Abstract
The T-cell is one of the main players in the mammalian immune response. It ensures antigen recognition at the surface of antigen-presenting cells in a complex and highly sensitive and specific process, in which the encounter of the T-cell receptor with the agonist peptide associated with the major histocompatibility complex triggers T-cell activation. While signaling pathways have been elucidated in increasing detail, the mechanism of TCR triggering remains highly controversial despite active research published in the past 10 years. In this paper, we present a short overview of pending questions on critical initial events associated with T-cell triggering. In particular, we examine biophysical approaches already in use, as well as future directions. We suggest that the most recent advances in fluorescence super-resolution imaging, coupled with the new classes of genetic fluorescent probes, will play an important role in elucidation of the T-cell triggering mechanism. Beyond this aspect, we predict that exploration of mechanical cues in the triggering process will provide new clues leading to clarification of the entire mechanism.
Using their receptors as sensors, T-cells patrol mammalian lymphoid organs and scan the surface of specialized cells, the antigen-presenting cells (APCs), for the presence of specific agonist peptides presented by major histocompatibility complexes (pMHCs) (Mazza et al., 1998). Binding of pMHCs, presented at the surface of APCs, to T-cell receptors (TCRs), triggers a signal that eventually leads to activation of T-cells and the T-cell immune response, i.e., cytokine secretion and∕or direct target cell killing. Generation of this adaptive immune response occurs with exquisite sensitivity and specificity. The T-cell is equipped with a unique TCR, which recognizes a very small number of specific agonist peptides presented at the surface of APCs by MHCs. These specific pMHCs are “lost” in a forest of nonspecific self and foreign-derived peptide MHCs (pMHCs), which share considerable structural similarity with the few pMHCs specific to a given TCR. How the T-cell achieves and regulates this function has prompted a tremendous amount of research and remains highly debated and scrutinized (Choudhuri and van der Merwe, 2007; Dustin, 2008; Krogsgaard et al., 2003). While signaling pathways involve intricate multimolecular complexes, and although feedback loops are now known in increasing detail (Smith-Garvin et al., 2009), significant aspects of the initiation process remain unresolved. (i) How do the low affinity and low number of receptors achieve sophisticated and accurate antigen recognition? (ii) How does TCR ligation by agonist pMHC end in kinase activation and immunoreceptor tyrosine-based activation motif (ITAM) phosphorylation? This is still not understood despite intense scrutinizing and speculation concerning the receptor conformational change hypothesis. (iii) How can receptor early aggregation—oligomerization, clustering or preclustering, pseudodimerization, and its effective role in T-cell triggering, which have generated several competing models, be further clarified? (iv) How can we learn more about immune synapse formation, driving forces, and their actual role in T-cell activation?
We will consider these issues from a biophysical point of view, providing the main approaches that have been attempted so as to gain insight into these aspects of T-cell activation. We will endeavor to propose further research directions based on the most recent advances in biophysics. We will focus our attention on the initial steps of the process in relation to surface receptor engagement. We will mainly concentrate on experimental approaches, although several theoretical models have also contributed to the current vision of the subject [see, for instance, Burroughs and van der Merwe (2007) and Wedagedera and Burroughs (2006)].
As molecular structures and biochemical events involved in T-cell activation have been increasingly well-described, biophysical approaches naturally tend to explore mechanisms governing the complex T-cell activation machinery using simplified experimental models with reduced biological assembly, well-defined molecular inputs, and physical organization for receptor engagement at the T-cell surface. Synthetic surfaces for modeling antigen presentation by APC were developed and have the advantage of respecting the two-dimensional (2D) nature of a genuine cell-cell interaction, thus, better reflecting the biological situation than injecting soluble ligands. Major contributions have been made and may continue due to advances in imaging techniques, as we suggest here. We will defend the idea that physical manipulation of cells at the molecular level using the most advanced nanotechnology, though extremely delicate, will enable significant progress in thoroughly understanding T-cell activation.
TCR SENSITIVITY AND AFFINITY
Sensitivity guarantees that intracellular pathogens, such as viruses and bacteria, which have evolved to avoid T-cell recognition by reducing the density of MHC molecules presenting pathogen-derived peptides, continue to be recognized. How this goal is achieved using low affinity and a small number of interactions is a major question.
Since sustained T-cell signaling is required to induce a functional T-cell response, and only a few agonist pMHC molecules are present on the APCs, it has been proposed that one pMHC molecule can serially engage several independent TCR molecules (Valitutti and Lanzavecchia, 1997; Valitutti et al., 1995). Consistent with this model is the observation that TCR-pMHC binding is most often of low affinity (in the micromolar range) and possesses rapid association and dissociation kinetics (Eisen et al., 1996; Wulfing and Davis, 1998). Indeed, the serial triggering model requires that pMHCs bind and dissociate from one TCR with kinetics compatible to engagement of another TCR, thereby inducing sustained TCR signaling. Thus, in the mid-1990s, at the same time as the serial triggering model was proposed (Valitutti et al., 1995), a proofreading model was also suggested (McKeithan, 1995; Rabinowitz et al., 1996). That model proposed that based on kinetic parameters, such as the dissociation constant, the cell performs a sort of evaluation of pMHC∕TCR binding to decide whether a threshold level for T-cell activation has been reached. Thus, serial triggering and kinetic proofreading models predict the existence of an optimal TCR-pMHC half-life for T-cell activation. Experimental data support this prediction (Kalergis et al., 2001). However, exceptions have been seen in which the efficacy of the pMHC at inducing T-cell activation does not correlate with the half-lives of their interactions with TCR. One caveat of all these modeling efforts is that interaction measurements have been made in solution with disembodied TCR and pMHC complexes. In contrast, T-cell activation takes place between surface-tethered ligands and receptors, which may significantly alter kinetic properties of the molecular bonds, the formation of which is regulated by diffusion of the molecules in the membrane. Moreover, in the 2D configuration, interactions may be subjected to mechanical forces exerted by the two cells interacting. An understanding of the binding properties of the TCR∕pMHC and of triggering of TCR-mediated signaling in T-cells must take into account constraints exerted by the actual 2D nature of the T-cell∕APC interaction. However, measurement in-situ of molecular kinetic rates is a real challenge. First, collective information, such as contact lifetime, is of no help in assessing kinetic rates within the interface, and an individual binding event is hard to monitor with high time resolution. Several years ago, Irvine et al. (2002) published impressive results using three-dimensional (3D) fluorescence microscopy to visualize a single pMHC complex in cell-cell contact. In those experiments, the bound state of the complex was revealed indirectly by an intracellular calcium concentration increase and no unbinding event could be observed in that way. Alternatively, Zhu et al. (2007) proposed a semitheoretical approach using fluorescence recovery after photobleaching experiments and a diffusion-coupled model to analyze recovery profiles (Tolentino et al., 2008; Wu et al., 2008). The model was based on various hypotheses, one of which is that of binding equilibrium in the contact, which might not be the case in this living dynamic system. To test serial triggering and proofreading kinetic models would require monitoring within cell-cell contact, individual molecular binding, diffusion, internalization, and possible recycling.
With additional improvements, the bimolecular fluorescence complementation (BiFC) approach [reviewed inKerppola (2008)] might possibly offer a track for monitoring molecular binding within the interface. The technique enables direct visualization of protein interactions in living cells. BiFC analysis is based on the association between two nonfluorescent fragments of a fluorescent protein when they are brought into proximity with each other by an interaction between proteins fused to these fragments. YFP fragments, appropriately truncated, have been used for this purpose. Up until now, the principal limitation to the technique lies in the stabilization of interaction partners by associations between fluorescent protein fragments, which freeze the interaction and significantly hamper dynamic monitoring. It also suffers from low temporal dynamics since, in already published systems, complex maturation takes minutes to hours (Kerppola, 2008), precluding the possibility of monitoring dynamics within seconds. Eventually, fluorescent fragments should not hamper receptor ligand recognition. This makes a serious list of potential sources of failure, but the general principle of fluorescence enhancement due to protein fragments brought in proximity by ligand-receptor association seems to be an interesting track to follow, and we believe that future efforts to engineer fragments of fluorescent proteins will produce significant improvements in BiFC analysis.
