Summary
Antibiotic resistance arises through mechanisms such as selection of naturally occurring resistant mutants and horizontal gene transfer. Recently, oxidative stress has been implicated as one of the mechanisms whereby bactericidal antibiotics kill bacteria. Here we show that sub-lethal levels of bactericidal antibiotics induce mutagenesis, resulting in heterogeneous increases in the minimum inhibitory concentration for a range of antibiotics, irrespective of the drug target. This increase in mutagenesis correlates with an increase in ROS, and is prevented by the ROS scavenger thiourea and by anaerobic conditions, indicating that sub-lethal concentrations of antibiotics induce mutagenesis by stimulating the production of ROS. We demonstrate that these effects can lead to mutant strains that are sensitive to the applied antibiotic but resistant to other antibiotics. This work establishes a radical-based molecular mechanism whereby sub-lethal levels of antibiotics can lead to multidrug resistance, which has important implications for the widespread use and misuse of antibiotics.
Introduction
There are a number of mechanisms whereby bacteria can develop antibiotic resistance (Dwyer et al., 2009; Hegreness et al., 2008; Livermore, 2003; McKenzie and Rosenberg, 2001), including horizontal transfer of resistance genes (Davies, 1994), drug-specific selection of naturally occurring resistant variants within a population, and increased mutagenesis in hypermutator strains (Andersson, 2003; Chopra et al., 2003). Quinolones, which are DNA damaging antibiotics, can stimulate the emergence of drug resistance via SOS-independent recombination (Lopez et al., 2007), and through the induction of RecA-mediated processes including homologous recombination (Drlica and Zhao, 1997) and SOS-regulated error-prone polymerases (Cirz et al., 2005). β-lactams can also induce the SOS response via RecA (Kohanski et al., 2007) and the DpiAB two-component system (Miller et al., 2004), and these drugs have been shown to induce DinB in an SOS-independent fashion resulting in increased frameshift mutations (Perez-Capilla et al., 2005).
Antibiotic treatment can also result in multidrug resistance (Cohen et al., 1989), which has been associated with mutations in multidrug efflux pumps, such as AcrAB (Ma et al., 1993). These drug efflux pumps can be regulated by a number of transcription factors, including the superoxide-responsive SoxRS system (Greenberg et al., 1990). In addition, there is evidence that low-level antibiotic treatment can lead to mutations that cause resistance (Girgis et al., 2009); however, the mechanisms underlying this effect are not well understood.
Bactericidal antibiotics, including β-lactams, quinolones and aminoglycosides, can stimulate bacteria to produce reactive oxygen species (ROS) (Dwyer et al., 2007; Kohanski et al., 2007; Kohanski et al., 2008), which are highly deleterious molecules that can interfere with the normal functions of oxygen-respiring organisms (Brumaghim et al., 2003; Fridovich, 1978; Imlay, 2006). Certain ROS, such as hydroxyl radicals, can directly damage DNA and lead to an accumulation of mutations (Demple and Harrison, 1994; Friedberg et al., 2006; Imlay et al., 1988). Oxidative DNA damage also activates the error-prone SOS response (Carlsson and Carpenter, 1980; Imlay and Linn, 1986, 1987), and error-correcting repair systems such as the “GO” repair system (Michaels and Miller, 1992; Miller, 1996). In this study, we hypothesized that ROS formation due to treatment with low levels of bactericidal antibiotics leads to an increase in mutation rates which can result in the emergence of multidrug resistance. We thus consider a possible molecular mechanism whereby bactericidal antibiotics act as active, reactive mutagens.
Results
To test the above hypothesis, we examined mutation rates in E. coli strain MG1655 following treatment with low levels of the bactericidal antibiotics, norfloxacin (quinolone), ampicillin (β-lactam) and kanamycin (aminoglycoside), respectively. Mutation rates were determined by plating aliquots of treated cultures onto rifampicin plates, counting rifampicin-resistant colonies, and using the MSS maximum likelihood method (Rosche and Foster, 2000) to estimate the number of mutation events per culture (see Experimental Procedures for additional details). The mutation rate for untreated wildtype E. coli was approximately 1.5×10−8 mutations/cell/generation.
