Abstract
Under mating conditions, yeast cells adopt a characteristic pear-shaped morphology, called a “shmoo,” as they project a cell extension toward their mating partners. Mating partners make contact at their shmoo tips, dissolve the intervening cell wall, and fuse their plasma membranes. We identified mutations in ERG4, encoding the enzyme that catalyzes the last step of ergosterol biosynthesis, that impair both shmoo formation and cell fusion. Upon pheromone treatment, erg4Δ mutants polarized growth, lipids, and proteins involved in mating but did not form properly shaped shmoos and fused with low efficiency. Supplementation with ergosterol partially suppressed the shmooing defect but not the cell fusion defect. By contrast, removal of the Erg4 substrate ergosta-5,7,22,24(28)-tetraenol, which accumulates in erg4Δ mutant cells and contains an extra double bond in the aliphatic chain of the sterol, restored both shmooing and cell fusion to wild-type levels. Thus, a two-atom change in the aliphatic moiety of ergosterol is sufficient to obstruct cell shape remodeling and cell fusion.
Fusion between cells underlies developmental events from fertilization to the genesis of syncytial tissues. Although cell fusion has been studied for >100 years, little is known about the molecular machineries that control it (1). Indeed, a currently prevailing view is that the molecular machineries that catalyze cell fusion may have evolved independently and thus differ fundamentally from one system to another (1). Different model systems, therefore, are invaluable for identifying basic principles that may be shared among diverse cell fusion machineries.
Yeast mating represents a powerful model to study cell fusion (2). Saccharomyces cerevisiae haploid cells exist in two mating types, a and α, which each secrete a pheromone that is sensed by a G protein-coupled receptor in the plasma membrane of the complementary cell type. On pheromone detection, a MAPK signaling cascade activates G1 cell cycle arrest, transcriptional induction of >100 genes, and polarized growth toward the mating partner (3). The polarized growth changes the cell’s shape from spheroid to a pear-like shape or “shmoo,” enabling partners to make contact. Once in contact, the cell walls of mating partners knit together, forming a mating pair. Cell wall material separating the two plasma membranes is then removed, allowing apposition of the plasma membranes, which fuse to form a dinucleate zygote (2).
Numerous genes have been identified whose products orchestrate distinct stages of the mating process. Remarkably, the protein(s) that mediates membrane bilayer fusion has eluded identification despite numerous experimental approaches (2, 4–6). Using a reverse genetics strategy, PRM1 was identified in our laboratory as the first gene shown to be required at the plasma membrane fusion step (7). Prm1 is a multispanning membrane protein that is expressed only in response to pheromone and localizes at the site of cell fusion. When both partners lack Prm1, cell fusion is drastically reduced (∼50% of mating partners fuse). Electron microscopic analysis of the resulting unfused mating pairs revealed that the cell wall was degraded normally and the plasma membranes were tightly apposed but unfused. Without fusion, the area of the two adhered plasma membranes increases and buckles into either cell, forming characteristic intrusions apparent by light and electron microscopy. These striking phenotypes place Prm1 function proximal to lipid bilayer fusion (7).
However, Prm1 is unlikely to be the sole factor required for fusion, because ∼50% of prm1Δ × prm1Δ mating pairs still fuse. Moreover, a closer inspection revealed that a significant fraction of mating pairs lyse while attempting to fuse (8, 9), suggesting that membrane fusion activity may persist in prm1Δ × prm1Δ mating pairs but acts in a dysregulated fashion (10). We aimed to identify additional genes acting in conjunction with Prm1 by isolating genetic enhancers of the prm1Δ × prm1Δ cell fusion defect. Such studies identified, for example, the Golgi-resident proteases Kex1 and Kex2 as novel mediators of cell fusion (11). However, even in the combined absence of Prm1 and Kex2, cell fusion proceeds, albeit with low efficacy (∼15% of mating pairs fused) (11).
In this study, we have taken this genetic enhancer approach to the next level. We performed a genetic screen to isolate mutations that, in the absence of both PRM1 and KEX2, abolish cell fusion. We identified mutations in ERG4, which encodes an enzyme that catalyzes the last step in ergosterol biosynthesis. Unexpectedly, these studies revealed an amazing and sophisticated complexity regarding structural requirements of sterols in polarized cell growth and cell fusion.
Results
ERG4 Facilitates Cell Fusion During Mating.
