Abstract
Large amounts of antibiotics used for human therapy, as well as for farm animals and even for fish in aquaculture, resulted in the selection of pathogenic bacteria resistant to multiple drugs. Multidrug resistance in bacteria may be generated by one of two mechanisms. First, these bacteria may accumulate multiple genes, each coding for resistance to a single drug, within a single cell. This accumulation occurs typically on resistance (R) plasmids. Second, multidrug resistance may also occur by the increased expression of genes that code for multidrug efflux pumps, extruding a wide range of drugs. This review discusses our current knowledge on the molecular mechanisms involved in both types of resistance.
Keywords: R plasmids, transposons, integrons, type IV secretion system, multidrug efflux pumps
INTRODUCTION
The discovery of penicillin in 1928 was followed by the discovery and commercial production of many other antibiotics. We now take for granted that any infectious disease is curable by antibiotic therapy. Antibiotics are manufactured at an estimated scale of about 100,000 tons annually worldwide, and their use had a profound impact on the life of bacteria on earth. More strains of pathogens have become antibiotic resistant, and some have become resistant to many antibiotics and chemotherapeutic agents, the phenomenon of multidrug resistance.
Indeed, some strains have become resistant to practically all of the commonly available agents. A notorious case is the methicillin-resistant Staphylococcus aureus (MRSA), which is resistant not only to methicillin (which was developed to fight against penicillinase-producing S. aureus) but usually also to aminoglycosides, macrolides, tetracycline, chloramphenicol, and lincosamides. Such strains are also resistant to disinfectants, and MRSA can act as a major source of hospital-acquired infections. An old antibiotic, vancomycin, was resurrected for treatment of MRSA infections. However, transferable resistance to vancomycin is now quite common in Enterococcus and found its way finally to MRSA in 2002, although such strains are still rare (1).
An even more serious threat may be the emergence of gram-negative pathogens that are resistant to essentially all of the available agents (2). Research had time to react against the threat by MRSA. Thus, there are newly developed agents that are active against vancomycin-resistant MRSA, such as linezolid and quinupristin/dalfopristin. However, the emergence of “pan-resistant” gram-negative strains, notably those belonging to Pseudomonas aeruginosa and Acinetobacter baumanii, occurred more recently, after most major pharmaceutical companies stopped the development of new antibacterial agents. Hence, there are almost no agents that could be used against these strains, in which an outer membrane barrier of low permeability and an array of efficient multidrug efflux pumps are combined with multitudes of specific resistance mechanisms.
Multidrug resistance in bacteria occurs by the accumulation, on resistance (R) plasmids or transposons, of genes, with each coding for resistance to a specific agent, and/or by the action of multidrug efflux pumps, each of which can pump out more than one drug type.
BIOCHEMICAL MECHANISMS OF RESISTANCE
It is first necessary for us to review briefly the commonly encountered mechanisms of resistance.
Mutational Alteration of the Target Protein
Man-made compounds, such as fluoroquinolones, are unlikely to become inactivated by the enzymatic mechanisms described below. However, bacteria can still become resistant through mutations that make the target protein less susceptible to the agent. Fluoroquinolone resistance is mainly (but not exclusively) due to mutations in the target enzymes, DNA topoisomerases (3). Whether resistance of this type is easily transferred to other cells on plasmids depends on the mode of the drug's action. With fluoroquinolones, which will kill the bacterial cell containing any drug-susceptible enzyme, the addition of the gene coding for a drug-resistant enzyme will not make the bacteria completely resistant, and plasmid-mediated transfer of the mutated target gene is unlikely to occur. Nevertheless, the mutants will become more and more prevalent by clonal selection in the presence of selective pressure.
When high-level streptomycin resistance is selected in Escherichia coli in the laboratory, mutation in one of the ribosomal proteins (RpsL) is usually selected. Again, bringing in this mutant gene on a plasmid will not make the recipient resistant because the drug action on the susceptible ribosome (containing the chromosomally coded intact RpsL) will result in cell death. This type of resistance is almost never found in the streptomycin-resistant E. coli of clinical origin, presumably because resistance caused by the drug modification (see below) is effectively expressed on plasmids. However, ribosomal resistance mutation is often found in the aminoglycoside-resistant clinical strains of Mycobacterium tuberculosis.
Another example of resistance attributable to target modification is that conferred by the erm gene, which is usually plasmid coded and produces the methylation of adenine at position 2058 of the 50S rRNA, causing resistance to macrolides (erythromycin and many others), lincosamide, and streptogramin of group B, the MLS phenotype (4). The molecular basis of this phenotype was elucidated through the crystal structure of 50S ribosomal subunit (5).
Sulfa drugs (synthetic competitors of p-aminobenzoic acids that inhibit dihydropteroate synthetase and trimethoprim, a synthetic inhibitor of dihydrofolate reductase) have been used in combination. They select for drug-resistant mutants of the respective enzymes. In this case, the high-level production of drug-resistant target enzymes from plasmids can make the bacteria resistant, and the resistant genes have spread widely on plasmids (6).
Enzymatic Inactivation of the Drug
This is a common resistance mechanism for antibiotics of natural origin, such as aminoglycosides (kanamycin, tobramycin, and amikacin), which are inactivated by enzymatic phosphorylation [by aminoglycoside phosphoryltransferase (APH)], acetylation [by aminoglycoside acetyltransferase (AAC)], or adenylation (by aminoglycoside adenyltransferase or nucleotidyltransferase), and β-lactams (penicillins, cephalosporins, and carbapenems such as imipenem), which are inactivated by enzymatic hydrolysis by β-lactamases, usually in the periplasm. Genes coding for these inactivating enzymes can easily produce resistance as additional genetic components on plasmids.
Aminoglycosides
Aminoglycosides are inactivated by modifications that reduce the net positive charges on these polycationic antibiotics (7, 8). There are now many dozens of aminoglycoside-modifying enzymes known; for example, AAC (3)-II designates an aminoglycoside acetyltransferase acting on position 3 of the substrate and belonging to the second phylogenic grouping among these enzymes (9). It was proposed in 1973 (10) that a major source of antibiotic-inactivating enzymes is the antibiotic-producing microorganisms, which must protect themselves. Indeed APH (3′)-I, a plasmid-coded aminoglycoside phosphorylase, which is present in 46% of the aminoglycoside-resistant gram-negative bacteria, is strongly related to a chromosomally coded enzyme in the producing strain Streptomyces fradiae (9). A similar situation also exists for AAC (3)-II, a plasmid-coded enzyme present in more than 60% of aminoglycoside-resistant gram-negative bacteria examined (9).
After these findings, the pharmaceutical industry developed either semisynthetic or natural aminoglycosides that are not the substrates of common inactivating enzymes. Nevertheless, these drugs are still susceptible to the action of some enzymes, and plasmids found in aminoglycoside-resistant strains in recent years often contain genes coding for several inactivating enzymes (11).
β-Lactams
Only a few years after the introduction of penicillin into clinical practice, S. aureus developed resistance caused by a β-lactamase coded for it by a plasmid gene. Although this problem was solved by the introduction of methicillin and similar compounds that resist the enzymatic hydrolysis, another enzyme, TEM β-lactamase, was reported in gram-negative bacteria in strains containing multiple-drug-resistant R plasmids that date from 1962 (12). This enzyme became widespread throughout the world, making penicillins with gram-negative activity, such as ampicillin, almost useless. (Methicillin and its relatives are inactive against gram-negative bacteria because they are pumped out efficiently by the multidrug efflux pump; see below.) β-Lactamases are classified into several phylogenetic families. Class A includes both the S. aureus and TEM enzymes, whereas Class C represents chromosomally coded enzymes (e.g., AmpC) that are present in many gram-negative bacteria. These two classes are both similar to serine proteases in their mechanism, whereas Class B enzymes are metalloenzymes that hydrolyze carbapenems efficiently.
In response to the spread of β-lactam resistance, various β-lactams were developed. Although the first-generation cephalosporins, such as cephaloridine and cefazolin, were rapidly hydrolyzed by both TEM and AmpC, both cephamycins (such as cefoxitin) and the third-generation cephalosporins containing an oxyimino side chain (such as cefotaxime) were initially reported to resist both types of enzymes. However, the former was inactive against some gram-negative bacteria such as Enterobacter and Serratia. Although the latter was capable of killing these organisms, their introduction into the clinics was followed by the emergence of resistant strains that over-produced the chromosomal AmpC enzyme. In fact, the AmpC enzymes have very low KM values for these compounds, and the values of Vmax/KM were quite high (13). The AmpC enzyme, however, needs to be induced, and the third-generation cephalosporins were effective against these bacteria simply because they were ineffective inducers of this enzyme. Thus, the third-generation cephalosporins selected for constitutive mutants of ampC. Furthermore, strong expression of plasmid-coded AmpC has been found recently in species that do not express the chromosomally coded ampC.
Subsequently, fourth-generation cephalosporins (cefepime, cefpirome) that are more resistant to hydrolysis by the AmpC enzyme have been developed. However, continued selective pressure resulted in the selection of plasmids that produced mutants of common enzymes, such as TEM or its relative SHV, which can now hydrolyze third- and sometimes even fourth-generation cephalosporins. These enzymes are called ESBL (extended spectrum β-lactamases) (14).
Especially troublesome among the ESBL enzymes are those called CTX-M (15). The genes coding for these enzymes appear to have originated from the chromosome of an infrequently encountered gram-negative bacterium Kluyvera and have transferred to R plasmids. This transfer or mobilization unusually appears to have occurred many times, and consequently the enzyme rapidly became widespread among R-plasmid-containing pathogenic bacteria (16).
β-Lactams with a new nucleus, such as carbapenems (e.g., imipenem), still remain quite effective, but their use may eventually result in the increased prevalence of enzymes capable of hydrolyzing these compounds (17).
Another example of new resistance mechanisms arising through mutations of prevalent resistance genes is a plasmid-coded ciprofloxacin resistance gene (18). Here an aminoglycoside acetylase, AAC (6′)-Ib, has mutated to acetylate the amino group of ciprofloxacin.
Acquisition of Genes for Less Susceptible Target Proteins from Other Species
Sequencing of the genes coding for the targets of penicillin, DD-transpeptidase or penicillin-binding proteins (PBPs), revealed that penicillin resistance among Streptococcus pneumoniae was due to the production of mosaic proteins, parts of which came from other organisms (19). We note that S. pneumoniae is an organism capable of natural transformation and may import foreign DNA. Interestingly, a similar mechanism of penicillin resistance was also found in another organism capable of natural transformation, Neisseria meningitidis.
An extreme case of this scenario is the generation of MRSA. MRSA strains contain a new methicillin-resistant PBP, called PBP-2A or 2′, whose expression is often induced by methicillin and other β-lactams. The gene for this new PBP is located in a large (30–60-kb) segment of DNA, which apparently came from an organism other than S. aureus (1, 20) and also contains other antibiotic resistance genes. S. aureus is not naturally transformable, and it is unclear how this horizontal transfer of a large DNA segment occurred.
Bypassing of the Target
Vancomycin, a fermentation product from streptomycetes, has an unusual mode of action. Instead of inhibiting an enzyme, it binds to a substrate, the lipid-linked disaccharidepentapeptide, a precursor of cell wall peptidoglycan. Because of this mechanism, many assumed that it would be impossible to generate resistance against vancomycin. However, vancomycin resistance is now prevalent among enterococci, normal inhabitants of our intestinal tract. Because enterococci are naturally resistant to β-lactams, aminoglycosides, macrolides, and tetracycline, these vancomycin-resistant strains of enterococci become prevalent in a hospital environment, colonize the patients, and cause infections that are difficult to treat.