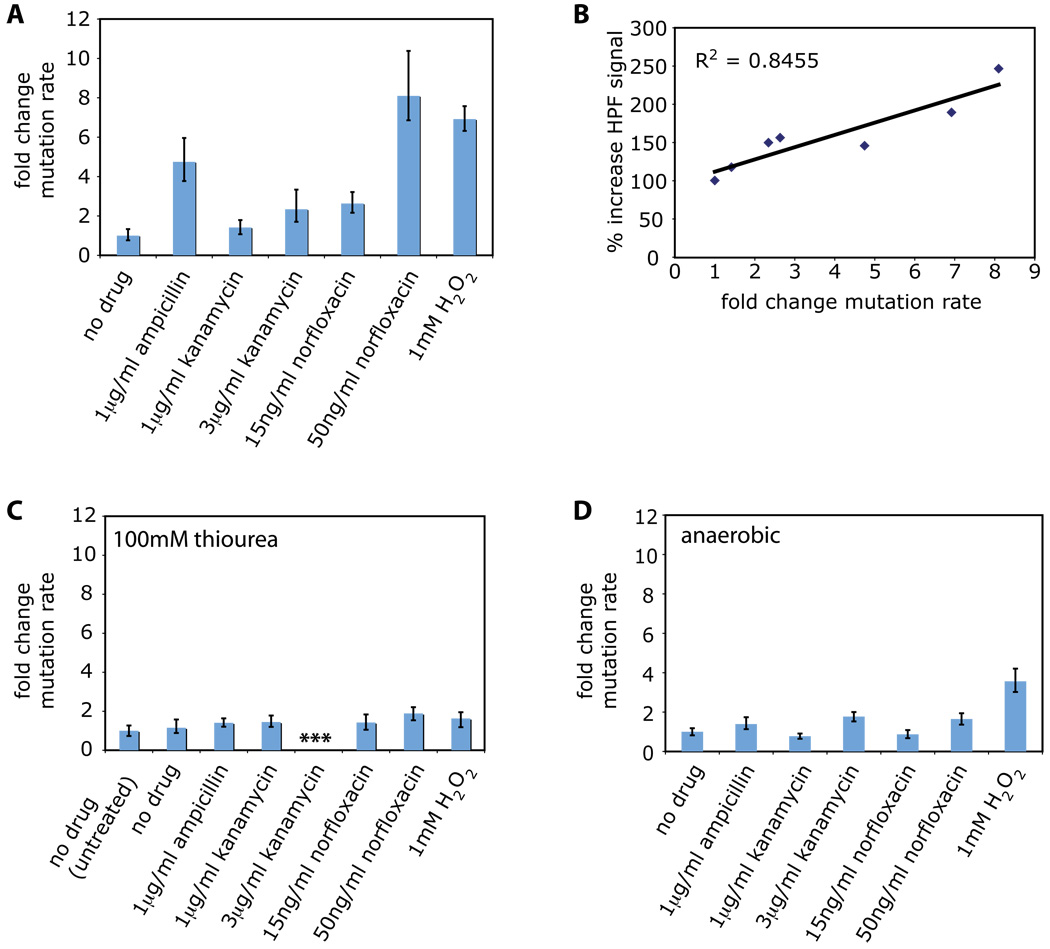
Treatment with 1µg/ml ampicillin, 3µg/ml kanamycin, 15ng/ml norfloxacin, or 50ng/ml norfloxacin resulted in significant increases in the mutation rate relative to an untreated control (Figure 1A). Treatment with 1µg/ml kanamycin resulted in a modest increase in mutation rate (Figure 1A). The largest increases in mutation rate were seen following treatment with ampicillin or 50ng/ml norfloxacin (Figure 1A). These changes were on par with the increase in mutation rate observed following treatment with 1mM hydrogen peroxide (Figure 1A), a concentration of hydrogen peroxide known to induce hydroxyl radical formation via Fenton chemistry (Imlay et al., 1988). To determine if there is a correlation between these changes in mutation rate and ROS formation, we examined radical levels using the radical-sensitive dye hydroxyphenyl fluorescein, HPF (Setsukinai et al., 2003) (see Experimental Procedures for more details). We found a significant correlation (R2 = 0.8455) between the fold change in mutation rate and peak HPF signal for the treatments described above (Figure 1B).
Figure 1. Low levels of bactericidal antibiotics increase mutation rate due to reactive oxygen species formation.
(A) Fold change in mutation rate (mean +/− 95% CI) relative to an untreated control (no drug) for wildtype E. coli (MG1655) following an overnight treatment with 1µg/ml ampicillin, 1µg/ml kanamycin, 3µg/ml kanamycin, 15ng/ml norfloxacin, 50ng/ml norfloxacin, or 1mM hydrogen peroxide (H2O2). (B) Correlation between oxidative stress levels (HPF fluorescence) and fold change in mutation rate for wildtype E. coli for the treatments described in A. (C–D) Fold change in mutation rate (mean +/− 95% CI) relative to an untreated control (no drug) for wildtype E. coli following an overnight treatment with 100mM thiourea and no drug, 1µg/ml ampicillin, 1µg/ml kanamycin, 3µg/ml kanamycin, 15ng/ml norfloxacin, 50ng/ml norfloxacin, or 1mM hydrogen peroxide (H2O2) under (C) aerobic growth conditions with 100mM thiourea or (D) anaerobic growth conditions.
The strong correlation between ROS formation and fold change in mutation rate following treatment with bactericidal antibiotics suggests that ROS actively contribute to bactericidal drug-induced mutagenesis. To test if this is indeed the case, we added 100mM thiourea to wildtype E. coli treated with antibiotics or hydrogen peroxide at the concentrations noted above (Figure 1C). Thiourea is a potent hydroxyl radical scavenger which mitigates the effects of hydroxyl radical damage in both prokaryotes and eukaryotes (Novogrodsky et al., 1982; Repine et al., 1981; Touati et al., 1995). We have previously shown that thiourea reduces hydroxyl radical formation and cell killing following treatment with bactericidal antibiotics (Kohanski et al., 2007).
The addition of thiourea significantly reduced the mutation rate to near untreated levels following the addition of 1mM hydrogen peroxide, norfloxacin, or ampicillin (Figure 1C). Interestingly, we were unable to detect any rifampicin-resistant colonies after plating up to 109 cells following treatment with both 3µg/ml kanamycin and thiourea (Figure 1C). However, we were able to detect rifampicin-resistant colonies after scaling up the system to 1L flasks and plating up to 1010 cells following treatment with both 3µg/ml kanamycin and thiourea (data not shown). These results suggest a role for kanamycin-mediated interference with ribosome function and translation, in the absence of oxidative stress, on significantly lowering mutation rate.
To further demonstrate that antibiotic-mediated ROS formation has a mutagenic component, we examined mutation rates under anaerobic growth conditions (see Experimental Procedures for additional details) following treatment of wildtype E. coli with antibiotics or hydrogen peroxide as described above (Figure 1D). We observed mutation rates near untreated levels for all antibiotic treatments tested (Figure 1D). Treatment with 1mM hydrogen peroxide, which results in direct addition of an oxidant, led to an increase in mutation rate relative to the no-drug control under anaerobic growth conditions (Figure 1D), but this increase was considerably smaller than that exhibited under aerobic growth conditions (Figure 1A).