To identify novel cell fusion factors, we performed a genetic screen for mutations that enhance the prm1Δ × prm1Δ kex2Δ mating defect. We subjected a prm1Δ MATα strain to random mutagenesis with ethylmethanosulfonate (EMS). The resulting mutants were grown as isolated colonies and mated to a prm1Δ kex2Δ MATa lawn. Diploid cells were selected, and the density of diploid papillae forming within each mating patch was used as an indicator of mating efficiency, as described in ref. 11. We considered mutants that gave rise to patches with a low density of diploid cells to be mating defective. We then mated these candidates to a wild-type MATa strain and discarded as sterile those mutants that failed to mate. We backcrossed the remaining mutants to the parental prm1Δ strain and, for the mutant with the most severe mating defect (Fig. S1), we identified ERG4 as the affected gene by complementation cloning. Sequencing of the genomic ERG4 locus in the mutant strain revealed the presence of a nonsense mutation at codon 144, predicted to result in a truncation of the Erg4 protein. Furthermore, deletion of ERG4 in prm1Δ MATα cells recapitulated the mutant mating defect. This defect was complemented by a plasmid bearing wild-type ERG4 but not by one bearing the ERG4 allele isolated from the mutant strain. We conclude that the prm1Δ kex2Δ mating defect-enhancing phenotype of the mutant strain is due to a loss-of-function mutation in ERG4, suggesting that ERG4 plays a role in cell fusion.
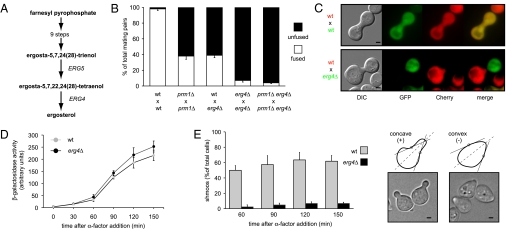
Erg4 catalyzes the last step of the yeast ergosterol biosynthetic pathway (Fig. 1A) (12). Two recent reports suggest a role for ERG4 in mating and cell fusion (13, 14). Whereas Jin et al. (13) report that deletion of ERG4 has no defect at the cell fusion step, Tiedje et al. (14) show that erg4Δ cells exhibit defects in both cell polarization and cell fusion. To test whether ERG4 acts in cell fusion, we used a quantitative cell fusion assay (8). Mating partners carrying deletions in PRM1, ERG4, or both were mixed and allowed to mate. One partner expressed soluble GFP and the other expressed a Pgk1-mCherry fusion protein as cytoplasmic markers. Mating pairs with mixed fluorescent signals throughout were scored as fused, and mating pairs in which fluorescence remained restricted to each partner were scored as unfused (Fig. 1B).
Fig. 1.
ERG4 mutant is defective in cell fusion. (A) Schematic representation of the ergosterol biosynthetic pathway highlighting the last two steps. (B) Quantitative cell fusion assays. All phenotypes were tested for both mating types, and the results were indistinguishable. Error bars indicate standard errors. (C) erg4Δ cells form morphologically abnormal mating pairs. MATα cells carrying cytoplasmic GFP were mixed with MATa cells carrying Pgk1-mCherry on nitrocellulose filters and incubated on YPD plates for 3 h at 30 °C. Fixed mating mixtures were then imaged by DIC and wide-field fluorescence microscopy. (Scale bars: 2 μm.) (D) erg4Δ cells preserve a normal pheromone response pathway. Strains harboring a FUS1-LACZ transcriptional reporter were grown and treated with 6 μM α-factor. At different time points, cells were harvested and assayed for β-galactosidase activity. Error bars indicate SEs. (E) erg4Δ cells are defective for shmoo formation. MATa cells were treated with α-factor, and aliquots were collected, fixed, and observed by bright-field microscopy at the indicated time points. For quantification (Left), cells that showed two convex edges along the axis of polarization (dotted lines) were scored as positive hits (Right). (Scale bars: 1 μm.)
In agreement with our previous results (7), deletion of PRM1 in both mating partners resulted in a substantial block to cell fusion. Deletion of ERG4 in only one partner reduced cell fusion efficiency [∼40% of mating pairs fused (Fig. 1B)], regardless of the mating type of the mutant (Fig. S1). When both mating partners lacked ERG4, cell fusion efficiency decreased to <10% and was further reduced in the absence of PRM1 [≤5% of mating pairs fused (Fig. 1B)]. The magnitude of these defects suggested an additive, rather than synergistic, effect of the double mutant, indicating that Prm1 and Erg4 act at different steps along the fusion pathway. Concordant with this notion, no prm1Δ-like cytoplasmic projections were observed in mating pairs derived from erg4Δ cells, suggesting that this mutant arrested at a step upstream of plasma membrane apposition. We also did not observe cell lysis in these mating pairs. Overall, the phenotypic differences between erg4Δ and prm1Δ and the additive defect of erg4Δ and prm1Δ mutations suggest that ERG4 and PRM1 participate in distinct processes during mating.
ERG4 Mutants Are Deficient in Shmoo Formation.
While scoring mating pairs in the quantitative cell fusion assay, we noticed that mating pairs that include at least one erg4Δ partner are morphologically distinct. In contrast to the elongated mating pairs formed by wild-type or prm1Δ cells, erg4Δ mating pairs have a shortened conjugal bridge, with mating partners displaying an ellipsoid cell shape reminiscent of unbudded, vegetatively growing cells (Fig. 1C). Analysis of cultures under mating conditions revealed that erg4Δ cells are defective in mating pair formation: ∼50% of erg4Δ cells in a mating mixture participated in recognizable mating pairs, compared to nearly 80% of cells in wild-type mating mixtures.
