuclei in the polyclonal cell line, the protein was undetectable in the monoclonal lines (Supplementary Figure S1d).
Generation of LEDGF hybrids
To retarget lentiviral integration, we substituted the LEDGF/p75 chromatin-binding region (amino acid 1–324, Figure 1) by alternative DNA-binding proteins. LEDGF325–530 was fused to linker histone 1 (H1; histone 1, H1F0) and heterochromatin protein 1β (CBX1, formerly HP1β). H1F0 binds to nucleosomes without apparent preference for the underlying DNA sequence,27 continuously shuttling among chromatin-binding sites.28 CBX1 is associated with pericentric heterochromatin. CBX1 has a single N-terminal chromodomain, which recognizes histone tails via methylated lysine residues, for example trimethylated histone H3 at lysine 9 (H3K9me3) (ref. 29). Both constructs, referred to as H1-LEDGF325–530 and CBX1-LEDGF325–530, were introduced in LEDGF/p75-depleted cell lines (A3, B5, and D11, respectively) using MLV-based viral vectors and selected with blasticidin. In parallel, control cell lines complemented with MLV-based vectors encoding RNAi-resistant LEDGF/p75 (LEDGF BC) or eGFP-LEDGF325–530 were generated.
Viability of the selected cell lines was similar to the parental HeLaP4-CCR5 cell line (data not shown). Expression of the fusion proteins in the A3 KD cell line was verified by western blot (Supplementary Figure S2). Although not all proteins were expressed to the same extent, the migration of all fusion proteins corresponded with their predicted molecular weight. Comparable data were obtained for the B5 and D11-derived cell lines (data not shown).
LEDGF hybrids colocalize with HIV-IN in the nucleus
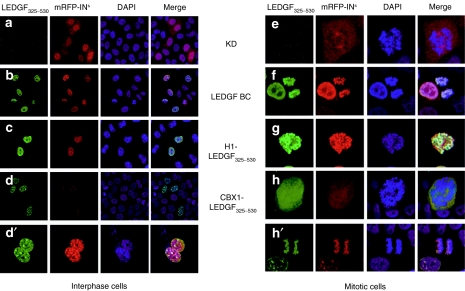
Endogenous LEDGF/p75 displays a characteristic pattern of dense fine speckles in the interphase nucleoplasm and localizes to condensed chromosomes during mitosis.3,30 Immunocytochemistry using antibodies recognizing the C-terminal portion of LEDGF/p75 showed no fluorescence in the KD cells (Figure 2a) corroborating depletion of LEDGF/p75. In contrast, KD cells complemented with RNAi-resistant LEDGF/p75 (LEDGF BC) displayed the typical dense fine speckled pattern of LEDGF/p75 (refs. 8,9,31) (Figure 2b). Complementation of KD cells with the H1-LEDGF325–530 fusion resulted in a nuclear distribution (Figure 2c). In contrast, CBX1-LEDGF325–530 was distributed in multiple irregularly shaped foci over the nuclear area during interphase (Figure 2d,d′), a pattern paralleling that of WT CBX1 (refs. 32,33). Similar to LEDGF/p75, the H1-LEDGF325–530 fusion colocalized with condensed chromatin during mitosis (Figure 2g). In contrast, CBX1-LEDGF325–530 diffused throughout the cytoplasm during late prophase (Figure 2h) and metaphase (not shown), but colocalized with condensed chromatin during anaphase (Figure 2h′), resembling the distribution pattern of WT CBX1 (refs. 32,33).
Figure 2.
Subcellular localization and HIV-IN interaction of stably expressed LEDGF hybrids in interphase and mitotic cells. Stable KD cell lines were complemented with LEDGF325–530 fusion proteins and transfected with mRFP-INs. Laser scanning microscopy (LSM) images of cells stained with anti-LEDGF325–530 antibody are shown (green). Nuclei were stained with DAPI (4',6-diamidino-2-phenylindole; blue). The respective constructs are indicated. Mitotic cells are displayed in the right panel. The data are representative for the vast majority of the imaged cells. CBX1, heterochromatin protein 1β (formerly HP1β); H1, histone H1; HIV, human immunodeficiency virus; IN, integrase; KD, knockdown; LEDGF, lens epithelium–derived growth factor; mRFP, monomeric red fluorescent protein; NLS, nuclear localization signal.
In order to reconstitute LEDGF function, LEDGF325–530 fusions should, in addition to nuclear localization, support chromatin tethering of HIV-IN. In accordance with previous data,3,4 transient expression of IN fused to the monomeric red fluorescent protein (mRFP-INs) in KD cells resulted in a diffuse fluorescent signal throughout the cytoplasm throughout the cell cycle (Figure 2a,e). Complementation with LEDGF/p75 relocated mRFP-INs to the nucleus and condensed chromatin (Figure 2b,f). Fusion of LEDGF325–530 to the linker histone H1 rescued the nuclear phenotype of mRFP-INs and the binding to condensed chromatin (Figure 2c,g). CBX1-LEDGF325–530 relocated mRFP-INs to the nucleus (Figure 2d,d′), in accordance with the distribution of WT CBX1 during late prophase (diffuse) and anaphase (condensed) (Figure 2h,h′).33 B5 and D11 KD lines complemented in parallel with the same viral vector constructs resulted in similar cellular phenotypes (data not shown).