Study of the resistance mechanism showed that the end of the pentapeptide, D-Ala-D-Ala, where vancomycin binds, was replaced in the resistant strain by an ester structure, D-Ala-D-lactic acid, which is not bound by vancomycin (160). Production of this altered structure requires the participation of several imported genes.
Preventing Drug Access to Targets
Drug access to the target can be reduced locally. It can be also reduced by an active efflux process. In gram-negative bacteria, the access can be reduced generally by decreasing the influx across the outer membrane barrier.
Local inhibition of drug access
Tet(M) or Tet(S) proteins, produced by plasmid-coded genes in gram-positive bacteria, bind to ribosomes with high affinity and apparently change the ribosomal conformation, thereby preventing the association of tetracyclines to ribosomes (21). Plasmid-coded Qnr proteins, which have become more prevalent in recent years, are thought to protect DNA topoisomerases from (fluoro)quinolones (22).
Drug-specific efflux pumps
Drug resistance owing to active efflux was discovered with the common tetracycline resistance protein TetA in gram-negative bacteria (23), which catalyze a proton-motive-force-dependent outward pumping of a tetracycine-Mg complex (24).
Nonspecific inhibition of drug access
In the laboratory where nutrient-rich medium is used, β-lactams often select for porin-deficient mutants (25). However, the generally decreased outer membrane permeability is somewhat detrimental to the bacterial growth because the nutrient influx is also reduced, and such mutants are not common among clinical specimens. Nevertheless, porin mutants are found in some species of Enterobacteriaceae (Enterobacter aerogenes, Klebsiella pneumoniae) as a means of last resort resistance to the more recent versions of β-lactams that withstand inactivation by common β-lactamases (26). Mutations within the coding sequences of the porin also have been reported, which possibly reduce the permeation rates of bulky β-lactams without affecting those of smaller nutrient molecules (26). The multidrug efflux pumps are discussed below.
SOURCES OF THE RESISTANCE GENES
There are so many resistance genes that rely on many different mechanisms. Where did they come from?
Producing Organisms
As described above, some of the aminoglycoside-resistant genes appear to be derived from streptomycetes producing these antibiotics. The genes coding for vancomycin resistance appear to originate in a similar way. Resistance here requires the production of several new enzymes, and it is unlikely that the genes coding for these enzymes evolved in the few decades after vancomycin was introduced. Indeed, the genes in the vancomycin-resistant clinical isolates of enterococci were found to be homologs of those found in the vancomycin-producing streptomycetes, organized in exactly the same manner (27), an observation that leaves no doubt on the origin of these resistance genes.
Microorganisms in the Environment, Especially Soil
Some resistance genes are found in the chromosome of environmental bacteria. A classical case is the ampC gene in the environmental genera of Enterobacteriaceae, such as Enterobacter, Serratia, and Proteus, and in the soil organism P. aeruginosa. These genes do not show signs that they have been imported in the recent past, and it is revealing that the ampC gene of the exclusive animal symbiont E. coli lacks the induction mechanism and that the pathogen Salmonella spp. lacks the ampC gene entirely. In this connection, examination of a random collection of soil-dwelling strains of Streptomyces and their relatives showed that 60% to 100% of them were resistant to several antibiotics tested, suggesting that antibiotic-resistant genes are abundantly present in this habitat (28). Although most antibiotics may be present in soil only at very low concentrations, the recent discovery of microorganisms that utilize antibiotics as nutrients (29) is suggestive of the evolutionary origin of some antibiotic degradation (resistance) genes.
ASSEMBLY, MAINTENANCE, AND TRANSFER OF RESISTANCE GENES
R plasmids often contain many resistance genes; they are maintained stably in the host strains of bacteria and are transferred very efficiently to neighboring drug-susceptible cells. How do all these processes occur?
Assembly of Resistance Genes in R Plasmids
We have seen that most drug resistance genes are effective when expressed from plasmids. Remarkably, many such genes are often present on a single R plasmid, so that multidrug resistance can be transferred to a susceptible bacterium in a single conjugation event. When the R plasmids were discovered in Japan in the 1950s, many of them already contained resistance genes for aminoglycosides, tetracycline, chloramphenicol, and sulfonamides. Sequencing of many plasmids has now shown how this clustering occurred.
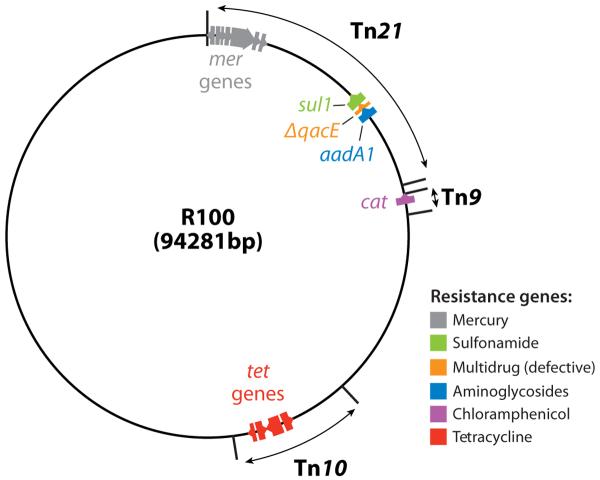
In the sequence of the early-generation R plasmids, we see that most of the resistance genes are components of transposons, which can deliver the genes to any piece of DNA. This is seen in the plasmid R100 (Figure 1).
Figure 1.
Map of an early R plasmid R100. Tetracycline resistance gene tetA is in the transposon Tn10, chloramphenicol acetyltransferase (cat) as a part of Tn9, and sulfonamide resistance gene sul1 and an aminoglycoside adenyltransferase gene aadA1 as a part of the large transposon Tn21. The resistance genes are color coded, and the direction of transcription is shown by the arrows. Much of the blank areas outside the transposons are occupied by the transfer genes, needed for the formation of relaxosomes, the coupling factor, and the mating-pair formation. This figure is based on the nucleotide sequence deposited by (30) and GenBank sequence NC 002134, submitted by G. Sempei and K. Mizobuchi. ΔqacE is a defective version of a gene coding for a multidrug efflux pump of Smr family (see text).
Tn21 is a particularly remarkable example of large, complex, multiply composite transposons (31). Interestingly, it contains mercury resistance genes. The sulfonamide resistance and aminoglycoside resistance genes in Tn21 were assembled as described below.
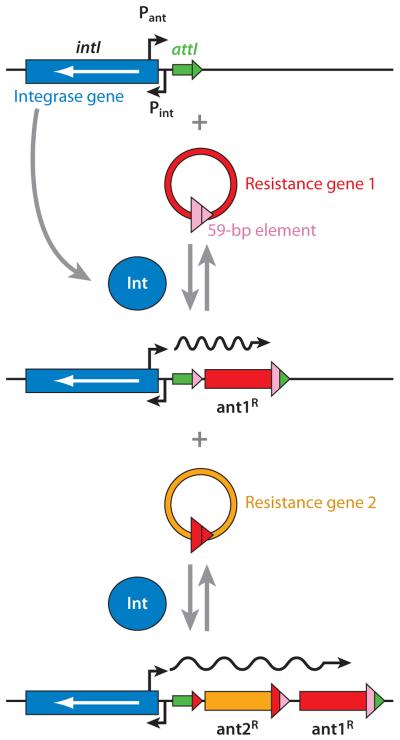
The discovery that many resistance genes in R plasmids contain a unique 59-base 3′-sequence tag led to the discovery of a remarkable apparatus called an integron (32). An integron contains a gene coding for an integrase, which catalyzes the insertion of resistance genes at a predetermined site downstream from a strong promoter (Figure 2). Once integrated, the resistance gene becomes marked by the tag, so that it can easily become integrated into another integron, perhaps containing a different set of resistance genes. In addition to this advantage of high mobility, the resistance genes when inserted into an integron become organized into a single operon, with the same orientation of transcription under a strong promoter supplied by the integron structure. In the example seen in Tn21, the integron already containing a sulfonamide resistance gene sul1 and a truncated version of a multidrug efflux gene qacE has integrated an aminoglycoside resistance gene aadA1 at the specific integration site attI. An integron may contain up to eight resistance genes (33).
Figure 2.
Presumed mechanism of resistance gene capture by integrons. An integron contains the gene for site-specific integrase (blue) and the specific integration site attI (green). When the resistance gene 1 (red) in the circular cassette form containing the 3′-terminal 59-bp element (pink) is available, the gene is integrated at the attI site, regenerating a slightly altered attI sequence (now in green and pink). This can then accept the second resistance gene casette, and the process can go on in this manner. The resistance genes, all in the same orientation, are transcribed by the powerful promoter provided by the integron (Pant). From Reference 159 with permission from Elsevier Ltd.
In addition, many of these integrons carry enzymatic machinery for transposing the entire integron structure to other places. Furthermore, the integron is usually inserted into a larger transposon such as Tn21, which allows the entire array of the resistance genes to hop between different plasmids and between plasmid and chromosome. It is unclear how the modern integrons evolved, but there is a rather similar mechanism that builds the assembly of many genes in Vibrio cholerae (34).
Recently, many integron structures were found to be associated with a downstream structure called ISCR element, containing a putative transposase gene (35). It apparently functions in an unusual, open-ended transposition event and recruits various resistance genes and delivers them close to the integron structure, resulting in the assembly of yet more resistance genes.
Maintenance of R Plasmids in the Host Cells
Recombinant plasmids derived from cloning vectors are frequently lost from the host cells even when they exist in relatively high copy numbers. In contrast, most natural R plasmids are remarkably stable and are rarely lost during the multiplication of host cells, even when the copy number is low. This is because R plasmids usually contain genes that ensure the correct partitioning of copies to daughter cells (36). In addition, some natural plasmids contain the “killer” elements composed of a stable killer protein or mRNA and an unstable inhibitor protein or antisense RNA, so that the loss of the plasmids will result in the death of the host cell (36).
Because of the presence of these regulatory networks, plasmids sharing a similar replication/partition machinery cannot stably coexist in the same host cell, and this phenomenon has been utilized for the classification of plasmids into “incompatibility groups.” In plasmids of enteric bacteria, this classification includes about two dozen such groups (37, 38).