The mating pair formation defect could be explained by an inability of erg4Δ cells to form shmoos due to an impaired pheromone response and/or a defect in cell shape remodeling. To distinguish between these possibilities, we quantified pheromone response and shmoo formation in MATa cells treated with the mating pheromone α-factor. After the addition of α-factor, erg4Δ MATa cells arrest bud formation, leading to a population composed mainly of unbudded cells. This indicates that erg4Δ cells properly detect the pheromone and undergo G1 cell cycle arrest. To test the integrity of the pheromone MAPK signaling pathway, we measured the activity of a FUS1-LACZ reporter gene (6) whose expression requires the MAPK-dependent activation of the transcription factor Ste12. We found that the pheromone-induced activity of the FUS1 promoter in erg4Δ cells closely parallels that of wild-type cells (Fig. 1D). We conclude that MATa erg4Δ cells transduce the pheromone signal normally. In this regard, Erg4 is distinct from the upstream biosynthetic enzymes Erg2, Erg3, and Erg6, which, when lost, disrupt pheromone signaling (13).
To quantify shmoo formation, we scored shmoo-positive cells as those that, by visual inspection, showed two concave inflection points along the axis of polarization and shmoo-negative cells as those that showed no concave features (Fig. 1E). After 2 h in the presence of pheromone, ∼65% of wild-type cells formed shmoos (Fig. 1E). By contrast, <10% of erg4Δ cells had formed shmoos during the same period. Prolonged incubation of erg4Δ cells with α-factor for 3 and 4 h did not improve shmoo formation (Fig. S1). Therefore, although erg4Δ cells can detect pheromone, arrest the cell cycle, and activate mating-specific genes normally, they are severely impaired in remodeling cell shape to form a normal mating projection.
Sterols and Shmoo Tip Proteins Properly Polarize in erg4Δ Cells.
Ergosterol is particularly enriched in the yeast plasma membrane (15) and accumulates at the shmoo tip with other proteins involved in cell fusion, such as Prm1 and Fig1 (8, 16). We wondered whether the shmoo formation and cell fusion defects of erg4∆ could be attributed to defects in such polarization, as has been shown in erg3Δ and erg6Δ mutants (13, 16). Therefore, we subjected erg4Δ cells to mating pheromone and analyzed the distribution of sterols and membrane-associated proteins.
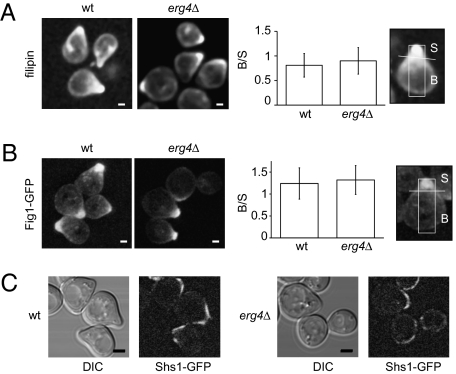
To visualize sterols, we used filipin staining (16). Qualitative observations showed an indistinguishable distribution in wild-type and erg4Δ cells (Fig. 2A). To quantitatively compare sterol polarization, we defined a shmoo area ‘‘S’’ and a body area ‘‘B’’ and then calculated the body/shmoo (B/S) fluorescence ratio (Materials and Methods). The resulting B/S ratios were indistinguishable for wild-type and erg4Δ cells, indicating that sterols polarize properly in the mutant.
Fig. 2.
erg4Δ cells show normal polarization of sterols and proteins. (A) Wild-type and erg4Δ MATa strains were treated with α-factor for 3 h, stained with filipin, and imaged by confocal microscopy (Left). (Scale bars: 1 μm.) Wild-type and erg4Δ MATa strains harboring either Fig1-GFP (B) or Shs1-GFP (C) were treated with α-factor for 3 h and then imaged by confocal microscopy. For A and B, fluorescence distribution along the longitudinal axis of cells was quantified as the fluorescence intensity ratio between the shmoo (S) and body (B) areas as described in Materials and Methods. Error bars indicate SEs. (Scale bars: B, 1 μm; C, 2 μm.)
We similarly examined the distribution of the mating-induced membrane proteins Fig1 and Prm1 by expressing GFP-fusions of these proteins. Fig1-GFP localization appeared qualitatively similar in erg4Δ and wild-type cells (Fig. 2B), as did the distribution of Prm1-GFP (Fig. S2). The B/S ratio confirmed that both proteins were polarized to a comparable extent.
Septins also become distributed in a polarized manner during shmoo formation and form a cortical filamentous collar around the neck of the growing shmoo (17), which is thought to function as a diffusion barrier (18). We found that the septin subunit Shs1-GFP localized around the neck of the growing shmoo and was excluded from its tip similarly in erg4∆ and wild-type cells (Fig. 2C).