Antibiotic-resistant strains can arise via drug-mediated selection of pre-existing antibiotic-resistant variants that occur naturally within a population (Livermore, 2003). Antibiotic-induced oxidative stress may be an additional mechanism that allows for the accumulation of mutations that increase resistance to drugs, irrespective of the drug target of the applied antibiotic. To test this, we measured changes in the MIC of wildtype E. coli over a period of 5 days of selective growth for the following antibiotics: norfloxacin, kanamycin, ampicillin, tetracycline and chloramphenicol. During the growth period, the cultures were exposed to no drug, norfloxacin, ampicillin, or kanamycin (see Experimental Procedures for more details). In all cases, growth in the absence of antibiotics did not change the MIC for any of the drugs tested (data not shown).
Treatment with 25ng/ml norfloxacin led to an increase in the MIC for norfloxacin and kanamycin, respectively (Figure S1A). The observed increases in MIC following treatment with norfloxacin were concentration dependent (see Supplemental Data for more details). Treatment of wildtype E. coli with 3µg/ml kanamycin led to an increase in the MIC for kanamycin and minimal increases in the MIC for norfloxacin and ampicillin, respectively (Figure S1C). The MIC for tetracycline and chloramphenicol, respectively, did not change (Figure S1C), indicating that kanamycin treatment may not lead to mutants resistant to other classes of ribosome inhibitors.
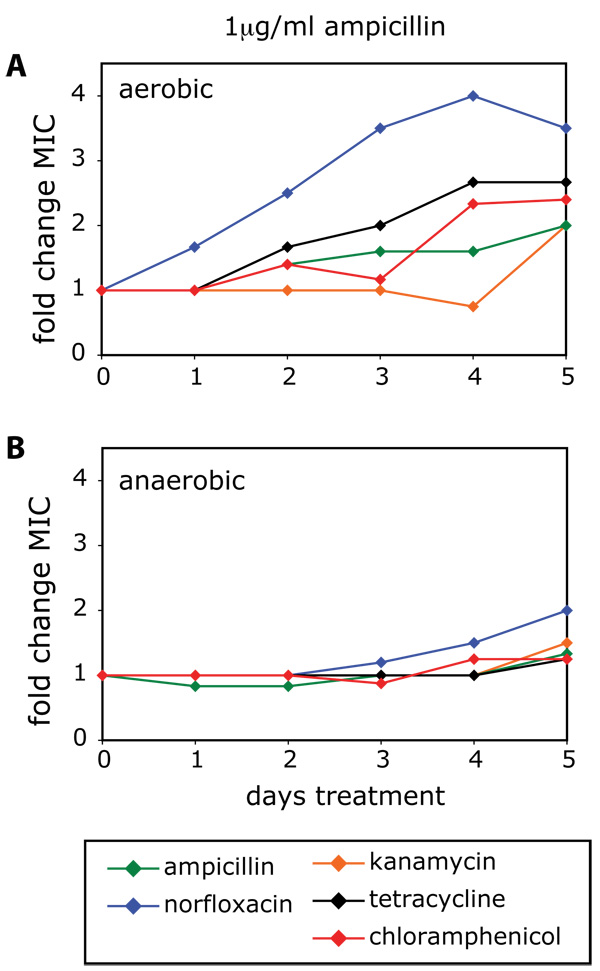
Treatment of wildtype E. coli with 1µg/ml ampicillin for 5 days led to an increase in the MIC to different levels for ampicillin, norfloxacin, kanamycin, tetracycline and chloramphenicol, respectively (Figure 2A). These results show that treatment with a β-lactam can stimulate formation of mutants that are potentially resistant to a wide range of antibiotics. Cultures that had been grown for 5 days in the presence of low levels of ampicillin were shifted to a drug-free environment, and grown without any ampicillin for 2 additional days. The MICs, which were increased after 5 days of ampicillin treatment (Figure 2A), remained elevated and did not change significantly following two days of growth in the absence of ampicillin (Figure S2). These findings demonstrate that the observed increases in MIC are stable and not due to a transient adaptation to growth in the presence of ampicillin.
Figure 2. Low levels of bactericidal antibiotics can lead to broad-spectrum increases in MIC due to ROS-mediated mutagenesis.
(A–B) Fold change in MIC relative to an aerobic no-drug control for ampicillin, norfloxacin, kanamycin, tetracycline and chloramphenicol, following 5 days of growth in the presence 1µg/ml ampicillin under (A) aerobic or (B) anaerobic growth conditions. See also Figure S1.
To determine if the observed increases in MIC were related to antibiotic-mediated ROS formation, we measured the MIC for ampicillin, norfloxacin, kanamycin, tetracycline and chloramphenicol, respectively, following treatment with no drug or 1µg/ml ampicillin under anaerobic growth conditions. Untreated anaerobic growth had no effect on MIC relative to untreated aerobic growth (data not shown). Following treatment with 1µg/ml ampicillin under anaerobic conditions, we observed almost no increase in MIC for ampicillin, kanamycin, tetracycline or chloramphenicol (Figure 2B). The MIC for norfloxacin exhibited a small increase by day 5 (Figure 2B); however, this change in MIC was much smaller than the increase in MIC for norfloxacin following ampicillin treatment under aerobic growth conditions (Figure 2A). These results suggest that ROS formation due to treatment with low levels of bactericidal antibiotics can lead to mutagenesis and the emergence of bacteria resistant to a wide range of antibiotics.
Drug resistance may not always be uniform throughout a population. Some cells within a population may remain susceptible to the antibiotic whereas other cells display varying degrees of drug resistance (de Lencastre et al., 1993), a phenomena referred to as heteroresistance. Antibiotic-stimulated, ROS-mediated mutagenesis could be a mechanism that stimulates the formation of a range of mutations that result in varying MICs within a population of cells. We sought to determine if the observed increases in population-level MIC for ampicillin following 5 days of treatment with 1µg/ml ampicillin (Figure 2A) exhibited heterogeneity in MIC at the single colony level.
