Abstract
Motivation
Genome-wide experiments only rarely show resounding success in yielding genes associated with complex polygenic disorders. We evaluate 49 obesity-related genome-wide experiments with publicly-available findings, including microarray, genetics, proteomics and gene knock-down from human, mouse, rat and worm, in terms of their ability to rediscover a comprehensive set of genes previously found to be causally associated or having variants associated with obesity.
Results
Individual experiments show poor predictive ability for rediscovering known obesity-associated genes. We show that intersecting the results of experiments significantly improves the sensitivity, specificity and precision of the prediction of obesity-associated genes. We create an integrative model that statistically significantly outperforms all 49 individual genome-wide experiments. We find that genes known to be associated with obesity are significantly implicated in more obesity-related experiments and use this to provide a list of genes that we predict to have the highest likelihood of association for obesity. The approach described here can include any number and type of genome-wide experiments and might be useful for other complex polygenic disorders as well.
1 INTRODUCTION
Multiple genome-wide technologies have been developed and experiments performed since the sequencing of the human genome (2005). As the amount of publicly available genome-wide data keeps increasing, and increasing amounts of funding goes towards the building of consortia such as the Programs for Genomic Applications (http://www.nhlbi.nih.gov/resources/pga), it is important to evaluate the different types of genome-wide experiments in terms of their relevance to complex, polygenic human disorders.
Many investigators have used an integrative approach to identify genes associated with complex disorders. Particularly, integration of genetic and gene expression experiments has been widely done to identify genes associated with type 1 diabetes (Eaves, et al., 2002) and obesity (Ghazalpour, et al., 2005; Mir, et al., 2003; Schadt, et al., 2005), among others. These studies demonstrate the utility of integrating data from genetic and microarray studies, but often depend on the specific characteristics of both. For instance, expression quantitative trait loci (eQTLs) and metabolic quantitative trait loci (mQTLs) may be used to find genetic loci associated with expression level differences of genes or metabolic abundance; however, these approaches can only be used when both gene expression levels (or metabolic measurements) and genetic markers have been measured from the same individuals (Fu, et al., 2007; Jansen and Nap 2001; Schadt, et al., 2003). This limits its applicability with much of the publicly-available genome-wide data sets. In addition, these methods have not yet been scaled to handle more than just two genome-wide modalities, and require one modality to be genetic.
Several systematic approaches have been developed for the prioritization of human disease genes by integrating multiple heterogeneous sources of molecular data (Tiffin, et al., 2006; Aerts, et al., 2006; Calvo, et al., 2006; Freudenberg and Propping, 2002; Perez-Iratxeta, et al., 2002; Turner, et al., 2003). Aerts and colleagues used a number of data sources to assemble characteristics for a set of disease or pathway related training genes (Aerts, et al., 2006). Their system learns the characteristics of true positive genes from a training set, then applies that set of characteristics to identify and rank candidate disease genes. Beyond its utility in finding additional genes, the set of characteristics learned from true positive genes is not otherwise studied, and may be unnecessarily complex and overfit to exactly match the true positive genes. In addition, the Aerts approach demonstrated better sensitivity across monogenic disorders, where known genes ranked higher, than for polygenic disorders; the performance of their approach on polygenic disorders without the assistance of prior literature was not given. Importantly, although these and other papers show the value of integrating molecular data of various types, many of these methods require integration with prior knowledge bases or are restricted to genes previously associated with Mendelian disorders, and cannot function with just prior data in predicting genes associated with complex polygenic disorders. This is a major disadvantage, as knowledge bases of functional annotations for genes are incomplete. Additionally, most of the previous methods use a “one set of data fits all” approach, in that the same prior data and knowledge sources are felt to be able to useful to identify genes across all genetic disorders; a customized approach in using disease-relevant data sets may offer advantages in sensitivity and specificity. Finally, the previous approaches do not suggest whether serially increasing the number of integrated datasets improves sensitivity, nor do they suggest whether precision (positive predictive value) is also increased with integration.
The present study involves a purely data-driven integration of primary data-sets of different types all related to a model complex human disorder. To our knowledge, we describe the first systematic study on the effectiveness of genome-scale experimentation in rediscovering genes known to be associated with a complex, polygenic condition, and the first systematic evaluation of how basic integration of primary experimental results can significantly improve sensitivity, specificity, and precision of rediscovering genes known to be associated with a complex disease, and by extension, performance in finding novel candidate gene associations. We use obesity as our model, as it reflects a polygenic condition, involving the interaction of multiple genes and the environment. Our study includes 49 publicly-available, obesity-related genome-wide experiments and our method of predicting disease genes is not biased towards prior knowledge of gene function.
2 METHODS
2.1 Calculations for sensitivity, specificity, precision
Performance of individual experiments and pair-wise intersections of experiments is evaluated by Positive Predictive Value (PPV or precision), sensitivity and specificity in rediscovering a set of genes known to be associated with obesity. The PPV of an experiment or an intersection of experiments is defined as the percentage of genes identified as positive in an experiment or intersection that are positive in the gold standard. Sensitivity = number of gold standard genes identified / 273 gold standard genes. False positive rate = number of positive genes identified not in gold standard / (16,186 human NCBI Gene identifiers – 273).
2.2 Obesity-related and control experiments used
Our study includes 49 obesity-related experiments, of which 39 show positive genes under our re-analysis. A positive result was defined as a significant gene expression change in a microarray or a proteomics experiment, the genes under the LOD peak for a genetics experiment, and genes that when knocked down show the phenotype of increased or decreased fat storage for an RNAi experiment. The experiment types evaluated include microarray, genetics, proteomics and RNAi knock-down, in human, mouse, rat and worm. A description of each experiment and details of analysis for each experiment type (or modality) and the gold standard is found in Supplementary Methods. Gene results for all experiments, experimental modalities, conditions, and species were integrated and stored in a single data-base as NCBI Gene identifiers.