LEDGF hybrids rescue lentiviral transduction
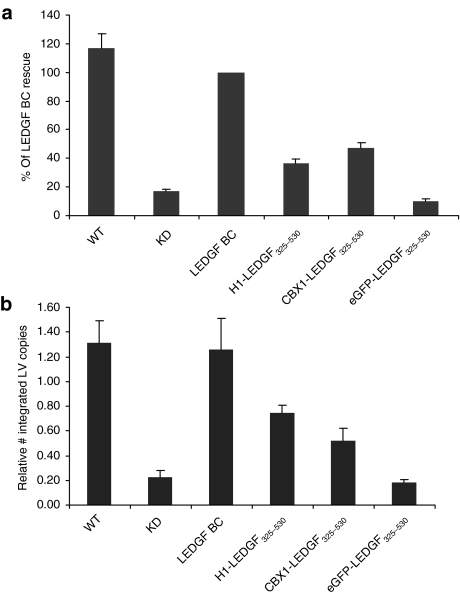
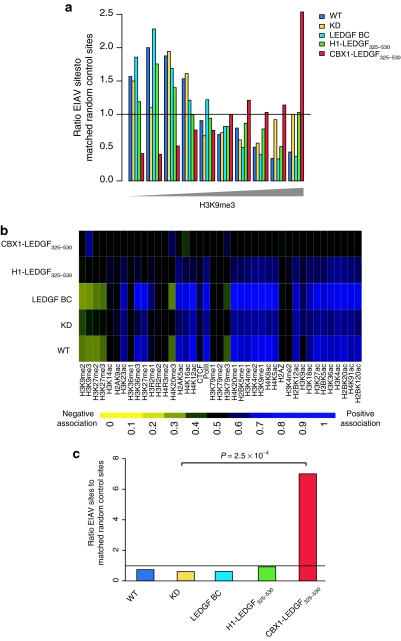
After demonstrating that the fusions were capable of interacting with HIV-1 IN and tethering IN to chromatin, we asked whether they could support efficient lentiviral transduction. We infected the engineered cell lines with an HIV vector expressing firefly luciferase (fLuc).34 Transduction efficiency was evaluated by assaying fLuc activity (relative light units/µg total protein). Transduction efficiency of KD cells was sevenfold lower than that of the parental cells (Figure 3a, WT). Back-complementation of KD with RNAi-resistant LEDGF/p75 (LEDGF BC) rescued lentiviral vector transduction to WT levels. Fusion of LEDGF325–530 to either the linker histone H1 or CBX1 partially rescued viral vector transduction (36.3 and 47.5%, respectively). Fusion of enhanced green fluorescent protein (eGFP) to LEDGF325–530 did not rescue transduction above the levels seen in KD cells. Similar data were obtained when evaluating transduction using eGFP as a reporter (Supplementary Figure S4) or using a near-complete HIV construct (HIVNL4-3.fLuc virus) in which fLuc is driven by the proviral long terminal repeat promoter (Supplementary Figure S5). In parallel, we quantified the number of integrated proviral vector copies in the complemented KD lines following transduction with HIV-based lentiviral vectors (Figure 3b). In KD cells, a 5.8-fold decrease in integrated copies was shown compared with WT cells, which was rescued completely upon back-complementation. Expression of fusion proteins partially rescued integration (60 and 41% of LEDGF BC integration for H1- and CBX1-LEDGF325–530, respectively).
Figure 3.
Rescue of HIV-based lentiviral vector transduction by LEDGF325–530 hybrids. WT and LEDGF BC cells are used as controls. (a) Relative luciferase activity (RLU/µg protein) following HIV-based vector transduction (LV CMV eGFP-T2A-fLuc). Data are compiled at least six independent experiments and are expressed as percentages relative to LEDGF BC cells (mean ± SD). Complemented B5 and D11 cells resulted in similar results (Supplementary Figure S3). (b) Vector integration measured by Q-PCR in WT cells and LEDGF BC cells and LEDGF325–530 fusions, data are represented as mean ± SD. CBX1, heterochromatin protein 1β (formerly HP1β); CMV, cytomegalovirus; eGFP, enhanced green fluorescent protein; H1, histone H1; HIV, human immunodeficiency virus; IN, integrase; KD, knockdown; LEDGF, lens epithelium–derived growth factor; LV, lentivirus; Q-PCR, quantitative PCR; RLU, relative light units; WT, wild type.
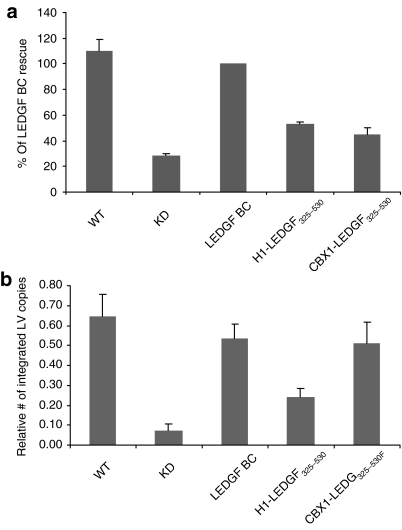
LEDGF/p75 is known to interact with other lentiviral INs in addition to HIV-IN.4,5,7 To investigate complementation of other lentiviruses, fusion-containing and control cells were transduced with an EIAV vector, engineered to encode eGFP. LEDGF325–530-hybrids were capable of rescuing EIAV transduction in the LEDGF/p75-depleted cells (Figure 4), paralleling the results obtained with HIV-1 based vectors. Whereas back-complementation of KD cells rescued vector transduction to WT levels (Figure 4a), fusion of LEDGF325–530 to linker histone H1 or CBX1 partially rescued viral vector transduction (53 and 45.1%, respectively). The number of integrated copies in KD cells was decreased 8.8-fold compared to WT cells (Figure 4b). Complementing the KD cells with H1-LEDGF325–530 resulted in a partial rescue of vector integration (3.3-fold increase compared to KD), and expression of CBX1-LEDGF325–530 resulted in a 6.9-fold increase. Thus, expression of the chimeric proteins rescued EIAV as well as HIV transduction.
Figure 4.
