Abstract
Protein synthesis has an overall error rate of approximately 10-4 for each mRNA codon translated. The fidelity of translation is mainly determined by two events: synthesis of cognate amino acid:tRNA pairs by aminoacyl-tRNA synthetases (aaRSs) and accurate selection of aminoacyl-tRNAs (aa-tRNAs) by the ribosome. To ensure faithful aa-tRNA synthesis, many aaRSs employ a proofreading (“editing”) activity, such as phenylalanyl-tRNA synthetases (PheRS) that hydrolyze mischarged Tyr-tRNAPhe. Eukaryotes maintain two distinct PheRS enzymes, a cytoplasmic (ctPheRS) and an organellar form. CtPheRS is similar to bacterial enzymes in that it consists of a heterotetramer in which the α-subunits contain the active site and the β-subunits harbor the editing site. In contrast, mitochondrial PheRS (mtPheRS) is an α-subunit monomer that does not edit Tyr-tRNAPhe, and a comparable transacting activity does not exist in organelles. Although mtPheRS does not edit, it is extremely specific as only one Tyr-tRNAPhe is synthesized for every ∼7,300 Phe-tRNAPhe, compatible with an error rate in translation of ∼10-4. When the error rate of mtPheRS was increased 17-fold, the corresponding strain could not grow on respiratory media and the mitochondrial genome was rapidly lost. In contrast, error-prone mtPheRS, editing-deficient ctPheRS, and their wild-type counterparts all supported cytoplasmic protein synthesis and cell growth. These striking differences reveal unexpectedly divergent requirements for quality control in different cell compartments and suggest that the limits of translational accuracy may be largely determined by cellular physiology.
Keywords: aminoacyl-tRNA synthetase, protein synthesis, tRNA
Typical error rates for individual steps in gene maintenance and expression range from 10-8 for DNA replication (1) to 10-5 for mRNA transcription (2) and 10-4 for mRNA translation (3). These low error rates are achieved through high substrate specificity augmented by monitoring and proofreading of erroneous product synthesis, ensuring a high level of quality control. Whereas each cellular quality control mechanism has optimized its own level of specificity, translation as a whole limits misincorporation of the incorrect amino acid to one per 10,000 mRNA codons (3, 4). The fidelity of translation is determined by multiple events including synthesis of cognate amino acid:tRNA pairs by aminoacyl-tRNA synthetases (aaRSs), binding and delivery of aminoacyl-tRNAs (aa-tRNAs) to the ribosome by elongation factors, and accurate selection of aa-tRNAs by the ribosome (5, 6). Despite the widely held notion that limits on quality control, and tolerable error rates, are a fundamental aspect of all cells, recent studies suggest that wide disparities exist between different cell types particularly during translation. For example, partial ablation of aaRS proofreading in mice had no discernible effect on early growth and development but specifically impacted neuronal cells leading to ataxia and neurodegeneration in older animals (7).
During translation of the genetic code, aaRSs provide a critical step in quality control by preferentially selecting cognate pairs of tRNAs and amino acids while discriminating against near- and noncognate molecules. The unique combinations of sequences and structures found in particular tRNAs allow cognate molecules to be specifically selected out of the large cellular pool of similar molecules without recourse to proofreading (5, 8). Amino acids, by contrast, present a much more challenging problem for their cognate aaRS. The 20 naturally occurring amino acids do not display a sufficiently diverse range of functional groups that would allow aaRSs to discriminate between them with a level of accuracy consistent with the error rate assigned to translation (9). To prevent degeneracy of the genetic code by the infiltration of near-cognate amino acids, a number of proofreading activities are employed by aaRSs. These editing activities can occur through the hydrolysis of misactivated aminoacyl-adenylates (pretransfer editing) and/or through the hydrolysis of mischarged aa-tRNAs (posttransfer editing) (10). For example, the editing activities of phenylalanyl-tRNA synthetase (PheRS) prevent the delivery of Tyr-tRNAPhe to the ribosome and protect against the mistranslation of Phe codons as Tyr (11, 12).
The eubacterial, archaeal, and eukaryotic cytoplasmic PheRSs are heterotetrameric proteins composed of two α/β-heterodimers in which the α-subunits contain the active site and the β-subunits contain the editing site (13, 14). In eukaryotic cells separate translational systems are maintained in the cytoplasm and organelles, and aaRSs from both are encoded in the nucleus, with organelle forms synthesized as preproteins, which are then imported and processed (15). The mitochondrial form of PheRS (mtPheRS), as well as the chloroplast form, is an α-subunit monomer that lacks a recognizable editing domain, consistent with the absence of editing in some other mitochondrial aaRSs (16–18). Whereas ctPheRS and eubacterial PheRS possess a posttransfer editing activity against misacylated species, mtPheRS does not have the ability to edit mischarged Tyr-tRNAPhe, nor do mitochondria contain any trans-editing activity able to compensate for this deficiency (19). We now show that in Saccharomyces cerevisiae mtPheRS relies solely on a high level of specificity for Phe over Tyr for quality control of aminoacylation. A decrease in the amino acid specificity of PheRS blocked mitochondrial biogenesis but did not affect normal growth when tested in either the yeast cytoplasm or a bacterial model system. These data reveal strikingly different requirements for aaRS-mediated translation quality control in various cellular environments.
Results
Amino Acid Specificity of PheRS.