Three experiments for brain cancer were also chosen as a study of an arbitrary control disease. All three modalities (microarray, RNAi, a knowledge base of brain cancer genes) were represented in the obesity-related experiments. The same comparisons were performed with the control and obesity-related experiments.
2.3 Gold standard list of obesity-related genes
A list of genes known to be involved in human obesity was obtained from the 2004 human Obesity Gene Map Database (OGMD; http://obesitygene.pbrc.edu), accessed in June, 2005 (Perusse, et al., 2005). The genes include 10 genes with single mutations resulting in human obesity, 59 genes associated with obesity-related Mendelian disorders and 113 genes that show associations between DNA sequence variations and human obesity phenotypes. There are 10 genes with mutations causally associated in mouse models of obesity and 164 genes that when mutated or expressed as transgenes in mouse, demonstrate body-weight and adiposity related phenotypes.
2.4 Receiver operating characteristic (ROC) curves
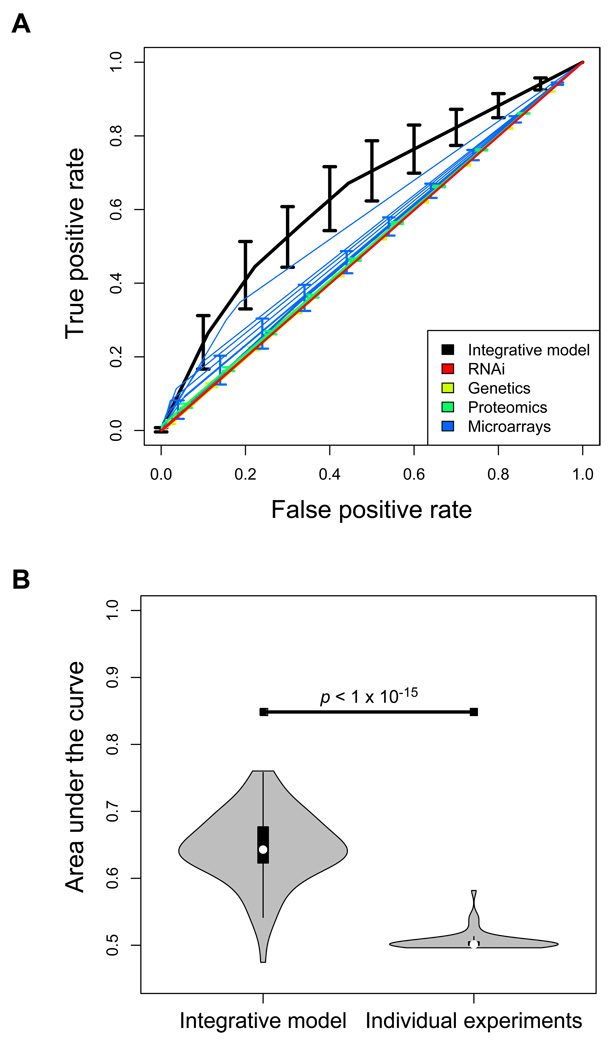
ROC curves were plotted for individual experiments, as well as average curves for each experimental modality. Microarray curves were constructed at threshold false discovery rates (FDRs) of 5% and 10%. Microarray experiments were re-analyzed using Significance Analysis of Microarrays (SAM) (Tusher, et al., 2001) at FDRs of 5% and 10%. The rest of the curves were constructed from a single point. ROC curves were also plotted for the integrative model across all experiments. This model calculated for each gene the number of individual experiments in which it was listed as positive. Each point on its ROC curve corresponds to a threshold for the number of experiments in which a gene appears as positive, which ranged from 0 to 8. The model was trained using 90% of all genes and tested on the remaining 10%, and this was repeated 100 times. The cross-validation trials were used to calculate average performance and standard deviation. The AUCs for the 100 cross-validated trials of the integrative model were compared to the AUCs for the individual experiments using a Wilcoxon rank sum test with continuity correction; results remained significant when using a t-test with Welch correction for unequal variances (Fig. 1).
Fig. 1.
An integrative model outperforms every one of its component obesity-related experiments. A. Receiver-operating characteristic curves are plotted for each of 49 obesity-related experiments and by experimental modality. An integrative model, considering genes by the number of obesity-related experiments in which they were positive, is shown in black. Each point on this curve indicates a different threshold number of positive experiments. Model error bars were constructed using 100 trials of 10-fold cross-validation, and indicate ± 1 standard deviation. B. Violin-plot showing the distribution of areas under the ROC curves for 100 cross-validated trials of the integrative model and the 49 individual obesity-related experiments. Significance was assessed using the Wilcoxon rank sum test. White dot indicates median, box covers between the 25% and 75% quantiles, whiskers cover extreme data points within 1.5 times the interquartile range from the box, and gray indicates the distribution of points.
2.5 Comparing individual and pair-wise intersections of experiments to the gold standard
Individual experiments and all possible pair-wise intersections were compared to the gold standard by using the PPV. Each pair of experiments was considered as a single experiment, in which genes positive in both experiments were considered as positive for the pair.