THE TCR TRIGGERING MYSTERY
TCR∕CD3 conformational changes
As TCR binds the agonist pMHC, information is transferred to the ITAMs of the associated CD3 subunits, which are subsequently phosphorylated by the Src kinases Lck and Fyn. This is the process referred to as TCR triggering (Trautmann and Randriamampita, 2003; van der Merwe, 2001). The central unresolved question here is how TCR ectodomain ligation results in phosphorylation of the cytoplasmic tails. The idea of TCR-CD3 conformational changes induced by pMHC binding is currently one of the most strongly debated hypotheses. Although many models naturally converge toward this hypothesis, experimental proof and localization of such an event are still scarce.
TCR and CD3 fragments, individual chains, intact TCR ectodomains and TCR∕pMHC complexes have been crystallized to gain insight into the structural basis for TCR complex signaling (Rudolph et al., 2006). Very recently, the group of Rossjohn used site-directed fluorescence labeling to follow the conformational changes of the TCR. Using this method, they showed that pMHC ligation led to a specific conformational change within the constant domain of the TCR, which is reversible upon removal of antigen stimulation (Beddoe et al., 2009). Though clarifying many aspects of antigen recognition, these studies have not revealed any relevant conformational changes in TCRs upon binding that would support the idea that TCRs must be triggered in this way. Yet no structural information is available on the entire T-cell receptor complex, including the transmembrane domain and cytoplasmic tail. Up until now, reconstitution of such a membrane receptor in an appropriate lipid system so as to obtain 3D well-ordered crystals for refining structural analysis has been an enormous challenge, likely requiring a few more years of technological development in the field of structural biology.
Using a thermodynamic approach, Krogsgaard et al. (2005) deduced from kinetics and calorimetric effects that a conformational and∕or flexibility adjustment must occur during TCR binding to MHC (Qi et al., 2006). They suggested a model whereby TCRs would twist with respect to CD3, thus, initiating a structural change within the TCR-CD3 complex.
Conformational change in the cytoplasmic tail of the CD3ε has been observed by Gil et al. (2002, 2005), showing that upon TCR ligation, a proline-rich region in CD3 is exposed and can then recruit the SH3 domain of the Nck adaptor protein. Modeling this conformational change in CD3ε using molecular dynamics, Alarcon’s group recently showed that mutation of two relevant amino acid residues expected to block transmission of conformational change within the ectodomains of CD3 also blocked T-cell activation. The decrease in T-cell activation was observed even in the presence of an excess of wild type CD3ε suggesting the existence of cooperativity between TCR complexes (Martinez-Martin et al., 2009). Yet this model is challenged by experiments that show that mutation of the proline-rich region of CD3ε exposed after conformational changes does not prevent T-cell activation (Mingueneau et al., 2008).
Another hypothesis targets a CD3 ITAM interaction with the membrane: the idea is that in resting T-cells, positively charged intracellular domains that contain ITAMs are tightly associated with the acidic lipids of the inner leaflet of the plasma membrane, leading to the burial of crucial tyrosines in the membrane and preventing their phosphorylation by Lck. This view is supported by the work of Aivazian and Stern (2000), who showed by circular dichroism that the cytoplasmic domain of the CD3 zeta subunit underwent a folding transition in the presence of lipid vesicles made of acidic phospholipids. In this conformation, the cytoplasmic tail could not be phosphorylated by src family tyrosine kinases, in contrast to the lipid-free unstructured form. Recently,Xu et al. (2008) transfected the CD3ε cytoplasmic domain in Jurkat cells and demonstrated a close interaction of the CD3ε cytoplasmic domain with the plasma membrane through fluorescence resonance energy transfer between a C-terminal fluorescent protein and a membrane fluorophore. Although these elegant results suggest a possible mechanism for T-cell signaling initiation, they do not explain what drives ITAM extraction from the membrane.
Alternatively, the permissive geometry model for TCR triggering predicts that dimeric pMHCs simultaneously bind two TCR∕CD3 receptors, forcing TCRαβ to rotate with respect to each of them and leading a scissorlike movement of the CD3 dimers that opens the cytoplasmic tail, enabling signaling inside the T-cell (Minguet and Schamel, 2008).
At this stage, it would be advantageous to be able to prove TCR∕CD3 conformational change upon agonist peptide ligation. For this purpose, it is tempting to examine recent developments in fluorescence resonance energy transfer (FRET) in order to dissect TCR internal molecular dynamics coupled with surface receptor engagement. Known for decades to detect molecular distances in the nanometer range, the technique initially introduced by Förster in the 1950s (Förster, 1946, 1960) has lately been the object of renewed interest due to outstanding progress in specific labeling of proteins in live cells (Giepmans et al., 2006; Schultz, 2007) and significant advances in adapting the strategy to microscopic imaging techniques (Jares-Erijman and Jovin, 2003). The principle of the measurement is based on resonant coupling of the dipole moment of excited states of a donor and an acceptor, which must both be in close spatial and energetic proximity. The effect is over a very short length range. Transfer efficiency is around 50% for a characteristic distance, R0, on the order of tens of nanometers depending on spectral overlapping between donor emission and acceptor excitation; it drops to almost zero for a separation distance of 2R0. Therefore, monitoring transfer efficiency within a well-chosen donor-acceptor pair provides a nanometric molecular rule, as Förster himself said. Needless to say, application of such a method in order to evidence molecular conformational change requires precise linking of the donor and the acceptor to appropriate sites on the targeted molecule. Small genetic encoded tags of fluorescent proteins expressed in-situ might be used as well. The available strategies are reviewed in the excellent paper by Giepmans et al., in which the fluorescent toolbox for protein assessment in live cells is given. Recently, Nguyen and Daugherty (2005) revealed an evolutionary strategy for optimizing FRET fluorophores. Using mutagenesis and screening, they evolved a FRET pair exhibiting a 20-fold ratiometric FRET signal change as compared to threefold for the parental classical cyan-yellow fluorescent protein pair. The pioneering work of Vilardaga et al. (2003) was published in 2003 and described a millisecond switch of G-protein-coupled receptors in living cells using FRET. They were able to show a conformational switch underlying receptor activation by inserting cyan and yellow fluorescent proteins at various positions of the third intracellular loop and∕or of the C-terminus of the transmembrane receptor, respectively. A similar strategy should be possible for exploring the potential TCR∕CD3 conformational change upon surface binding and activation triggering. For instance, tagged Nck and CD3ε could be introduced in T-cells to follow, via FRET, the conformational change in CD3ε that has been shown by Gil et al. (2002) to reveal a binding site for Nck. Up until now, FRET has been used only to examine the interaction of TCRs or CD3 with coreceptor implications (Zal and Gascoigne, 2004; Zal et al., 2002) or ITAM interactions with membranes, as mentioned above (Xu et al., 2008), but not to assess TCR-CD3 internal dynamics. Moreover, while classical FRET is measured by the decrease in donor fluorescence intensity and acceptor-sensitized fluorescence upon donor excitation, which might present some pitfalls, transfer efficiency may also be measured based on donor lifetime in the presence and absence of the acceptor, which is concentration- and intensity-independent (Levitt et al., 2009; Wallrabe and Periasamy, 2005).
T-cell activation mechanics
However, whether or not a more accurate evaluation of the assumed conformational changes would help to clarify the question of T-cell initiation, it will not necessarily disclose the driving force behind conformational change. Lately, the idea has begun to take root in our minds and that of others that both theoretical and experimental models are missing a key aspect of T-cell activation: the potential role of mechanical forces in the formation of cell-cell contact, molecular bonds, and ensuing signaling. Initially, this hypothesis was simply evoked as a possible working hypothesis (Dustin and Zhu, 2006; van der Merwe, 2001). Then, it was put forward in several models involving conformational change, as, for instance, the model shown by Sun et al. (2001), who proposed that one of the chains of the CD3 complex, i.e., CD3γ because of its rigid transmembrane domain, could, after ligand binding, mediate a pistonlike movement of the TCR complex inside the plasma membrane using external forces that would be provided by the cell-to-cell interaction.
Recently, a TCR deformation model was detailed in two successive dedicated papers by Ma et al. (2007, 2008), wherein they proposed TCR deformation induced by mechanical forces as a T-cell triggering hypothesis, reconciling available experimental data and explaining both T-cell activation specificity and sensitivity. They pinpointed the fact that T-cell∕APC interactions take place under constant mechanical stress due to the dynamic nature of cell-cell interactions and confinement encountered in the lymphoid organs. In their model, pMHC-TCR binding per se does not trigger TCR, but rather provides anchorage for a pulling force originating from the cytoskeleton, either to detach the contact in the case of too weak a binding or to exert conformational changes expected to be transmitted to Lck kinases and then to trigger signaling by increased access to and phosphorylation of ITAMs of the CD3 complex.