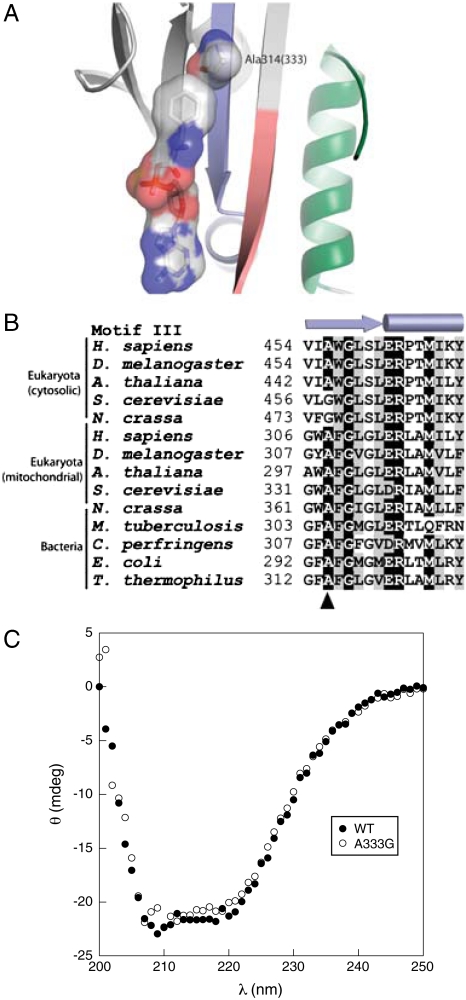
Previous studies of Escherichia coli PheRS showed that the replacement αA294G enlarged the amino acid binding pocket of the active site of the enzyme, allowing tRNA aminoacylation with para-halogenated Phe analogs (20). Steady-state kinetic analyses confirmed the role of this residue in E. coli PheRS quality control, the αA294G variant showing almost a 100-fold loss in specificity for Phe over Tyr (Table 1). To investigate the conservation of this specificity determinant, 1,179 PheRS α-subunit sequences (877 from eubacteria, 23 from archaea, and 160 and 119 cytosolic and mitochondrial eukaryotic, respectively) were aligned with ClustalX (21), guided by the three-dimensional structure of Thermus thermophilus PheRS [Fig. 1A (22)], and refined manually. αAla294 was found conserved at the equivalent position in all sequences except for 48 cytoplasmic PheRSs from 33 genera of eukaryotes that contained a Gly residue (Fig. 1B). This natural substitution of Ala by Gly showed no obvious phylogenetic distribution and was observed in all fungi (not in microsporidia), in the primitive eukaryote Trichoplax adhaerens, and also in some higher eukaryotes. The lack of conservation of the αAla294 specificity determinant among several PheRSs prompted us to compare the substrate specificity of the S. cerevisiae mitochondrial (αAla333) and cytoplasmic (αGly458) PheRSs, the latter of which contain a natural Ala to Gly substitution. The ctPheRS was 5 times more efficient (as reflected in kcat/KM) than the mtPheRS with respect to Phe activation and 80 times more efficient with respect to Tyr activation (Table 1). These differences in amino acid activation kinetics revealed that the specificity for Phe over Tyr is 15-fold higher for mtPheRS than it is for ctPheRS. To further investigate the ability of the αAla294 equivalent residues to confer specificity during Phe/Tyr discrimination in S. cerevisiae, mtPheRS was engineered through an A333G replacement whereas the converse change, G458A, was made in ctPheRS. MtPheRS A333G showed a 17-fold reduced specificity for Tyr in vitro compared to wild-type mtPheRS, whereas ctPheRS αG458A displayed a 20-fold increase compared to wild-type, confirming the critical role of this residue in quality control during amino acid activation (Table 1).
Table 1.
Steady-state kinetic constants for ATP-[32P]PPi exchange for cytosolic and mitochondrial wild-type and variant PheRS from S. cerevisiae and E. coli
| Phe |
Tyr |
||||||
|
KM (μM) |
kcat (s-1) |
kcat/KM (s-1/μM-1) |
KM (μM) |
kcat (s-1) |
kcat/KM (s-1/μM-1) |
Specificity (kcat/KM)Phe/(kcat/KM)Tyr | |
| mtPheRS | |||||||
| Wild-type | 5 ± 0.3 | 180 ± 7.6 | 35 ± 0.1 | 1,155 ± 160 | 5 ± 0.3 | 0.005 ± 0.0005 | 7,300 |
| A333G | 17 ± 1 | 140 ± 18 | 8 ± 0.1 | 660 ± 53 | 12.5 ± 0.5 | 0.02 ± 0.001 | 426 |
| ctPheRS | |||||||
| Wild-type | 3 ± 0.1 | 603 ± 41 | 194 ± 14 | 637 ± 308 | 184 ± 36 | 0.4 ± 0.2 | 485 |
| G458A | 233 ± 27 | 464 ± 37 | 2 ± 0.3 | 3,000 ± 1,200 | 0.6 ± 0.25 | 0.0002 ± 0.0001 | 10,000 |
| E. Coli | |||||||
| Wild-type | 2 ± 0.8 | 199 ± 25 | 110 ± 40 | 2,200 ± 700 | 35 ± 7 | 0.016 ± 0.002 | 6,800 |
| A294G | 4.5 ± 1.6 | 185 ± 13 | 45 ± 17 | 320 ± 80 | 185 ± 23 | 0.6 ± 0.08 | 75 |
Fig. 1.