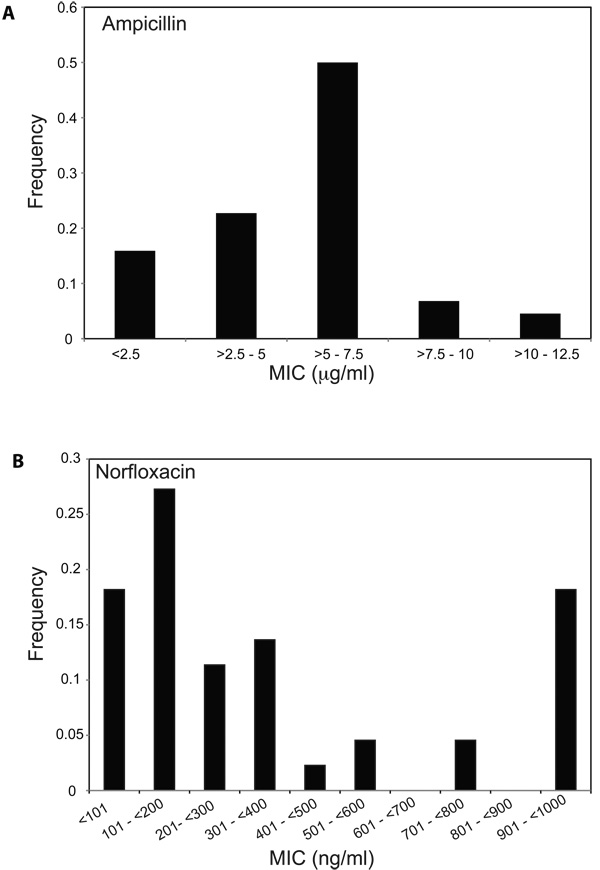
We isolated individual colonies following ampicillin treatment and measured the MIC of each clone to ampicillin. We found that these isolates exhibited a range of resistance to ampicillin (>2.5–12.5µg/ml), with some isolates remaining completely susceptible (≤ 2.5µg/ml) to treatment with this drug (Figure 3A). We also found that the MICs for these isolates to norfloxacin ranged from <100ng/ml (completely susceptible) to ≥ 1000ng/ml (Figure 3B). Although levels of resistance from clinical isolates are typically quite high (with MICs in the range of 10–1000µg/ml for norfloxacin (Becnel Boyd et al., 2009)), the upper ranges of the MICs for ampicillin or norfloxacin observed here (Figure 3) are near the peak serum concentrations for these drugs (Bryskier, 2005), indicating that these MICs might be near the limit for the amount of drug a human can tolerate. These data show that heterogeneous increases in MIC to ampicillin arise in E. coli following treatment with low-levels of ampicillin, and treatment with one drug class can lead to heterogeneous increases in MIC against other classes of antibiotics.
Figure 3. Ampicillin treatment of E. coli results in heterogeneous increases in MIC for ampicillin and norfloxacin.
(A–B) Shown are the distributions of (A) ampicillin or (B) norfloxacin MICs for 44 ampicillin-treated isolates. The maximum growth inhibitory concentration tested for norfloxacin was 1000ng/ml, and the MICs for these isolates may be ≥ 1000ng/ml.
Resistance to multiple antibiotics has been linked to mutations in drug-efflux systems such as the AcrAB multidrug (MDR) efflux pump (George and Levy, 1983; Ma et al., 1993), as well as mutations in transcription factors controlling these systems, such as MarA (Alekshun and Levy, 1997), Rob (Ariza et al., 1995) and SoxS (Greenberg et al., 1990). Our results suggest that ROS-mediated DNA damage induced by low levels of bactericidal antibiotics can result in mutations in a wider range of genes, potentially in some unrelated to the applied antibiotic and drug efflux systems. This implies that treatment with ampicillin, for example, may generate mutants that are not ampicillin resistant but are resistant to other antibiotics.
To determine if these types of resistant strains arise, we examined multidrug resistance following 5 days of treatment with 1µg/ml ampicillin or no treatment. Mutants from ampicillin-treated or untreated cultures were selected on plates containing norfloxacin, ampicillin, kanamycin, tetracycline and chloramphenicol, respectively. From this primary selection, we determined cross-resistance to the other four antibiotics via replica plating (see Experimental Procedures for additional details). We found substantially more primary resistant colonies and higher rates of cross-resistance following ampicillin treatment as compared to no treatment (Table 1). Ampicillin-selected mutants displayed a range of cross-resistance to the other classes of antibiotics, and showed a strong correlation (89% cross-resistance) with norfloxacin resistance (Table 1). We also found that ampicillin-treated cells selected originally on the basis of norfloxacin or kanamycin resistance were only 75% and 63% cross-resistant to ampicillin, respectively (Table 1). Interestingly, primary resistance selection with the static drugs tetracycline and chloramphenicol yielded isolates that were always (100%) cross-resistant to ampicillin (Table 1); this effect deserves further study. Also of note, ampicillin-treated, kanamycin-resistant strains were found to have very low cross-resistance to tetracycline (7%) and no cross-resistance with chloramphenicol (Table 1). This is consistent with previous work showing a lack of cross-resistance to tetracycline or chloramphenicol following selective treatment with aminoglycosides (Grassi, 1979). While the majority of these multidrug cross-resistant strains exhibit resistance against the treatment drug, ampicillin, our results demonstrate that treatment with ampicillin can also generate mutants that are not resistant to ampicillin, yet are resistant to other classes of antibiotics.