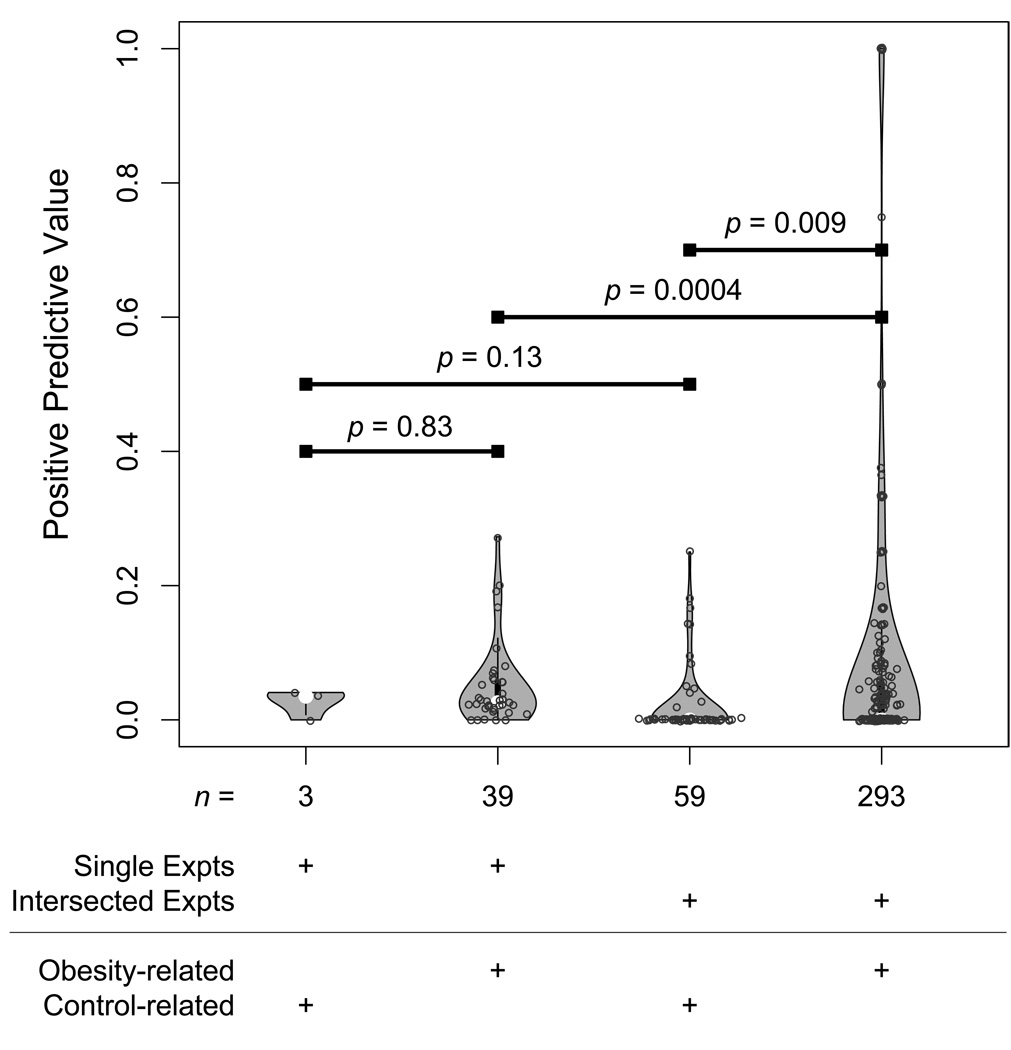
The PPVs of individual obesity experiments, control experiments, inter-sections of obesity-related experiments and intersections containing at least one control experiment were compared using Wilcoxon rank sum tests with continuity correction (Fig. 2), excluding experiments and intersections yielding no positive genes. Microarrays analyzed at both 5% FDR and 10% FDR were treated separately. The same result was obtained for both FDRs, and when a t-test with Welch correction for unequal variances was used.
Fig. 2.
Pair-wise intersections of experiments significantly outperforms individual experiments in rediscovering known associated genes. A violin-plot shows the distributions of positive predictive values (PPV) for individual obesity-related and control experiments, as well as the pair-wise intersections of obesity-related experiments and pair-wise intersections involving at least one control experiment. Individual experiments and intersections with no positive genes were excluded, as PPV cannot be calculated. After significance was assessed using the Wilcoxon rank sum test, a slight scatter was added to the graphical x- and y-axis positioning of points, to separate overlapping points.
We calculated the number of non-informative intersections when inter-sections were performed across genetics, microarray, and proteomics types of measurement modalities and when intersections were performed within the same type of experiment. The two groups were compared using the Fisher exact test. Again, the values were calculated separately for microarrays analyzed at both 5% and 10% FDR, and the same result was obtained.
2.6 Prediction of novel obesity-related genes
There are 16,186 human NCBI Gene identifiers in Homologene corresponding to ortholog family identifiers with a single gene (no significant human paralogs). Of these, 273 genes are gold standard genes. Each of the 16,186 genes was considered measured/negative, measured/positive, or absent in an experiment. A gene was considered as absent for an experiment if no ortholog was specified in Homologene for the experiment species, or if it corresponded to a gene ortholog family identifier containing more than one gene for the experiment species (paralogs). Each gene was assigned a score equal to the number of experiments in which it appeared as positive. We then measured the number of experiments in which each gold standard gene was implicated and did the same for non-gold standard genes. The two groups were compared by a Fisher exact test and a two-tailed t-test with Welch correction for unequal variances. The genes with the highest scores were the most likely to be obesity-related.
3 RESULTS
The results of 49 publicly-available obesity-related genome-wide experiments (Supplementary Methods and Table 1) were compared to a gold standard, the Obesity Gene Map, an annual well-cited listing of genes previously associated with obesity published by Perusse, Bouchard, and colleagues. All experimental datasets are demonstrably obesity-related, as indicated by their associated MeSH terms (Supplementary Methods). The experiments were performed across different organisms and modalities and examine different aspects of obesity. Although we do not expect an overall similarity between the datasets, we do expect each to identify a subset of genes important in the development of obesity. Some experiments directly compared obese and non-obese samples from human, mouse and rat by microarrays and proteomics technologies. Human and rat genome scan studies involved obesity-related traits. Other studies directly examined adipogenesis in human and mouse using microarrays or proteomics. Many studied this process in stimulated 3T3-L1 fibroblasts, a standard cellular model. We also include a minority of experiments showing a phenotype of altered fat mass or adipogenesis under conditions such as loss of insulin signaling, a genome-wide RNAi experiment in worm examining fat storage, and a human microarray study of a complex genetic syndrome with a fat cell phenotype.