Some plasma membrane proteins, like the SNARE Snc1, polarize through dynamic partitioning driven by localized secretion combined with slow diffusion and spatially separated endocytosis (19). We tested whether this mechanism was affected in erg4Δ by monitoring Snc1-GFP localization and found that it was indistinguishable from that in wild-type cells (Fig. S2). Thus, the defects of erg4Δ cells in shmoo formation and cell fusion are not readily explained by underlying defects in the tested mechanisms of lipid or membrane protein polarization.
Shmoo Formation, but Not Cell Fusion, Is Rescued by Exogenously Supplied Ergosterol.
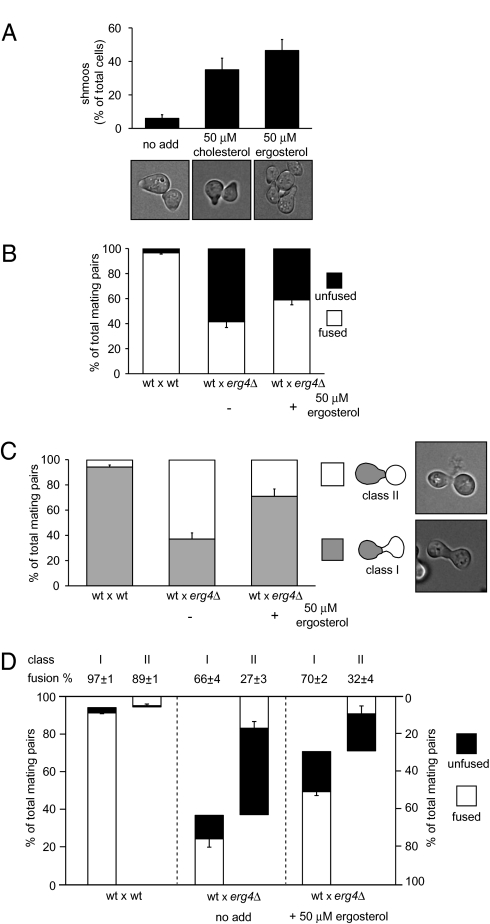
If the mating defects observed in erg4Δ cells are due to lack of ergosterol, then sterol supplementation in the growth medium would be expected to complement these erg4Δ deficiencies. Because yeast cells take up sterols only under anaerobic conditions (20), we added ergosterol or cholesterol to erg4Δ and wild-type cultures grown anaerobically and then assayed the cultures for shmoo formation and cell fusion (Fig. 3). Indeed, ergosterol or cholesterol suppressed the erg4Δ shmooing defect almost completely [from 6% of cells forming shmoos to 47% (Fig. 3A), compared to 60% in wild-type cells (Fig. 1E)]. By contrast, cell fusion efficiency increased only modestly from 41% to 59% after ergosterol supplementation (Fig. 3B), indicating that ergosterol depletion is sufficient to explain the shmoo formation defect but not the cell fusion defect.
Fig. 3.
Sterol supplementation partially suppresses erg4Δ shmooing and cell fusion defects. (A) erg4Δ cells were grown in anaerobic conditions in the presence of ergosterol and cholesterol. Cells were then harvested and treated with α-factor for 3 h, fixed, and observed under the microscope for shmoo formation quantification. Error bars indicate SEs. (B–D) Cell fusion assay carried out with erg4Δ cells grown anaerobically either in the presence or absence of ergosterol and then mated with wild-type MATα cells bearing cytoplasmic GFP. Total cell fusion scores are shown in B. Different classes of mating pairs were classified depending on either the presence (Class I) or absence (Class II) of proper shmoo morphology in the erg4Δ partner, as shown in C. A breakdown of cell fusion scores for each class of mating pair, genotype, and growing conditions is shown in D. Error bars indicate standard errors.
Analysis of the morphology of mating pairs revealed further insights into the relationship between shmoo formation and cell fusion efficiency. We grouped mating pairs in two classes: Class I mating pairs, which showed elongated, normal projections contributed by both partners; and Class II mating pairs, in which at least one partner showed no projection (Fig. 3C). For erg4Δ × wild-type mating pairs (in which the wild-type mating partner expressed cytoplasmic GFP), we scored exclusively the morphologies of the erg4Δ partners.
In almost all wild-type × wild-type mating pairs both partners display well defined projections (Fig. 3C; 94% Class I mating pairs). Rare Class II wild-type × wild-type mating pairs are likely due to asynchronous mating events in which one partner completed the previous cell cycle earlier. The cell fusion score is slightly higher for wild-type × wild-type mating pairs in Class I (97% of mating pairs fused) than Class II (89% of mating pairs fused) (Fig. 3D), consistent with the idea that the latter class arises from more recently formed mating pairs.