Table 1. Cross-resistance following ampicillin treatment and primary resistance selection with 5 different classes of antibiotics.
Wildtype E. coli were treated with 1mg/ml ampicillin or no drug for 5 days. These ampicillin-treated or untreated cells were spread on plates containing norfloxacin, ampicillin, kanamycin, tetracycline or chloramphenicol, and mutants resistant to the individual drugs were isolated. Resistance to the other 4 classes of antibiotics was determined by replica plating of the primary-selected strains onto plates containing the respective antibiotic. Shown is percent resistance (resistant colonies/total primary resistant colonies). Note: Double the volume of no-drug control cells were plated for primary resistance selection for E. coli as compared to the ampicillin-treated cells.
| E. coli control strain | % cross-resistant following ampicillin treatment | |||||
|---|---|---|---|---|---|---|
| Norfloxacin | Ampicillin | Kanamycin | Tetracycline | Chloramphenicol | ||
| primary selection | Norfloxacin | 100% (40/40) |
75% (30/40) |
25% (10/40) |
23% (9/40) |
23% (9/40) |
| Ampicillin | 89% (77/87) |
100% (87/87) |
20% (17/87) |
54% (47/87) |
21% (18/87) |
|
| Kanamycin | 20% (17/83) |
63% (52/83) |
100% (83/83) |
7% (6/83) |
0% (0/83) |
|
| Tetracycline | 79% (63/80) |
100% (80/80) |
14% (11/80) |
100% (80/80) |
78% (62/80) |
|
| Chloramphenicol | 87% (67/77) |
100% (77/77) |
35% (27/77) |
100% (77/77) |
100% (77/77) |
|
| % cross-resistant following no-drug treatment | ||||||
| Norfloxacin | Ampicillin | Kanamycin | Tetracycline | Chloramphenicol | ||
| primary selection | Norfloxacin | 100% (10/10) |
0% (0/10) |
0% (0/10) |
0% (0/10) |
0% (0/10) |
| Ampicillin | 0% (0/10) |
100% (10/10) |
0% (0/10) |
0% (0/10) |
0% (0/10) |
|
| Kanamycin | 0% (0/15) |
0% (0/15) |
100% (15/15) |
0% (0/15) |
0% (0/15) |
|
| Tetracycline | 100% (1/1) |
0% (0/1) |
0% (0/1) |
100% (1/1) |
0% (0/1) |
|
| Chloramphenicol | 100% (1/1) |
0% (0/1) |
0% (0/1) |
0% (0/1) |
100% (1/1) |
|
We sought to determine if some of the ampicillin-treated, cross-resistant isolates had acquired mutations in specific antibiotic targets or in genes making up the common oxidative damage cell death pathway induced by bactericidal antibiotics (Kohanski et al., 2007; Kohanski et al., 2008), or if the observed cross-resistance (Table 1) was solely a function of altered drug efflux. We examined 6 norfloxacin-resistant isolates, 6 kanamycin-resistant isolates, and the untreated control strain. We sequenced gyrA and gyrB, which code for the subunits of DNA Gyrase, the known target of quinolones, rpsL, which codes for a component of the 30S subunit of the ribosome and has been associated with aminoglycoside resistance, ampC, which has been associated with ampicillin resistance, icdA, arcA, cpxA, sdhB, and iscR which are genes involved in the common mechanism of cell death, as well as tolC, marRA and its promoter region, and acrA and its promoter region, which are involved in multidrug efflux.
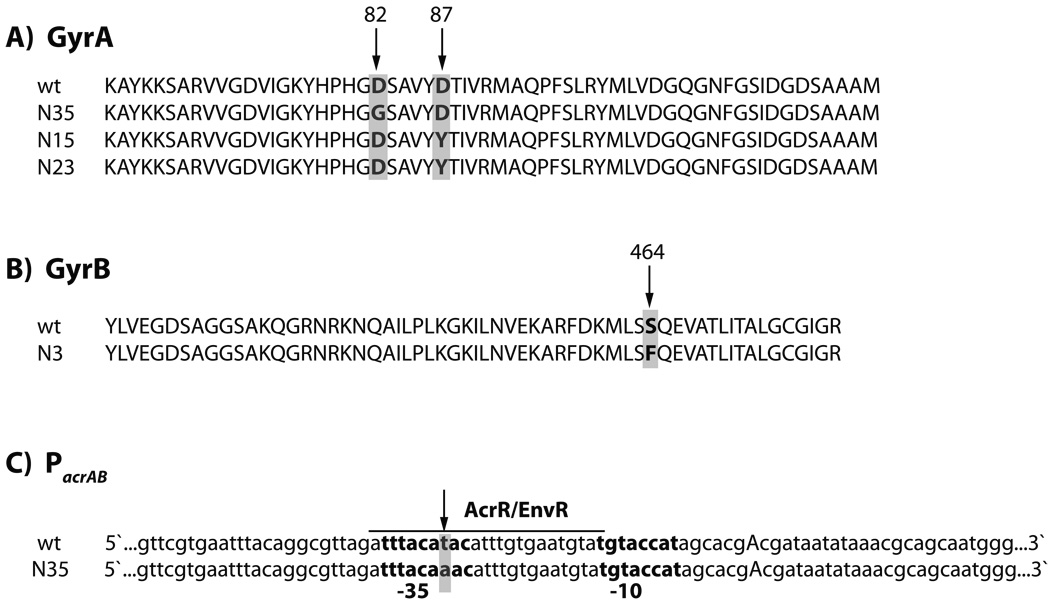
We found that 3 of the 6 norfloxacin-resistant isolates contained point mutations in gyrA that resulted in a substitution of glycine for aspartic acid at amino acid 82 in one isolate, and a substitution of tyrosine for aspartic acid at amino acid 87 in two other isolates (Figure 4A). We also found that one of these 6 norfloxacin-resistant isolates, which did not have a mutation in gyrA, had a point mutation resulting in the conversion of serine to phenylalanine at residue 464 of GyrB (Figure 4B). Interestingly, the point mutations we found in gyrA and gyrB are all in the quinolone resistance determining regions of GyrA and GyrB, respectively, and these mutations have been observed in clinical isolates of Bacteroides fragilis (Oh et al., 2001), Salmonella enterica (Weill et al., 2006) and Pseudomonas aeruginosa (Mouneimne et al., 1999).