Cell-to-Cell Transfer of R Plasmids
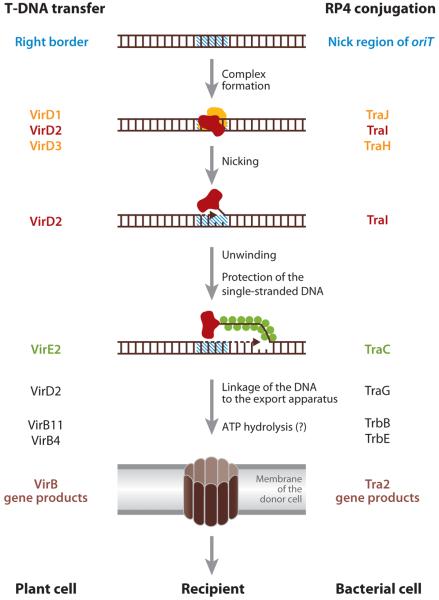
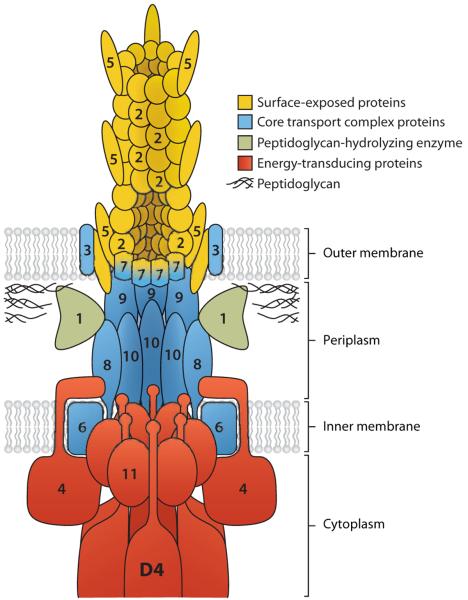
R plasmids are not only stably maintained, but also usually transferred between bacterial cells at a very high efficiency, in many cases approaching 100%. The molecular mechanism of this cell-to-cell transfer has been studied mainly in E. coli and its relatives and in the plasmids of incompatibility group FI (F or sex plasmid), P (RP4), and W. In recent years, the plasmid transfer genes were found to be homologs of the virB genes of Agrobacterium tumefaciens, which transfer a piece of bacterial DNA into a plant nucleus, and several effector protein injection systems of human and animal pathogens (39, 40). These systems are now called Type IV Secretion Systems. A specific sequence in the circular, double-stranded DNA of plasmids is recognized, and a single-stranded cut is made by the VirD2/TraI enzyme (Figure 3), with the enzyme protein remaining covalently attached to the 5′-end of the cut strand. This complex is sometimes called a relaxosome, as the nicking of one strand results in the relaxation of supercoiling of the plasmid DNA. The protein-(single-stranded) DNA complex is then exported by the “mating-pair formation” complex, composed of about a dozen different proteins (Figure 4). The export of relaxosomes, rather than protein-free DNA, explains the observation that some Type IV Secretion Systems export pure proteins. Before this happens, however, the leading 5–-end of the exported DNA, with the VirD2/TraI protein attached, must be taken up by an ATPase (VirD4 or TraG). The latter is called a coupling protein because of its role in coupling the export substrate to the export apparatus (Figure 4). Although one of the original strands of the R plasmid DNA is transferred completely into the recipient cells, in the donor cell this strand is replaced by a rolling-circle replication mechanism, so that the complete R plasmids now exist both in donor and recipient cells. One would suspect that the entry of the DNA of foreign origin would be recognized by the strain-specific restriction endonuclease of the recipient cells. However, the initial DNA piece that enters is single stranded (although the complementary strand will be synthesized soon) and thus may escape this mechanism. In addition, some R plasmids are equipped with functions that antagonize the attack by restriction endonucleases (41).
Figure 3.
Steps in T-DNA transfer in Agrobacterium tumefaciens (annotation on the left) and conjugative plasmid DNA transfer in enteric bacteria (annotation on the right). From Reference 41 with permission from Elsevier Ltd.
Figure 4.
A model of the mating-pair formation complex with the coupling protein (VirD4). The proteins are named after the VirB components in Agrobacterium tumefaciens, but homologs for most of these proteins are found in the conjugative R plasmid transfer systems. Number n denotes the product of virBn gene. The virD4 product is shown as D4. From Reference 39 with permission from Elsevier Ltd.
Some R plasmids are very small and do not contain the genes needed for the construction of a mating-pair formation complex. However, these plasmids can be mobilized by the transfer genes of other plasmids and can be transferred efficiently into recipient cells. Finally, it should be emphasized that conjugational transfer of plasmids is not a novel mechanism invented for multidrug resistance; indeed, horizontal transfer of genes, to a large extent through the conjugational mechanism, has been the main mechanism used for the evolution of many groups of bacteria (42).
Gram-positive bacteria also carry out cell-to-cell transfer of plasmids (43). In addition, a large transposon of a special class, called conjugative transposons, play a prominent role in these bacteria (44). They use a λ-phage-like mechanism of transposition, which generates a circular DNA as an intermediate. This intermediate can insert at a predetermined location in the plasmid or chromosomal DNA, but it can also behave like a conjugative plasmid and can transfer its copy into another bacterium by conjugation. Remarkably, in the transposons containing the tetracycline resistance gene, the transfer function is induced strongly by the presence of tetracycline. Thus, in these transposons, the drug resistance genes are not just passive passengers but are a part of an integrated regulatory mechanism of conjugational transfer. Such transfer also occurs easily between distantly related organisms, for example between gram-positive and gram-negative bacteria (45).
MULTIDRUG EFFLUX PUMPS
The active efflux of drugs has been known to play a predominant role in the resistance to certain individual drugs, such as tetracycline, as described above. Analysis of S. aureus strains that were resistant to multiple cationic bacteriocides and were causing hospital-acquired infections showed, however, that these strains contained plasmids coding for a multidrug efflux transporter QacA (or QacB), belonging to the Major Facilitator superfamily (MFS) (46), the first multidrug efflux pump identified in bacteria. Since then, multidrug efflux pumps belonging to various families have been discovered, and the contribution of such pumps to multidrug resistance became clear (47, 48). Structures and possible mechanisms of these transporters have been reviewed recently (49)
Multidrug Efflux Pumps Belonging to the Major Facilitator Superfamily
MFS is one of the largest families of transporters and contains many important efflux pumps.
MFS Pumps with 14 transmembrane segments
QacA and QacB were the first examples of this class (46). These pumps actively extrude monocationic biocides and dyes, such as benzalkonium chloride, cetyltrimethylammonium bromide, and ethidium bromide, and in addition, QacA pumps out dicationic biocides, such as chlorhexidine and pentamidine isethionate. Each protein contains 14 transmembrane segments (TMSs) with several acidic amino acid residues in the transmembrane region. Comparison of these two protein sequences and site-directed mutagenesis showed that the extrusion of dicationic compounds by QacA, but not by QacB, depended upon the presence of an aspartate residue in the TMS 10.
The expression of QacA pump is negatively regulated by the QacR repressor, and binding of some of the QacA substrates to QacR relieves this repression, resulting in the increased expression of the pump. Although QacA has not been crystallized, crystallographic studies of the QacR-inducer complex (50) gave us insights on how a binding site can accommodate diverse ligands. Thus, the QacR drug-binding pocket is large (1100Å3), and many drugs bind to one of the “minipockets” within this cavity. Unlike the binding of most ligands to conventional enzymes, the binding involves few hydrogen bonds and relies mostly on stacking and van der Waals interactions, with the charges of the ligands usually neutralized by acidic residues. These features are similar to the binding of ligands to the human pregnane X receptor, a transcriptional regulator that binds to a number of xenobiotics (51). QacR can bind two ligands simultaneously to different minipockets of the binding site (52), and a given ligand can be bound in different ways within the binding cavity (53). Another regulatory protein, TtgR, belonging to the same family as QacR but regulating a transporter in the Resistance-Nodulation-Division (RND) family (see below), was crystallized with several antibiotics and plant secondary metabolites (54). The binding pocket here is even larger (1500Å3), and some ligands utilize the broad hydrophobic area of the pocket, whereas phloretin in its high-affinity mode binds to the deep valley within the pocket, utilizing several H-bonding interactions, consistent with the extremely wide range of ligands that regulate the expression of the cognate RND pump (54).
Another multidrug transporter belonging to this branch is EmrB of E. coli, which confers resistance to uncouplers such as carbonyl cyanide m-chlorophenylhydrazone and to antibiotics such as nalidixic acid and thiolactomycin (46, 55). EmrB is coded for by a chromosomal gene, together with EmrA, a periplasmic adaptor protein that connects the pump to an outer membrane channel, TolC, so that the drugs can be exported directly into the medium, as discussed below in the section on RND family pumps.
MFS Pumps with 12 TMSs
The chromosomally coded NorA of S. aureus, in contrast, belongs to a branch with 12 TMS transporters (46). NorA produces resistance to fluoroquinolones, as well as to cationic dyes and cationic inhibitors including puromycin and tetraphenylphosphonium (56). The S. aureus chromosome contains at least two other homologs of NorA, NorB and NorC, which produce a similar phenotype (57). Because all these efflux pumps are inhibited by reserpine, sensitization of bacteria to substrate drugs in the presence of reserpine can be used as an effective tool to assess the contribution of the efflux process to resistance. Thus, in about one-half of S. aureus strains isolated from the bloodstream of patients, efflux made a strong contribution to resistance, and more than half of these strains overexpressed the three chromosomally coded pumps just mentioned (58).
Some MFS pumps have been studied as model systems, although they have little relevance in the clinic. LmrP of Lactococcus lactis pumps out cationic dyes, daunomycin, tetracyclines, and macrolides (59), and MdfA of E. coli can confer increased resistance to cationic dyes, chloramphenicol, and fluoroquinolones when overexpressed from plasmids in a mutant strain lacking the constitutive RND pump AcrB (60), although mdfA deletion produces no effect on drug susceptibility (61). Study of the LmrP pump showed that fluorescent dyes are captured from the inner leaflet of the bilayer, the pump thus acting as a “vacuum cleaner” of the membrane. The MdfA pump is remarkably versatile, for example, pumping out neutral substrates in an electrogenic manner while treating cationic substrates in an electroneutral way. Another unexpected finding was that the mdfA deletion mutants were hypersensitive to alkaline pH, presumably because MdfA functions as a K+/H+ antiporter (161). This is reminiscent of the similar function for a chromosomally coded “monodrug efflux pump,” TetB(L), of Bacillus subtilis (62).
Among MFS multidrug efflux pumps, the crystal structure was solved for EmrD of E. coli (63), which pumps out uncouplers, quaternary ammonium biocides, as well as sodium dodecylsulfate (60). The central cavity, a likely substrate-binding site, is lined with side chains of both an aliphatic and an aromatic nature.
Multidrug Efflux Pumps of the Small Multidrug Resistance Family
Efflux pumps of a very different structure, belonging to the Small Multidrug Resistance (SMR) family (64), were first discovered coded on S. aureus plasmids and were subsequently found to be coded also on the chromosomes of gram-negative bacteria. These proteins, which extrude cationic compounds such as quaternary ammonium biocides or ethidium, represent the smallest transport proteins known, containing only four TMS in a 110-residue sequence in the EmrE of E. coli. EmrE has been studied intensively (65). There is only one charged residue in the transmembrane segments, Glu14, which has an unusually high pKa of about 8.5. When the transporter encounters the substrate, this residue becomes deprotonated and instead binds the substrate, as shown with the transporter solution in detergents. Thus, the inward flux of proton(s) and the outward flux of the substrate appear to be coupled by the sharing of a common binding site. In terms of the three-dimensional structure, EmrE seems to exist primarily as a symmetrical dimer, according to the biochemical (66) and the electron cryomicroscopy data (67). Although much confusion ensued following the publications suggesting that the dimer is asymmetric, with one protomer facing the outside and the other facing inside, these papers have now been retracted as a result of calculations using a faulty software.
Multidrug Efflux Pumps of Resistance-Nodulation-Division Family
Transporters of this family play, by far, a predominant role in the multidrug resistance of gram-negative bacteria.