Active site specificity of mitochondrial and cytosolic PheRS. (A) Structure of active site of the α-subunit of T. thermophilus PheRS in complex with phenylalanyl-adenylate (22). The optimal size of the Phe binding pocket is provided by residue Ala314 (residue A333 of S. cerevisiae mitochondrial PheRS). Motifs 1, 2, and 3 forming the active site of PheRS are colored in green, red, and blue, respectively. (B) Alignment summary of PheRS motif 3. The secondary structure elements of motif 3 are indicated on the top of the alignment, and position Ala314 is indicated by the arrowhead. Residues displaying more than 85% identity or similarity are depicted on black or gray background, respectively. (C) Secondary structural analysis of mtPheRS. Circular dichroism was measured at 25 °C with 5 μM samples of S. cerevisiae wild-type mtPheRS or the A333G variant.
Mitochondrial PheRS Specificity is Essential for Respiratory Function.
MtPheRS A333G displayed comparable substrate specificity to wild-type ctPheRS but lacked the latter protein’s ability to edit Tyr-tRNAPhe, which provided a means to investigate the importance of specificity for translational quality control in vivo by replacing wild-type mtPheRS with the A333G variant. To exclude possible indirect effects of the A333G replacement resulting from changes in secondary structure, CD spectral analysis was performed. Wild-type and mutant mtPheRS proteins showed nearly identical far-UV CD spectra, suggesting A333G does not induce major changes in global secondary structure (Fig 1C).
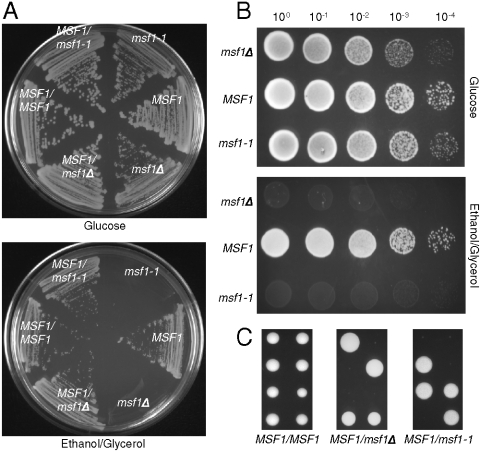
To test the effect of changing amino acid specificity in vivo, a chromosomal A333G replacement in MSF1 (encoding mtPheRS) was constructed in a haploid yeast strain resulting in the msf1-1 allele, and this strain was then crossed to a haploid wild-type to ensure the presence of fully functional mitochondria with an intact genome. Dissection of the heterozygous diploid MSF1/msf1-1 strain on fermentative medium (glucose) resulted in growth of all spores in each tetrad and replica plating onto media requiring respiratory function (ethanol plus glycerol) resulted in a no-growth phenotype that segregated 2∶2. On fermentative media both the msf1-1 and msf1Δ strains showed a reduced level of growth compared to MSF1 (Fig. 2A and B). However, on respiratory media neither the msf1-1 nor msf1Δ strain showed any growth (Fig 2B). To determine if the mitochondria are unable to respire from the onset or the ability to respire is lost with time, MSF1/msf1-1 was sporulated and dissected directly onto respiratory medium (Fig. 2C). While msf1-1 spores were able to germinate, growth was quickly arrested. It is possible that the limited growth seen is a result of the presence of both wild-type and mutant mtPheRS in the mitochondria at the onset of germination. Thus, the mitochondria may be able to respire for a very short time with the mitochondrial genome becoming unstable as a result of protein turnover and the increase in population of mutant mtPheRS present.
Fig. 2.
Growth of S. cerevisiae msf1-1. (A) Streak plates showing growth of diploid and haploid strains on YPDA and ethanol plus glycerol medium. (B) Serial dilutions of strains obtained from dissection of MSF1/msf1-1 on YPDA; MSF1 msf1-1, and msf1Δ on YPDA and ethanol plus glycerol medium. Plates were incubated at 30 °C for 3 days. (C) Dissection of tetrads from MSF1/MSF1, MSF1/msf1Δ, and MSF1/msf1-1 strains on ethanol plus glycerol medium. Plates were incubated at 30 °C for 3 (MSF1/MSF1) or 4 days (MSF1/msf1Δ and MSF1/msf1-1).
To investigate the presence or absence of the mitochondrial genome, msf1-1 was crossed with a MSF1 rho0 strain that encodes a wild-type mtPheRS but is devoid of the mitochondrial genome. If the msf1-1 strain is limited in respiratory activity because of the mutant mtPheRS but retains an intact mitochondrial genome, crossing with a MSF1 rho0 strain would result in a diploid with fully functional mitochondria, imparting the ability to grow on media requiring respiration. When tetrads from MSF1/msf1-1 were dissected, the spores allowed to germinate for 36 h, and then crossed with the MSF1 rho0 strain to monitor the loss of the mitochondrial genome, 84 out of 100 msf1-1 spores maintained their mitochondrial genome, whereas 16 of 100 spores had lost their mitochondrial genome at the time of crossing. However, after germination, if msf1-1 was grown in an overnight liquid culture before crossing, the MSF1 rho0 strain was unable to complement msf1-1, indicating a complete loss of the mitochondrial genome from msf1-1. These results demonstrate that reducing the specificity of mtPheRS is sufficient to destabilize the mitochondrial genome, which is then lost over time. This result is in agreement with previous findings in yeast where mutations inactivating mitochondrial translation result in mitochondrial genome instability (23).
PheRS Quality Control Mechanisms are not Essential in E. coli or S. cerevisiae Cytoplasm.