In contrast to wild-type × wild-type mating pairs, only 37% of wild-type × erg4Δ mating pairs fell into Class I (Fig. 3C). Although both classes displayed a pronounced defect in cell fusion, the morphologically normal Class I wild-type × erg4Δ mating pairs fused more efficiently than the Class II mating pairs (66% and 27%, respectively; Fig. 3D), suggesting that the population of erg4Δ partners that can form a mature mating projection is more fusion-competent, albeit still less so than wild-type cells.
After ergosterol supplementation, the proportion of Class I wild-type × erg4Δ mating pairs almost doubled from 37% to 71% (Fig. 3C). By contrast, cell fusion efficiency within each class was not substantially changed (66% and 70% of mating pairs fused for Class I, 27% and 32% for Class II, without and with ergosterol supplementation, respectively). Thus, the observed increase in cell fusion after ergosterol supplementation is due mainly to the increase in the proportion of properly formed Class I mating pairs. However, supplemental ergosterol was unable to rescue the decreased cell fusion efficiency of even these morphologically normal mating pairs.
Sterol Analysis of erg Mutants.
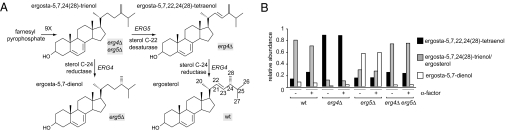
We reasoned that the defects observed in the erg4Δ mutant could be attributed, in principle, to lack of ergosterol, inappropriate accumulation of the intermediate that serves as the Erg4 substrate [ergosta-5,7,22,24(28)-tetraenol], or both. Ergosta-5,7,22,24(28)-tetraenol would be predicted not to accumulate in a mutant that lacks the enzyme that produces it, Erg5. Importantly, elimination of late steps of ergosterol biosynthesis usually does not block the pathway because the remaining enzymes can use improperly modified precursors as substrates (15, 21); thus, as depicted in Fig. 4A, erg4Δ and erg5Δ mutants should accumulate sterols that differ from each other and from ergosterol by the presence or absence of a double bond in the aliphatic chain of the sterol.
Fig. 4.
Accumulation of different sterols in erg mutants. (A) Structures of the expected predominant sterols for each mutant strain. In the double mutant erg4Δ erg5Δ, the remaining sterol biosynthetic route ends in the production of ergosta-5,7,24(28)-trienol, whereas in the single mutant erg4Δ, the lack of sterol C-24 reductase activity produces the accumulation of ergosta-5,7,22,24(28)-tetraenol. As Erg4 sterol C-24 reductase activity can use ergosta-5,7,24(28)-trienol as a substrate, the predominant sterol in the erg5Δ strain should be ergosta-5,7-dienol. (B) Semiquantitative analysis of accumulated sterols. Free sterols were further purified and analyzed by mass spectrometry. The normalized peak areas of the protonated sterols in the MS1 are shown. Original spectra are depicted in Figs. S3–S5.
We determined the sterol composition of erg4Δ, erg5Δ, and erg4Δ erg5Δ mutants during vegetative growth and mating conditions. To this end, we purified free sterols by fractionation of total lipid extracts by TLC. Sterols were detected as ammonia adducts and protonated species by tandem mass spectrometry (MS/MS). MS analysis confirmed that erg4Δ mutants accumulate ergosta-5,7,22,24(28)-tetraenol, whereas erg5Δ and erg4Δ erg5Δ mutants accumulate ergosta-5,7-dienol and ergosta-5,7,24(28)-trienol, respectively (Fig. 4 and Figs. S3 and S4). Semiquantitative analysis of sterol composition of the different strains incubated in the presence of α-factor showed no differences when compared with normal growth conditions (Fig. 4B and Fig. S5), indicating that no global changes in sterol composition occur during shmoo formation.
Thus, taken together, our results confirm the identity of the predicted species as the major sterols that accumulate in each mutant. These sterols only differ by number and placement of double bonds in their aliphatic chains. We note in particular that ergosta-5,7,22,24(28)-tetraenol, which accumulates in the erg4Δ mutant, is the only sterol among this set that contains two conjugated double bonds in its aliphatic moiety, which would add conformational rigidity.
ERG5 Deletion Suppresses the erg4Δ Phenotype.
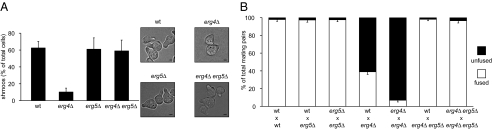
Next, we analyzed shmoo formation and cell fusion when ergosta-5,7,22,24(28)-tetraenol production is impaired. Unlike erg4Δ cells, pheromone-treated erg5Δ cells efficiently formed shmoos, which were morphologically indistinguishable from those formed by wild-type cells (Fig. 5A). Furthermore, both erg5Δ × wild-type and erg5Δ × erg5Δ mating pairs fuse with almost 100% efficiency (Fig. 5B).