Resistance-nodulation-division pump usually exists as a part of a tripartite complex
Efflux pumps of this superfamily (such as AcrB of E. coli and MexB of P. aeruginosa) (68) play an important role in producing multidrug resistance in gram-negative bacteria. This is because these pumps become associated with two other classes of proteins, the outer membrane channel exemplified by TolC of E. coli, belonging to the OMF (outer membrane factor) family of proteins (69) and the periplasmic “adapter” protein such as AcrA of E. coli and MexA of P. aeruginosa, classified into the MFP (membrane fusion protein) family (70) (Figure 5a). The association of these proteins was confirmed by chemical cross-linking (71) and by the isolation of the complex containing all three proteins (72). As was emphasized already in 1996 (73), this construction allows the direct export of drugs into the external medium, rather than into the periplasmic space. This is a huge advantage because once exported into the external space drug molecules must traverse the outer membrane barrier to reenter bacterial cells. Thus, these pumps work synergistically with the outer membrane barrier. Wild-type strains of most gram-negative bacteria are resistant to most lipophilic antibiotics (for E. coli, they include penicillin G, oxacillin, cloxacillin, nafcillin, macrolides, novobiocin, linezolid, and fusidic acid), and this “intrinsic resistance” was often thought to be caused by the exclusion of drugs by the outer membrane barrier. Indeed, breaching the outer membrane barrier does sensitize E. coli cells to the drugs just mentioned (74). However, the inactivation of the major RND pump AcrB of E. coli makes the bacteria almost completely susceptible to these agents [the minimal inhibitory concentration (MIC) of a lipophilic penicillin, cloxacillin, goes down from 512 μg/ml in the wild type to only 2 μg/ml (75)] even in the presence of the intact outer membrane barrier. Thus, the characteristic intrinsic resistance of gram-negative bacteria owes as much to the RND pumps as to the outer membrane barrier.
Figure 5.
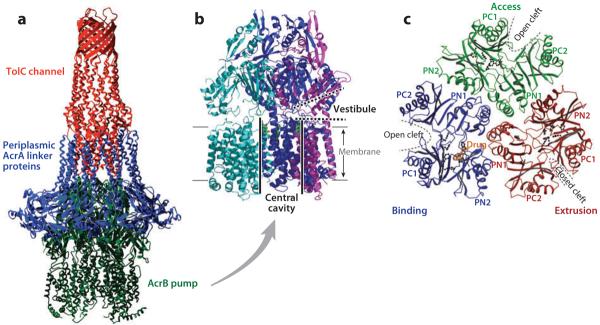
(a) The model of the AcrB-AcrA-TolC tripartite complex that exports drugs directly into the medium. The transmembrane domain of the AcrB pump trimer is embedded in the cytoplasmic membrane, whereas its periplasmic domain is connected to the TolC channel through a number of periplasmic AcrA linker proteins. From Reference 105 with permission from Elsevier Ltd. (b) AcrB trimer. Each protomer is shown in a different color. The large central cavity (thick black lines) is connected to the periplasm through vestibules (thick dotted lines) between protomers. The proximal portion of the structure was cut away to reveal the presence of vestibule. Drawn by using PyMol with Protein Data Bank coordinate file 1OYE. (c) The periplasmic domain of the asymmetric AcrB trimer viewed from the top. The conformation of each protomer is characteristic, with open or closed external clefts. A drug molecule is seen bound to the binding protomer. From Reference 113 with permission from Nature Publishing Group.
Some RND pumps show extremely wide substrate specificity. E. coli AcrB can pump out not only most of the common antibiotics but also dyes, detergents, and even solvents (Table 1). Although AcrB cannot pump out aminoglycosides, there are homologs such as AcrD that carry out this function. Most gram-negative bacteria contain several chromosomal genes coding for such pumps, and their expression may become increased through regulatory responses or mutations, making the bacteria more resistant to practically all antimicrobial agents in one single step.
Table 1.
Properties of some Resistance-Nodulation-Division (RND) multidrug efflux pumpsa
| Organism | RND pump | Associates with: | Substratesb | Regulator | |
|---|---|---|---|---|---|
| MFP | OMF | ||||
| Escherichia coli | AcrB | AcrA | TolC | AC, BS, CM, CP, CV, EB, FA, FQ, ML, NO, OS, PN, SDS, TC, TM, TR, TX | AcrR, MarA, SoxS, Rob, SdiA |
| E. coli | AcrD | AcrA | TolC | AG, DC, FU, NO | BaeR |
| Pseudomonas aeruginosa | MexB | MexA | OprM | AC, AG, CM, CP, CV, EB, ML, NO, OS, PN, SDS, SF, TC, TM, TR | MexR, NalC (PA3721), NalD |
| P. aeruginosa | MexD | MexC | OprJ | CM, CP, FQ, ML, NO, TC, TR | NfxB |
| P. aeruginosa | MexF | MexE | OprN | CM, FQ, TC | MexT |
| P. aeruginosa | MexY | MexX | OprM | AG, FQ, ML, TC | MexZ |
Properties of well-studied pumps are shown together with the periplasmic adaptor protein (MFP) and the outer membrane channel (OMF) they interact with. Sources for the data can be found in (47, 79) as well as in the text.
Abbreviations for substrates: AC, acriflavine; AG, aminoglycosides; BS, bile salts; CM, chloramphenicol; CP, cephalosporins; CV, crystal violet; DC, deoxycholate; EB, ethidium bromide; FA, fatty acids; FQ, fluoroquinolines; FU, fusidic acid: ML, macrolides; NO, novobiocin; OS, organic solvents; PN, penicillins; SDS, sodium dodecylsulfate; SF, sulfonamides: TC, tetracyclines; TM, trimethoprim; TR, triclosan; TX, Triton X-100.
Finally, at least some RND pumps appear to capture the drugs either in the periplasmic space or from a location that is in equilibrium with the periplasm. This was predicted (73) because carbenicillin, which cannot cross the inner (cytoplasmic) membrane owing to the presence of two carboxylate groups, is still a good substrate for these pumps. This allows the pumps not only to prevent the entry of drugs into the cytoplasm, but also to extrude agents, such as β-lactams, that have targets in the periplasm. Consistent with this concept, the substrate specificity of RND pumps is primarily determined by their large periplasmic, extramembrane domains, as shown by study of chimeric transporters (76) or mutants with altered substrate specificity (77). Importantly, the periplasmic capture mode allows the RND tripartite pumps to collaborate synergistically with simple efflux pumps that extrude substrate drugs only into the periplasm (78). A review describes pumps in P. aeruginosa (79).
Regulation of RND pump expression
Expression of many RND pumps is repressed by specific repressor proteins (47, 80). Sometimes, the pump expression is increased by substrates, for example, MexXY, an aminoglycoside efflux pump of P. aeruginosa, is induced by its substrates (81). However, the mechanism of induction is more complex than with that of QacA, described above, where the repressor QacR is inactivated by the binding of the inducer drug. Thus, induction requires the inhibition of ribosomal function, which is not necessarily caused by aminoglycosides (82), and apparently involves another protein (83).
With most other systems, the pump gene expression is not regulated by the substrates, and it is not known what ligands, if any, become bound to the local repressors. Thus, the efflux pump overproduction in clinical isolates occurs frequently as a result of mutation in the repressor gene, as with mexR mutations leading to overproduction of MexAB-OprM system in P. aeruginosa. A fraction of such MexAB-overproducing strains has no changes in the mexR gene and has mutations in the gene PA3721, the so-called nalC phenotype (84). Finally, the nalD gene, located upstream from the mexAB-oprM operon, also is involved in regulation (83). The mexCD-oprJ operon is again under the control of the NfxB repressor, coded by an upstream gene, and the pump overproducers have mutations in the repressor gene (85). The mexEF-oprN operon is exceptional in being regulated by an upstream activator, MexT. MexEF-OprN-overproducing strains do not contain mutations in the mexT gene, and instead have mutations in PA2491, a gene coding for an enzyme-like molecule (86).
The expression of acrAB operon in E. coli is repressed by its cognate repressor AcrR, but the small effector molecules that bind to this repressor are not known. Instead, the transcription of the acrAB operon is mainly controlled by the AraC family global activators MarA, SoxS, and Rob (47, 80). MarA and SoxS are exceptionally small proteins containing only the DNA-binding domain, and thus, regulation occurs at the level of production of these proteins. The MarA production is regulated by a repressor MarR, which becomes inactivated by salicylate and plumbagin. Interestingly, transketolase A, whose production is elevated under oxidative stress, binds to MarR and relieves its repression of MarA production (87). The level of active SoxS repressor in the cell is determined by its own repressor SoxR, which contains Fe-S centers and becomes inactivated by superoxide. Rob is constitutively expressed and is also much larger than MarA and SoxS, containing a domain outside the DNA-binding domain. Indeed, Rob becomes inactivated by binding substrate-like molecules, such as α,α′-dipyridyl (88), fatty acids, and bile salts (89).
Expression of some RND-type transporters, such as MdtBC (90, 91), is regulated by a two-component system, BaeSR. Indole upregulates MdtBC as well as AcrD through BaeSR (92), although it is unclear if indole is directly sensed by the sensor kinase BaeS.
Plasmid-driven overproduction of SdiA, the soluble receptor for the quorum-sensing signal acyl-homoserine lactone, increased expression of AcrAB (93). However, growth of E. coli in the presence of hexanoyl-homoserine lactone did not upregulate the acrAB operon (94).
Biochemical and crystallographic studies
Biochemical studies of the RND-type efflux pumps were hampered by the fact that the pumps function as multiprotein complexes spanning two membranes. Nevertheless, the basic kinetic constants of the AcrB pump were recently determined (162). When β-lactams are added to intact E. coli cells, they diffuse across the outer membrane and, in periplasm, are either hydrolyzed by the periplasmic β-lactamase or extruded by the AcrAB-TolC pump. Because the kinetic constants of the β-lactamase are known, we can calculate the periplasmic drug concentration from the hydrolysis rate. The efflux rate at that periplasmic concentration is obtained as the difference between the rate of influx (following Fick's first law of diffusion) and the hydrolysis rate. Data with nitrocefin suggests saturation kinetics of efflux with a half-saturation concentration of ~5 μM and a turnover number about 10. Although nitrocefin did not show positive cooperativity, such a behavior was evident with cephalothin, cephamandole, and cephaloridine, which showed much higher K0.5 values than nitrocefin did. The positive cooperativity is consistent with the functionally rotating mechanism of AcrB function, described below.
Another biochemical approach used was the reconstitution of purified RND transporter into proteoliposomes. Because most of the substrates of these pumps are lipophilic and spontaneously cross the bilayer, the assay used the movement of labeled phospholipid from the exporter-containing vesicle to acceptor vesicles that were devoid of proteins (95). This study showed that the AcrB pump functions as a proton antiporter and also showed that, among the substrates, conjugated bile salts had the highest affinity to AcrB. Another result was that the addition of AcrA increased very strongly the rate of transport (95). Although this was interpreted as the result of connection between vesicles mediated by AcrA, AcrA is essential in producing the aminoglycoside pumping activity of reconstituted AcrD (96) where such juxtaposition of vesicles is not needed, and AcrA probably activates directly the pumping activity of AcrB.