Figure 4. Ampicillin treatment leads to the formation of norfloxacin-resistant isolates with mutations in gyrA, gyrB or the acrAB promoter (PacrAB) and kanamycin-resistant isolates with mutations in rpsL or arcA.
(A–B) Isolates with point mutation resulting in (A) a D82G or D87Y substitution in GyrA, or (B) a S464F substitution in GyrB. (C) T-to-A DNA base pair mutation in the AcrR/EnvR binding site of the -35 region of PacrAB. PacrAB is partially annotated to show the -10 and -35 regions (bold), the transcription start site (capitalized A) and the AcrR/EnvR binding site (underlined). (D) Isolates with insertion between basepair 92 and 93 (K58) and between basepair 78 and 79 (K62) resulting in truncation of RpsL. (E) Isolate with a single basepair insertion between basepair 211 and 212 resulting in a truncated ArcA protein missing the majority of the helix-turn-helix (HTH) DNA binding domain.
As noted above, mutations in rpsL have been associated with aminoglycoside resistance. We found that 2 of the 6 kanamycin-resistant isolates had point mutations in rpsL. These mutations led to a frame shift and truncated form of RpsL in both isolates (Figure 4D). It is possible that these mutations contribute to kanamycin resistance in these isolates.
Among the ampicillin-treated, drug-resistant mutants, we did not find any mutations in ampC (data not shown), a gene associated with ampicillin resistance. We also did not find any mutations in icdA, cpxA, sdhB, or iscR (data not shown). Interestingly, we did find a single insertion mutation in arcA in one of the drug-resistant isolates. ArcA is a two-component system transcription factor containing a sensor domain and a DNA binding domain, and the mutation we found results in a truncated ArcA protein that is missing the DNA-binding element of the protein (Figure 4E). We have previously shown that two-component systems are important elements in the common mechanism of cell death, and a knockout of arcA is more tolerant to treatment with ampicillin and kanamycin compared to norfloxacin (Kohanski et al., 2008). This isolate is resistant to ampicillin and kanamycin, but not to norfloxacin. This result suggests that mutations leading to low-level antibiotic resistance can occur in genes that are involved in the common mechanism of cell death.
We did not find any mutations in tolC, marRA, the marRA promoter, or acrA (data not shown); however, we did find a T-to-A conversion in the promoter upstream of acrA (Figure 4C) in one of the norfloxacin-resistant isolates that also had a mutation in gyrA (Figure 4A). This promoter mutation occurs within the annotated -35 site of the promoter and the binding site for the repressor transcription factors AcrR and EnvR (Keseler et al., 2005; Miller et al., 2002). The observed mutations could reduce the ability of these repressors to bind to the acrAB promoter, which would result in increased pump expression and drug resistance. These sequencing results demonstrate that ampicillin treatment can lead to the formation of norfloxacin-resistant strains with mutations in DNA Gyrase and/or mutations that can affect drug efflux pump activity, which likely contribute to the emergence of multidrug resistance.
To demonstrate that sub-lethal levels of bactericidal antibiotics can lead to an increase in multidrug cross-resistance in Gram-positive as well as Gram-negative bacteria, we also examined multidrug cross-resistance in Staphylococcus aureus (S. aureus), following treatment with low-levels of ampicillin (35ng/ml) for 5 days. Previously, we demonstrated antibiotic-mediated ROS formation in S. aureus (Kohanski et al., 2007). In the present study, we found substantially more primary resistant S. aureus colonies and higher rates of cross-resistance following ampicillin treatment as compared to no treatment (Table 2). Interestingly, we were unable to enrich for tetracycline- or chloramphenicol-resistant S. aureus isolates following treatment with low-levels ampicillin as compared with the no-drug treatment. This may be due to the lower level of ROS formation we have observed with S. aureus (Kohanski et al., 2007).
Table 2. Cross-resistance for S. aureus following ampicillin treatment and primary resistance selection with 5 different classes of antibiotics.
Wildtype S. aureus were treated with 35ng/ml ampicillin or no drug for 5 days. These ampicillin-treated or untreated cells were spread on plates containing norfloxacin, ampicillin, kanamycin, tetracycline or chloramphenicol, and mutants resistant to the individual drugs were isolated. Resistance to the other 4 classes of antibiotics was determined by replica plating of the primary-selected strains onto plates containing the respective antibiotic. Shown is percent resistance (resistant colonies/total primary resistant colonies).