Fig. 5.
erg5Δ suppresses erg4Δ. (A) Cells growing on YPD were treated with α-factor for 3 h, fixed, and examined under the microscope for quantification of shmooing efficiency. Error bars indicate standard errors. Bright-field microscopy representative images are shown at Right. (Scale bars: 1 μm.) (B) Quantitative cell fusion assays were performed testing both mating types for all mutants, and the results were indistinguishable. Error bars indicate SEs.
To our surprise, erg4Δ erg5Δ double mutant cells also behaved identically to wild-type cells. Deletion of ERG5 in erg4Δ cells restored shmoo formation to wild-type levels (Fig. 5A). Also, erg4Δ erg5Δ double mutants mated and fused with wild-type cells and with erg4Δ erg5Δ cells with efficiencies indistinguishable from wild type (Fig. 5B). Ectopic expression of ERG5 in the erg4Δ erg5Δ double mutant confirmed the role of erg5Δ as an erg4Δ suppressor (Fig. S6).
The ability of erg5∆ to suppress erg4∆ indicates that neither ergosterol nor the Erg4 protein per se is required for shmoo formation and cell fusion. Rather, the aberrantly high levels of ergosta-5,7,22,24(28)-tetraenol that accumulate in the absence of Erg4 may explain both these defects.
Discussion
We have shown that ERG4 is required for proper cell shape remodeling and cell fusion during yeast mating and that, surprisingly, deletion of ERG5 suppresses these defects. These results can be explained in terms of sterol composition: wild-type, erg5Δ, and erg4Δ erg5Δ cells contain sterol derivatives with zero or one double bonds in their aliphatic chains and these cells form shmoos and fuse normally; by contrast, erg4Δ cells contain a predominant sterol with two conjugated double bonds in its aliphatic chain, and these cells can neither form shmoos nor fuse robustly. Addition of supplemental ergosterol or cholesterol (one or zero double bonds in the aliphatic chain, respectively) restores shmoo formation in erg4Δ cells, indicating a requirement for the more flexible sterol aliphatic chain. In contrast, supplemental ergosterol did not rescue cell fusion, indicating that the more rigid aliphatic chain of ergosta-5,7,22,24(28)-tetraenol interferes with a later step in the mating pathway. It is remarkable that such subtle modifications in the aliphatic moiety of ergosterol can exert such pronounced consequences on cell physiology.
What role could ERG4 play in shmoo formation and mating? One possibility is that the aliphatic chain of ergosterol, or the Erg4 protein itself, could act as a cofactor to proteins important in cell remodeling or cell wall breakdown. For example, the PAK kinase Ste20, which affects mating and cell polarity, has recently been suggested to interact physically with Erg4 (14). Changes in sterol composition may also act indirectly; for example, by leading to a change in sphingolipid metabolism (22). However, because both shmooing and cell fusion defects in erg4Δ cells are suppressed by ERG5 deletion, it seems unlikely that a physical interaction with Erg4 or with ergosterol per se is required.
Alternatively, by affecting sterol structure, Erg4 could alter the biophysical properties of the lipid bilayer, particularly membrane curvature and lipid phase separation (23, 24). Cholesterol is thought to be a major modulator of membrane shape changes, including SNARE-mediated and viral membrane fusion (25, 26). Empirical evidence as well as simulations show that the double bond in the aliphatic chain of ergosterol increases its ability to pack tightly with surrounding lipids, and promotes phase separation within the membrane (27, 28). By extension, the second, conjugated double bond of ergosta-5,7,22,24(28)-tetraenol would further restrict the mobility of the aliphatic chain and so would be predicted to lead to even higher condensation properties. Thus, it is plausible that erg4Δ cells, owing to the accumulation of ergosta-5,7,22,24(28)-tetraenol, have more tightly packed membranes, in which lateral diffusion of proteins and lipids may be suboptimal.
Importantly, the erg4∆ defects are distinct from those previously reported for cells with altered sterol composition (29). First, the lack of a cone of negative curvature in the shmoo is a phenotypic trait of erg4Δ cells that has not been observed in other mating-defective erg mutants. Second, a recent study shows that erg3Δ and erg6Δ mutants display a “prm1Δ-like” phenotype (15), while we showed that erg4∆ does not. In contrast to our results, this study reports only a mild cell fusion defect for the erg4Δ mutant (15). Possibly, the difficulty of recognizing erg4Δ mating pairs of the aberrant Class II morphology led to an oversampling of morphologically normal Class I mating pairs, which fuse more efficiently.
Our results suggest that the roles of ERG4 in shmooing and cell fusion are distinct: (i) erg4Δ × wild-type Class I mating pairs are morphologically normal but fuse with low efficiency and, (ii) ergosterol supplementation rescues the shmooing defect but does not improve cell fusion. Perhaps low levels of ergosterol are sufficient to promote growth polarization, whereas higher levels are required for efficient cell fusion. However, in light of our finding that removal of ergosta-5,7,22,24(28)-tetraenol by ERG5 deletion completely suppressed the erg4Δ phenotypes, a more likely hypothesis is that cell fusion is obstructed by accumulation of this tetraenol—a remarkable consequence of a two-atom chemical change in a lipid.