Most important pieces of knowledge concerning the RND pump complex came from crystallography. Thus, the structure of the outer membrane channel TolC was solved in 2000 (97, 98) (shown in red in Figure 5a), followed by the structure of its P. aeruginosa homolog, OprM (99). These proteins exist as a tightly woven trimer, containing a single 12-stranded β-barrel traversing the outer membrane and a remarkably long (~70-Å) periplasmic extension of the channel in the form of long α-helical bundles. Murakami et al. (100) solved the crystallographic structure of AcrB trimer in 2002, which was a first for a proton antiporter. The periplasmic portion is at least as large as the transmembrane portion, and the top of the periplasmic domain, the TolC-binding domain, has a dimension that is similar to the tip of the α-helical bundle of TolC (Figure 5a,b). This was followed by the crystallographic elucidation of the structures of the central portion (about two-thirds) of the adaptor proteins MexA (101, 102) and AcrA (103) (blue in Figure 5a). These are elongated proteins, as shown in 1999 (104), and in crystals show a strong tendency to become packed side by side to form hexamers and heptamers. Thus, the structures of all three component classes of the tripartite assembly are known, and it is possible to propose how these proteins are assembled together (105) (Figure 5a). In such models, the coiled-coil domain of the adaptor is assumed to interact with the outer membrane channel protein, a hypothesis that is supported by biochemical data (106, 107). Studies with chimeric constructs of AcrA homologs showed that the C-terminal domain, whose structure is not known from crystallography, is essential for interaction with AcrB (108). Mutant studies also suggest that the domain close to this end of AcrA, containing the α + β structure, is the main site of its interaction with the periplasmic domain of RND pumps (109).
The outer membrane component TolC and its homologs are first synthesized with the signal sequence and then are exported into the outer membrane. The periplasmic adaptors are either lipoproteins with the covalently bound lipid at its N terminus or contain a hydrophobic N-terminal helix and are thought to become bound to the outer surface of the inner membrane. The RND pump itself, as a classic example of polytopic inner membrane protein, requires the signal-recognition particle and the SecY apparatus for insertion into the membrane (110). The interaction between the component proteins of the AcrB-AcrA-TolC complex was studied in vitro by titration calorimetry (105).
Above, I mentioned that AcrB likely captures some of its substrates from the periplasm. Indeed the AcrB trimer structure showed that there was a small opening (vestibule) between subunits at the bottom of the periplasmic domain, close to the external surface of the membrane bilayer (100), and this led to the top of the large central cavity in the transmembrane domain (Figure 5b). It was thus hypothesized that the drugs diffused through the vestibule into the central cavity, where they were captured. Indeed, cocrystallization of AcrB with various drugs produced crystals with drug molecules within this central cavity (111). However, there was no proof that the bound drugs were on their correct path to extrusion, and the extrusion pathways were not clear in the crystal structure. We have attempted to solve this problem by crystallizing AcrB mutants in which one of the residues putatively involved in proton translocation was altered; we assumed that such proteins would assume a conformation mimicking that of a transient intermediate during the process of drug extrusion (112). There was an extensive conformational alteration in the transmembrane domain, but only minimal alterations were seen in the periplasmic domain (112), which is likely the main site of substrate binding and extrusion (see above). The reason became clear from the papers describing the asymmetric trimer structure of AcrB (113–115), especially as the Murakami group succeeded in solving the structure in which one of the protomers contained a substrate (minocycline or doxorubicin) (Figure 5c). Each protomer takes a conformation that is unique. Indeed the transmembrane domain of our mutant AcrB was similar in conformation to that of one of the protomers, called the extrusion protomer. However, extensive conformational changes in the periplasmic domain require complementary accommodation by the neighboring protomers (113–115). This was not possible in our construct, in which all protomers corresponded to the mutant protein. In the binding protomer, the periplasmic domain encloses an expanded binding pocket containing several aromatic residues, and there is an open pathway between the binding pocket and the vestibule. In the extrusion protomer, the binding pocket becomes much narrower, and the pathway to the vestibule becomes closed (Figure 6). These crystallographic observations suggest that the protomers within the trimeric AcrB go through a cyclic conformational change, from the open conformation through the ligand-bound one to finally the extrusion conformer, whose transmembrane domain shows signs of a disrupted network among proton-translocating residues. This cyclic change involves the opening and closing of the large external cleft in the periplasmic domain, and the forced closing of this cleft by fast-acting disulfide cross-linking agents stops the function of the pump instantaneously, thus providing biochemical support for this hypothesis (116). A similar result was obtained with a somewhat different approach (117). More recently, we succeeded in producing a giant gene containing three copies of the acrB genes connected together by linker sequences and, by using this method, showed conclusively that inactivation of just one protomer in the trimeric complex completely abolishes the pumping activity (118), a result supporting the functionally rotating mechanism mentioned above.
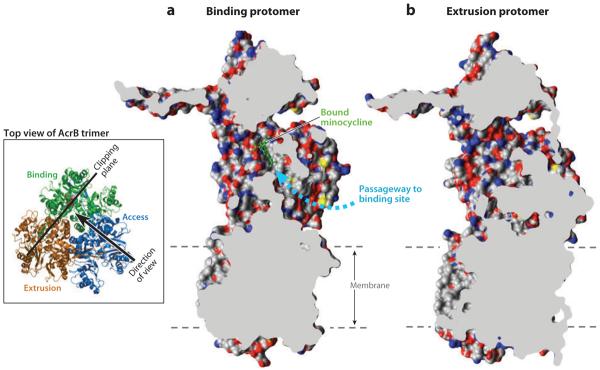
Figure 6.
Cut out view of the binding protomer with the bound minocycline (in a ball-and-stick representation in green) (a) and the extruding protomer (b), both from Protein Data Bank file 2DRD. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081) (119). The wide passageway from the external surface to the binding site (dashed arrow) seen in (a) appears to be completely closed in (b). The dashed lines show the approximate limits of the membrane bilayer. Modified after (120) with permission from Elsevier. The inset shows the location and direction of the clipping plane as well as the direction of the view in (a). The binding, extrusion, and access protomers are indicated.
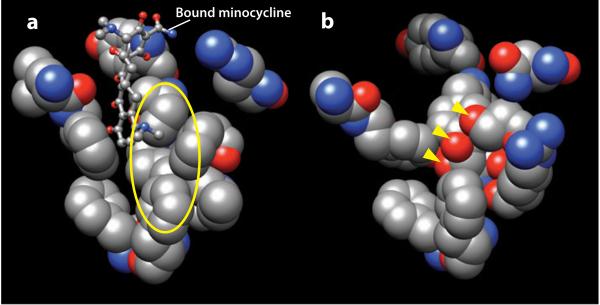
The binding pocket identified in the asymmetric structure (113–115) is large and flexible. In fact, the two substrates, which were cocrystalized with AcrB, minocycline, and doxorubicin, predominantly occupy somewhat different parts of the cavity; this explains the extremely wide substrate specificity of this pump. The pocket in AcrB, which favors lipophilic substrates, has a remarkably hydrophobic surface (Figure 7a). When AcrD, which pumps out basic and hydrophilic aminoglycosides (96), is homology modeled following the binding protomer of AcrB, its hypothetical binding pocket is seen to contain many electronegative atoms (Figure 7b). This observation increases our confidence that this is the genuine substrate binding site in these transporters.
Figure 7.
Substrate binding pockets of AcrB and a homology-modeled AcrD. (a) Residues facing the bound minocycline (in a ball-and-stick model), i.e., residues 177, 178, 275–278, 610, 612, 615, 620, 626, and 628 of the binding protomer of AcrB in Protein Data Bank file 2DRD are shown in space-filling models. The walls are remarkably hydrophobic, as seen for example in the area covered by the yellow ellipse. (b) Hypothetical structure of the corresponding region in AcrD. AcrD structure was built by homology modeling, using the binding protomer of AcrB as the template. The area covered by the ellipse in (a) is now seen to be studded with oxygen atoms. The oxygen atoms shown by the arrowheads belong to the side chains of, from top, Ser replacing Phe617 of AcrB, Tyr replacing Ile277, and Thr replacing Phe611. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081) (119).
Finally, we note that inhibitors of RND multidrug efflux pumps have been developed (for example, see Reference 121).
“Natural” substrates for multidrug efflux RND pumps
The idea that the multidrug efflux pumps may have other, more physiological functions than the extrusion of exogenous chemicals began with the finding that an MFS pump in B. subtilis, Blt, is actually a component of an operon that detoxifies spermidine (122). Clearly the RND-type multidrug pumps have not appeared in response to the widespread use of antibiotics, as most of these pumps are coded by chromosomal genes that are present in strains isolated long before the antibiotic era. For AcrB of E. coli, its main physiological function is to protect the bacteria against bile salts, detergents that are abundant in the environment where E. coli lives, the intestinal tract of vertebrates. This notion is supported by the observation that bile salts appear to have the highest affinity, among substrates, to the AcrB transporter (95). The AcrAB-TolC system was reported to be important in the pathogenicity of Salmonella spp. (123). However, this conclusion was reached by orally infecting experimental animals with bacteria and seems less convincing because the infecting bacteria will obviously encounter bile acids where the AcrAB pump will be needed for their survival. With plant pathogens, again RND pumps may be needed to protect the bacteria against plant secondary metabolites. In Pseudomonas syringae, the inactivation of one RND pump was reported to decrease the secretion of lipopeptide phytotoxins (124), but the decrease was only modest (41% to 67%).
With soil organisms such as P. aeruginosa, it seems likely that most of the RND pumps are involved in excluding toxic compounds produced by other soil microorganisms. The reports that they are involved in the export of homoserine lactone autoinducers are not convincing, as these compounds should diffuse across membranes spontaneously, and as the assay involves cells that are secreting these compounds and at the same time are exposed to the same compound secreted by other cells. However, the loss of some RND pumps was shown to result in the loss of invasiveness in the cell culture, through mechanisms that are unclear (125).
Contributions of RND efflux pumps to the resistance in clinical strains
RND pumps are making major contributions both to the intrinsic and elevated resistance of clinically relevant pathogens (48). For example, although fluoroquinoline resistance is often attributed to the mutational alteration of the target topoisomerases, high-level resistance seems to require an added contribution by increased efflux (126). Fluoroquinolone resistance in P. aeruginosa is rapidly increasing; efflux makes a major contribution in most such cases (48). A significant portion of aminoglycoside-resistant clinical isolates of gram-negative bacteria was previously classified as being caused by “decreased permeability.” These strains are now known to owe their resistance to increased active efflux (127, 128). In view of the important roles multidrug efflux plays in resistance, many scientists are concerned with the trend to increasingly add disinfectants to household products like soaps, because such compounds may select for pump overproduction mutants (129).
RND transporters that pump out nondrug substrates
Some of the RND transporters are involved in the export of compounds other than the antimicrobials. The “Nodulation” part of its name came from the putative NodGHI proteins (130), which are now fused into one RND protein (131), involved in the secretion of “nodulation factors” or lipochitin oligosaccharides by Rhizobium (132).
As mentioned above, typical multidrug exporters such as AcrB and MexB also export some simple solvents. Organisms that resist high concentrations of solvents, such as Pseudomonas putida, contain RND transporters optimized for the efflux of solvents, some of which can also pump out antimicrobials (133). Among the human proteins, the Niemann-Pick C1 disease protein, a member of RND family, attracted attention because mutations in the protein cause cholesterol accumulation in some organelles (134). Indeed, a close relative of this protein, Niemann-Pick C1-like protein, is involved in cholesterol transport in the intestinal epithelial cells (135).
There are many examples of RND transporters in the genome of M. tuberculosis. One of them (MmpL7) is involved in the export of complex, apolar lipid, phthiocerol dimycocerosate (136) and another one (MmpL8) in that of 2,3-diacyl-α,α′-trehalose-2′-sulfate, a precursor of sulfatides (137).