| S. aureus | % cross-resistant following ampicillin treatment | |||||
|---|---|---|---|---|---|---|
| Norfloxacin | Ampicillin | Kanamycin | Tetracycline | Chloramphenicol | ||
| primary selection | Norfloxacin | 100% (59/59) |
64% (38/59) |
56% (33/59) |
36% (21/59) |
19% (11/59) |
| Ampicillin | 41% (29/71) |
100% (71/71) |
18% (13/71) |
25% (18/71) |
14% (10/71) |
|
| Kanamycin | 13% (9/68) |
60% (41/68) |
100% (68/68) |
18% (12/68) |
15% (10/68) |
|
| Tetracycline | 0% (0/2) |
100% (2/2) |
0% (0/2) |
100% (2/2) |
0% (0/2) |
|
| Chloramphenicol | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | |
| % cross-resistant following no-drug treatment | ||||||
| Norfloxacin | Ampicillin | Kanamycin | Tetracycline | Chloramphenicol | ||
| primary selection | Norfloxacin | 100% (19/19) |
5% (1/19) |
26% (5/19) |
0% (0/19) |
5% (1/19) |
| Ampicillin | 0% (0/13) |
100% (13/13) |
0% (0/13) |
8% (1/13) |
0% (0/13) |
|
| Kanamycin | 2.6% (1/38) |
2.6% (1/38) |
100% (38/38) |
0% (0/38) |
8% (3/38) |
|
| Tetracycline | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | |
| Chloramphenicol | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | |
To demonstrate that these effects are not limited to lab strains, we considered a clinical isolate of E. coli from a patient with diarrhea (NCDC C771). We examined multidrug cross-resistance in the clinical isolate following treatment with 1µg/ml ampicillin (see Experimental Procedures for more details). As with the wildtype strains, we found substantially more primary resistant colonies and higher rates of cross-resistance in the clinical isolates following ampicillin treatment as compared to no treatment. We also found that ampicillin-treated cells selected originally on the basis of norfloxacin or kanamycin resistance were only 11.5% and 21.5% cross-resistant to ampicillin, respectively (Table 3). This further affirms that treatment with ampicillin can generate mutants that are not resistant to ampicillin, yet are resistant to other classes of antibiotics.
Table 3. Cross-resistance for E. coli clinical isolate NCDC C771 following ampicillin treatent and primary resistance selection with 5 different classes of antibiotics.
E. coli strain NCDC C771 was treated with 1 µg/ml ampicillin or no drug for 5 days. These ampicillin-treated or untreated cells were spread on plates containing norfloxacin, ampicillin, kanamycin, or chloramphenicol, and mutants resistant to the individual drugs were isolated. Resistance to the other 3 classes of antibiotics was determined by replica plating of the primary-selected strains onto plates containing the respective antibiotic. Shown is percent resistance (resistant colonies/total primary resistant colonies). Tetracycline cross-resistance was not quantified for NCDC C771 as this strain is resistant to tetracycline (MIC >35 µg/ml).
| E. coli clinical isolate | % cross resistant following ampicillin treatment | ||||
|---|---|---|---|---|---|
| Norfloxacin | Ampicillin | Kanamycin | Chloramphenicol | ||
| primary selection | Norfloxacin | 100% (78/78) |
11.5% (9/78) |
1.3% (1/78) |
10.3% (8/78) |
| Ampicillin | 13.2% (5/38) |
100 (38/38) |
2.6% (1/38) |
23.9% (9/38) |
|
| Kanamycin | 15.2% (12/79) |
21.5% (17/79) |
100% (79/79) |
7.6% (6/79) |
|
| Chloramphenicol | 41.4% (29/70) |
45.7% (32/70) |
22.9% (16/70) |
100% (70/70) |
|
| % cross resistant following no-drug treatment | |||||
| Norfloxacin | Ampicillin | Kanamycin | Chloramphenicol | ||
| primary selection | Norfloxacin | 0/0 | 0/0 | 0/0 | 0/0 |
| Ampicillin | 0/0 | 0/0 | 0/0 | 0/0 | |
| Kanamycin | 2.8% (1/36) |
11.1% (4/36) |
100% (36/36) |
2.8% (1/36) |
|
| Chloramphenicol | 0/3 | 0/3 | 0/3 | 100% (3/3) |
|
Discussion
Here we establish a radical-based molecular mechanism whereby sub-lethal levels of antibiotics can lead to multidrug resistance. This occurs via bactericidal antibiotic-mediated radical formation that results in the formation of mutations, some of which confer antibiotic resistance. Low-level resistance likely provides a first step toward clinically significant resistance (Goldstein, 2007), and the mechanism we propose and validate here establishes an antibiotic-stimulated mutagenic effect that likely works in conjunction with SOS-induced mutagenesis in the emergence of mutations that confer drug resistance.
Clinical situations where bacteria are exposed to low levels of antibiotics can occur with incomplete treatment of an infection, noncompliance with antibiotic treatment (e.g., a missed pill), and reduced or limited drug accessibility to certain tissues (e.g., bone or cerebrospinal fluid (Bryskier, 2005)). It is possible that mutations arising via antibiotic-mediated oxidative stress could be maintained in the normal bacterial flora of the body and transferred to virulent bacteria via horizontal gene transfer, a mechanism which can be induced by DNA damage (Beaber et al., 2004). Novel therapeutics targeting ROS-forming systems or error-prone DNA damage repair systems may help reduce and contain the spread of new antibiotic-resistant bacteria.
Experimental Procedures
Strains, media and antibiotics
All experiments were performed with wildtype E. coli strain MG1655 (ATCC 700926), in Luria-Bertani (LB) medium (Fisher Scientific). For all treatment conditions, we used 1mM hydrogen peroxide (VWR) and the bactericidal antibiotics: norfloxacin (Sigma), ampicillin and kanamycin (Fisher Scientific). Bactericidal antibiotics were used at concentrations of 15ng/ml norfloxacin, 50ng/ml norfloxacin, 1µg/ml ampicillin, 1µg/ml kanamycin or 3µg/ml kanamycin. Tetracycline (MP Biomedical) and chloramphenicol (Fluka) were used for MIC assays, rifampicin (Sigma) for determination of antibiotic resistant rates, and thiourea (Fluka) for radical-quenching experiments. Anaerobic media was made by heating LB in 17ml Bellco glass hungate tubes (Fisher) under anaerobic conditions in a Coy anaerobic chamber (Coy Lab Products Inc) to drive out dissolved oxygen (Norris and Ribbons, 1969). 10mM Resazurin (Sigma), which turns clear in the absence of oxygen, was used as an indicator for anaerobic conditions. Multidrug resistance was also determined in wildtype S. aureus (ATCC 25923) and the E. coli clinical isolate NCDC C771 (ATCC 23985).