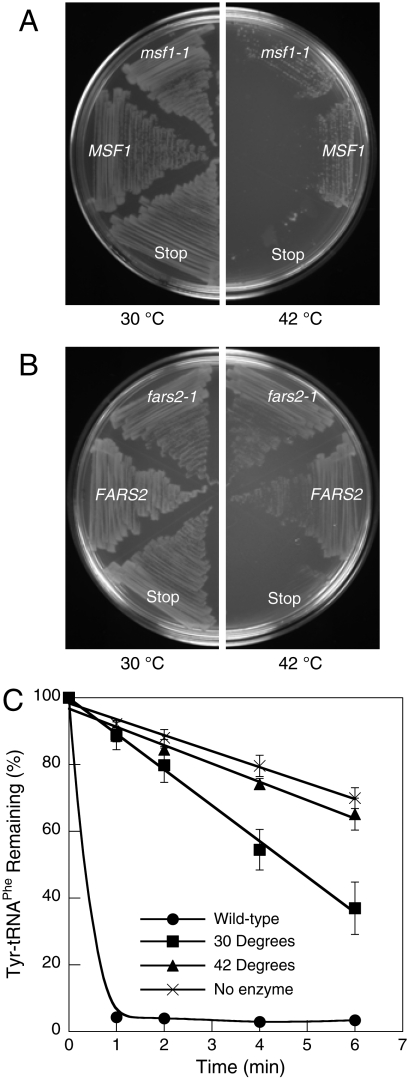
Loss of amino acid specificity encoded by the MSF1 gene resulted in ablation of mitochondrial biogenesis in S. cerevisiae, prompting us to investigate the requirements for this quality control mechanism in other cell types. In order to provide a direct comparison to mitochondria, E. coli was chosen as a model system because it has previously been demonstrated that mtPheRS can efficiently aminoacylate E. coli tRNAPhe (24, 25). To determine if loss of specificity in PheRS has a similar impact on cellular physiology in mitochondria and bacteria, we attempted to complement the E. coli strain NP37, which encodes a temperature-sensitive PheRS variant (26). NP37 was transformed with plasmids expressing MSF1, msf1-1, FARS2 (encoding human mtPheRS), or fars2-1 (A308G) and the resulting transformants grown at permissive or restrictive temperature (Fig. 3A and B). Both yeast and human wild-type mtPheRS, and the corresponding compromised specificity mutants, rescued the growth phenotype at a restrictive temperature. To determine if growth at the restrictive temperature resulted from retention of E. coli PheRS editing activity at 42 °C, Tyr-tRNAPhe hydrolysis activity was measured in cell-free extracts (Fig. 3C). NP37/fars2-1 extracts displayed editing activity when prepared from cells grown at 30 °C but not at 42 °C, confirming that E. coli NP37 lacks both PheRS editing and aminoacylation activities at the restrictive temperature.
Fig. 3.
Function of A333G mtPheRS in E. coli. Rescue of the growth phenotype of E. coli NP37 transformed with (A) yeast MSF1, msf1-1, or msf1 with a stop codon at A333 and (B) human FARS2, fars2-1, or fars2 with a stop codon at A308. (C) Transediting activity of E. coli wild-type and NP37 FARS2 complemented cell extracts grown at 30 or 42 °C. Reactions were carried out at 42 °C. Data points are an average of three independent experiments with errors bars representing 1 SD.
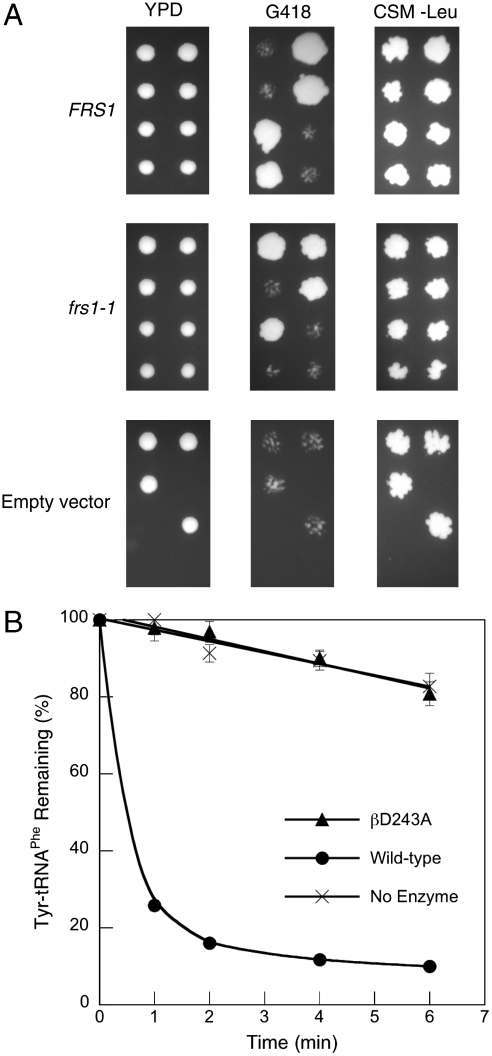
To further compare requirements for quality control in different cellular compartments, the effect of ablating the Tyr-tRNAPhe editing activity of ctPheRS was investigated in vivo. The yeast ctPheRS variant βD243A, which lacks editing activity (19), was inserted into the low copy-number centromeric plasmid pFL36, resulting in the plasmid pFL36-frs1-1, which was then used to transform the diploid strain YLR060W BY4743 (FRS1/frs1Δ). The resulting strains were sporulated, dissected onto yeast extract/peptone/dextrose (YPD), and replica plated onto G418 and complete supplement media minus leucine (CSM-Leu). The 36 resulting complete tetrads all showed 2∶2 segregation on G418 and showed growth on CSM-Leu, confirming the viability of the frs1Δ strain complemented with frs1-1 (Fig. 4A). Enzymatic Tyr-tRNAPhe hydrolysis activity was not observed in extracts prepared from frs2Δ pFL36-frs1-1 cells, indicating that ctPheRS editing activity is absent in this strain (Fig. 4B). Taken together, these findings suggest that neither high amino acid specificity nor near-cognate amino acid editing by PheRS are required for E. coli or S. cerevisiae cytoplasmic growth under normal conditions.