Rescue of EIAV-based vector transduction by LEDGF hybrids. LEDGF325–530 fusions are tested for their ability to rescue EIAV-eGFP transduction. (a) eGFP fluorescence was evaluated by fluorescence-activated cell-sorting analysis. Data are compiled at least six independent experiments and are expressed relative to LEDGF BC complemented cells (mean ± SD). (b) EIAV integration measured by Q-PCR. CBX1, heterochromatin protein 1β (formerly HP1β); eGFP, enhanced green fluorescent protein; EIAV, equine infectious anemia virus; H1, histone H1; KD, knockdown; LEDGF, lens epithelium–derived growth factor; LV, lentivirus; Q-PCR, quantitative PCR; WT, wild type.
Sequencing of proviral integration sites
We next asked whether the LEDGF325–530 fusions retargeted integration to genomic sites bound by the fusion partner. As HeLaP4 cells contain integrated HIV long terminal repeats that would interfere with the isolation of HIV provirus, we used the EIAV vector for distribution analysis. Both EIAV and HIV-IN interact with the LEDGF/p75 IN-binding domain7 and show the same integration site preferences in WT35 or LEDGF/p75-depleted13 cells. Integration sites were analyzed as described previously,13 yielding a total of 2,769 integration sites. Random control sites were generated computationally, and matched to experimental sites with respect to the distance to the nearest MseI cleavage site (matched random control, MRC). In the analyses that follow, the distribution of experimental EIAV sites is normalized to that of the MRC sites, as a control for recovery bias due to cleavage by restriction enzymes.36,37
Retroviral INs show weak but detectable target sequence specificity at the local site of integration. In line with previous reports,13,14 LEDGF/p75 depletion did not affect the consensus sequence flanking the integration site (Supplementary Figure S6). Likewise, expression of LEDGF325–530 fusions did not alter the consensus sequence, consistent with the idea that IN binding to local target DNA determines the sequence preference, independent of the tethering mechanism.
CBX1 fusion directs integration to intergenic regions
Lentiviruses favor integration in transcription units and gene-dense regions.15,35 In the absence of LEDGF/p75, this preference is reduced, and a preference for CpG islands and gene 5′-ends emerges.12,13,14 As an initial survey of the proviral integration site distribution, we examined the frequency of integration in these features. In KD cells, a reduction in the integration frequency in RefSeq transcription units from 67.2 to 51.2% was observed (Table 1), as previously reported for LEDGF/p75-depleted cells.13 Although this reduction was statistically significant (P < 0.0001, Fisher's exact test), integration events in the KD cells were still significantly favored in transcription units over random (P = 4.8 × 10‐6). In accordance with previous reports, we found that integration sites in the KD cells were favored near CpG islands. Both trends were reversed by LEDGF/p75 back-complementation. In contrast, expression of H1-LEDGF325–530 did not rescue integration in transcription units. However, upon expression of CBX1-LEDGF325–530, integration was significantly disfavored in transcription units compared with random (P = 0.026, Fisher's exact test), consistent with the distribution pattern of CBX1 in heterochromatic regions, which are generally gene-poor. Correlation of integration sites with the expression level of the targeted genes showed a slight shift towards genes with lower expression (P < 0.0001, χ2-test to trend comparing CBX1-LEDGF325–530 and WT cells) (Supplementary Figure S7). In addition, we analyzed the distribution of integration sites relative to transcription start sites (7.5-kb window around 5′-end of gene; Supplementary Figure S8). No significant differences were found between the positioning of integration sites in the different cell lines (χ2-test to trend).
Table 1.
Integration frequency near mapped genomic features in the human genome
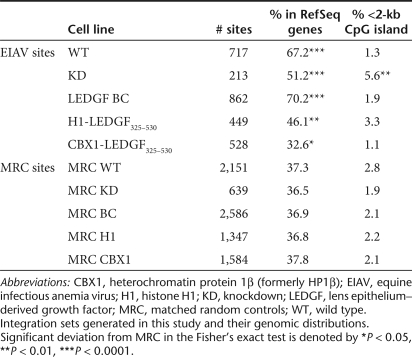
CBX1 is known to bind H3 di- or trimethylated at K9 (H3K9me2 and H3K9me3, respectively) via its chromodomain,29,38,39 so we investigated integration near the sites of these histone modifications.40,41 The H3K9me3 density near the sites of EIAV integration is summarized in Figure 5a. In WT cells, integration was disfavored in the areas high in H3K9me3 (P = 2.9 × 10‐29), consistent with the role of H3K9me3 in transcriptional repression and establishment of silent heterochromatin, features generally disfavored by lentiviral integration. In the KD cells, the same negative correlation remained, though its magnitude was reduced (P = 0.0012). Complementation with LEDGF/p75 restored the negative effect of H3K9me3 to WT levels. Integration site distribution in H1-LEDGF325–530 cells paralleled that seen in KD cells. In cells expressing CBX1-LEDGF325–530, however, the correlation was reversed, with integration sites showing a clear preference for regions denser in H3K9me3 (P = 1.3 × 10‐13).
Figure 5.
Expression of the CBX1-LEDGF325–530 retargets EIAV integration into CBX1-rich heterochromatin regions. (a) Relationship of integration frequency to sites of H3K9me3 in the human genome. (b) Integration frequency relative to density of histone methylation and acetylation. (c) Integration frequency in human chromosome 19 near CBX1-binding sites. CBX1, heterochromatin protein 1β (formerly HP1β); EIAV, equine infectious anemia virus; H1, histone H1; KD, knockdown; LEDGF, lens epithelium–derived growth factor; WT, wild type.