Materials and Methods
Media and Yeast Strains.
Synthetic complete (SC) and complex (YPD) media were prepared and supplemented with 2% glucose by using reagents from Difco and Sigma-Aldrich. All strains used in this study (Table S1) are derivatives of wild-type strain W303. For description of strain constructions, see SI Methods. For growth in anaerobic conditions, cells were first grown under aeration at 30 °C in YPD media up to OD600 = 0.3–0.5. Cultures were then diluted 20-fold in YPD with or without sterol supplementation, placed on an anaerobic jar system (BD BBL GasPak), and incubated at 30 °C overnight. Five millimolar (100×) ergosterol and cholesterol stock solutions were prepared in 50% ethanol/50% Tween 80 (Sigma-Aldrich).
Genetic Screening for Enhancers of prm1Δ.
Mutagenesis and genetic screening were performed as described in ref. 11. For a description, see SI Methods.
Quantitative Cell Fusion and Shmooing Assays.
Quantitative cell fusion assay was performed as described in ref. 8. For a description, see SI Methods. For shmooing assays, MATa strains were grown at 30 °C to mid-logarithmic growth phase and treated with 10 μg/mL α-factor. At indicated times, aliquots were taken and then fixed in 4% paraformaldehyde before inspection by microscopy. For each strain and condition, three independent experiments were done, and at least 200 cells were scored.
β-Galactosidase Assays.
β-Galactosidase assays were performed as described in ref. 11. For a description, see SI Methods.
Microscopy.
Visualization of sterol-rich domains was performed as described in ref. 16. For visualization of GFP fusion proteins, live cells were grown and directly mounted for microscopy. For a description, see SI Methods.
Quantification of Fluorescence Polarization.
Analysis of cell fluorescence intensity was performed by using ImageJ 1.40g (http://rsb.info.nih.gov/ij). Between 10 and 18 stacks spaced at 0.4-μm intervals spanning the entire cell were acquired and then processed for background subtraction and z-projected using a sum intensity projection. For each cell, we defined a rectangular area centered along the longitudinal axis of the cell with 1/3 of the cell width and cell length as the rectangular dimensions. The line that encompasses the two inflection points defined by the change of membrane curvature of the shmoo tips was used to separate the shmoo area "S" and the body area "B." Because the majority of pheromone-treated erg4Δ cells have no concave curvature along their membranes, we determined the average shmoo/cell axis length ratio of the subset of erg4Δ cells that showed shmoos with inflections and applied this ratio to the rest of the population. For each strain, two independent experiments were done where at least 10 cells were scored.
Sterol Purification and Mass Spectrometry.
Yeast total lipids were extracted as described, and free sterols were purified by silica chromatography (for a description, see SI Methods). Sterol fractions were diluted in CHCl3/MeOH/2-propanol 1/2/4 (vol/vol/vol) containing 5 mM ammonium acetate. All measurements were performed on a modified QSTAR Pulsar i quadrupole time-of-flight mass spectrometer (MDS Sciex) equipped with an automated nanospray chip ion source (NanoMate HD; Advion BioSciences). Acquired spectra were interpreted with Analyst QS 1.1 (MDS Sciex). Theoretical and experimentally determined m/z values for the [M+H], [M+NH4], and [M+H-H2O] ions are displayed in Table S2. MS/MS experiments with low collision energy were performed on [M+NH4] precursor ions for detection of the characteristic [M+H-H2O] fragment ions. For semiquantitative interpretation, peak areas of the [M+H] of the identified sterol species were extracted, and afterward a correction for overlapping isotopic peaks was performed manually (Figs. S4 and S5). The resulting corrected values were normalized according to the sum of all detected sterols.
Supplementary Material
Acknowledgments
We thank members of the Walter laboratory for valuable suggestions and discussion, and Sean O′Rourke and Ira Herskowitz for kindly providing the CEN/ARS yeast genomic DNA library. This work was supported by a Damon Runyon Cancer Research Fund postdoctoral fellowship (to P.S.A.), a Human Frontiers Science Project (to T.C.W.), and grants from the National Institute of Health (to P.W.). P.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Freely available online through the PNAS open access option.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914094107/DCSupplemental.