RND transporters also play a major role in resistance of gram-negative bacteria against toxic metals (138). CzrA, an RND transporter in Ralstonia that pumps out Co2+, Ni2+, Cd2+, and Zn2+, has been studied intensively, and an early successful reconstitution of an RND pump has been achieved in this system (139).
Other Multidrug Efflux Pumps Energized by Ionic Gradients
Several multidrug efflux transporters belong to the Multidrug and Toxin Extrusion (MATE) family (140). NorM of Vibrio parahaemolyticus, a protein with 12 transmembrane helices, pumps out fluoroquinolones and ethidium in exchange for the influx of Na+ (141). A similar pump was found in Bacteroides fragilis (142).
Multidrug Efflux Pumps of the ATP-Binding Cassette Superfamily
Transporters of ATP-Binding Cassette (ABC) superfamily play a major role in the multidrug resistance of cancer cells (143). Their role in drug resistance, however, seems to be more limited in bacteria (144). LmrA of L. lactis (145) is homologous to one-half of the mammalian MDR1 protein. Although its overproduction in E. coli confers resistance to cationic dyes, daunomycin, and triphenylphosphonium, it does not seem to play a significant role in the drug resistance of L. lactis. Nevertheless, biochemical studies of this pump led to a proposed mechanism for the coupling of drug extrusion and ATP hydrolysis (146) and to the discovery that the transmembrane domain of this pump can alone catalyze a proton-gradient-dependent flux of drugs (147). Studies were made on other ABC drug exporters of gram-positive bacteria, such as BmrA of B. subtilis (148, 149) and Sav1866 of S. aureus, whose X-ray crystallographic structure (150) had a major impact in the studies of ABC transporters.
Another ABC transporter, MacB in E. coli, occurs together with the structural gene for a periplasmic adaptor protein MacA and confers resistance to macrolides when overexpressed (151). In vitro reconstitution showed that the ATPase activity of MacB was stimulated by MacA, showing that the membrane fusion protein has functions other than the structural one (152).
MULTIDRUG RESISTANCE CAUSED BY ALTERED PHYSIOLOGICAL STATES
The antibiotic susceptibility of bacterial cells is affected by their physiological states. One important consequence of this phenomenon is the occurrence of “persister” cells. Thus, it was discovered early that even high concentrations of antibiotics do not kill all of the bacterial population, leaving behind a persister population that is genetically identical with the susceptible cells (153). When biofilms were found more resistant to most antibiotics, there were initial attempts to interpret this finding on the basis of a more limited diffusion of drugs through the biofilm structure. However, this mechanism cannot produce large increases in resistance. Although there are interesting data that link drug resistance in P. aeruginosa biofilms to the production of periplasmic β-(1–3)-glucans (154) and an efflux system (155), it is difficult, at least at present, to explain the extensive antibiotic resistance of biofilms on the basis of alteration of all cells in the population.
It is therefore attractive to explain such resistance as the result of the presence of a large number of persister cells in the biofilm population (153). The presence of persisters is now thought to be an example of the strategy whereby bacteria naturally generate mixtures of phenotypically different populations, so that one of them can be advantageous to a changing environmental demand (156). Persisters limit the efficacy of antibiotic therapy, and we note that a recent single-cell study has identified an antibiotic-susceptible phase in the life cycle of typical persister cells (157).
SUMMARY POINTS.
Multidrug resistance in bacteria is often caused by the accumulation of genes, each coding for resistance to a single drug, on R plasmids. The assembly of resistance genes on a single R plasmid is achieved by mechanisms provided by transposons, integrons, and ISCR elements. Integrons, for example, are especially powerful in producing multidrug resistance because they assemble several resistance genes in a correct orientation and supply a strong promoter for their expression. Furthermore, the resistance gene once incorporated into an integron becomes tagged, so that it could easily become a part of another integron.
Many of the resistance genes apparently have their evolutionary origins in the antibiotic-producing microbes, which must defend themselves from the antibiotics produced. Many also come from the environmental organisms, especially soil microorganisms, which have been exposed to various antibiotics throughout their evolutionary history. An alarming trend is the recent selection of mutants of widespread resistance genes that resulted in the broadening of the substrate range of the inactivated drugs, as in the extended-spectrum β-lactamases from the common TEM β-lactamase.
R plasmids are maintained extremely well and are often transferred efficiently from cell to cell.
Another mechanism of multidrug resistance is the active pumping out of drugs by multidrug efflux pumps. The RND superfamily pumps in gram-negative bacteria are especially important because they are usually coded by chromosomal genes and can be overexpressed easily and because some of them can pump out most of the antibiotics currently in use.
In some gram-negative species, these mechanisms may become augmented by the decrease in outer membrane permeability through mutations in porin genes.
Finally, persistence of pathogenic microorganisms may occur in an antibiotic-treated patient because they may get into a physiologically resistant state without any genetic changes.
FUTURE ISSUES.
There is a continuing need to define the molecular details of the resistance mechanism. This may lead, for example, to useful inhibitors of multidrug efflux pumps or to inhibitors of the R plasmid transfer process.
There are also practical issues of preventing the further increase in multidrug resistant bacteria. Because the usage of antibiotics was the cause of selection of these bacteria, one logical conclusion is to minimize antibiotic usage. Indeed, there is a striking correlation between the usage of β-lactams and the frequency of occurrence of penicillin-resistant pneumococci in the member countries of European Union (158).
Simultaneous administration of more than one agent (as is done with tuberculosis) may be considered. For example, an inhibitor of multidrug efflux pumps lowers the MICs of various drugs strongly in gram-negative bacteria and may prevent the emergence of resistant organisms (121).
ACKNOWLEDGMENTS
I thank colleagues for reading parts of this manuscript and for giving me the original figures for reproduction. I apologize to authors whose studies could not be cited because of size limitations. The work in the author's laboratory has been supported by a research grant from U.S. National Institutes of Health (AI-09644).
Glossary
- MRSA
methicillin-resistant Staphylococcus aureus
- Aminoglycosides
bactericidal antibiotics that are active against those gram-negative bacteria, resisting most other antibiotics owing to their low permeability outer membrane
- Transposons
discrete DNA sequences that can move to another sites. “Composite” transposons contain antibiotic resistance genes between two simple transposons, i.e., IS elements
- β-Lactams
antibiotics, including penicillins, cephalosporins, and carbapenems, which irreversibly inactivate DD-transpeptidases involved in the cross-linking of bacterial peptidoglycan, often causing cell lysis
- PBP
penicillin-binding proteins, or DD-transpeptidases, which cross-link the newly made peptidoglycan and correspond to the targets of β-lactam action
- Porins
proteins that produce, in the outer membrane of gram-negative bacteria, nonspecific diffusion channels that allow the influx of most of the effective antibiotics
- Major Facilitator superfamily (MFS) of secondary transporters
proton symporters or antiporters; some are involved in drug efflux, both single drug and multidrug
- TMS
transmembrane segment or the section of amino acid sequence that traverses the membrane bilayer
- Resistance-Nodulation-Division (RND) superfamily of transporters
many are antibiotic/proton antiporters, often with very wide substrate specificities
- ABC
a superfamily of ATP-dependent transporters, containing the transmembrane domain and the ATP-binding cassette (ABC) domain
Footnotes
DISCLOSURE STATEMENT The author is not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.de Lencastre H, Oliveira D, Tomasz A. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr. Opin. Microbiol. 2007;10:428–35. doi: 10.1016/j.mib.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Livermore DM. The need for new antibiotics. Clin. Microbiol. Infect. 2004;10(Suppl 4):1–9. doi: 10.1111/j.1465-0691.2004.1004.x. [DOI] [PubMed] [Google Scholar]
- 3.Hooper DC. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis. 2000;31(Suppl 2):S24–28. doi: 10.1086/314056. [DOI] [PubMed] [Google Scholar]
- 4.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 1995;39:577–85. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 2005;3:870–81. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 6.Huovinen P, Sundström L, Swedberg G, Sköld O. Trimethoprim and sulfonamide resistance. Antimicrob. Agents Chemother. 1995;39:279–89. doi: 10.1128/aac.39.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies J, Wright GD. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 1997;5:234–40. doi: 10.1016/S0966-842X(97)01033-0. [DOI] [PubMed] [Google Scholar]
- 8.Wright GD. Aminoglycoside-modifying enzymes. Curr. Opin. Microbiol. 1999;2:499–503. doi: 10.1016/s1369-5274(99)00007-7. [DOI] [PubMed] [Google Scholar]
- 9.Shaw KJ, Rather PN, Hare RS, Miller GH. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 1993;57:138–63. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benveniste R, Davies J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA. 1973;70:2276–80. doi: 10.1073/pnas.70.8.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Över U, Gür D, Ünal S, Miller GH. The changing nature of aminoglycoside resistance mechanisms and prevalence of newly recognized resistance mechanisms in Turkey. Clin. Microbiol. Infect. 2001;7:470–78. doi: 10.1046/j.1198-743x.2001.00284.x. [DOI] [PubMed] [Google Scholar]
- 12.Datta N, Kontomichalou P. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature. 1965;208:239–41. doi: 10.1038/208239a0. [DOI] [PubMed] [Google Scholar]
- 13.Vu H, Nikaido H. Role of β-lactam hydrolysis in the mechanism of resistance of a β-lactamase-constitutive Enterobacter cloacae strain to expanded-spectrum β-lactams. Antimicrob. Agents Chemother. 1985;27:393–98. doi: 10.1128/aac.27.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacoby GA, Medeiros AA. More extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 1991;35:1697–704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonnet R. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barlow M, Reik RA, Jacobs SD, Medina M, Meyer MP, et al. High rate of mobilization for blaCTX-Ms. Emerg. Infect. Dis. 2008;14:423–28. doi: 10.3201/eid1403.070405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 2007;20:440–58. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, et al. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006;12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 19.Spratt BG. Resistance to antibiotics mediated by target alterations. Science. 1994;264:388–93. doi: 10.1126/science.8153626. [DOI] [PubMed] [Google Scholar]
- 20.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–40. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 21.Connell SR, Tracz DM, Nierhaus KH, Taylor DE. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob. Agents Chemother. 2003;47:3675–81. doi: 10.1128/AAC.47.12.3675-3681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 2006;6:629–40. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 23.Levy SB. Active efflux mechanisms for antimicrobial resistance. Antimicrob. Agents Chemother. 1992;36:695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura N, Konishi S, Yamaguchi A. Mechanisms of drug/H+ antiport: complete cysteine-scanning mutagenesis and the protein engineering approach. Curr. Opin. Chem. Biol. 2003;7:570–79. doi: 10.1016/j.cbpa.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 25.Harder KJ, Nikaido H, Matsuhashi M. Mutants of Escherichia coli that are resistant to certain β-lactam compounds lack the ompF porin. Antimicrob. Agents Chemother. 1981;20:549–52. doi: 10.1128/aac.20.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Achouak W, Heulin T, Pages JM. Multiple facets of bacterial porins. FEMS Microbiol. Lett. 2001;199:1–7. doi: 10.1111/j.1574-6968.2001.tb10642.x. [DOI] [PubMed] [Google Scholar]