We carried out the same analysis using genome-wide ChIP-seq data for a panel of 39 histone modifications.41 Figure 5b shows correlations between integration sites and the density of these modifications. Each correlation is represented as a tile on the heat map, with the color denoting the strength and direction of the correlation. Histone modifications are grouped into clusters, reported to colocalize and associate with classes of functional genomic elements.41 In WT cells, EIAV sites positively correlated with histone modifications generally associated with active transcription, such as all acetylations, and some histone methylations (shown in blue). Integration sites in WT cells negatively correlated (shown in yellow) with H3K9me3 and other markers reported to be associated with transcriptionally silent regions (e.g., H3K27me3) and heterochromatin (e.g., H4K20me3 and H3K79me3) (refs. 40,42,43). In KD cells, most of the correlations persisted, though they were less pronounced. Complementation with LEDGF restored correlations to WT levels. In cells expressing CBX1-LEDGF325–530, however, most of the correlations were reversed, suggesting a dramatic redistribution of integration sites. In addition to H3K9me3, the modification bound by CBX1, regions high in H4K20me3 and H3K79me3 became favored for EIAV integration. The latter two modifications have also been associated with pericentric heterochromatin.
As CBX1 is enriched around centromeres, we compared the frequency of integration sites in pericentric regions. Integration sites in WT, KD, or LEDGF BC cells did not differ from random (Supplementary Figure S9). In contrast, in cells expressing CBX1-LEDGF325–530, these regions contained 2.7-fold as many integration sites as MRC sites (P = 0.0052, Fisher's exact test), significantly higher than KD cells (P = 0.0236, Fisher's exact test). Sites from H1-LEDGF325–530 cells also showed a preference for these regions, but this was not significantly higher than in KD cells (P = 0.0851, Fisher's exact test).
Finally, we used CBX1-binding sites mapped by DamID (ref. 44) to calculate the average number of CBX1-binding sites around integration sites. CBX1 occupancy around EIAV integration sites on chromosome 19 did not differ from random in WT cells and KD cells, and was not altered in H1-LEDGF325–530-expressing cells (Figure 5c). However, in cells complemented with the CBX1 fusion, 10-kb windows around integration sites contain seven times as many CBX1-binding sites as random (P = 2.5 × 10‐4). The same pattern held when integration sites across the genome were compared to CBX1-binding sites mapped genome-wide (P = 0.015, not shown). Thus, the CBX1-LEDGF/p75 fusion redirected integration to sites known to bind CBX1 and a collection of associated features.
Reporter gene expression remains efficient over time
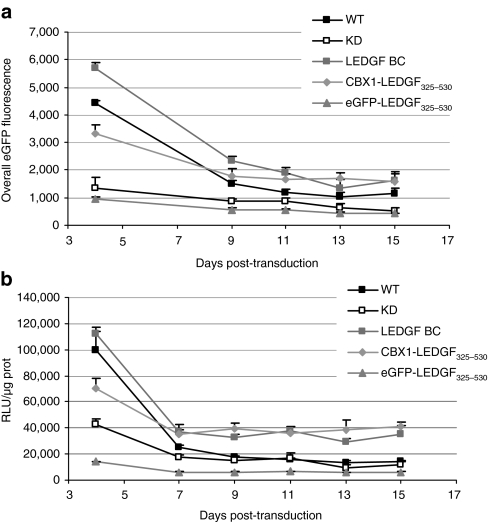
Having shown that the CBX1-LEDGF325–530 fusion retargets lentiviral integration to sites bound by CBX1, we wondered whether gene expression from the vector remained efficient, despite integration in regions rich in epigenetic marks associated with gene silencing. Vector encoded reporter activity was assessed over time following transduction of CBX1-LEDGF325–530 cells and compared to WT cells, KD cells, or KD cells complemented with LEDGF BC or eGFP- LEDGF325–530. Engineered cell lines were transduced with an HIV-based vector expressing both eGFP and fLuc (multiplicity of infection < 1) (ref. 34), and reporter expression was measured in cells over 2 weeks. Mean fluorescence intensity gradually decreased in all cell lines over time (Figure 6a). The relative difference in overall eGFP fluorescence (fold difference to the first measurement at day 4) reached 3.7- and 3.3-fold in WT cells or LEDGF BC cells, respectively, and 2.1-fold in the KD or eGFP-LEDGF325–530 cells. Surprisingly, eGFP reporter activity also decreased only twofold in the CBX1-LEDGF325–530 cells. In parallel, the same cells were analyzed for luciferase activity. Likewise, fLuc reporter activity decreased during the first week post-transduction and remained more or less constant thereafter (Figure 6b). The relative difference in luciferase activity (relative to day 4) showed the most prominent effect in the WT cells (6.1-fold), followed by the KD cells, the LEGDF BC cells and eGFP-LEDGF325–530 cells (3.1-, 3.3-, and 2.4-fold, respectively), whereas KD cells complemented with CBX1-LEDGF325–530 showed a 1.9-fold decrease. Taken together, these data demonstrate that despite retargeting to CBX1-binding regions transgene expression from HIV-based vectors remains efficient.
Figure 6.
Effect of retargeting by CBX1-LEDGF325–530 on transgene expression over time. WT and KD cells, together with LEDGF BC and eGFP-LEDGF325–530 cells were used as controls. All cells were transduced with an HIV-based vector carrying both eGFP and fLuc as reporter genes (LV CMV eGFP-T2A-fLuc) as in Supplementary Figure S3. Reporter activity was determined at the indicated time point following HIV-based vector (LV CMV eGFP-T2A-fLuc) transduction (days post-transduction). (a) Overall eGFP fluorescence over time is calculated as MFI × % gated cells and displayed as mean ± SD (n = 6). (b) Relative luciferase activity (RLU/µg protein) over time; data are expressed as mean ± SD (n = 6). CBX1, heterochromatin protein 1β (formerly HP1β); CMV, cytomegalovirus; eGFP, enhanced green fluorescent protein; fLuc, firefly luciferase; H1, histone H1; HIV, human immunodeficiency virus; KD, knockdown; LEDGF, lens epithelium–derived growth factor; MFI, mean fluorescence intensity; RLU, relative light units; WT, wild type.