Fig. 4.
Function of βD243A ctPheRS in S. cerevisiae. (A) Dissection of tetrads from S. cerevisiae FRS1/frsΔ complemented with pFL36-FRS1, pFL36-frs1-1, or pFL36 onto YPD and replica plated onto G418 and CSM-Leu. In liquid YPD doubling times for frsΔ pFL36-FRS1 and pFL36-frs1-1 were 1.37 ± 0.05 and 1.34 ± 0.09 h, respectively. (B) Posttransfer editing activity of S. cerevisiae frs1Δ pFL36-FRS1 and frs1Δ pFL36-frs1-1 extracts at 37 °C. Data points are an average of at least three independent experiments, with errors bars representing 1 SD.
Discussion
Amino Acid Selectivity of aaRSs.
Early studies on protein synthesis suggested overall error rates of around one misincorporated valine per 3,000 isoleucine codons translated (4). Upon the basis of this and other studies, it was proposed that aaRSs would generally need to achieve at least 3,000-fold selectivity for cognate over near-cognate amino acids to avoid elevating the error rate of protein synthesis (27). Amino acid selectivity takes into account both aaRS specificity and the relative cellular concentrations of the corresponding cognate and near-cognate substrates. aaRSs which did not show an overall selectivity above 1 in 3,000 were predicted to require some form of proofreading to maintain sufficient accuracy during aa-tRNA synthesis, as borne out by the discovery of numerous editing mechanisms (10, 28). Biochemical characterization of the yeast PheRSs is consistent with this model; ctPheRS, an editing enzyme, displays 480-fold specificity for Phe over Tyr, which corresponds to a selectivity ranging from 1 in 400–2,000 depending on growth conditions (29). MtPheRS, which does not edit, displays 7,300-fold specificity for Phe over Tyr, which corresponds to a selectivity of 11,700 upon the basis of mammalian mitochondrial amino acid pools (30). E. coli PheRS displays a comparable predicted selectivity of 1 in 14,400 for Phe under standard conditions (31) while also retaining a robust editing activity (Table 2). The three PheRSs characterized here are all predicted to be equally effective in providing a level of cognate amino acid selectivity consistent with protein synthesis error rates lower than 1 in 3,000; the significant divergence in exactly how each enzyme prevents Tyr-tRNAPhe synthesis may reflect adaptations to specific cellular conditions, which could place greatly different demands on the accuracy of translation. How amino acid pools vary under different conditions in diverse cellular environments, and how this changes the demands on aaRS quality control, remains to be determined.
Table 2.
Predicted selectivity of PheRS in different cells and compartments
| Cell type |
PheRS |
Cellular Phe∶Tyr* |
Specificity Phe/Tyr† |
Selectivity Phe/Tyr‡ |
Tyr-tRNAPhe editing |
Viable in vivo |
| Yeast mitochondria | ||||||
| mtPheRS | 1.6∶1§ | 7,300 | 11,700 | No | Yes | |
| A333G mtPheRS | 1.6∶1 | 420 | 690 | No | No | |
| Yeast cytoplasm | ||||||
| ctPheRS | 4.6∶1¶ | 485 | 2,200 | Yes | Yes | |
| D243A ctPheRS | 4.6∶1 | 485 | 2,200 | No | Yes | |
| E. coli | ||||||
| E. coli PheRS | 1.9∶1∥ | 6,800 | 14,400 | Yes | Yes | |
| A333G mtPheRS | 1.9∶1 | 420 | 890 | No | Yes | |
*Ratio of concentrations of free Phe and Tyr.
†See Table 1.
‡Selectivity was defined as (specificity) × ([Phe]/[Tyr]), as applied to amino acid activation, but does not take into account possible posttransfer editing.
§Estimate based on rat mitochondria (30).
¶Total cellular amino acid pools (29).
∥(31).
Divergent Cellular Requirements for Quality Control During Translation.
The mitochondrial form of PheRS does not contain the β domain responsible for posttransfer editing and relies instead on a high level of amino acid specificity to maintain the fidelity of aminoacylation. This reliance on specificity alone is similar to mtLeuRS and certain prolyl-tRNA synthetases, all of which are highly specific and do not require editing, in contrast to their respective counterparts in E. coli, which both display robust editing activities (17, 18, 32, 33). The detrimental effects on the cell of reducing specificity for Phe confirmed the reliance of mtPheRS function on accurate amino acid recognition. Introduction of the A333G mtPheRS variant reduced the selectivity for Phe over Tyr to ∼700∶1, below the quality control “threshold” of 3,000∶1, a level of accuracy that proved to be too low to sustain mitochondrial biogenesis. Several proteins of the respiratory chain complex are synthesized within mitochondria (34, 35), and posttranslational quality control is used extensively to prevent dysfunction of the organelle (36). Our data now indicate that aaRS specificity is also an indispensable component of the quality control machinery in mitochondria.