- 27.Marshall CG, Lessard IA, Park I, Wright GD. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 1998;42:2215–20. doi: 10.1128/aac.42.9.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–77. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 29.Dantas G, Sommer MO, Oluwasegun RD, Church GM. Bacteria subsisting on antibiotics. Science. 2008;320:100–3. doi: 10.1126/science.1155157. [DOI] [PubMed] [Google Scholar]
- 30.Williams LE, Detter C, Barry K, Lapidus A, Summers AO. Facile recovery of individual high-molecular-weight, low-copy-number natural plasmids for genomic sequencing. Appl. Environ. Microbiol. 2006;72:4899–906. doi: 10.1128/AEM.00354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liebert CA, Hall RM, Summers AO. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 1999;63:507–22. doi: 10.1128/mmbr.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall RM, Stokes HW. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica. 1993;90:115–32. doi: 10.1007/BF01435034. [DOI] [PubMed] [Google Scholar]
- 33.Rowe-Magnus DA, Mazel D. The role of integrons in antibiotic resistance gene capture. Int. J. Med. Microbiol. 2002;292:115–25. doi: 10.1078/1438-4221-00197. [DOI] [PubMed] [Google Scholar]
- 34.Mazel D. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 2006;4:608–20. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 35.Toleman MA, Bennett PM, Walsh TR. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nordstrom K, Austin SJ. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- 37.Novick RP, Clowes RC, Cohen SN, Curtiss R, 3rd, Datta N, Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol. Rev. 1976;40:168–89. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacob AE, Shapiro JA, Yamamoto L, Smith DI, Cohen SN, Berg D. Plasmids studied in Escherichia coli and in other enteric bacteria. In: Bukhari AI, Shapiro JA, Adhya SL, editors. DNA Insertion Elements, Plasmids, and Episomes. Cold Spring Harb. Lab.; Cold Spring Harbor, NY: 1977. pp. 607–38. [Google Scholar]
- 39.Schröder G, Lanka E. The mating pair formation system of conjugative plasmids—A versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54:1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 2005;59:451–85. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lessl M, Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994;77:321–24. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 42.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 43.Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rice LB. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 1998;42:1871–77. doi: 10.1128/aac.42.8.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Courvalin P. The Garrod Lecture: evasion of antibiotic action by bacteria. J. Antimicrob. Chemother. 1996;37:855–69. doi: 10.1093/jac/37.5.855. [DOI] [PubMed] [Google Scholar]
- 46.Paulsen IT, Brown MH, Skurray RA. Proton-dependent multidrug efflux systems. Microbiol. Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria. Drugs. 2004;64:159–204. doi: 10.2165/00003495-200464020-00004. [DOI] [PubMed] [Google Scholar]
- 48.Poole K. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 49.Higgins CF. Multiple molecular mechanisms for multidrug resistance transporters. Nature. 2007;446:749–57. doi: 10.1038/nature05630. [DOI] [PubMed] [Google Scholar]
- 50.Schumacher MA, Brennan RG. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol. Microbiol. 2002;45:885–93. doi: 10.1046/j.1365-2958.2002.03039.x. [DOI] [PubMed] [Google Scholar]
- 51.Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, et al. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292:2329–33. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- 52.Schumacher MA, Miller MC, Brennan RG. Structural mechanism of the simultaneous binding of two drugs to a multidrug-binding protein. EMBO J. 2004;23:2923–30. doi: 10.1038/sj.emboj.7600288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brooks BE, Piro KM, Brennan RG. Multidrug-binding transcription factor QacR binds the bivalent aromatic diamidines DB75 and DB359 in multiple positions. J. Am. Chem. Soc. 2007;129:8389–95. doi: 10.1021/ja072576v. [DOI] [PubMed] [Google Scholar]
- 54.Alguel Y, Meng C, Teran W, Krell T, Ramos JL, et al. Crystal structures of multidrug binding protein TtgR in complex with antibiotics and plant antimicrobials. J. Mol. Biol. 2007;369:829–40. doi: 10.1016/j.jmb.2007.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lomovskaya O, Lewis K. emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA. 1992;89:8938–42. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neyfakh AA, Borsch CM, Kaatz GW. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob. Agents Chemother. 1993;37:128–29. doi: 10.1128/aac.37.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Truong-Bolduc QC, Strahilevitz J, Hooper DC. NorC, a new efflux pump regulated by MgrA of Staphylococcus aureus. Antimicrob. Agents Chemother. 2006;50:1104–7. doi: 10.1128/AAC.50.3.1104-1107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeMarco CE, Cushing LA, Frempong-Manso E, Seo SM, Jaravaza TA, Kaatz GW. Efflux-related resistance to norfloxacin, dyes, and biocides in bloodstream isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 2007;51:3235–39. doi: 10.1128/AAC.00430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mazurkiewicz P, Driessen AJ, Konings WN. Energetics of wild-type and mutant multidrug resistance secondary transporter LmrP of Lactococcus lactis. Biochim. Biophys. Acta. 2004;1658:252–61. doi: 10.1016/j.bbabio.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 60.Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 2001;183:5803–12. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sulavik MC, Houseweart C, Cramer C, Jiwani N, Murgolo N, et al. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 2001;45:1126–36. doi: 10.1128/AAC.45.4.1126-1136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng J, Guffanti AA, Krulwich TA. The chromosomal tetracycline resistance locus of Bacillus subtilis encodes a Na+/H+ antiporter that is physiologically important at elevated pH. J. Biol. Chem. 1994;269:27365–71. [PubMed] [Google Scholar]
- 63.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312:741–44. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paulsen IT, Skurray RA, Tam R, Saier MH, Jr, Turner RJ, et al. The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol. Microbiol. 1996;19:1167–75. doi: 10.1111/j.1365-2958.1996.tb02462.x. [DOI] [PubMed] [Google Scholar]
- 65.Schuldiner S, Granot D, Mordoch SS, Ninio S, Rotem D, et al. Small is mighty: EmrE, a multidrug transporter as an experimental paradigm. News Physiol. Sci. 2001;16:130–34. doi: 10.1152/physiologyonline.2001.16.3.130. [DOI] [PubMed] [Google Scholar]
- 66.Schuldiner S. When biochemistry meets structural biology: the cautionary tale of EmrE. Trends Biochem. Sci. 2007;32:252–58. doi: 10.1016/j.tibs.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 67.Fleishman SJ, Harrington SE, Enosh A, Halperin D, Tate CG, Ben-Tal N. Quasi-symmetry in the cryo-EM structure of EmrE provides the key to modeling its transmembrane domain. J. Mol. Biol. 2006;364:54–67. doi: 10.1016/j.jmb.2006.08.072. [DOI] [PubMed] [Google Scholar]
- 68.Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, et al. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1999;1:107–25. [PubMed] [Google Scholar]
- 69.Paulsen IT, Park JH, Choi PS, Saier MH., Jr. A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol. Lett. 1997;156:1–8. doi: 10.1111/j.1574-6968.1997.tb12697.x. [DOI] [PubMed] [Google Scholar]
- 70.Dinh T, Paulsen IT, Saier MH., Jr. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J. Bacteriol. 1994;176:3825–31. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zgurskaya HI, Nikaido H. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J. Bacteriol. 2000;182:4264–67. doi: 10.1128/jb.182.15.4264-4267.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tikhonova EB, Zgurskaya HI. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J. Biol. Chem. 2004;279:32116–24. doi: 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- 73.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 1996;178:5853–59. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vaara M. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium. Antimicrob. Agents Chemother. 1993;37:2255–60. doi: 10.1128/aac.37.11.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nikaido H, Basina M, Nguyen V, Rosenberg EY. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those β-lactam antibiotics containing lipophilic side chains. J. Bacteriol. 1998;180:4686–92. doi: 10.1128/jb.180.17.4686-4692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elkins CA, Nikaido H. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J. Bacteriol. 2002;184:6490–98. doi: 10.1128/JB.184.23.6490-6498.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mao W, Warren MS, Black DS, Satou T, Murata T, et al. On the mechanism of substrate specificity by resistance nodulation division (RND)-type multidrug resistance pumps: the large periplasmic loops of MexD from Pseudomonas aeruginosa are involved in substrate recognition. Mol. Microbiol. 2002;46:889–901. doi: 10.1046/j.1365-2958.2002.03223.x. [DOI] [PubMed] [Google Scholar]
- 78.Lee A, Mao W, Warren MS, Mistry A, Hoshino K, et al. Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J. Bacteriol. 2000;182:3142–50. doi: 10.1128/jb.182.11.3142-3150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poole K, Srikumar R. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem. 2001;1:59–71. doi: 10.2174/1568026013395605. [DOI] [PubMed] [Google Scholar]
- 80.Grkovic S, Brown MH, Skurray RA. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 2002;66:671–701. doi: 10.1128/MMBR.66.4.671-701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. Contribution of the MexX-MexY-oprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2000;44:2242–46. doi: 10.1128/aac.44.9.2242-2246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jeannot K, Sobel ML, El Garch F, Poole K, Plesiat P. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J. Bacteriol. 2005;187:5341–46. doi: 10.1128/JB.187.15.5341-5346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morita Y, Sobel ML, Poole K. Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product. J. Bacteriol. 2006;188:1847–55. doi: 10.1128/JB.188.5.1847-1855.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cao L, Srikumar R, Poole K. MexAB-OprM hyperexpression in NalC-type multidrug-resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 2004;53:1423–36. doi: 10.1111/j.1365-2958.2004.04210.x. [DOI] [PubMed] [Google Scholar]
- 85.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, et al. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 1996;21:713–24. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 86.Sobel ML, Neshat S, Poole K. Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J. Bacteriol. 2005;187:1246–53. doi: 10.1128/JB.187.4.1246-1253.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Domain F, Bina XR, Levy SB. Transketolase A, an enzyme in central metabolism, derepresses the marRAB multiple antibiotic resistance operon of Escherichia coli by interaction with MarR. Mol. Microbiol. 2007;66:383–94. doi: 10.1111/j.1365-2958.2007.05928.x. [DOI] [PubMed] [Google Scholar]
- 88.Rosner JL, Dangi B, Gronenborn AM, Martin RG. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J. Bacteriol. 2002;184:1407–16. doi: 10.1128/JB.184.5.1407-1416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 2003;48:1609–19. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 90.Nagakubo S, Nishino K, Hirata T, Yamaguchi A. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 2002;184:4161–67. doi: 10.1128/JB.184.15.4161-4167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baranova N, Nikaido H. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 2002;184:4168–76. doi: 10.1128/JB.184.15.4168-4176.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol. Microbiol. 2005;55:1113–26. doi: 10.1111/j.1365-2958.2004.04449.x. [DOI] [PubMed] [Google Scholar]
- 93.Rahmati S, Yang S, Davidson AL, Zechiedrich EL. Control of the AcrAB multidrug efflux pump by quorum-sensing regulator SdiA. Mol. Microbiol. 2002;43:677–85. doi: 10.1046/j.1365-2958.2002.02773.x. [DOI] [PubMed] [Google Scholar]
- 94.Van Houdt R, Aertsen A, Moons P, Vanoirbeek K, Michiels CW. N-acyl-L-homoserine lactone signal interception by Escherichia coli. FEMS Microbiol. Lett. 2006;256:83–89. doi: 10.1111/j.1574-6968.2006.00103.x. [DOI] [PubMed] [Google Scholar]