Nevertheless, forces are increasingly being recognized as integral parts of biological signaling, and transduction of mechanical cues into biochemical events have been described in several cell models, including epithelial, endothelial, and muscle cells, using various biophysical approaches [see for instance Janmey and McCulloch (2007)]. Recently, del Rio et al. (2009) demonstrated that mechanical stretching of talin induces vinculin binding and activation, leading to several intracellular responses.
Another means of envisaging mechanical stress on molecular bonds is to think in terms of bond survival. Pulling on a molecular bond increases the frequency of bond dissociation and regulates the likelihood of rebinding. Bond survival effectively decreases exponentially with the level of the pulling force (Evans, 2001; Evans and Ritchie, 1997). This too might be a potential mechanism impacting the cell response to surface binding.
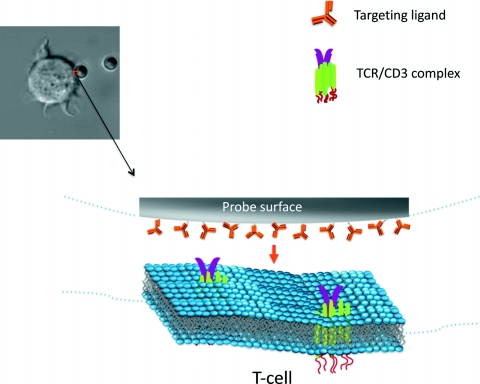
In order to gain insight into these questions, we recently undertook experimental exploration of the potential role of mechanical stress in T-cell activation. We implemented an electromagnetic tip to pull on micrometric magnetic particles devised to artificially engage TCR or CD3 receptors at the cell surface, as displayed in Fig. 1 (Carpentier et al., 2009a, 2009b). The particles were fixed on the Jurkat cell surface several hours before applying forces, allowing the cell to regain full responsiveness after receptor engagement. As constant force was applied to cell-particle-specific contact, the intracellular calcium concentration was monitored in order to detect the onset of intracellular signaling. Our preliminary results show that a calcium signal can be obtained in response to TCR or CD3 pulling in about 20% of tested cells. T-cell activation was obtained for a narrow force ranging from 200 pN to 600 pN. The calcium wave appeared after force application with a time delay of several tens of seconds depending on the applied force amplitude (Fig. 2). No such effect was obtained when equivalent mechanical stress was applied at the cell surface through nonspecific binding of a cationic magnetic particle. These results suggest, as proposed in several models, that mechanical forces play a role in the initial activation machinery and participate in the deciphering and transduction of the message contained in the TCR∕CD3-pMHC interaction. However, additional investigations are needed to further interpret the observed effect. Our efforts are currently focusing on refinement of magnetic actuation through better control of the cell-particle contact area and of the number of receptors engaged by the magnetic particle.
Figure 1. Artificial TCR/CD3 engagement.
Micrometric particle grafted with TCR/CD3-specific ligands brought into contact with the T-cell enables simplified and defined molecular engagement of surface receptors in a 2D configuration mimicking cell-cell interaction. The bound particle also provides a physical handle, which should enable actuating receptors using several types of micro- or nanomanipulation (see Fig. 2).
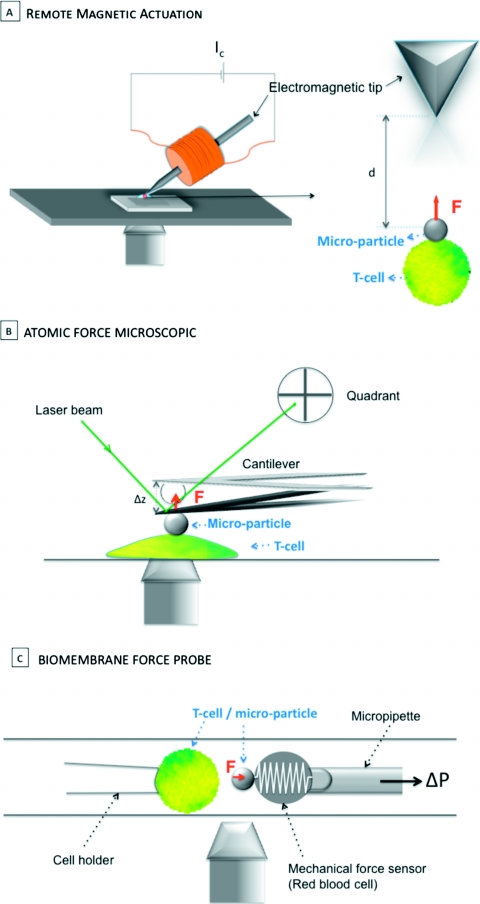
Figure 2. Pulling on TCR.
Three different means of TCR physical actuation: (a) remote magnetic actuation, which consists of using a grafted superparamagnetic particle to engage surface receptors, and a magnetic field gradient to generate a magnetic force. Here and in our experiments, the magnetic gradient is created by an electromagnetic tip made of a soft iron tip at the center of a copper coil supplied with direct current. The force exerted on the particle grows like the magnetic gradient amplitude, particle volume and current intensity in the coil, and like the inverse of the particle-tip distance. The applied force amplitude ranges from tens of piconewtons to a few nanonewtons. Constant force is applied. (b) Atomic force microscope lifting. AFM cantilever can be grafted with the appropriate ligands or chemically coupled with a particle itself grafted with the desired molecules. The setup enables use of various loading rates and measurement of rupture forces. Forces range from a few to a few hundred piconewtons. Force sensor size may be reduced to nanoscopic range (2–50 nm), enabling precise localization of the contact. (c) The biomembrane force probe uses micropipette aspiration to create the pulling force and a red blood cell as a force sensor. The contact is operated through a grafted micrometric particle bound to the red blood cell. This nanomechanical method, initially developed to probe unbinding kinetics of a single molecular bond under stress, might be well-adapted to exploring the effect of nanoforces on the cell response, provided an adequate fluorescence setup can be coupled with the force probe instruments. This experiment targets low forces in the piconewton range. Loading rate is precisely controlled in the range of 0.1–1 pN/nm.
On the other hand, a significant advance in this field would consist of exploring TCR mechanics using a single-molecule-force spectroscopy (SMFS) approach coupled with fluorescent detection of cell biological outcome. In principle, the experiment is quite feasible, since SMFS, and especially atomic force microscopy (AFM), probe a wide force range from 5 pN to tens of nN using a nanoscopic sensor enabling single-molecule binding (Muller et al., 2009). Yet, although interesting force results have been obtained on single molecules in vitro [see, for instance, titin unfolding (Linke and Grutzner, 2008)], much less information has been reported on entire cells, and still less on mammalian cells, involving mostly imaging [e.g., Xiong et al. (2009)] rather than force experiments. For pulling on a specific receptor in its native environment, the membrane of an entire living cell requires that the AFM stylus be functionalized with the appropriate ligand. This is not a limiting step, since an extended tool box for covalently attaching biomolecules to the AFM tip is currently available (Muller et al., 2009). Thus, nonspecific interactions must be avoided, which constitutes a real challenge at the surface of an entire cell. Next, the strength of the attachments—tip-ligand and ligand-receptor—has to be greater than the force expected to deform or unfold TCR∕CD3. But the main difficulty that can be anticipated from these experiments lies in cell membrane intrinsic mechanical properties that may be difficult to discriminate from TCR∕CD3 properties. Yet it is very likely that overcoming these difficulties will provide exciting knowledge about cell surface receptor engagement mechanical issues. An ideal configuration would couple the AFM setup with advanced fluorescence imaging, which is now becoming available on custom AFM instruments.