The need for a high level of substrate specificity in order for PheRS to function properly in translation was not observed in bacteria or the yeast cytoplasm. Despite a specificity for Phe over Tyr of only ∼420∶1, A333G mtPheRS was able to support growth of E. coli on complete media. βD243A ctPheRS has a similar specificity, cannot edit mischarged tRNAPhe, and also supports cytoplasmic protein synthesis and growth. This ability to tolerate low specificity is in sharp contrast to the requirement for high amino acid specificity in mitochondria and provides direct evidence that certain cell types differ with respect to their requirements for quality control during translation. The notion that translation quality control requirements are cell-specific is supported by other recent in vivo studies using a mouse model. Lee et al. found that a missense mutation in the editing site of AlaRS resulted in the accumulation of misfolded proteins and cell death in terminally differentiated Purkinje neuronal cells. Only these nondividing cells, which contain an extremely high concentration of protein, show this phenotype while all other cells appear normal. When taken together with these findings, our data now clearly indicate that the requirements for quality control during translation vary greatly depending on cellular physiology. Given that both E. coli and the yeast cytoplasm can tolerate a low specificity for Phe over Tyr yet contain proofreading PheRSs, it is also apparent that the true role of aaRS editing in the cell still remains to be fully elucidated.
Materials and Methods
Protein Preparation and Analysis.
Proteins were prepared as described previously (12, 19). E. coli BL21-RIL/pET16b producing His6-tagged mtPheRS encoded by the MSF1 gene was a gift from R.A. Zimmermann (University of Massachusetts, Amherst, MA). The ctPheRS α and β subunits, encoded by the FRS2 and FRS1 genes, respectively, were expressed in tandem from pQE31-FRS-sc (producing His6-tagged WT ctPheRS) in E. coli BL21-RIL. E. coli pET-21c(+) encoding human mtPheRS (FARS2), producing mature His6-tagged wild-type PheRS, was a gift from L.L. Spremulli (University of North Carolina, Chapel Hill, NC). His6-tagged proteins were purified on nickel-nitrilotriacetic acid-agarose by standard procedures. Point mutations were introduced by site-directed mutagenesis using the Quikchange procedure (Stratagene). CD spectra were measured at 25 °C in an Aviv 62A DS spectropolarimeter (Aviv). The protein concentration was 5 μM in 50 mM Tris-HCl, pH 7.5, and 5% glycerol. CD spectra were measured from 200 to 250 nm (five scans per sample) with a step size of 1 nm in a 1 mm path length cuvette with 1 nm bandwidth and 5 sec averaging time. Protein-only spectra were obtained by subtracting the CD signal for buffer alone.
ATP-PPi exchange reactions were performed at 37 °C as described (19) with the exception that the amounts of amino acids used were 1.3 μM–1 mM Phe or 170 μM–6.8 mM Tyr, and the concentration of enzymes used was 5–150 nM ctPheRS, 150 nM mtPheRS, or 100 nM E. coli PheRS. To ensure the absence of Phe contamination, Tyr was subjected to several cycles of heating and cooling to remove any trace amounts of Phe (37). Posttransfer editing reactions contained 100 mM Na-Hepes, pH 7.2, 30 mM KCl, 10 mM MgCl2, 2 μM [14C]Tyr-tRNAPhe, and 74 μg (OD595) of E. coli crude extract or 0.006 U of S. cerevisiae extract, where one unit of PheRS corresponds to the amount of protein necessary to catalyze the formation of 1 nmol of Phe-tRNAPhe min-1 at 37 °C. E. coli crude extracts were preincubated at 42 °C before addition to the reaction mixture. After the addition of crude extract, reaction mixtures were incubated at 37 or 42 °C, and the deacylation reaction followed by measuring the [14C]Tyr-tRNAPhe remaining in aliquots of 7 μL removed after 0–6 min of incubation.
Construction, Manipulation, and Growth of Yeast Strains.
Strains derived from S. cerevisiae W303 (MATa, ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100) were used to carry out all in vivo experiments with the exception of those used in test crosses. In vivo site-directed mutagenesis was performed by the delitto perfetto method (38). A counter selectable reporter (CORE) cassette was inserted into the wild- type MSF1 gene of wild-type diploid S. cerevisiae near the A333 codon through homologous recombination resulting in the msf1 ∷ CORE strain. Upon introduction of the CORE cassette, cells were sporulated, resulting in 1∶1 segregation of the cassette into the daughter spores. Haploid cells containing the msf1 ∷ CORE strain were rho0. Recombinant MSF1 with mutations created by quick-change site-directed mutagenesis was used to transform msf1 ∷ CORE rho0 haploid cells. Recombination of the mutated gene with the msf1 ∷ CORE led to excision of the CORE and insertion of the A333G mutation (msf1-1) into the chromosome. To recover mitochondria, the haploid W303 msf1-1 strain was mated to a W303 wild-type haploid strain, resulting in a heterozygous rho+ strain. The resulting diploid strain was sporulated to obtain haploid msf1-1 cells. Insertion of the A333G mutation in W303 msf1-1 was confirmed by sequencing. Haploid strains are referred to as W303 MSF1 or W303 msf1-1 depending on the genotype. W303 msf1Δ was created through the replacement of the MSF1 open reading frame with a KanMX4 cassette by homologous recombination in a W303 MSF1 homozygous diploid. W303 msf1Δ was then obtained by sporulation and dissection.
The presence of mitochondrial DNA was determined by crossing W303 haploid strains with KL14-4A/60 (MATa, his1, trp2, rho0) or D27310B/50 (MATα, ade5, rho0) (39) on minimal glycerol medium followed by replica plating onto minimal ethanol/glycerol medium. To determine the initial state of the mitochondria W303 diploid cells were grown in either yeast extract/peptone/dextrose/adenine (YPDA), sporulated on minimal sporulation medium (1% CH3COOK, 0.1% yeast extract, 0.05% glucose), and dissected directly onto ethanol plus glycerol medium and grown for 3–4 days at 30 °C.