Discussion
In this study, we present evidence that LEDGF/p75 can be engineered to target lentiviral integration to new positions in the genome. Alternative chromatin-binding domains (linker histone H1 or the heterochromatin protein 1β, CBX1, were fused to the C-terminal portion of LEDGF/p75 (amino acid 325–530, LEDGF325–530). CBX1 was selected to target sites of H3K9 di- and trimethylation, which are mapped in the genome and usually disfavored for lentiviral integration, so retargeting would be readily identifiable. H1 was used as a control, as it has no known preference for the underlying DNA sequence. Fusing a new chromatin-binding module to LEDGF325–530 changed the behavior of this protein from an integration-inhibitor into an efficient cofactor. Upon challenge by lentiviral vectors, LEDGF325–530-fusions supported efficient lentiviral transduction and integration compared to KD cells. Similar data were recently reported by Meehan et al.24—albeit using LEDGF hybrids that only lack the PWWP- and AT-hook domain (amino acid 1–199).
In addition, we characterized proviral integration sites using 454 pyrosequencing. Analysis of the EIAV integration distribution demonstrated that the CBX1 fusion retargeted lentiviral integration away from RefSeq genes (Table 1), to regions high in H3K9me3 (Figure 5a) and CBX1 binding (Figure 5c). The observation that integration can be retargeted away from genes and into heterochromatin using LEDGF hybrids raises hope for the development of safer lentiviral vectors for gene therapy. Before this study, attempts to retarget HIV integration employed fusions of IN with DNA-binding proteins.45,46,47,48 Some of these showed retargeting as purified enzymes, but until now this approach had limited effect on the distribution of integration sites in cells.
The CBX1 hybrid provides the first example of global redistribution of lentiviral integration sites in the cellular genome, and the first instance of manipulation of a host tethering factor to do so. The success of the CBX1 fusion may be due to the abundance in the genome of its target ligand compared with site-specific DNA-binding domains previously employed, or perhaps its level of occupancy. Even though integration is targeted toward regions in the genome that are generally associated with gene silencing, transgene expression remained efficient over time (Figure 6). Whether new classes of genes are activated as a result is at present unknown and remains to be investigated.
Our findings open possibilities to engineer viral vectors that incorporate LEDGF hybrids to target integration into safe landing sites, thereby reducing the risk of insertional mutagenesis. Hare et al. have recently reported49 a set of amino-acid substitutions in HIV-IN that abolish LEDGF/p75 binding, together with mutations in the LEDGF/p75 protein that restore binding. Gene delivery vectors could thus use an altered IN/LEDGF pair to direct integration, even in the presence of WT LEDGF/p75. To date, the altered IN does not show WT integration activity, but this may be improved with further engineering.
Our data also address issues in HIV biology. Our findings strengthen the idea that LEDGF/p75 is the dominant tether for lentiviral integration. Moreover, we show that chromatin-binding proteins with multiple specificities can successfully replace the LEDGF/p75 DNA-binding elements and rescue HIV infection in an LEDGF/p75 KD model. Still, the hybrids did not mediate rescue to WT levels, which leaves open the question of whether some portions of the N-terminus of LEDGF/p75 absent from our fusions stimulate IN activity or reporter gene expression. The fact that integration can be retargeted to genomic regions usually disfavored for integration indicates that integration in these areas in WT cells is disfavored due to the lack of a tether, rather than to an inherent integration barrier such as steric hindrance resulting from the condensed chromatin structure.
In conclusion, these results establish that LEDGF/p75 is the dominant targeting factor for lentiviral integration and that its interaction with lentiviral INs can be exploited to develop safe and target-specific lentiviral vectors for gene therapy.
Materials and Methods
Generation of LEDGF/p75 depleted cell lines. Stable KD cells were generated using MLV-based vectors encoding miRNA-based short-hairpin RNAs26 against LEDGF/p75 with a zeocin resistance cassette (Supplementary Figure S1a). HeLaP4-CCR5 cells (gift from P. Charneau, Institut Pasteur, Paris, France) were transduced and selected with zeocin (200 µg/ml; Invitrogen, Merelbeke, Belgium), resulting in polyclonal cells with 93% suppression of LEDGF/p75 mRNA compared to WT cells (Supplementary Figure S1b). Monoclonal cells were selected having <3% of WT LEDGF/p75 mRNA (referred to as A3, B5, and D11 in Supplementary Figure S1b). LEDGF protein was undetectable in polyclonal or monoclonal cells by western blot (Supplementary Figure S1c). Whereas LEDGF/p75 was not depleted from all nuclei in the polyclonal cell line, no protein could be detected in the monoclonals (Supplementary Figure S1d).
Construction of MLV-based retroviral vectors. MLV vector constructs were cloned in pLNCX (Clontech, Saint-Germain-en-Laye, France). pLNC-2x miRNA_L3 ZeoR was constructed by cloning zeocin cDNA and an artificial miR30-based shRNA-dimer into pLNC_MCS (primer sequences are included in Supplementary Table S1). Artificial miR30-based hairpin structures were cloned as described by Sun et al.26 Briefly, Hs LEDGF/p75_L3_miRNA was amplified using UNI_miRNAi_s and UNI_miRNAi_as primers and a template with the LEDGF/p75 specific L3 siRNA sequence.3,10,26 The product was cloned twice in peGFP-N3 (Clontech), resulting in peGFP-N3_2x L3mir.