3.1 Evaluation of sensitivity and specificity
Receiver-Operating Characteristic curves illustrate test performance by sensitivity and specificity, summarized by the area under the curve (AUC). The average ROC curves for each experimental modality, as well as most individual experiment curves, show poor sensitivity and specificity in the recall of the gold standard list of genes (Fig. 1A). Most experiments show a performance very close to the diagonal line of non-discrimination. Some microarray experiments do slightly better, with higher sensitivities likely due to the larger number of genes measured and yielded as positive.
We created a simple meta-experimental model, considering genes based on the number of 49 experiments in which they were positive. Even this simple integrative model outperforms any one of the experimental modalities or individual experiments in terms of sensitivity and specificity (Fig. 1A), as shown by the AUCs of the cross-validated trials of the integrative model being significantly higher than the AUCs for the individual experiments (Wilcoxon rank sum test, p < 1×10−15; Fig. 1B). In the integrative model, the maximum sensitivity of 66% at a specificity of 56% is achieved by the set of genes positive in any one of the 49 experiments, while the set of genes positive in any two experiments yields a sensitivity of 38% at a specificity of 83%. In other words, just building a repository of related genome-wide experiments to identify genes associated with a complex trait, such as obesity, may enable a more complete discovery of disease-related genes than any one of those experiments.
3.2 Performance of individual experiments
A common assumption made by labs and investigators is that individual genome-wide experiments can be used to identify genes important in the development of disease. We tested this assumption by comparing individual experiments to the gold standard using the PPV, which is computable for experiments yielding any positive findings. Ten of the 49 experiments are non-informative, identifying no positive genes; all are microarray experiments that show no positive genes under the stringent conditions of our re-analysis.
Most individual experiments show poor predictive ability for the gold standard genes (Supplementary Table 1). The mean PPV of the remaining 39 informative obesity-related experiments is 5% with a standard deviation of 6.1% and median 3.0%. In fact, there is no statistically significant difference between the PPVs of obesity-related and control experiments (Wilcoxon rank sum test p = 0.83; Fig. 2).
The PPV for individual experiments is statistically significantly affected by both the measurement modality and the species (two-way ANOVA with p = 0.001 and 0.002, respectively), though we acknowledge that certain measurement modalities were used only in model organisms (such as the proteomics studies in mouse). Three proteomics experiments show PPVs greater than three standard deviations from the mean (range 19% to 27%) yielding 5 to 21 genes in each.
The poorest performers, with PPVs equal to zero, include the worm genome-wide RNAi experiment, one proteomics experiment, and three genome scans. While worm genes may be less relevant to human obesity because of evolutionary distance, it is more likely that we are limited by the paucity of orthologous human genes currently related to worm genes in Homologene. It is not surprising that one proteomics experiment shows a zero PPV, given the few proteins identified in a proteomics experiment. Most of the genome scans used in this study contain a single linkage peak or QTL over numerous genes, while only a single gene under any one peak might be expected to be involved in obesity. Without prior knowledge, one true positive can be lost when surrounded by hundreds of other genes in a large region of linkage disequilibrium.
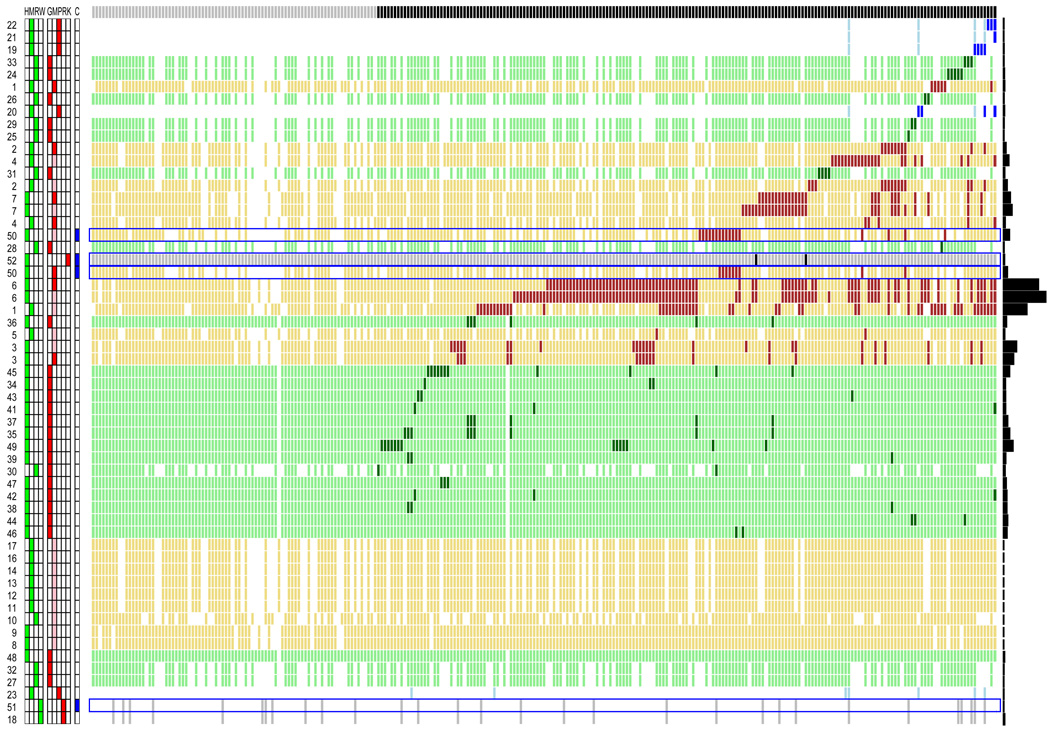
Fig. 3 illustrates the performance of each experiment in predicting the 273 gold standard genes by type of experiment and organism. Most microarray experiments identify a very large number of genes as positive. Although some of these experiments, such as those studying human and mouse adipogenesis, identify the greatest number of gold standard genes, they also yield the largest number of positive genes, many of which are not currently associated with obesity, potentially decreasing their PPV. In addition, while a microarray experiment measures thousands of genes and thus most of the gold standard genes, a proteomics 2D gel identifies fewer proteins and far fewer of the obesity gold standard genes.