Likewise, the biomembrane force probe (Evans and Kinoshita, 2007; Evans et al., 1995) approach should be useful for exploring T-cell triggering mechanical questions; although it is not quite clear how the technique could be coupled with high-performance fluorescence detection to monitor cell outcome. The technique pioneered by E. Evans in the late 1990s enables fine-tuned analysis of a single molecular bond under applied force at a wide range of amplitudes (0.1 pN to 1 nN) (Evans and Ritchie, 1997; Evans et al., 1995; Merkel et al., 1999). The principle consists of using a biotinylated erythrocyte as a force transducer. It is maintained with a glass micropipette, which also tunes nanospring stiffness by controlled aspiration. A streptavidin glass bead is attached to the red blood cell and decorated with appropriate ligands to target defined receptors on the facing cell, which is itself maintained by another glass micropipette. When the decorated glass bead is brought into contact with the target cell, one or more bonds form. The ligand density must be adjusted to obtain high probability of a single molecular bond. When the two surfaces are separated, the bond is submitted to a traction force that induces red blood cell deformation. Knowledge of nanospring stiffness enables deriving the applied force. Here too, implementation of a technique whose capacity toward receptors and ligands immobilized on a solid substrate has been clearly demonstrated will be more delicate in the context of an entire living cell (Evans and Kinoshita, 2007; Evans and Calderwood, 2007). Nevertheless, this approach should enable a description of the impact of force on bond dissociation kinetics.
These potential approaches to TCR mechanics are summarized in Fig. 2.
CLUSTERING∕RECEPTOR DYNAMICS
The idea of TCR oligomerization, aggregation and clustering has long been advanced to explain TCR triggering. This view was initially supported by observations that soluble multimeric, but not monomeric, pMHCs can trigger TCR activation (Abastado et al., 1995; Boniface et al., 1998) but was countered by the fact that TCR triggering can occur at low density of pMHC in a biological situation, rendering highly unlikely the possibility of simultaneously engaging two adjacent TCRs as a general T-cell mechanism. Binding-induced self- association was suggested as a solution to this difficulty (Reich et al., 1997), but three-dimensional fluorescence microscopy experiments aimed at visualizing the individual pMHC complex showed that even a single agonist ligand could be detected by the T-cell and trigger transient calcium signaling (Irvine et al., 2002; Sykulev et al., 1996). Preclustering of multivalent TCR complexes was also evoked as an advantageous configuration for conformational change achievement upon ligand-receptor binding (Fernandez-Miguel et al., 1999).
The kinetic segregation model suggests that molecules presenting short extracellular domains in the interface between the T cell and the APC initiate close contact zones between the two cells, whereas inhibitory molecules with long extracellular domains such as CD45 are excluded because of their size (Choudhuri et al., 2005). TCR complexes are then segregated from inhibitory molecules and can thus initiate signaling. This model was supported by experiments showing that shortening of the extracellular domains of inhibitory molecules could abrogate TCR signaling (Irles et al., 2003). But this finding is subjected to controversy, since overexpression of chimeras could inhibit basal phosphorylation even when induced by soluble anti-CD3 antibodies (Trautmann and Randriamampita, 2003).
Another view involving coreceptors and endogenous pMHC complexes was proposed, in which the signal induced by a few specific pMHC complexes was increased due to TCR aggregation by endogenous pMHC. This model was supported by experiments showing that specific pMHC∕TCR can recruit, via the CD4 coreceptor, a second TCR that binds endogeneous pMHC complexes, forming a pseudodimer that induces Ca2+ signaling in T-cells (Krogsgaard et al., 2005).
In a recent review, Varma (2008) suggested that the need for multivalency of TCR engagement might depend on the affinity of the TCR∕pMHC complex.
The clustering we are talking about here is on a much smaller scale than the large-scale segregation of cell surface molecules occurring later to form the so-called immune synapse that we will discuss below. However, microcluster formation has been repeatedly described prior to the formation of the immune synapse (Grakoui et al., 1999b; Krummel et al., 2000).
Despite these numerous models, the role, extent, and timing of receptor aggregation have not been clarified. It would help to confirm not only the existence of TCR preclustering on resting lymphocytes, but also the role of clusters in antigen recognition and their dynamics in terms of receptor engagement. Current progress in fluorescence imaging techniques, though still presenting difficulties for use in living material, should be of significant value.
Indeed, new technologies are now emerging for achieving spatial resolution beyond the diffraction limit of emitted light. This limitation occurs because light traveling through a lens cannot be focused on a point, but only upon an airy disk with a diameter of about half the wavelength of the emitted light. Since wavelengths of visible light range from 400 nm to 700 nm, objects closer than 200–350 nm apart cannot be resolved but appear to merge into one.
Recently, Gustafson and co-workers introduced a 3D-structured illumination microscopy method that increases the spatial resolution of wide-field fluorescence microscopy beyond its classical limit by a factor of 2 (Gustafsson, 2008; Gustafsson et al., 2008). The principle consists of illuminating the specimen with multiple interfering beams of light. The emitted light then contains higher-resolution image information, encoded through spatial frequency mixing. This new information is computationally extracted and used to generate a 3D reconstruction with twice the resolution, in all three dimensions, as is possible in a conventional wide-field microscope. This technique has recently been implemented by Schermelleh et al. (2008) to resolve the single nuclear pore complex, localizing distinct components, and detecting double-layered invaginations, as previously seen only by electron microscopy. However, the images were obtained on formaldehyde fixed cells.
Other approaches for attaining spatial super-resolution consist of making use of single-molecule fluorescence. As mentioned above, in conventional optical fluorescence microscopy, a point source is imaged by a diffraction limit spot. However, localization of the point source coincides with the center of the diffraction spot and can be precisely determined by fitting a Gaussian to it. Thus, imaging of a single-molecule distant by more that 200 nm enables very good localization with a precision <20 nm. To exploit this property, a method has been found to isolate single-molecule fluorescence at high densities (up to ∼105∕μm2). The strategy, so-called photoactivation localization microscopy (PALM), consists of applying serial photoactivation and subsequent photobleaching of sparse subsets of photoactivatable fluorescent protein in a sample. On the single-molecule level, both photobleaching and photoactivation are stochastic rapid one-step events. Fluorescent protein molecules are thus localized one by one. The procedure is repeated approximately 10,000 times until all photoactivatable protein molecules in the sample have been photoactivated, localized, and photobleached. The position information from many molecular subsets is then assembled into a super-resolution image (Betzig et al., 2006; Hess et al., 2006).
Although very promising, this technique requires rapid imaging speed relative to the phenomenon being imaged, along with efficient labeling of the target protein with a suitable photoactivable probe; moreover, it involves multiple cell illuminations. First demonstrated on fixed whole cells, it has recently been applied to live-cell super-resolution imaging to investigate nanoscale dynamics within individual adhesion complexes with a PALM frame rate of 25 s and spatial resolution down to 60 nm, i.e., around 20 min imaging time (Ji et al., 2008). The relatively low time resolution of the technique is the main limitation, yet it may be expected to be overcome in the coming months by improving both the acquisition speed and probe chemistry.
IMMUNE SYNAPSE FORMATION
A seminal object in the field of T-cell activation during the past decade is the so-called immune synapse—the tight junction formed at the interface between the T-cell and the APC (Paul and Seder, 1994). Formation of this large-scale spatial organization upon cell-cell contact provided an attractive mechanism by which T-cells could engage APCs, scan the repertoire of available pMHCs, and respond to low-abundance antigenic stimuli. In the late 1990s, a landmark study (Monks et al., 1998) employing fluorescence imaging demonstrated that transmembrane receptors and signaling molecules segregated in a concentric patterning involving both surface receptors and intracellular signaling molecules. The TCR and protein kinase C were found in the central cluster, which was subsequently defined as the central supramolecular activation cluster (cSMAC). Integrins and their associated adapter talin were found in a peripheral ring surrounding the TCR. This domain was referred to as the peripheral SMAC (pSMAC).