pLNC_LEDGF BC-Ires-Bsd was constructed by digesting LEDGF BC-Ires-Bsd from pCHMWS-LEDGF BC-Ires-Bsd with BamHI–MluI and cloning in pLNC_MCS digested with BglII–MluI. pCHMWS-LEDGF BC-Ires-Bsd was constructed by replacing eGFP in pCHMWS-eGFP-Ires-Bsd with LEDGF BC (kind gift from M. Llano4) using LEDGF_KZ and LEDGF_as SalI primers. The androgen receptor DNA-binding domain (AR-DBD) (gift from F. Claessens) was amplified using Flag_s BglII and AR DBD.CTE15_as primers and cloned in pCHMWS_eGFP-LEDGF325–530 BC-Ires-Puro.19 Next, flag AR-DBD-LEDGF325–530 was amplified (Flag_s AgeI, StuI 325_as primers), generating pLNC_flag AR-DBD-LEDGF325–530-Ires-Bsd. The latter plasmid was used to generate all fusions. pLNC_H1-LEDGF325–530-Ires-Bsd was constructed by amplifying human H1F0 (NM_005318) from a HeLa cDNA. The PCR fragment was XmaI–XhoI digested to replace flag AR-DBD in pLNC_flag AR-DBD-LEDGF325–530-Ires-Bsd. For pLNC_CBX1-LEDGF325–530-Ires-Bsd, human CBX1 (NM_006807) was amplified with HsCBX1_s AgeI and HsCBX1_as XhoI primers from pLgwCbx1-V5-EcoDam (gift from B. Van Steensel44).
Retroviral vector production and transduction. Lentiviral vector production was performed as described earlier.34,50 Briefly, vesicular stomatitis virus glycoprotein pseudotyped HIV-based particles were produced by PEI transfection using pCHMWS_eGFP-T2A-fLuc as a transfer plasmid.34 EIAV-vector particles were produced likewise using p6.1G3CeGFPw (M. Patel and J. Olsen, unpublished results, University of North Carolina, Chapel Hill) and pEV53B and vesicular stomatitis virus glycoprotein encoding pMD.G. HIV_NL4-3.fLuc single round virus was prepared by transient transfection with pNL4-3.LucR‐E‐ (National Institutes of Health AIDS Research and Reference Reagent Program) and pMD.G. MLV-based viral vectors were essentially produced as described for HIV-based vectors,50 except that 293T producer cells were transfected with pCMVgagpol, the respective transfer plasmids (see higher) and pMD.G in a 12.5/32/7 ratio.
For lentiviral transduction experiments, cells were typically plated at 20,000 cells/well in a 96-well plate and transduced overnight. After 73 hours, 90% of cells were reseeded into two plates (FACS analysis and Luc-assay). The remainder was cultured for quantitative PCR or integration site analysis for at least 20 days to eliminate nonintegrated DNA. Stable cell lines were generated by transduction of the monoclonal LEDGF/p75 KD cells with retroviral vectors and subsequent selection with blasticidin (3 µg/ml; Invitrogen, Merelbeke, Belgium).
Quantitative PCR. Integrated proviral copies were quantified by real-time quantitative PCR on genomic DNA as reported earlier.13 To determine LEDGF mRNA levels, total RNA was used for reverse transcription using the High-Capacity cDNA Archive kit (Applied Biosystems, Nieuwerkerk a/d. Ijssel, the Netherlands). Samples corresponding to 400 ng RNA were used for analysis using iQ5 Multicolor RT PCR detection system (BioRad, Nazareth, Belgium). Each reaction contains 12.5 µl 2× iQ Supermix (Biorad, Nazareth, Belgium), 40 nmol/l of forward and reverse primer, and 40 nmol/l of probe in a final volume of 25 µl. LEDGF/p75 primer/probe set: LEDGF Fwd4, 5′-GAA CTT GCT TCA CTT CAG GTC-3′, LEDGF Rev4, 5′-TCG CCG TAT TTT TTT CAG TGT-3′, LEDGF probe4, 5′-FAM-TGC AAC AAG CTC AGA AAC ACA CAG AGA TGA-TAMRA-3′. In all cases, RNaseP was used as endogenous house-keeping control (TaqMan RnaseP Control Reagent; Applied Biosystems). All samples were run in quadruplet for 3 minutes at 95 °C followed by 50 cycles of 10 seconds at 95 °C and 30 seconds at 55 °C. Data were analyzed with iQ5 Optical System Software (BioRad, Nazareth, Belgium).
Luciferase activity assay. Cells were lysed with 70 µl of lysis buffer (50 mmol/l Tris pH 7.5, 200 mmol/l NaCl, 0.2% NP40, 10% glycerol). The lysate was assayed according to the manufacturer's protocol (ONE-Glo; Promega, Madison, WI). Luciferase activity was normalized for total protein (BCA; Pierce, Rockford, IL). All conditions were run at least in triplicate in each experiment.
Western blot analysis. SDS (1%) protein extracts were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. LEDGF fusions were detected using A300-848a antibody (1:2,000 dilution; Bethyl Laboratories, Montgomery, TX) and visualized by chemiluminescence (ECL+; Amersham Biosciences, Uppsala, Sweden). Equal loading was verified with β-tubulin (1:4,000 dilution, T-4026; Sigma-Aldrich, St Louis, MO).
Laser scanning microscopy. Cells were transfected with pmRFP-INs and visualized as described earlier.19 LEDGF325–530 fusion proteins were detected as described previously using a polyclonal rabbit LEDGF325–530-specific antibody (A300-848a, 1/500 dilution; Bethyl Laboratories) and Alexa-488-labeled goat anti-rabbit secondary antibodies (Molecular Probes, Invitrogen, Merelbeke, Belgium). All images were acquired using an LSM 510 META imaging unit (Carl Zeiss, Zaventem, Belgium). Alexa-488 was excited at 488 nm (AI laser), mRFP at 543 nm (HeNe laser) and DAPI (4′,6-diamidino-2-phenylindole) at 790 nm (Spectra-Physics Mai Tai laser; Spectra Physics, Mountain View, CA). After the main beam splitter (HFT KP 700/543 for mRFP, HFT UV/488/543/633 for eGFP, and HFT KP650 for DAPI) the fluorescence signal was divided by a secondary dichroic beam splitter (NFT 490 for eGFP, NFT 545 for mRFP).