Fig. 3.
Performance of each experiment in rediscovering the 273 gold standard positives (columns). Each row is a single experiment, sorted by positive predictive value, with highest at the top. Left-most column indicates the experiment number (Supplementary Table 1). Left grid (green) indicates species, middle (red) measurement type, and right (blue) control or obesity-related. Abbreviations: H, Human; M, Mouse; R, Rat; W, Worm; G, Genetics; M, Microarray; P, Proteomics; R, RNAi; K, Knowledge; C, Control. Microarray experiments are shown at both 5% (red) and 10% (pink) false-discovery rates. White elements are genes unmeasured in an experiment. In each row, darkest elements are gold standard genes positive in an experiment, lighter elements are gold standard genes measured in an experiment, but not positive. Four control experiments have a blue border. Bar chart at extreme right visually indicates total number of positives identified in each experiment, ranging from 0 to 3067. All 273 genes were measured in at least one experiment, but 89 genes are not positive in any experiment.
3.3 Performance of intersections of experiments
To test our hypothesis that intersections of genome-wide experiments perform better than individual experiments in identifying obesity-related genes, we performed comprehensive pair-wise intersections of all experiments. Importantly, the average PPV for intersections of pairs of obesity-related experiments was significantly higher as compared to individual obesity-related experiments (mean PPV 10% versus 5%; Wilcoxon rank sum test p = 0.0004; Fig. 2), with some pairs having PPVs of 50% or higher. Also, unlike the individual experiments, the pair-wise intersections of obesity-related experiments have significantly higher PPVs than pair-wise intersections containing a control experiment (Wilcoxon rank sum test p = 0.009; Fig. 2). Both results remained significant when using a t-test with Welch correction for unequal variances, though both results must be interpreted in the context of non-independence of the single and paired experiments.
While the average pair showed a higher PPV than single experiments, the median pair PPV (0%) did not improve, as more pairs of experiments yielded a zero PPV than single experiments. However, the average pair of experiments with a zero PPV yielded 6.2 positive genes, while the average single experiments with a zero PPV yielded 24.6 genes. In other words, pairs of experiments yielding intersecting genes can yield a high PPV, but can also often yield a zero PPV, though when this occurs, there are fewer genes on average for (potentially mistaken) follow up.
In the PPV calculation above, we exclude non-informative individual experiments and non-informative pair-wise intersections, as no PPV can be calculated from an experiment or pair of experiments with no positive findings. Interestingly, we find that intersecting the 39 informative obesity-related experiments across measurement type (e.g. intersecting a microarray with a proteomics experiment) yields significantly fewer non-informative intersections, as compared to pair-wise intersections within the same measurement type (Fisher exact test p < 1×10−15). Of the 385 intersections across measurement type, 149 (39%) are non-informative, while 299 of the 356 intersections within the same measurement type (84%) are non-informative.
The above calculation of the distribution of PPVs is empirically based on the 741 ways 39 experiments may be paired, excluding the 448 pairings with no positive findings. This distribution is most useful when serially adding data from a second experiment. PPV may also be calculated retrospectively given an increasing thresh-old of positive experiments across the 39 studies. Of the 2,868 genes positive in 2 or more experiments, 106 were in the gold-standard, yielding a PPV of 3.7%. PPV increases to 4.1%, 5.0%, 9.6%, 6.3%, 25%, to a maximum of 33%, when considering genes positive in 3 through 8 or more experiments, respectively.
3.4 Prediction of novel obesity-related genes
We find that the number of experiments in which a gene appears as positive is significantly higher for gold standard genes as com-pared to non-gold standard genes (Fisher exact test for count data with simulation based on 2×106 replicates p = 5×10−7; two tailed t-test p = 6.1×10−13). This observation led to a simple regression model that predicts the likelihood of a gene being obesity-related just given the number of experiments in which it was positive.
We found 52 genes positive in 5 or more of the 49 experiments; those positive in 6 or more are listed in Table 1. We predict these 52 genes have the highest likelihood of association with obesity. Most of these genes do not yet have variants associated with obesity. Five of the genes are gold standard genes. The highest scoring genes IL1R1, ADIPOQ and CYP1B1, appear as positive in 8 experiments. ADIPOQ is a gold standard positive gene. IL1R1 (interleukin receptor 1, type I) variants are associated with the metabolic syndrome (McCarthy, et al., 2003) and type 1 diabetes (Bergholdt, et al., 2000), but not obesity. In addition, CYP1B1 is induced when C3H10T1/2 cells are stimulated by an adipogenic hormonal mixture (Cho, et al., 2005).
Table 1.
52 genes were positive in 5 or more experiments; those 16 positive in 6 or more are listed here. The third column indicates the number of positive experiments for each gene. Star indicates gold standard gene.
| Symbol | Name | + |
|---|---|---|
| IL1R1 | interleukin 1 receptor, type I | 8 |
| *ADIPOQ | adiponectin, C1Q and collagen domain containing | 8 |
| CYP1B1 | Cyto. P450, fam.1, subfam. B, polypep. 1 | 8 |
| TRIB2 | tribbles homolog 2 (Drosophila) | 7 |
| HADHA | hydroxyacyl-CoA dehydrogenase /3-ketoacyl- Coenzyme A thiolase/enoyl-Coenzyme A hydra- tase, alpha subunit |
6 |
| GAS6 | growth arrest-specific 6 | 6 |
| CYB5A | cytochrome b5 type A (microsomal) | 6 |
| KCNK3 | potassium channel, subfam. K, memb. 3 | 6 |
| KLF11 | Kruppel-like factor 11 | 6 |
| ARL4A | ADP-ribosylation factor-like 4A | 6 |
| MDH1 | malate dehydrogenase 1, NAD (soluble) | 6 |
| PRSS23 | protease, serine, 23 | 6 |
| NCOA1 | nuclear receptor coactivator 1 | 6 |
| IGSF4 | immunoglobulin superfamily, member 4 | 6 |
| SLC1A4 | solute carrier family 1 , member 4 | 6 |
| PPP1CB | Prot. phosphatase 1, cat. sub., β isoform | 6 |
There are also several other non-gold standard genes that are directly or indirectly implicated in fat metabolism. GAS6, HADHA, and NCOA1 were implicated in 6 of the 49 experiments. Importantly, GAS6 and NCOA1, a co-activator for steroid and nuclear hormone receptors are sufficient to cause obesity when knocked out (Maquoi, et al., 2005; Picard, et al., 2002). GAS6 is also necessary for the development of diabetic nephropathy in certain models (Nagai, et al., 2005). Deficiency of HADHA, the alpha subunit of the mitochondrial trifunctional protein, in humans leads to hypoketotic hypoglycemia and fatty liver (Ibdah, et al., 1999).