- 95.Zgurskaya HI, Nikaido H. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1999;96:7190–95. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aires JR, Nikaido H. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol. 2005;187:1923–29. doi: 10.1128/JB.187.6.1923-1929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–19. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 98.Koronakis V, Eswaran J, Hughes C. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 2004;73:467–89. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- 99.Akama H, Kanemaki M, Yoshimura M, Tsukihara T, Kashiwagi T, et al. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J. Biol. Chem. 2004;279:52816–19. doi: 10.1074/jbc.C400445200. [DOI] [PubMed] [Google Scholar]
- 100.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–93. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 101.Akama H, Matsuura T, Kashiwagi S, Yoneyama H, Narita S, et al. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 2004;279:25939–42. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 102.Higgins MK, Bokma E, Koronakis E, Hughes C, Koronakis V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. USA. 2004;101:9994–99. doi: 10.1073/pnas.0400375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14:577–87. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zgurskaya HI, Nikaido H. AcrA is a highly asymmetric protein capable of spanning the periplasm. J. Mol. Biol. 1999;285:409–20. doi: 10.1006/jmbi.1998.2313. [DOI] [PubMed] [Google Scholar]
- 105.Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V. Three's company: component structures bring a closer view of tripartite drug efflux pumps. Curr. Opin. Struct. Biol. 2004;14:741–47. doi: 10.1016/j.sbi.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 106.Stegmeier JF, Polleichtner G, Brandes N, Hotz C, Andersen C. Importance of the adaptor (membrane fusion) protein hairpin domain for the functionality of multidrug efflux pumps. Biochemistry. 2006;45:10303–12. doi: 10.1021/bi060320g. [DOI] [PubMed] [Google Scholar]
- 107.Lobedanz S, Bokma E, Symmons MF, Koronakis E, Hughes C, Koronakis V. A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc. Natl. Acad. Sci. USA. 2007;104:4612–17. doi: 10.1073/pnas.0610160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Elkins CA, Nikaido H. Chimeric analysis of AcrA function reveals the importance of its C-terminal domain in its interaction with the AcrB multidrug efflux pump. J. Bacteriol. 2003;185:5349–56. doi: 10.1128/JB.185.18.5349-5356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Krishnamoorthy G, Tikhonova EB, Zgurskaya HI. Fitting periplasmic membrane fusion proteins to inner membrane transporters: mutations that enable Escherichia coli AcrA to function with Pseudomonas aeruginosa MexB. J. Bacteriol. 2007;190:691–98. doi: 10.1128/JB.01276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee HC, Bernstein HD. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl. Acad. Sci. USA. 2001;98:3471–76. doi: 10.1073/pnas.051484198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu EW, McDermott G, Zgurskaya HI, Nikaido H, Koshland DE., Jr. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science. 2003;300:976–80. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
- 112.Su CC, Li M, Gu R, Takatsuka Y, McDermott G, et al. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J. Bacteriol. 2006;188:7290–96. doi: 10.1128/JB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–79. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 114.Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–98. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 115.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grütter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Takatsuka Y, Nikaido H. Site-directed disulfide cross-linking shows that cleft flexibility in the periplasmic domain is needed for the multidrug efflux pump AcrB of Escherichia coli. J. Bacteriol. 2007;189:8677–84. doi: 10.1128/JB.01127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Seeger MA, von Ballmoos C, Eicher T, Brandstatter L, Verrey F, et al. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat. Struct. Mol. Biol. 2008;15:199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 118.Takatsuka Y, Nikaido H. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J. Bacteriol. 2009 doi: 10.1128/JB.01441-08. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 120.Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta Prot. Proteom. 2009 doi: 10.1016/j.bbapap.2008.10.004. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 2001;45:105–16. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Neyfakh AA. Natural functions of bacterial multidrug transporters. Trends Microbiol. 1997;5:309–13. doi: 10.1016/S0966-842X(97)01064-0. [DOI] [PubMed] [Google Scholar]
- 123.Piddock LJV. Multidrug-resistance efflux pumps—not just for resistance. Nat. Rev. Microbiol. 2006;4:629–36. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 124.Kang H, Gross DC. Characterization of a resistance-nodulation-cell division transporter system associated with the syr-syp genomic island of Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 2005;71:5056–65. doi: 10.1128/AEM.71.9.5056-5065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hirakata Y, Srikumar R, Poole K, Gotoh N, Suematsu T, et al. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 2002;196:109–18. doi: 10.1084/jem.20020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mazzariol A, Tokue Y, Kanegawa TM, Cornaglia G, Nikaido H. High-level fluoroquinolone-resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrob. Agents Chemother. 2000;44:3441–43. doi: 10.1128/aac.44.12.3441-3443.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ziha-Zarifi I, Llanes C, Köhler T, Pechere JC, Plesiat P. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 1999;43:287–91. doi: 10.1128/aac.43.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.El'Garch F, Jeannot K, Hocquet D, Llanes-Barakat C, Plesiat P. Cumulative effects of several nonenzymatic mechanisms on the resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 2007;51:1016–21. doi: 10.1128/AAC.00704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aiello AE, Larson EL, Levy SB. Consumer antibacterial soaps: effective or just risky? Clin. Infect. Dis. 2007;45(Suppl 2):S137–47. doi: 10.1086/519255. [DOI] [PubMed] [Google Scholar]
- 130.Baev N, Endre G, Petrovics G, Banfalvi Z, Kondorosi A. Six nodulation genes of nod box locus 4 in Rhizobium meliloti are involved in nodulation signal production: nodM codes for D-glucosamine synthetase. Mol. Gen. Genet. 1991;228:113–24. doi: 10.1007/BF00282455. [DOI] [PubMed] [Google Scholar]
- 131.Barnett MJ, Fisher RF, Jones T, Komp C, Abola AP, et al. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA. 2001;98:9883–88. doi: 10.1073/pnas.161294798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Spaink HP. Root nodulation and infection factors produced by rhizobial bacteria. Annu. Rev. Microbiol. 2000;54:257–88. doi: 10.1146/annurev.micro.54.1.257. [DOI] [PubMed] [Google Scholar]
- 133.Ramos JL, Duque E, Gallegos MT, Godoy P, Ramos-Gonzalez MI, et al. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 2002;56:743–68. doi: 10.1146/annurev.micro.56.012302.161038. [DOI] [PubMed] [Google Scholar]
- 134.Chang TY, Chang CC, Ohgami N, Yamauchi Y. Cholesterol sensing, trafficking, and esterification. Annu. Rev. Cell Dev. Biol. 2006;22:129–57. doi: 10.1146/annurev.cellbio.22.010305.104656. [DOI] [PubMed] [Google Scholar]
- 135.Altmann SW, Davis HR, Jr, Zhu LJ, Yao X, Hoos LM, et al. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 2004;303:1201–4. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- 136.Cox JS, Chen B, McNeil M, Jacobs WR., Jr. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 137.Domenech P, Reed MB, Dowd CS, Manca C, Kaplan G, Barry CE., 3rd The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J. Biol. Chem. 2004;279:21257–65. doi: 10.1074/jbc.M400324200. [DOI] [PubMed] [Google Scholar]
- 138.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003;27:313–39. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 139.Goldberg M, Pribyl T, Juhnke S, Nies DH. Energetics and topology of CzcA, a cation/proton antiporter of the resistance-nodulation-cell division protein family. J. Biol. Chem. 1999;274:26065–70. doi: 10.1074/jbc.274.37.26065. [DOI] [PubMed] [Google Scholar]
- 140.Hvorup RN, Winnen B, Chang AB, Jiang Y, Zhou XF, Saier MH., Jr. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur. J. Biochem. 2003;270:799–813. doi: 10.1046/j.1432-1033.2003.03418.x. [DOI] [PubMed] [Google Scholar]
- 141.Morita Y, Kataoka A, Shiota S, Mizushima T, Tsuchiya T. NorM of Vibrio parahaemolyticus is an Na+-driven multidrug efflux pump. J. Bacteriol. 2000;182:6694–97. doi: 10.1128/jb.182.23.6694-6697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Miyamae S, Ueda O, Yoshimura F, Hwang J, Tanaka Y, Nikaido H. AMATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob. Agents Chemother. 2001;45:3341–46. doi: 10.1128/AAC.45.12.3341-3346.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE, Gottesman MM. P-glycoprotein: from genomics to mechanism. Oncogene. 2003;22:7468–85. doi: 10.1038/sj.onc.1206948. [DOI] [PubMed] [Google Scholar]
- 144.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008;72:317–64. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lubelski J, Konings WN, Driessen AJ. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol. Mol. Biol. Rev. 2007;71:463–76. doi: 10.1128/MMBR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.van Veen HW, Higgins CF, Konings WN. Multidrug transport by ATP binding cassette transporters: a proposed two-cylinder engine mechanism. Res. Microbiol. 2001;152:365–74. doi: 10.1016/s0923-2508(01)01208-6. [DOI] [PubMed] [Google Scholar]
- 147.Venter H, Shilling RA, Velamakanni S, Balakrishnan L, Van Veen HW An ABC transporter with a secondary-active multidrug translocator domain. Nature. 2003;426:866–70. doi: 10.1038/nature02173. [DOI] [PubMed] [Google Scholar]
- 148.Chami M, Steinfels E, Orelle C, Jault JM, Di Pietro A, et al. Three-dimensional structure by cryoelectron microscopy of YvcC, an homodimeric ATP-binding cassette transporter from Bacillus subtilis. J. Mol. Biol. 2002;315:1075–85. doi: 10.1006/jmbi.2001.5309. [DOI] [PubMed] [Google Scholar]
- 149.Steinfels E, Orelle C, Fantino JR, Dalmas O, Rigaud JL, et al. Characterization of YvcC (BmrA), a multidrug ABC transporter constitutively expressed in Bacillus subtilis. Biochemistry. 2004;43:7491–502. doi: 10.1021/bi0362018. [DOI] [PubMed] [Google Scholar]
- 150.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–85. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 151.Kobayashi N, Nishino K, Yamaguchi A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 2001;183:5639–44. doi: 10.1128/JB.183.19.5639-5644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tikhonova EB, Devroy VK, Lau SY, Zgurskaya HI. Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol. Microbiol. 2007;63:895–910. doi: 10.1111/j.1365-2958.2006.05549.x. [DOI] [PubMed] [Google Scholar]
- 153.Lewis K. Persister cells and the riddle of biofilm survival. Biochemistry. 2005;70:267–74. doi: 10.1007/s10541-005-0111-6. [DOI] [PubMed] [Google Scholar]
- 154.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–10. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 155.Zhang L, Mah TF. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J. Bacteriol. 2008;190:4447–52. doi: 10.1128/JB.01655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dhar N, McKinney JD. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr. Opin. Microbiol. 2007;10:30–38. doi: 10.1016/j.mib.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 157.Gefen O, Gabay C, Mumcuoglu M, Engel G, Balaban NQ. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc. Natl. Acad. Sci. USA. 2008;105:6145–49. doi: 10.1073/pnas.0711712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bronzwaer SL, Cars O, Buchholz U, Molstad S, Goettsch W, et al. A European study on the relationship between antimicrobial use and antimicrobial resistance. Emerg. Infect. Dis. 2002;8:278–82. doi: 10.3201/eid0803.010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rowe-Magnus DA, Mazel D. Resistance gene capture. Curr. Opin. Microbiol. 1999;2:483–88. doi: 10.1016/s1369-5274(99)00004-1. [DOI] [PubMed] [Google Scholar]
- 160.Courvalin P. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 2006;42(Supp 1):S25–34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 161.Lewinson O, Padan E, Bibi E. Alkalitolerance: a biological function for a multidrug transporter in pH homeostasis. Proc. Natl. Acad. Sci. USA. 2004;101:14073–78. doi: 10.1073/pnas.0405375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nagano K, Nikaido H. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2009;106:5854–58. doi: 10.1073/pnas.0901695106. [DOI] [PMC free article] [PubMed] [Google Scholar]