Integration site amplification. Integration sites were amplified by linker-mediated PCR as described previously.13 Genomic DNA was digested using MseI and linkers were ligated (Supplementary Table S2). Proviral–host junctions were amplified by nested PCR using barcoded primers. This enabled pooling of PCR products into one sequencing reaction. Products were gel-purified and sequenced on the 454 GS-FLX instrument at the University of Pennsylvania.
Bioinformatic analysis. For integration sites to be authentic, sequences needed a best unique hit when aligned to the human genome (hg18 draft) using BLAT, the alignment began within 3 bp of the viral long terminal repeat end, and had >98% sequence identity. Statistical methods are detailed in Berry et al.37 Integration site counts were compared with MRCs by a Fisher's exact test (where stated), or by multiple regression models for integration intensity and a c-logit test for significance.37 Analysis was carried out using R (http://www.r-project.org). Histone modification data from Barski et al.40 and Wang et al.41 were used. The number of sequence tags from the ChIP-Solexa data sets in a defined window around each EIAV integration site or MRC, was calculated. CBX1-binding sites were analyzed using data from Vogel et al.44 For each DamID probe set available, probes were aligned onto the hg18 draft using BLAT, and their associated log2-binding ratios used to select the top 5% of sites. For each integration site or MRC, the average number of high-affinity probes within a defined window around the site was calculated. Pericentric regions were defined as 1-Mb upstream or downstream of the unsequenced gap on each chromosome.
SUPPLEMENTARY MATERIALFigure S1. Generation of LEDGF/p75 depleted HeLaP4 cell lines.Figure S2. Western blot analysis of LEDGF hybrids.Figure S3. Rescue of HIV-based LV transduction by LEDGF hybrids in B5 and D11 monoclonal LEDGF/p75 KD lines.Figure S4. Rescue of HIV-based LV transduction by LEDGF hybrids using eGFP fluorescence as a read-out.Figure S5. Rescue of HIVNL4.3-fLuc infection by LEDGF hybrids.Figure S6. Alignment of 20 bp surrounding EIAV integration sites from each cell type.Figure S7. Distribution of EIAV integration sites relative to gene expression level.Figure S8. Distribution of EIAV integration sites relative to transcription start sites.Figure S9. Integration frequency of EIAV near centromeres.Table S1. Sequences of primers used in this study to clone hybrid LEDGF constructs.Table S2. Sequences of primers and linkers used in this study to isolate EIAV integration sites.
Supplementary Material
Generation of LEDGF/p75 depleted HeLaP4 cell lines.
Western blot analysis of LEDGF hybrids.
Rescue of HIV-based LV transduction by LEDGF hybrids in B5 and D11 monoclonal LEDGF/p75 KD lines.
Rescue of HIV-based LV transduction by LEDGF hybrids using eGFP fluorescence as a read-out.
Rescue of HIVNL4.3-fLuc infection by LEDGF hybrids.
Alignment of 20 bp surrounding EIAV integration sites from each cell type.
Distribution of EIAV integration sites relative to gene expression level.
Distribution of EIAV integration sites relative to transcription start sites.
Integration frequency of EIAV near centromeres.
Sequences of primers used in this study to clone hybrid LEDGF constructs.
Sequences of primers and linkers used in this study to isolate EIAV integration sites.
Acknowledgments
We thank the K.U. Leuven Cell Imaging Core for use of the confocal microscope. R.G. is a postdoctoral fellow of the Flemish Fund for Scientific Research (FWO Vlaanderen). K.R. was supported by National Institutes of Health (NIH) training grant T32 AI-07324-17. S.V. is funded by the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen). J.D. has a Mathilde-Krim postdoctoral fellowship from Amfar. M.M. is funded by the Affiliation Française contre les Maladies Musculaire. Research was funded by the CellCoVir SBO grant (60813) of the IWT; FWO grant G.0530.08; EC grant THINC (HEALTH-F3-2008-201032) to Z.D. and by NIH grant AI52845, the University of Pennsylvania Center for AIDS Research, and the Penn Genome Frontiers Institute with a grant with the Pennsylvania Department of Health to F.D.B.
REFERENCES
- Dietz F, Franken S, Yoshida K, Nakamura H, Kappler J., and , Gieselmann V. The family of hepatoma-derived growth factor proteins: characterization of a new member HRP-4 and classification of its subfamilies. Biochem J. 2002;366 Pt 2:491–500. doi: 10.1042/BJ20011811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara T, Singh DP., and , Fatma N. LEDGF, a survival factor, activates stress-related genes. Prog Retin Eye Res. 2002;21:341–358. doi: 10.1016/s1350-9462(02)00007-1. [DOI] [PubMed] [Google Scholar]
- Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, et al. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, et al. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busschots K, Vercammen J, Emiliani S, Benarous R, Engelborghs Y, Christ F, et al. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J Biol Chem. 2005;280:17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Devroe E, Silver PA., and , Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J Biol Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- Cherepanov P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007;35:113–124. doi: 10.1093/nar/gkl885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens G, Cherepanov P, Debyser Z, Engelborghs Y., and , Engelman A. Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase interactor LEDGF/p75. J Biol Chem. 2004;279:33421–33429. doi: 10.1074/jbc.M404700200. [DOI] [PubMed] [Google Scholar]