Very soon after this discovery, the pioneering work of Grakoui et al. (1999a) using T-cells in an interaction with artificial surfaces, demonstrated that TCR central cluster formation could be achieved with receptor engagement restricted to TCR and LFA-1. Supported planar bilayers created by fusion of lipid vesicles, including lipid-anchored proteins with solid substrates (Grakoui et al., 1999a), were used to model the display of ligand to T-cell receptors. Planar geometry provided the possibility of using evanescent wave technology, which enabled greater spatial resolution of events occurring during T-cell recognition than with genuine cell-cell contact. This elegant optical technique available on internal reflection fluorescence microscopes is based on sample excitation at a high incident angle through the slide in which the sample is placed. At a specific critical angle, the beam of light is totally reflected from the glass∕water interface. The reflection generates a very thin electromagnetic field (usually less than 200 nm) in the aqueous medium, with frequency identical to that of the incident light, enabling imaging of the sample on a thin thickness close to the glass slide. Using this technique, Grakoui et al., 1999a revealed spatial organization very close to that described on entire cells by Monks et al. (1998). Using two different TCR systems, they correlated clustering properties with soluble molecule kinetic constants and concluded that spatiotemporal dynamics provided machinery for integrating cell surface events into T-cell activation. Following these experiments, Qi et al. (2001) calculated spontaneous evolution of synaptic patterns, which could be expected considering receptor-ligand binding∕dissociation rates, protein mobility, and the membrane shape itself coupled to protein size, elasticity of tethering to the membrane, and local separation between membranes. They found that binding of more than one type of receptor-ligand pair, membrane shape changes and protein transport could lead to spontaneous formation of multiring patterns similar to those shown by Monks et al. (1998). The bull’s-eye structure then became a widely spread paradigm of T-cell activation. Additional studies identified a number of transmembrane and cytoskeletal proteins that are actively removed from the contact region (Allenspach et al., 2001; Delon et al., 2001; Roumier et al., 2001) in the so-called distal SMAC (dSMAC). This patterning of receptors and signaling molecules was soon thought to drive T-cell signaling.
However, there were substantial difficulties with this hypothesis. Detailed comparison of the kinetics of signaling versus synapse formation in T-cells ruled out the hypothesis of a role for synapse in early TCR signaling (Bromley et al., 2001; Delon and Germain, 2000). It was shown, for example, that TCR-mediated tyrosine kinase signaling in naive murine T-cells occurred primarily at the periphery of the synapse and was largely abated before organization into cSMAC and pSMAC could be observed (Lee et al., 2002). Moreover, dynamic imaging showed that T-cells underwent massive morphological rearrangements, as well as intracellular calcium changes within seconds of their encounter with an antigen-bearing APC (Negulescu et al., 1996). Recently, Kaizuka et al. (2007) confirmed TCR actin-driven centripetal flow, but in the meantime they observed early TCR-LFA-1 segregation well before initiation of synapse formation. In addition, studies of T-cell∕dendritic cell (DC) contacts showed that efficient signaling and activation of T-cells occurred without the well-segregated pattern characteristic of mature T-cell∕B cell synapse (Blanchard et al., 2004). Instead of a well-defined cSMAC, T-cells interacting with DC accumulated the TCR within multiple small clusters distributed throughout the contact site, referred to as “multifocal” synapses (Brossard et al., 2005). All together, these results raise doubts as to the exact role of the bull’s-eye structure in T-cell activation. Indeed, a direct causal relationship between changes in the synaptic pattern and signaling remains to be established. Actually, when questioning how the physical organization and composition of cell-cell contact control T-cell activation, it was necessary not only to monitor but to control the physical organization of cell surface receptors. This was achieved by recent introduction of micro∕nanofabrication techniques enabling control of the physical pattern of ligands and fixing barriers against molecular diffusion (Abbott et al., 1992; Falconnet et al., 2006; Groves et al., 1997; Voldman et al., 1999). The impact of predetermined physical organization of ligands on T-cell triggering, synapse formation, and overall signaling could then be studied (DeMond et al., 2008; Irvine et al., 2007; Kaizuka et al., 2007; Mossman et al., 2005; Torres et al., 2008). By imposing geometric constraints and restricted transport of pMHC or costimulatory molecules, Mossman et al. first observed that impeding TCR cluster translocation to the central zone of the contact might correlate with T-cell activation augmentation, as observed through intracellular calcium monitoring and TCR-specific phosphotyrosine colocalization. Using similar nanopatterned-supported membranes to redirect TCR spatial organization, DeMond et al. (2008) recently showed a TCR centripetal translocation coupled to actin flow through a viscous friction mode. Currently, the immune synapse does not seem to play a major role in the triggering phase of T-cell activation, but rather to control triggering extinction and to support further activation expansion. Yet its specific function and well-identified driving forces remain to be established. This still leaves the field open for new ideas aimed at artificial manipulation of the immune synapse in parallel with refined cell response monitoring. Coupling artificial membrane technologies with caging approaches may provide interesting investigative routes for that purpose (DeMond et al., 2006).
CONCLUSION
Evolution has developed refined mechanisms to defend mammalian organisms against pathogens. T-cell triggering and activation upon agonist peptide encounter involve a sophisticated machinery leading from pMHC binding at the cell surface to immune responses like cytokine production and direct target cell killing. This process occurs with exquisite sensitivity and specificity, and while the elicited intracellular biochemical cascade is increasingly well understood, the initial triggering steps remain highly debated. Biophysical approaches using artificial membranes, simplified molecular constructions, and refined fluorescence imaging have helped to better describe initial interactions, minimal requirements, and associated molecular dynamics. Ongoing work aimed at overcoming light diffraction limits to obtain higher spatial resolution will be extremely valuable in gaining further insights into key questions such as that of TCR clustering (Hess et al., 2009). Likewise, how initial binding events are coupled with cytoskeleton reorganization is an important question that we have not treated in this paper, but which must be elucidated so as to gain a clearer vision of the T-cell activation process [reviewed in Gomez and Billadeau (2008)]. Super-resolution techniques associated with recent advances in the field of genetically encoded fluorescent proteins should be extremely helpful in this research. In addition, it should be kept in mind that T-cell activation is, in real life, triggered by a true cell—the antigen-presenting cell—the role of which may have been underestimated in our current understanding of T-cell triggering. This partner might indeed play more than a passive role: among other things, it might strongly influence the mechanical context in which TCR is engaged.
Yet we feel that a biophysical approach to the fascinating question of T-cell activation must also take into account the role of mechanical cues in this process. For this purpose, micro- or nanomechanical actuation of key receptors involved in cell activation must be achieved concomitantly with monitoring of the cell response, through well-adapted biological fluorescent probes. This requires precise and controlled receptor engagement, which remains to be developed. Hopefully, investigating this piece of the puzzle will help to unveil a new mechanism capable of integrating existing models into a unified image of the T-cell.