IL6ST, IL18RAP, CXCL12, JUN, DBI and HES1 were implicated in 5 of the 49 experiments. IL6ST (interleukin 6 signal transducer) transduces signals for multiple cytokines with anti-obesity effects, including interleukin 6, ciliary neurotrophic factor, and leukemia inhibitory factor (Jansson, et al., 2006; Wallenius, et al., 2002; Watt, et al., 2006). IL18RAP is an accessory subunit for the receptor for IL18, elevated levels of which are associated with the metabolic syndrome (Hung, et al., 2005), type 2 diabetes (Thorand, et al., 2005) and insulin resistance (Fischer, et al., 2005). CXCL12 (stromal cell-derived factor 1) has been associated with complications of type 2 diabetes (Butler, et al., 2005). Mice with defective JUN show poor weight gain (Behrens, et al., 1999). Both DBI, also known as diazepam binding inhibitor and acyl coenzyme A binding protein, and HES1, a transcriptional repressor, have been shown to be necessary for adipogenesis (Mandrup, et al., 1998; Ross, et al., 2004). DBI is regulated by SREBP and PPARs, including PPARG (Neess, et al., 2006; Sandberg, et al., 2005).
It is important to note that while literature evidence exists for the plausible role for these genes in human obesity, none had yet been positively genetically or causally associated with human obesity or a specific phenotype of obesity in murine knockout or transgenic models enough to be in the 2004 gold standard.
4 DISCUSSION
We find that the sensitivity of individual obesity-related experiments in finding known obesity-associated genes is quite low, but one can maximize the sensitivity of finding obesity-related genes by incorporating the results of a multitude of experiments (Fig 1). One of the reasons for low experimental sensitivity is that the range of measurable genes in genome-wide experiments varies depending on the type and generation of technology being used. For example, current generation microarrays measure about 94% of the genes known to be associated with obesity, while commonly used proteomics technologies are limited by resolution and the few proteins typically chosen for identification. Another factor is that 89 of the 273 gold standard genes known to be associated with obesity are not positive in any of the 49 experiments. Of these, 21 genes are candidates associated with complex Mendelian disorders, for which obesity is just one of many symptoms.
While this work is not the first to integrate genome-wide experimental data to yield candidate genes associated with disease, we feel the strongest point of this work is that such a simple method for integration works. It is easy to imagine how a complex model, such as data fusion (Aerts, et al., 2006) or a Bayesian method (Calvo, et al., 2006), can be built to learn how to prioritize disease genes. Here, we show that a model -- as simple as counting the number of experiments in which a gene is implicated -- is significantly predictive. Prior literature or pathway knowledge is not needed. Considering genes by the number of related experiments in which they were implicated offers significantly better sensitivity and specificity than any individual experiment. Simply put, more is better. This makes the case for the value of building repositories of disease-related genome-wide experiments, a practice that is increasingly supported and funded by NIH and others.
Consistent with the known challenge of applying genome-wide experiments to complex disorders, most individual experiments show lower predictive ability for genes with known sequence variants or functional changes associated with obesity. But, on average, intersecting two genome-wide experiments shows better predictive ability than individual experiments. This is important, because individual scientists are more likely to desire fewer false positives at the expense of sensitivity. It is also of strategic interest to minimize non-informative intersections. We find that intersecting experiments across measurement technologies, rather than within the same technologies, significantly reduces the number of non-informative intersections.
We do not make the claim of exclusively identifying obesity-causative genes. We are interested in genes likely to be predictive for the development of human obesity. Without further biological validation, it would be difficult to claim “causality”. The 52 genes we predict as highly likely to be obesity-associated include non-gold standard genes with independent functional evidence suggestive of involvement in obesity. These are excellent candidates for future genotyping studies.
The proteomics experiments show a trend towards higher precision. In a single case, we have proteomics and microarray data from the same experiment: a study of the effect of impaired insulin signaling and adipocyte size. Unlike the proteomics experiment, the microarray experiment shows no significantly changed genes under the conditions of our re-analysis. This suggests that proteomics might outperform microarrays because of its intrinsic technology rather than the biological process studied.
There are a few limitations to our approach. Importantly, although the gold standard genes represent the current state of knowledge on obesity, this knowledge is dynamic and will always be incomplete. This analysis is dependent on the quality of the gold standard, and may be sensitive to the addition of a single functional category (Myers, et al., 2006). Another limitation is that our intersections are cis-intersections, identifying genes positive in more than one experiment. The next step would be to consider genes by pathway co-involvement or genes that may be upstream or down-stream of a given gene. We acknowledge that some of the experiments incorporated in our analysis used older technologies, which have improved considerably since the studies we used were published. We also acknowledge that our study may exclude other obesity-related experiments, though many of these are not publicly-available. However, automated extraction of all obesity-related experiments from all repositories and laboratory web-sites is currently intractable and beyond the scope of this study.
In spite of these limitations, we have several significant findings:
The 49 individual obesity-related experiments and 4 types of experiments demonstrate poor sensitivity and specificity in rediscovering known obesity-related genes.
Known obesity-related genes are implicated in significantly more genome-wide experiments than unrelated genes.