- Vanegas M, Llano M, Delgado S, Thompson D, Peretz M., and , Poeschla E. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J Cell Sci. 2005;118 Pt 8:1733–1743. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove L, Christ F, Van Maele B, De Rijck J, Gijsbers R, Van den Haute C, et al. Transient and stable knockdown of the integrase cofactor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J Virol. 2006;80:1886–1896. doi: 10.1128/JVI.80.4.1886-1896.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, et al. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, et al. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, et al. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS ONE. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR., and , Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Ciuffi A., and , Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC., and , Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombrouck A, De Rijck J, Hendrix J, Vandekerckhove L, Voet A, De Maeyer M, et al. Virus evolution reveals an exclusive role for LEDGF/p75 in chromosomal tethering of HIV. PLoS Pathog. 2007;3:e47. doi: 10.1371/journal.ppat.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijck J, Vandekerckhove L, Gijsbers R, Hombrouck A, Hendrix J, Vercammen J, et al. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. J Virol. 2006;80:11498–11509. doi: 10.1128/JVI.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F., and , Ponting CP. The Tudor domain ‘Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Turlure F, Maertens G, Rahman S, Cherepanov P., and , Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006;34:1653–1665. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S., and , Poeschla EM. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J Mol Biol. 2006;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- Shun MC, Botbol Y, Li X, Di Nunzio F, Daigle JE, Yan N, et al. Identification and characterization of PWWP domain residues critical for LEDGF/p75 chromatin binding and human immunodeficiency virus type 1 infectivity. J Virol. 2008;82:11555–11567. doi: 10.1128/JVI.01561-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan AM, Saenz DT, Morrison JH, Garcia-Rivera JA, Peretz M, Llano M, et al. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PLoS Pathog. 2009;5:e1000522. doi: 10.1371/journal.ppat.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffi A, Diamond TL, Hwang Y, Marshall HM., and , Bushman FD. Modulating target site selection during human immunodeficiency virus DNA integration in vitro with an engineered tethering factor. Hum Gene Ther. 2006;17:960–967. doi: 10.1089/hum.2006.17.960. [DOI] [PubMed] [Google Scholar]
- Sun D, Melegari M, Sridhar S, Rogler CE., and , Zhu L. Multi-miRNA hairpin method that improves gene knockdown efficiency and provides linked multi-gene knockdown. BioTechniques. 2006;41:59–63. doi: 10.2144/000112203. [DOI] [PubMed] [Google Scholar]
- Bustin M, Catez F., and , Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Catez F, Ueda T., and , Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol. 2006;13:305–310. doi: 10.1038/nsmb1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C., and , Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- Ochs RL, Muro Y, Si Y, Ge H, Chan EK., and , Tan EM. Autoantibodies to DFS 70 kd/transcription coactivator p75 in atopic dermatitis and other conditions. J Allergy Clin Immunol. 2000;105 6 Pt 1:1211–1220. doi: 10.1067/mai.2000.107039. [DOI] [PubMed] [Google Scholar]
- Llano M, Delgado S, Vanegas M., and , Poeschla EM. Lens epithelium-derived growth factor/p75 prevents proteasomal degradation of HIV-1 integrase. J Biol Chem. 2004;279:55570–55577. doi: 10.1074/jbc.M408508200. [DOI] [PubMed] [Google Scholar]
- Minc E, Allory Y, Courvalin JC., and , Buendia B. Immunolocalization of HP1 proteins in metaphasic mammalian chromosomes. Methods Cell Sci. 2001;23:171–174. doi: 10.1007/978-94-010-0330-8_18. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Haraguchi T, Masumoto H., and , Hiraoka Y. Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J Cell Sci. 2003;116:3327–3338. doi: 10.1242/jcs.00635. [DOI] [PubMed] [Google Scholar]
- Ibrahimi A, Vande Velde G, Reumers V, Toelen J, Thiry I, Vandeputte C, et al. Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Hum Gene Ther. 2009;20:845–860. doi: 10.1089/hum.2008.188. [DOI] [PubMed] [Google Scholar]
- Hacker CV, Vink CA, Wardell TW, Lee S, Treasure P, Kingsman SM, et al. The integration profile of EIAV-based vectors. Mol Ther. 2006;14:536–545. doi: 10.1016/j.ymthe.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J., and , Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K., and , Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S, et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008;4:e1000190. doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MJ, Guelen L, de Wit E, Peric-Hupkes D, Lodén M, Talhout W, et al. Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome Res. 2006;16:1493–1504. doi: 10.1101/gr.5391806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD. Tethering human immunodeficiency virus 1 integrase to a DNA site directs integration to nearby sequences. Proc Natl Acad Sci USA. 1994;91:9233–9237. doi: 10.1073/pnas.91.20.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes-Son ML., and , Chow SA. Correct integration mediated by integrase-LexA fusion proteins incorporated into HIV-1. Mol Ther. 2002;5:360–370. doi: 10.1006/mthe.2002.0559. [DOI] [PubMed] [Google Scholar]
- Bushman FD., and , Miller MD. Tethering human immunodeficiency virus type 1 preintegration complexes to target DNA promotes integration at nearby sites. J Virol. 1997;71:458–464. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, Dong Z, Wilkinson TA, Barbas CF., 3rd, and , Chow SA. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Shun MC, Gupta SS, Valkov E, Engelman A., and , Cherepanov P. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 2009;5:e1000259. doi: 10.1371/journal.ppat.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraerts M, Michiels M, Baekelandt V, Debyser Z., and , Gijsbers R. Upscaling of lentiviral vector production by tangential flow filtration. J Gene Med. 2005;7:1299–1310. doi: 10.1002/jgm.778. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation of LEDGF/p75 depleted HeLaP4 cell lines.
Western blot analysis of LEDGF hybrids.
Rescue of HIV-based LV transduction by LEDGF hybrids in B5 and D11 monoclonal LEDGF/p75 KD lines.
Rescue of HIV-based LV transduction by LEDGF hybrids using eGFP fluorescence as a read-out.
Rescue of HIVNL4.3-fLuc infection by LEDGF hybrids.
Alignment of 20 bp surrounding EIAV integration sites from each cell type.
Distribution of EIAV integration sites relative to gene expression level.
Distribution of EIAV integration sites relative to transcription start sites.
Integration frequency of EIAV near centromeres.
Sequences of primers used in this study to clone hybrid LEDGF constructs.
Sequences of primers and linkers used in this study to isolate EIAV integration sites.