REFERENCES
- Abastado, J P, Lone, Y C, Casrouge, A, Boulot, G, and Kourilsky, P (1995). “Dimerization of soluble major histocompatibility complex-peptide complexes is sufficient for activation of T-cell hybridoma and induction of unresponsiveness.” J. Exp. Med. 182, 439–447. 10.1084/jem.182.2.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott, N L, Folkers, J P, and Whitesides, G M (1992). “Manipulation of the wettability of surfaces on the 0.1- to 1-micrometer scale through micromachining and molecular self-assembly.” Science 257, 1380–1382. 10.1126/science.257.5075.1380 [DOI] [PubMed] [Google Scholar]
- Aivazian, D, and Stern, L J (2000). “Phosphorylation of T-cell receptor zeta is regulated by a lipid dependent folding transition.” Nat. Struct. Biol. 7, 1023–1026. 10.1038/80930 [DOI] [PubMed] [Google Scholar]
- Allenspach, E J, Cullinan, P, Tong, J, Tang, Q, Tesciuba, A G, Cannon, J L, Takahashi, S M, Morgan, R, Burkhardt, J K, and Sperling, A I (2001). “ERM-dependent movement of CD43 defines a novel protein complex distal to the immunological synapse.” Immunity 15, 739–750. 10.1016/S1074-7613(01)00224-2 [DOI] [PubMed] [Google Scholar]
- Beddoe, T, et al. (2009). “Antigen ligation triggers a conformational change within the constant domain of the alphabeta T-cell receptor.” Immunity 30, 777–788. 10.1016/j.immuni.2009.03.018 [DOI] [PubMed] [Google Scholar]
- Betzig, E, Patterson, G H, Sougrat, R, Lindwasser, O W, Olenych, S, Bonifacino, J S, Davidson, M W, Lippincott-Schwartz, J, and Hess, H F (2006). “Imaging intracellular fluorescent proteins at nanometer resolution.” Science 313, 1642–1645. 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- Blanchard, N, Decraene, M, Yang, K, Miro-Mur, F, Amigorena, S, and Hivroz, C (2004). “Strong and durable TCR clustering at the T/dendritic cell immune synapse is not required for NFAT activation and IFN-gamma production in human CD4+ T cells.” J. Immunol. 173, 3062–3072. [DOI] [PubMed] [Google Scholar]
- Boniface, J J, Rabinowitz, J D, Wulfing, C, Hampl, J, Reich, Z, Altman, J D, Kantor, R M, Beeson, C, McConnell, H M, and Davis, M M (1998). “Initiation of signal transduction through the T-cell receptor requires the multivalent engagement of peptide/MHC ligands.” Immunity 9, 459–466. 10.1016/S1074-7613(00)80629-9 [DOI] [PubMed] [Google Scholar]
- Bromley, S K, Burack, W R, Johnson, K G, Somersalo, K, Sims, T N, Sumen, C, Davis, M M, Shaw, A S, Allen, P M, and Dustin, M L (2001). “The immunological synapse.” Annu. Rev. Immunol. 19, 375–396. 10.1146/annurev.immunol.19.1.375 [DOI] [PubMed] [Google Scholar]
- Brossard, C, Feuillet, V, Schmitt, A, Randriamampita, C, Romao, M, Raposo, G, and Trautmann, A (2005). “Multifocal structure of the T-cell—dendritic cell synapse.” Eur. J. Immunol. 35, 1741–1753. 10.1002/eji.200425857 [DOI] [PubMed] [Google Scholar]
- Burroughs, N J, and van der Merwe, P A (2007). “Stochasticity and spatial heterogeneity in T-cell activation.” Immunol. Rev. 216, 69–80. [DOI] [PubMed] [Google Scholar]
- Carpentier, B, Hivroz, C, and Henry, N (2009a). “Mechanical forces in T-cell triggering.” Biophys. J. 96, 368a. 10.1016/j.bpj.2008.12.1982 [DOI] [Google Scholar]
- Carpentier, B, Pierobon, P, Hivroz, C, and Henry, N (2009b). “T-cell artificial focal triggering tools: linking surface interactions with cell response.” PLoS ONE 4, e4784. 10.1371/journal.pone.0004784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri, K, and van der Merwe, P A (2007). “Molecular mechanisms involved in T-cell receptor triggering.” Semin. Immunol. 19, 255–261. 10.1016/j.smim.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Choudhuri, K, Wiseman, D, Brown, M H, Gould, K, and van der Merwe, P A (2005). “T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand.” Nature (London) 436, 578–582. 10.1038/nature03843 [DOI] [PubMed] [Google Scholar]
- del Rio, A, Perez-Jimenez, R, Liu, R, Roca-Cusachs, P, Fernandez, J M, and Sheetz, M P (2009). “Stretching single talin rod molecules activates vinculin binding.” Science 323, 638–641. 10.1126/science.1162912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delon, J, and Germain, R N (2000). “Information transfer at the immunological synapse.” Curr. Biol. 10, R923–933. 10.1016/S0960-9822(00)00870-8 [DOI] [PubMed] [Google Scholar]
- Delon, J, Kaibuchi, K, and Germain, R N (2001). “Exclusion of CD43 from the immunological synapse is mediated by phosphorylation-regulated relocation of the cytoskeletal adaptor moesin.” Immunity 15, 691–701. 10.1016/S1074-7613(01)00231-X [DOI] [PubMed] [Google Scholar]
- DeMond, A L, Mossman, K D, Starr, T, Dustin, M L, and Groves, J T (2008). “T-cell receptor microcluster transport through molecular mazes reveals mechanism of translocation.” Biophys. J. 94, 3286–3292. 10.1529/biophysj.107.119099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMond, A L, Starr, T, Dustin, M L, and Groves, J T (2006). “Control of antigen presentation with a photoreleasable agonist peptide.” J. Am. Chem. Soc. 128, 15354–15355. 10.1021/ja065304l [DOI] [PubMed] [Google Scholar]
- Dustin, M L (2008). “T-cell activation through immunological synapses and kinapses.” Immunol. Rev. 221, 77–89. 10.1111/j.1600-065X.2008.00589.x [DOI] [PubMed] [Google Scholar]
- Dustin, M L, and Zhu, C (2006). “T cells like a firm molecular handshake.” Proc. Natl. Acad. Sci. U.S.A. 103, 4335–4336. 10.1073/pnas.0600899103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, H N, Sykulev, Y, and Tsomides, T J (1996). “Antigen-specific T-cell receptors and their reactions with complexes formed by peptides with major histocompatibility complex proteins.” Adv. Protein Chem. 49, 1–56. 10.1016/S0065-3233(08)60487-8 [DOI] [PubMed] [Google Scholar]
- Evans, E (2001). “Probing the relation between force—lifetime—and chemistry in single molecular bonds.” Annu. Rev. Biophys. Biomol. Struct. 30, 105–128. 10.1146/annurev.biophys.30.1.105 [DOI] [PubMed] [Google Scholar]
- Evans, E, and Kinoshita, K (2007). “Using force to probe single-molecule receptor-cytoskeletal anchoring beneath the surface of a living cell.” Methods Cell Biol. 83, 373–396. 10.1016/S0091-679X(07)83016-0 [DOI] [PubMed] [Google Scholar]
- Evans, E, and Ritchie, K (1997). “Dynamic strength of molecular adhesion bonds.” Biophys. J. 72, 1541–1555. 10.1016/S0006-3495(97)78802-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, E, Ritchie, K, and Merkel, R (1995). “Sensitive force technique to probe molecular adhesion and structural linkages at biological interfaces.” Biophys. J. 68, 2580–2587. 10.1016/S0006-3495(95)80441-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, E A, and Calderwood, D A (2007). “Forces and bond dynamics in cell adhesion.” Science 316, 1148–1153. 10.1126/science.1137592 [DOI] [PubMed] [Google Scholar]
- Falconnet, D, Csucs, G, Grandin, H M, and Textor, M (2006). “Surface engineering approaches to micropattern surfaces for cell-based assays.” Biomaterials 27, 3044–3063. 10.1016/j.biomaterials.2005.12.024 [DOI] [PubMed] [Google Scholar]
- Fernandez-Miguel, G, Alarcon, B, Iglesias, A, Bluethmann, H, Alvarez-Mon, M, Sanz, E, and de la Hera, A (1999). “Multivalent structure of an alphabeta T-cell receptor.” Proc. Natl. Acad. Sci. U.S.A. 96, 1547–1552. 10.1073/pnas.96.4.1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster, T (1946). “Energiewanderung und fluoreszenz.” Naturwiss. 33, 166–175. 10.1007/BF00585226 [DOI] [Google Scholar]
- Förster, T (1960). “Transfer mechanisms of electronic excitation energy.” Radiat. Res. Suppl. 2, 326–339. 10.2307/3583604 [DOI] [Google Scholar]
- Giepmans, B N, Adams, S R, Ellisman, M H, and Tsien, R Y (2006). “The fluorescent toolbox for assessing protein location and function.” Science 312, 217–224. 10.1126/science.1124618 [DOI] [PubMed] [Google Scholar]