Based on this, we created a simple integrative model that statistically significantly outperforms each of the 49 individual experiments in sensitivity and specificity.
Individual obesity-related experiments show poor precision in rediscovering known obesity-related genes, but intersecting the results of pairs of experiments statistically significantly improves the precision.
Putting all of our findings together, our analysis suggests a two-pronged strategy; we can now recommend to individual investigators and collaborators that they use two or more types of genome-wide measurement and intersect the results of these to maximally yield known and potentially novel disease-related genes. We recommend that consortia and associations support the building of databases and repositories to hold genome-wide experimental results, and to require data-sharing policies that enable multi-experiment analyses.
Note added during review: GAS6 is newly listed in the 2005 Obesity Gene Map, as we predicted (Rankinen, et al., 2006).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Russ Altman for critical comments and suggestions. The work was supported by grants from the Lucile Packard Foundation for Children's Health, National Library of Medicine (K22 LM008261), Howard Hughes Medical Institute, Pharmaceutical Research and Manufacturers of America Foundation, National Institute of Diabetes and Digestive and Kidney Diseases (K12 DK63696, R01 DK62948, and R01 DK060837), Harvard-MIT Division of Health Sciences and Technology, and Lawson Wilkins Pediatric Endocrine Society.
Footnotes
Supplementary Information: Available Online and at http://buttelab.stanford.edu/doku.php?id=public:obesityintegration
REFERENCES
- A decade of genome-wide biology. Nature genetics. 2005;37:3–3. 1p. [Google Scholar]
- Aerts S, Lambrechts D, Maity S, Van Loo P, Coessens B, De Smet F, Tranchevent LC, De Moor B, Marynen P, Hassan B, Carmeliet P, Moreau Y. Gene prioritization through genomic data fusion. Nature biotechnology. 2006;24:537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- Behrens A, Sibilia M, Wagner EF. Amino-terminal phosphorylation of c-Jun regulates stress-induced apoptosis and cellular proliferation. Nature genetics. 1999;21:326–329. doi: 10.1038/6854. [DOI] [PubMed] [Google Scholar]
- Bergholdt R, Larsen ZM, Andersen NA, Johannesen J, Kristiansen OP, Mandrup-Poulsen T, Nerup J, Pociot F. Characterization of new polymorphisms in the 5' UTR of the human interleukin-1 receptor type 1 (IL1R1) gene: linkage to type 1 diabetes and correlation to IL-1RI plasma level. Genes and immunity. 2000;1:495–500. doi: 10.1038/sj.gene.6363719. [DOI] [PubMed] [Google Scholar]
- Butler JM, Guthrie SM, Koc M, Afzal A, Caballero S, Brooks HL, Mames RN, Segal MS, Grant MB, Scott EW. SDF-1 is both necessary and sufficient to promote proliferative retinopathy. The Journal of clinical investigation. 2005;115:86–93. doi: 10.1172/JCI22869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, Spinazzola A, Zeviani M, Carr SA, Mootha VK. Systematic identification of human mitochondrial disease genes through integrative genomics. Nature genetics. 2006;38:576–582. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- Cho YC, Zheng W, Yamamoto M, Liu X, Hanlon PR, Jefcoate CR. Differentiation of pluripotent C3H10T1/2 cells rapidly elevates CYP1B1 through a novel process that overcomes a loss of Ah Receptor. Archives of biochemistry and biophysics. 2005;439:139–153. doi: 10.1016/j.abb.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Eaves IA, Wicker LS, Ghandour G, Lyons PA, Peterson LB, Todd JA, Glynne RJ. Combining mouse congenic strains and microarray gene expression analyses to study a complex trait: the NOD model of type 1 diabetes. Genome research. 2002;12:232–243. [PubMed] [Google Scholar]
- Fischer CP, Perstrup LB, Berntsen A, Eskildsen P, Pedersen BK. Elevated plasma interleukin-18 is a marker of insulin-resistance in type 2 diabetic and non-diabetic humans. Clinical immunology (Orlando, Fla. 2005;117:152–160. doi: 10.1016/j.clim.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Fu J, Swertz MA, Keurentjes JJ, Jansen RC. Meta-Network: a computational protocol for the genetic study of metabolic networks. Nature protocols. 2007;2:685–694. doi: 10.1038/nprot.2007.96. [DOI] [PubMed] [Google Scholar]
- Freudenberg J, Propping P. A similarity-based method for genome-wide prediction of disease-relevant human genes. Bioinformatics (Oxford, England) 2002;18 Suppl 2:S110–S115. doi: 10.1093/bioinformatics/18.suppl_2.s110. [DOI] [PubMed] [Google Scholar]
- Ghazalpour A, Doss S, Sheth SS, Ingram-Drake LA, Schadt EE, Lusis AJ, Drake TA. Genomic analysis of metabolic pathway gene expression in mice. Genome biology. 2005;6:R59. doi: 10.1186/gb-2005-6-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung J, McQuillan BM, Chapman CM, Thompson PL, Beilby JP. Elevated interleukin-18 levels are associated with the metabolic syndrome independent of obesity and insulin resistance. Arteriosclerosis, thrombosis, and vascular biology. 2005;25:1268–1273. doi: 10.1161/01.ATV.0000163843.70369.12. [DOI] [PubMed] [Google Scholar]
- Ibdah JA, Bennett MJ, Rinaldo P, Zhao Y, Gibson B, Sims HF, Strauss AW. A fetal fatty-acid oxidation disorder as a cause of liver disease in pregnant women. The New England journal of medicine. 1999;340:1723–1731. doi: 10.1056/NEJM199906033402204. [DOI] [PubMed] [Google Scholar]
- Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- Jansson JO, Moverare-Skrtic S, Berndtsson A, Wernstedt I, Carlsten H, Ohlsson C. Leukemia inhibitory factor reduces body fat mass in ovariec-tomized mice. European journal of endocrinology / European Federation of Endocrine Societies. 2006;154:349–354. doi: 10.1530/eje.1.02082. [DOI] [PubMed] [Google Scholar]
- Mandrup S, Sorensen RV, Helledie T, Nohr J, Baldursson T, Gram C, Knudsen J, Kristiansen K. Inhibition of 3T3-L1 adipocyte differentiation by expression of acyl-CoA-binding protein antisense RNA. The Journal of biological chemistry. 1998;273:23897–23903. doi: 10.1074/jbc.273.37.23897. [DOI] [PubMed] [Google Scholar]
- Maquoi E, Voros G, Carmeliet P, Collen D, Lijnen HR. Role of Gas-6 in adipogenesis and nutritionally induced adipose tissue development in mice. Arteriosclerosis, thrombosis, and vascular biology. 2005;25:1002–1007. doi: 10.1161/01.ATV.0000160611.68791.c6. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Meyer J, Moliterno DJ, Newby LK, Rogers WJ, Topol EJ. Evidence for substantial effect modification by gender in a large-scale genetic association study of the metabolic syndrome among coronary heart disease patients. Human genetics. 2003;114:87–98. doi: 10.1007/s00439-003-1026-1. [DOI] [PubMed] [Google Scholar]
- Mir AA, Myakishev MV, Polesskaya OO, Moitra J, Petersen D, Miller L, Orosz A, Vinson C. A search for candidate genes for lipodystrophy, obesity and diabetes via gene expression analysis of A-ZIP/F-1 mice. Genomics. 2003;81:378–390. doi: 10.1016/s0888-7543(03)00024-7. [DOI] [PubMed] [Google Scholar]
- Myers CL, Barrett DR, Hibbs MA, Huttenhower C, Troyanskaya OG. Finding function: evaluation methods for functional genomic data. BMC Genomics. 2006;7:187. doi: 10.1186/1471-2164-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K, Matsubara T, Mima A, Sumi E, Kanamori H, Iehara N, Fukatsu A, Yanagita M, Nakano T, Ishimoto Y, Kita T, Doi T, Arai H. Gas6 induces Akt/mTOR-mediated mesangial hypertrophy in diabetic nephropathy. Kidney international. 2005;68:552–561. doi: 10.1111/j.1523-1755.2005.00433.x. [DOI] [PubMed] [Google Scholar]
- Neess D, Kiilerich P, Sandberg MB, Helledie T, Nielsen R, Mandrup S. ACBP--a PPAR and SREBP modulated housekeeping gene. Molecular and cellular biochemistry. 2006;284:149–157. doi: 10.1007/s11010-005-9039-9. [DOI] [PubMed] [Google Scholar]
- Perez-Iratxeta C, Bork P, Andrade MA. Association of genes to genetically inherited diseases using data mining. Nature genetics. 2002;31:316–319. doi: 10.1038/ng895. [DOI] [PubMed] [Google Scholar]
- Perusse L, Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Snyder EE, Bouchard C. The human obesity gene map: the 2004 update. Obesity research. 2005;13:381–490. doi: 10.1038/oby.2005.50. [DOI] [PubMed] [Google Scholar]
- Picard F, Gehin M, Annicotte J, Rocchi S, Champy MF, O'Malley BW, Chambon P, Auwerx J. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: the 2005 update. Obesity (Silver Spring, Md. 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- Ross DA, Rao PK, Kadesch T. Dual roles for the Notch target gene Hes-1 in the differentiation of 3T3-L1 preadipocytes. Molecular and cellular biology. 2004;24:3505–3513. doi: 10.1128/MCB.24.8.3505-3513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg MB, Bloksgaard M, Duran-Sandoval D, Duval C, Staels B, Mandrup S. The gene encoding acyl-CoA-binding protein is subject to metabolic regulation by both sterol regulatory element-binding protein and peroxisome proliferator-activated receptor alpha in hepatocytes. The Journal of biological chemistry. 2005;280:5258–5266. doi: 10.1074/jbc.M407515200. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, Linsley PS, Mao M, Stoughton RB, Friend SH. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, Lum PY, Leonardson A, Thieringer R, Metzger JM, Yang L, Castle J, Zhu H, Kash SF, Drake TA, Sachs A, Lusis AJ. An integrative genomics approach to infer causal associa-tions between gene expression and disease. Nature genetics. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorand B, Kolb H, Baumert J, Koenig W, Chambless L, Meisinger C, Illig T, Martin S, Herder C. Elevated levels of interleukin-18 predict the development of type 2 diabetes: results from the MONICA/KORA Augsburg Study 1984–2002. Diabetes. 2005;54:2932–2938. doi: 10.2337/diabetes.54.10.2932. [DOI] [PubMed] [Google Scholar]
- Tiffin N, Adie E, Turner F, Brunner HG, van Driel MA, Oti M, Lopez-Bigas N, Ouzounis C, Perez-Iratxeta C, Andrade-Navarro MA, et al. Computational disease gene identification: a concert of methods prioritizes type 2 diabetes and obesity candidate genes. Nucleic Acids Res. 2006;34:3067–3081. doi: 10.1093/nar/gkl381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner FS, Clutterbuck DR, Semple CA. POCUS: mining genomic sequence annotation to predict disease genes. Genome biology. 2003;4:R75. doi: 10.1186/gb-2003-4-11-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallenius V, Wallenius K, Ahren B, Rudling M, Carlsten H, Dickson SL, Ohlsson C, Jansson JO. Interleukin-6-deficient mice develop mature-onset obesity. Nature medicine. 2002;8:75–79. doi: 10.1038/nm0102-75. [DOI] [PubMed] [Google Scholar]
- Watt MJ, Dzamko N, Thomas WG, Rose-John S, Ernst M, Carling D, Kemp BE, Febbraio MA, Steinberg GR. CNTF reverses obesity-induced insulin resistance by activating skeletal muscle AMPK. Nature medicine. 2006;12:541–548. doi: 10.1038/nm1383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.