S. cerevisiae strain YLR060W BY4743 (MATa/MATα, his3Δ1/his3Δ1, leu2Δ0/leu2Δ0, lys2Δ0/LYS2, MET15/met15Δ0, ura3Δ0/ura3Δ0, FRS1/frs1::kanMX4) (ATCC) was used to carry out all in vivo experiments on yeast cytosolic pheRS (FRS1). A 3-kb genomic region including FRS1 and its native regulators was cloned into the centromeric shuttle vector pFL36 and the D243A mutation (frs1-1) introduced through site-directed mutagenesis. YLR060W BY4743 was transformed with pFL36-FRS1, pFL36-frs1-1, or pFL36. Resulting strains were sporulated, dissected on YPD, and replica plated onto YPD with 200 μg/mL geneticin (G418) and complete supplement media minus leucine (CSM -Leu; Sunrise Science Products). Growth rates of haploid frsΔ strains complemented with pFL36-FRS1 and pFL36-frs1-1 were determined in duplicate in 250-mL flasks with 50 mL of YPD. Cultures were shaken at 225 rpm at 30 °C. Samples of 1 mL were taken every hour with growth monitored spectrophotometrically at an absorbance of 660 nm. Cell-free extracts were prepared the same as E. coli cell-free extracts with the exception the cells were grown in 100 mL YPD overnight, washed, and resuspended in 5 mL 100 mM Tris-HCl, pH 8.0, 5 mM 2-mercaptoethanol, 500 μM diisopropyl fluorophosphate, 500 μM phenylmethylsulfonyl fluoride. Cells were opened with 1 mL glass beads by vortexing for 2 min, 6 times.
E. Coli pheSts Complementation and Preparation of Cell-Free Extracts.
E. coli NP37 (26) was transformed with the mature human mtPheRS (FARS2) cloned in pET-21c(+) and pRARE, which expresses six rare tRNAs (Novagene). Point mutations were introduced by quick-change site-directed mutagenesis with the Quikchange kit (Stratagene). Transformants were plated on LB supplemented with 100 μg/mL ampicillin, 30 μg/mL chloramphenicol, 0.4 mM IPTG at 30 or 42 °C for 48 h. Prior to preparation of cell-free extracts, revertants of NP37 were removed from plates grown at 42 °C for 48 h. The remaining cells were then removed from the plates and resuspended in 100 mM Tris-HCl, pH 8.0, 5 mM 2-mercaptoethanol, 500 μM diisopropyl fluorophosphate, 500 μM phenylmethylsulfonyl fluoride, washed once, and resuspended in the same buffer. Cells were sonicated at 70% output with a Sonifier 450 (Branson) equipped with a microprobe. The resulting extract was centrifuged at 100,000 × g for 1 h. The soluble extracts were dialyzed overnight at 4 °C against 100 mM Tris-HCl, pH 8.0, 5 mM 2-mercaptoethanol, 1 mM ethylenediaminetetraacetic acid and concentrated in the same buffer plus 50% glycerol.
Acknowledgments.
We thank K. Musier-Forsyth, B.R. So, and C. Hausmann for critical reading of the manuscript and I. Artsimovitch for invaluable assistance. This work was supported by Grant MCB-744791 from the National Science Foundation and a Binational Science Foundation grant (to M.I.), a predoctoral fellowship from the American Heart Association (to J.L.), and an Ohio State University Center for RNA Biology fellowship and a Grant-in-Aid of Research from the Ohio State University Chapter of Sigma Xi (to N.M.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 2.Blank A, Gallant JA, Burgess RR, Loeb LA. An RNA polymerase mutant with reduced accuracy of chain elongation. Biochemistry. 1986;25:5920–5928. doi: 10.1021/bi00368a013. [DOI] [PubMed] [Google Scholar]
- 3.Kurland CG. Translational accuracy and the fitness of bacteria. Annu Rev Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 4.Loftfield RB, Vanderjagt D. The frequency of errors in protein biosynthesis. Biochem J. 1972;128:1353–1356. doi: 10.1042/bj1281353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ibba M, Söll D. Quality control mechanisms during translation. Science. 1999;286:1893–1897. doi: 10.1126/science.286.5446.1893. [DOI] [PubMed] [Google Scholar]
- 6.Dale T, Uhlenbeck OC. Amino acid specificity in translation. Trends Biochem Sci. 2005;30:659–665. doi: 10.1016/j.tibs.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 8.Guth EC, Francklyn CS. Kinetic discrimination of tRNA identity by the conserved motif 2 loop of a class II aminoacyl-tRNA synthetase. Mol Cell. 2007;25:531–542. doi: 10.1016/j.molcel.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pauling L. Festschrift für Prof. Dr. Arthur Stoll. Basel, Switzerland: Birkhauser Verlag; 1958. [Google Scholar]
- 10.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 11.Ling J, Yadavalli SS, Ibba M. Phenylalanyl-tRNA synthetase editing defects result in efficient mistranslation of phenylalanine codons as tyrosine. RNA. 2007;13:1881–1886. doi: 10.1261/rna.684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling J, et al. Resampling and editing of mischarged tRNA prior to translation elongation. Mol Cell. 2009;33:654–660. doi: 10.1016/j.molcel.2009.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy H, Ling J, Irnov M, Ibba M. Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J. 2004;23:4639–4648. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Safro M, et al. Aminoacyl-tRNA Synthetases. Georgetown, Texas: Landes Bioscience; 2005. Phenylalanyl-tRNA synthetases; pp. 250–265. [Google Scholar]