- Gil, D, Schamel, W W, Montoya, M, Sanchez-Madrid, F, and Alarcon, B (2002). “Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T-cell receptor signaling and synapse formation.” Cell 109, 901–912. 10.1016/S0092-8674(02)00799-7 [DOI] [PubMed] [Google Scholar]
- Gil, D, Schrum, A G, Alarcon, B, and Palmer, E (2005). “T-cell receptor engagement by peptide-MHC ligands induces a conformational change in the CD3 complex of thymocytes.” J. Exp. Med. 201, 517–522. 10.1084/jem.20042036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, T S, and Billadeau, D D (2008). “T-cell activation and the cytoskeleton: you can’t have one without the other.” Adv. Immunol. 97, 1–64. 10.1016/S0065-2776(08)00001-1 [DOI] [PubMed] [Google Scholar]
- Grakoui, A, Bromley, S K, Sumen, C, Davis, M M, Shaw, A S, Allen, P M, and Dustin, M L (1999a). “The immunological synapse: a molecular machine controlling T-cell activation.” Science 285, 221–227. 10.1126/science.285.5425.221 [DOI] [PubMed] [Google Scholar]
- Grakoui, A, Donermeyer, D L, Kanagawa, O, Murphy, K M, and Allen, P M (1999b). “TCR-independent pathways mediate the effects of antigen dose and altered peptide ligands on T-cell polarization.” J. Immunol. 162, 1923–1930. [PubMed] [Google Scholar]
- Groves, J T, Ulman, N, and Boxer, S G (1997). “Micropatterning fluid lipid bilayers on solid supports.” Science 275, 651–653. 10.1126/science.275.5300.651 [DOI] [PubMed] [Google Scholar]
- Gustafsson, M G (2008). “Super-resolution light microscopy goes live.” Nat. Methods 5, 385–387. 10.1038/nmeth0508-385 [DOI] [PubMed] [Google Scholar]
- Gustafsson, M G, Shao, L, Carlton, P M, Wang, C J, Golubovskaya, I N, Cande, W Z, Agard, D A, and Sedat, J W (2008). “Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination.” Biophys. J. 94, 4957–4970. 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, S T, Girirajan, T P, and Mason, M D (2006). “Ultra-high resolution imaging by fluorescence photoactivation localization microscopy.” Biophys. J. 91, 4258–4272. 10.1529/biophysj.106.091116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, S T, Gould, T J, Gunewardene, M, Bewersdorf, J, and Mason, M D (2009). “Ultrahigh resolution imaging of biomolecules by fluorescence photoactivation localization microscopy.” Methods Mol. Biol. 544, 483–522. 10.1007/978-1-59745-483-4_32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irles, C, Symons, A, Michel, F, Bakker, T R, van der Merwe, P A, and Acuto, O (2003). “CD45 ectodomain controls interaction with GEMs and Lck activity for optimal TCR signaling.” Nat. Immunol. 4, 189–197. 10.1038/ni877 [DOI] [PubMed] [Google Scholar]
- Irvine, D J, Doh, J, and Huang, B (2007). “Patterned surfaces as tools to study ligand recognition and synapse formation by T cells.” Curr. Opin. Immunol. 19, 463–469. 10.1016/j.coi.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Irvine, D J, Purbhoo, M A, Krogsgaard, M, and Davis, M M (2002). “Direct observation of ligand recognition by T cells.” Nature (London) 419, 845–849. 10.1038/nature01076 [DOI] [PubMed] [Google Scholar]
- Janmey, P A, and McCulloch, C A (2007). “Cell mechanics: integrating cell responses to mechanical stimuli.” Annu. Rev. Biomed. Eng. 9, 1–34. 10.1146/annurev.bioeng.9.060906.151927 [DOI] [PubMed] [Google Scholar]
- Jares-Erijman, E A, and Jovin, T M (2003). “FRET imaging.” Nat. Biotechnol. 21, 1387–1395. 10.1038/nbt896 [DOI] [PubMed] [Google Scholar]
- Ji, N, Shroff, H, Zhong, H, and Betzig, E (2008). “Advances in the speed and resolution of light microscopy.” Curr. Opin. Neurobiol. 18, 605–616. 10.1016/j.conb.2009.03.009 [DOI] [PubMed] [Google Scholar]
- Kaizuka, Y, Douglass, A D, Varma, R, Dustin, M L, and Vale, R D (2007). “Mechanisms for segregating T-cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells.” Proc. Natl. Acad. Sci. U.S.A. 104, 20296–20301. 10.1073/pnas.0710258105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalergis, A M, Boucheron, N, Doucey, M A, Palmieri, E, Goyarts, E C, Vegh, Z, Luescher, I F, and Nathenson, S G (2001). “Efficient T-cell activation requires an optimal dwell-time of interaction between the TCR and the pMHC complex.” Nat. Immunol. 2, 229–234. 10.1038/85286 [DOI] [PubMed] [Google Scholar]
- Kerppola, T K (2008). “Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells.” Ann. Rev. Biophys. 37, 465–487. 10.1146/annurev.biophys.37.032807.125842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogsgaard, M, Huppa, J B, Purbhoo, M A, and Davis, M M (2003). “Linking molecular and cellular events in T-cell activation and synapse formation.” Semin Immunol. 15, 307–315. 10.1016/j.smim.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Krogsgaard, M, Li, Q J, Sumen, C, Huppa, J B, Huse, M, and Davis, M M (2005). “Agonist/endogenous peptide-MHC heterodimers drive T-cell activation and sensitivity.” Nature (London) 434, 238–243. 10.1038/nature03391 [DOI] [PubMed] [Google Scholar]
- Krummel, M F, Sjaastad, M D, Wulfing, C, and Davis, M M (2000). “Differential clustering of CD4 and CD3zeta during T-cell recognition.” Science 289, 1349–1352. 10.1126/science.289.5483.1349 [DOI] [PubMed] [Google Scholar]
- Lee, S J, Hori, Y, Groves, J T, Dustin, M L, and Chakraborty, A K (2002). “Correlation of a dynamic model for immunological synapse formation with effector functions: two pathways to synapse formation.” Trends Immunol. 23, 492–499. 10.1016/S1471-4906(02)02285-8 [DOI] [PubMed] [Google Scholar]
- Levitt, J A, Matthews, D R, Ameer-Beg, S M, and Suhling, K (2009). “Fluorescence lifetime and polarization-resolved imaging in cell biology.” Curr. Opin. Biotechnol. 20, 28–36. 10.1016/j.copbio.2009.01.004 [DOI] [PubMed] [Google Scholar]
- Linke, W A, and Grutzner, A (2008). “Pulling single molecules of titin by AFM—recent advances and physiological implications.” Pfluegers Arch. Eur. J. Physiol. 456, 101–115. 10.1007/s00424-007-0389-x [DOI] [PubMed] [Google Scholar]
- Ma, Z, Janmey, P A, and Finkel, T H (2007). “The receptor deformation model of TCR triggering.” FASEB J. 22, 1002–1008. 10.1096/fj.07-9331hyp [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Z, Sharp, K A, Janmey, P A, and Finkel, T H (2008). “Surface-anchored monomeric agonist pMHCs alone trigger TCR with high sensitivity.” PLoS Biol. 6, e43. 10.1371/journal.pbio.0060043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Martin, N, Risueno, R M, Morreale, A, Zaldivar, I, Fernandez-Arenas, E, Herranz, F, Ortiz, A R, and Alarcon, B (2009). “Cooperativity between T-cell receptor complexes revealed by conformational mutants of CD3epsilon.” Sci. Signal. 2, ra43. 10.1126/scisignal.2000402 [DOI] [PubMed] [Google Scholar]
- Mazza, G, Housset, D, Piras, C, Gregoire, C, Lin, S Y, Fontecilla-Camps, J C, and Malissen, B (1998). “Glimpses at the recognition of peptide/MHC complexes by T-cell antigen receptors.” Immunol. Rev. 163, 187–196. 10.1111/j.1600-065X.1998.tb01197.x [DOI] [PubMed] [Google Scholar]
- McKeithan, T W (1995). “Kinetic proofreading in T-cell receptor signal transduction.” Proc. Natl. Acad. Sci. U.S.A. 92, 5042–5046. 10.1073/pnas.92.11.5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel, R, Nassoy, P, Leung, A, Ritchie, K, and Evans, E (1999). “Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy.” Nature (London) 397, 50–53. 10.1038/16219 [DOI] [PubMed] [Google Scholar]
- Mingueneau, M, Sansoni, A, Gregoire, C, Roncagalli, R, Aguado, E, Weiss, A, Malissen, M, and Malissen, B (2008). “The proline-rich sequence of CD3epsilon controls T-cell antigen receptor expression on and signaling potency in preselection CD4+CD8+ thymocytes.” Nat. Immunol. 9, 522–532. 10.1038/ni.1608 [DOI] [PubMed] [Google Scholar]
- Minguet, S, and Schamel, W W (2008). “A permissive geometry model for TCR-CD3 activation.” Trends Biochem. Sci. 33, 51–57. 10.1016/j.tibs.2007.10.008 [DOI] [PubMed] [Google Scholar]