References
- 1.Chen EH, Grote E, Mohler W, Vignery A. Cell-cell fusion. FEBS Lett. 2007;581:2181–2193. doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 2.Ydenberg CA, Rose MD. Yeast mating: a model system for studying cell and nuclear fusion. Methods Mol Biol. 2008;475:3–20. doi: 10.1007/978-1-59745-250-2_1. [DOI] [PubMed] [Google Scholar]
- 3.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 4.Gammie AE, Brizzio V, Rose MD. Distinct morphological phenotypes of cell fusion mutants. Mol Biol Cell. 1998;9:1395–1410. doi: 10.1091/mbc.9.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philips J, Herskowitz I. Osmotic balance regulates cell fusion during mating in Saccharomyces cerevisiae. J Cell Biol. 1997;138:961–974. doi: 10.1083/jcb.138.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trueheart J, Boeke JD, Fink GR. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguilar PS, Engel A, Walter P. The plasma membrane proteins Prm1 and Fig1 ascertain fidelity of membrane fusion during yeast mating. Mol Biol Cell. 2007;18:547–556. doi: 10.1091/mbc.E06-09-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin H, Carlile C, Nolan S, Grote E. Prm1 prevents contact-dependent lysis of yeast mating pairs. Eukaryot Cell. 2004;3:1664–1673. doi: 10.1128/EC.3.6.1664-1673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engel A, Walter P. Membrane lysis during biological membrane fusion: collateral damage by misregulated fusion machines. J Cell Biol. 2008;183:181–186. doi: 10.1083/jcb.200805182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heiman MG, Engel A, Walter P. The Golgi-resident protease Kex2 acts in conjunction with Prm1 to facilitate cell fusion during yeast mating. J Cell Biol. 2007;176:209–222. doi: 10.1083/jcb.200609182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zweytick D, Hrastnik C, Kohlwein SD, Daum G. Biochemical characterization and subcellular localization of the sterol C-24(28) reductase, erg4p, from the yeast saccharomyces cerevisiae. FEBS Lett. 2000;470:83–87. doi: 10.1016/s0014-5793(00)01290-4. [DOI] [PubMed] [Google Scholar]
- 13.Jin H, McCaffery JM, Grote E. Ergosterol promotes pheromone signaling and plasma membrane fusion in mating yeast. J Cell Biol. 2008;180:813–826. doi: 10.1083/jcb.200705076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiedje C, Holland DG, Just U, Höfken T. Proteins involved in sterol synthesis interact with Ste20 and regulate cell polarity. J Cell Sci. 2007;120:3613–3624. doi: 10.1242/jcs.009860. [DOI] [PubMed] [Google Scholar]
- 15.Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 16.Bagnat M, Simons K. Cell surface polarization during yeast mating. Proc Natl Acad Sci USA. 2002;99:14183–14188. doi: 10.1073/pnas.172517799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gladfelter AS. Control of filamentous fungal cell shape by septins and formins. Nat Rev Microbiol. 2006;4:223–229. doi: 10.1038/nrmicro1345. [DOI] [PubMed] [Google Scholar]
- 18.Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- 19.Valdez-Taubas J, Pelham HR. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr Biol. 2003;13:1636–1640. doi: 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Sturley SL. Conservation of eukaryotic sterol homeostasis: new insights from studies in budding yeast. Biochim Biophys Acta. 2000;1529:155–163. doi: 10.1016/s1388-1981(00)00145-1. [DOI] [PubMed] [Google Scholar]
- 21.Heese-Peck A, et al. Multiple functions of sterols in yeast endocytosis. Mol Biol Cell. 2002;13:2664–2680. doi: 10.1091/mbc.E02-04-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan XL, et al. Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol Biol Cell. 2009;20:2083–2095. doi: 10.1091/mbc.E08-11-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bacia K, Schwille P, Kurzchalia T. Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proc Natl Acad Sci USA. 2005;102:3272–3277. doi: 10.1073/pnas.0408215102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumgart T, Hess ST, Webb WW. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- 25.Qiang W, Weliky DP. HIV fusion peptide and its cross-linked oligomers: efficient syntheses, significance of the trimer in fusion activity, correlation of beta strand conformation with membrane cholesterol, and proximity to lipid headgroups. Biochemistry. 2009;48:289–301. doi: 10.1021/bi8015668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang J, et al. Fusion step-specific influence of cholesterol on SNARE-mediated membrane fusion. Biophys J. 2009;96:1839–1846. doi: 10.1016/j.bpj.2008.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czub J, Baginski M. Comparative molecular dynamics study of lipid membranes containing cholesterol and ergosterol. Biophys J. 2006;90:2368–2382. doi: 10.1529/biophysj.105.072801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X, et al. Effect of the structure of natural sterols and sphingolipids on the formation of ordered sphingolipid/sterol domains (rafts). Comparison of cholesterol to plant, fungal, and disease-associated sterols and comparison of sphingomyelin, cerebrosides, and ceramide. J Biol Chem. 2001;276:33540–33546. doi: 10.1074/jbc.M104776200. [DOI] [PubMed] [Google Scholar]
- 29.Tomeo ME, Fenner G, Tove SR, Parks LW. Effect of sterol alterations on conjugation in Saccharomyces cerevisiae. Yeast. 1992;8:1015–1024. doi: 10.1002/yea.320081204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.