- 15.Sissler M, Pütz J, Fasiolo F, Florentz C. Mitochondrial aminoacyl-tRNA synthetases. In: Ibba M, Francklyn C, Cusack S, editors. The Aminoacyl-tRNA Synthetases. Georgetown, Texas: Landes Bioscience; 2005. pp. 271–284. [Google Scholar]
- 16.Duchene AM, et al. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lue SW, Kelley SO. An aminoacyl-tRNA synthetase with a defunct editing site. Biochemistry. 2005;44:3010–3016. doi: 10.1021/bi047901v. [DOI] [PubMed] [Google Scholar]
- 18.Karkhanis VA, Boniecki MT, Poruri K, Martinis SA. A viable amino acid editing activity in the leucyl-tRNA synthetase CP1-splicing domain is not required in the yeast mitochondria. J Biol Chem. 2006;281:33217–33225. doi: 10.1074/jbc.M607406200. [DOI] [PubMed] [Google Scholar]
- 19.Roy H, Ling J, Alfonzo J, Ibba M. Loss of editing activity during the evolution of mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem. 2005;280:38186–38192. doi: 10.1074/jbc.M508281200. [DOI] [PubMed] [Google Scholar]
- 20.Ibba M, Kast P, Hennecke H. Substrate specificity is determined by amino acid binding pocket size in Escherichia coli phenylalanyl-tRNA synthetase. Biochemistry. 1994;33:7107–7112. doi: 10.1021/bi00189a013. [DOI] [PubMed] [Google Scholar]
- 21.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fishman R, Ankilova V, Moor N, Safro M. Structure at 2.6 è resolution of phenylalanyl-tRNA synthetase complexed with phenylalanyl-adenylate in the presence of manganese. Acta Crystallogr D. 2001;57:1534–1544. doi: 10.1107/s090744490101321x. [DOI] [PubMed] [Google Scholar]
- 23.Contamine V, Picard M. Maintenance and integrity of the mitochondrial genome: A plethora of nuclear genes in the budding yeast. Microbiol Mol Biol Rev. 2000;64:281–315. doi: 10.1128/mmbr.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aphasizhev R, et al. Conservation in evolution for a small monomeric phenylalanyl-tRNA synthetase of the tRNAPhe recognition nucleotides and initial aminoacylation site. Biochemistry. 1996;35:117–123. doi: 10.1021/bi9517998. [DOI] [PubMed] [Google Scholar]
- 25.Bullard JM, Cai YC, Demeler B, Spremulli LL. Expression and characterization of a human mitochondrial phenylalanyl- tRNA synthetase. J Mol Biol. 1999;288:567–577. doi: 10.1006/jmbi.1999.2708. [DOI] [PubMed] [Google Scholar]
- 26.Kast P, Keller B, Hennecke H. Identification of the pheS 5 mutation, which causes thermosensitivity of Escherichia coli mutant NP37. J Bacteriol. 1992;174:1686–1689. doi: 10.1128/jb.174.5.1686-1689.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fersht AR. Enzymic editing mechanisms and the genetic code. Proc R Soc Lond B. 1981;212:351–379. doi: 10.1098/rspb.1981.0044. [DOI] [PubMed] [Google Scholar]
- 28.Beuning PJ, Musier-Forsyth K. Hydrolytic editing by a class II aminoacyl-tRNA synthetase. Proc Natl Acad Sci USA. 2000;97:8916–8920. doi: 10.1073/pnas.97.16.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Force E, Benitez T. Effects of varying media, temperature, and growth rates on the intracellular concentrations of yeast amino acids. Biotechnol Prog. 1995;11:386–392. doi: 10.1021/bp00034a004. [DOI] [PubMed] [Google Scholar]
- 30.Ross-Inta C, Tsai CY, Giulivi C. The mitochondrial pool of free amino acids reflects the composition of mitochondrial DNA-encoded proteins: Indication of a post-translational quality control for protein synthesis. Biosci Rep. 2008;28:239–249. doi: 10.1042/BSR20080090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taymaz-Nikerel H, et al. Development and application of a differential method for reliable metabolome analysis in Escherichia coli. Anal Biochem. 2009;386:9–19. doi: 10.1016/j.ab.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 32.Beuning PJ, Musier-Forsyth K. Species-specific differences in amino acid editing by class II prolyl-tRNA synthetase. J Biol Chem. 2001;276:30779–30785. doi: 10.1074/jbc.M104761200. [DOI] [PubMed] [Google Scholar]
- 33.Sternjohn J, Hati S, Siliciano PG, Musier-Forsyth K. Restoring species-specific posttransfer editing activity to a synthetase with a defunct editing domain. Proc Natl Acad Sci USA. 2007;104:2127–2132. doi: 10.1073/pnas.0611110104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzagoloff A, Myers AM. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- 35.Foury F, Roganti T, Lecrenier N, Purnelle B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998;440:325–331. doi: 10.1016/s0014-5793(98)01467-7. [DOI] [PubMed] [Google Scholar]
- 36.Tatsuta T, Langer T. Quality control of mitochondria: Protection against neurodegeneration and ageing. EMBO J. 2008;27:306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin SX, Baltzinger M, Remy P. Fast kinetic study of yeast phenylalanyl-tRNA synthetase: An efficient discrimination between tyrosine and phenylalanine at the level of the aminoacyladenylate-enzyme complex. Biochemistry. 1983;22:681–689. doi: 10.1021/bi00272a024. [DOI] [PubMed] [Google Scholar]
- 38.Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- 39.Groudinsky O, Dujardin G, Slonimski PP. Long range control circuits within mitochondria and between nucleus and mitochondria. II. Genetic and biochemical analyses of suppressors which selectively alleviate the mitochondrial intron mutations. Mol Gen Genet. 1981;184:493–503. doi: 10.1007/BF00352529. [DOI] [PubMed] [Google Scholar]