- Monks, C R, Freiberg, B A, Kupfer, H, Sciaky, N, and Kupfer, A (1998). “Three-dimensional segregation of supramolecular activation clusters in T cells.” Nature (London) 395, 82–86. 10.1038/25764 [DOI] [PubMed] [Google Scholar]
- Mossman, K D, Campi, G, Groves, J T, and Dustin, M L (2005). “Altered TCR signaling from geometrically repatterned immunological synapses.” Science 310, 1191–1193. 10.1126/science.1119238 [DOI] [PubMed] [Google Scholar]
- Muller, D J, Helenius, J, Alsteens, D, and Dufrene, Y F (2009). “Force probing surfaces of living cells to molecular resolution.” Nat. Chem. Biol. 5, 383–390. 10.1038/nchembio.181 [DOI] [PubMed] [Google Scholar]
- Negulescu, P A, Krasieva, T B, Khan, A, Kerschbaum, H H, and Cahalan, M D (1996). “Polarity of T-cell shape, motility, and sensitivity to antigen.” Immunity 4, 421–430. 10.1016/S1074-7613(00)80409-4 [DOI] [PubMed] [Google Scholar]
- Nguyen, A W, and Daugherty, P S (2005). “Evolutionary optimization of fluorescent proteins for intracellular FRET.” Nat. Biotechnol. 23, 355–360. 10.1038/nbt1066 [DOI] [PubMed] [Google Scholar]
- Paul, W E, and Seder, R A (1994). “Lymphocyte responses and cytokines.” Cell 76, 241–251. 10.1016/0092-8674(94)90332-8 [DOI] [PubMed] [Google Scholar]
- Qi, S, Krogsgaard, M, Davis, M M, and Chakraborty, A K (2006). “Molecular flexibility can influence the stimulatory ability of receptor-ligand interactions at cell-cell junctions.” Proc. Natl. Acad. Sci. U.S.A. 103, 4416–4421. 10.1073/pnas.0510991103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, S Y, Groves, J T, and Chakraborty, A K (2001). “Synaptic pattern formation during cellular recognition.” Proc. Natl. Acad. Sci. U.S.A. 98, 6548–6553. 10.1073/pnas.111536798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz, J D, Beeson, C, Lyons, D S, Davis, M M, and McConnell, H M (1996). “Kinetic discrimination in T-cell activation.” Proc. Natl. Acad. Sci. U.S.A. 93, 1401–1405. 10.1073/pnas.93.4.1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich, Z, Boniface, J J, Lyons, D S, Borochov, N, Wachtel, E J, and Davis, M M (1997). “Ligand-specific oligomerization of T-cell receptor molecules.” Nature (London) 387, 617–620. 10.1038/42500 [DOI] [PubMed] [Google Scholar]
- Roumier, A, Olivo-Marin, J C, Arpin, M, Michel, F, Martin, M, Mangeat, P, Acuto, O, Dautry-Varsat, A, and Alcover, A (2001). “The membrane-microfilament linker ezrin is involved in the formation of the immunological synapse and in T-cell activation.” Immunity 15, 715–728. 10.1016/S1074-7613(01)00225-4 [DOI] [PubMed] [Google Scholar]
- Rudolph, M G, Stanfield, R L, and Wilson, I A (2006). “How TCRs bind MHCs, peptides, and coreceptors.” Annu. Rev. Immunol. 24, 419–466. 10.1146/annurev.immunol.23.021704.115658 [DOI] [PubMed] [Google Scholar]
- Schermelleh, L, et al. (2008). “Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy.” Science 320, 1332–1336. 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, C (2007). “Molecular tools for cells and systems biology.” HFSP J. 1(4), 230–248. 10.2976/1.2812442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Garvin, J E, Koretzky, G A, and Jordan, M S (2009). “T-cell activation.” Annu. Rev. Immunol. 27, 591–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z J, Kim, K S, Wagner, G, and Reinherz, E L (2001). “Mechanisms contributing to T-cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer.” Cell 105, 913–923. 10.1016/S0092-8674(01)00395-6 [DOI] [PubMed] [Google Scholar]
- Sykulev, Y, Joo, M, Vturina, I, Tsomides, T J, and Eisen, H N (1996). “Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T-cell response.” Immunity 4, 565–571. 10.1016/S1074-7613(00)80483-5 [DOI] [PubMed] [Google Scholar]
- Tolentino, T P, Wu, J, Zarnitsyna, V I, Fang, Y, Dustin, M L, and Zhu, C (2008). “Measuring diffusion and binding kinetics by contact area FRAP.” Biophys. J. 95, 920–930. 10.1529/biophysj.107.114447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, A J, Wu, M, Holowka, D, and Baird, B (2008). “Nanobiotechnology and cell biology: micro- and nanofabricated surfaces to investigate receptor-mediated signaling.” Ann. Rev. Biophys. 37, 265–288. 10.1146/annurev.biophys.36.040306.132651 [DOI] [PubMed] [Google Scholar]
- Trautmann, A, and Randriamampita, C (2003). “Initiation of TCR signalling revisited.” Trends Immunol. 24, 425–428. 10.1016/S1471-4906(03)00182-0 [DOI] [PubMed] [Google Scholar]
- Valitutti, S, and Lanzavecchia, A (1997). “Serial triggering of TCRs: a basis for the sensitivity and specificity of antigen recognition.” Immunol. Today 18, 299–304. 10.1016/S0167-5699(97)01075-X [DOI] [PubMed] [Google Scholar]
- Valitutti, S, Muller, S, Cella, M, Padovan, E, and Lanzavecchia, A (1995). “Serial triggering of many T-cell receptors by a few peptide-MHC complexes.” Nature (London) 375, 148–151. 10.1038/375148a0 [DOI] [PubMed] [Google Scholar]
- van der Merwe, P A (2001). “The TCR triggering puzzle.” Immunity 14, 665–668. 10.1016/S1074-7613(01)00155-8 [DOI] [PubMed] [Google Scholar]
- Varma, R (2008). “TCR triggering by the pMHC complex: valency, affinity, and dynamics.” Sci. Signal. 1, pe21. 10.1126/stke.119pe21 [DOI] [PubMed] [Google Scholar]
- Vilardaga, J P, Bunemann, M, Krasel, C, Castro, M, and Lohse, M J (2003). “Measurement of the millisecond activation switch of G protein-coupled receptors in living cells.” Nat. Biotechnol. 21, 807–812. 10.1038/nbt838 [DOI] [PubMed] [Google Scholar]
- Voldman, J, Gray, M L, and Schmidt, M A (1999). “Microfabrication in biology and medicine.” Annu. Rev. Biomed. Eng. 1, 401–425. 10.1146/annurev.bioeng.1.1.401 [DOI] [PubMed] [Google Scholar]
- Wallrabe, H, and Periasamy, A (2005). “Imaging protein molecules using FRET and FLIM microscopy.” Curr. Opin. Biotechnol. 16, 19–27. 10.1016/j.copbio.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Wedagedera, J R, and Burroughs, N J (2006). “T-cell activation: a queuing theory analysis at low agonist density.” Biophys. J. 91, 1604–1618. 10.1529/biophysj.105.066001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J, Fang, Y, Zarnitsyna, V I, Tolentino, T P, Dustin, M L, and Zhu, C (2008). “A coupled diffusion-kinetics model for analysis of contact-area FRAP experiment.” Biophys. J. 95, 910–919. 10.1529/biophysj.107.114439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfing, C, and Davis, M M (1998). “A receptor/cytoskeletal movement triggered by costimulation during T-cell activation.” Science 282, 2266–2269. 10.1126/science.282.5397.2266 [DOI] [PubMed] [Google Scholar]
- Xiong, Y, Lee, A C, Suter, D M, and Lee, G U (2009). “Topography and nanomechanics of live neuronal growth cones analyzed by atomic force microscopy.” Biophys. J. 96, 5060–5072. 10.1016/j.bpj.2009.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C, Gagnon, E, Call, M E, Schnell, J R, Schwieters, C D, Carman, C V, Chou, J J, and Wucherpfennig, K W (2008). “Regulation of T-cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif.” Cell 135, 702–713. 10.1016/j.cell.2008.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zal, T, and Gascoigne, N R (2004). “Using live FRET imaging to reveal early protein-protein interactions during T-cell activation.” Curr. Opin. Immunol. 16, 674–683. [PubMed] [Google Scholar]
- Zal, T, Zal, M A, and Gascoigne, N R (2002). “Inhibition of T-cell receptor-coreceptor interactions by antagonist ligands visualized by live FRET imaging of the T-hybridoma immunological synapse.” Immunity 16, 521–534. 10.1016/S1074-7613(02)00301-1 [DOI] [PubMed] [Google Scholar]
- Zhu, D M, Dustin, M L, Cairo, C W, and Golan, D E (2007). “Analysis of two-dimensional dissociation constant of laterally mobile cell adhesion molecules.” Biophys. J. 92, 1022–1034. 10.1529/biophysj.106.089649 [DOI] [PMC free article] [PubMed] [Google Scholar]