Determination of mutation rate
Mutation rates were examined following 24 hours of growth in the presence of a bactericidal antibiotic. Drug levels were chosen such that there was an observable effect on growth or survival within the first 6 hours after drug addition (Figure S3), followed by “recovery” of the culture to near untreated colony density 24 hours after treatment. All treatment conditions exhibited recovery to near untreated colony density levels, with the exception of 50ng/ml norfloxacin. This allowed us to compare mutation frequencies for cultures of similar densities following treatment with an antibiotic.
Mutation rates were determined using a rifampicin-based selection method (Giraud et al., 2001). Briefly, an overnight culture of E. coli was diluted 1:10000 into 50ml LB in a 250ml baffled flask and grown for 3.5 hours at 37°C and 300RPM. Cultures were grown at high shaking speeds and in baffled flasks to maximize aeration and ROS formation. The culture was diluted 1:3 into fresh LB containing no drug, an antibiotic or hydrogen peroxide at the concentrations described above. For experiments with thiourea, thiourea in solid form was added to each diluted culture for a final concentration of 100mM. 1ml aliquots (ten replicates) of these diluted cultures were grown in 14ml tubes for 24 hours at 37°C and 300RPM. Aliquots of each treatment were serially diluted and plated on LB-agar plates for colony forming unit per ml (cfu/ml) determination. Aliquots of each treatment were also plated on LB-agar plates containing 100µg/ml rifampicin and grown for 48 hours at 37°C. Colonies were counted at 24 and 48 hours, and the colony count from the 48-hour time point was used to estimate mutation rates. For experiments in anaerobic conditions, cells were diluted 1:10000 into 15ml of anaerobic LB in sealed hungate tubes to minimize exposure to oxygen. Antibiotic treatments, growth temperature, shaking speed, and sample collection were as described above for the aerobic growth conditions.
The colony counts from the 10 replicates were then used in the MSS maximum-likelihood method (Rosche and Foster, 2000; Sarkar et al., 1992) to estimate the number of mutational events per culture. The MSS maximum likelihood method is a recursive algorithm based on the Lea-Coulson function for solving the Luria-Delbruck distribution for a given number of mutational events (Sarkar et al., 1992); its utility has been demonstrated in vitro (Rosche and Foster, 2000). The mutation rate was determined by dividing the number of mutational events per culture by the total number of bacteria plated on the rifampicin plates (Rosche and Foster, 2000). Fold change in mutation rate was determined for all treatments and conditions relative to an untreated MG1655 control. Three biological replicates were run for each treatment condition and the averages are shown in Figure 1.
ROS detection using HPF
To detect ROS formation, we used the fluorescent reporter dye, 3’-(p-hydroxyphenyl) fluorescein (HPF, Invitrogen), and flow cytometry as previously described (Kohanski et al., 2007). Average fluorescence was determined at 0 (baseline), 1, 3 and 6 hours (normalized to a no-dye control) following antibiotic treatment at the concentrations described above, and peak fluorescence levels were used to determine the change in mean fluorescence relative to baseline (Figure 1B).
Determination of MIC
For wildtype E. coli, MICs for norfloxacin, ampicillin, kanamycin, tetracycline and chloramphenicol were measured over 5 days of treatment with no drug, 25ng/ml norfloxacin, 50ng/ml norfloxacin, 1µg/ml ampicillin, or 3µg/ml kanamycin. Briefly, an overnight culture of E. coli was diluted 1:10000 into 50ml LB in a 250ml baffled flask and grown for 3.5 hours at 37°C and 300RPM. The culture was diluted 1:3 into fresh LB containing no drug or antibiotics at the above concentrations. 1ml aliquots of these diluted cultures were grown in 14ml tubes for 24 hours at 37°C and 300RPM. Each day thereafter for 5 days, in order to avoid mutations arising due to evolution during stationary phase (GASP mutants) (Zinser and Kolter, 2004), cells were diluted 1:1000 into 1ml of LB in a 14ml tube containing the respective antibiotic and grown for 24 hours at 37°C and 300RPM.
MICs were also measured for anaerobically grown E. coli over 5 days of treatment with no drug or 1µg/ml ampicillin. Briefly, an overnight culture of E. coli was diluted 1:1000 into 15ml anaerobic LB in sealed hungate tubes containing no drug or 1µg/ml ampicillin. These cultures were grown in the sealed hungate tubes for 24 hours at 37°C and 300RPM. Each day thereafter for 5 days, cells were diluted 1:1000 into 15ml of anaerobic LB in a sealed hungate tube containing the respective antibiotic and grown for 24 hours at 37°C and 300RPM.
To determine the MIC on each day, an aliquot of cells from each treatment condition was diluted 1:10000 into LB and dispensed into 96-well plates (100µl total volume per well) containing various concentrations (10 replicates per drug concentration) of norfloxacin, ampicillin, kanamycin, tetracycline or chloramphenicol. Plates were incubated at 37°C and 300RPM for 24 hours after which time, the optical density at 600nm (OD600) was measured using a SPECTRAFluor Plus (Tecan). The median OD600 was calculated for each drug concentration and the MIC was determined as the concentration that inhibited 90% of growth based on OD600. Fold change in MIC was determined by dividing the treated MIC on each day by its respective MIC from day 0.
Determination of MIC variability and multidrug resistance
Wildtype E. coli were grown for 5 days in the presence of 1µg/ml ampicillin or no drug (untreated) as described above. These long-term treated cultures were diluted 1:1000 into 25ml LB in 250ml flasks and grown for 3 hours at 37°C and 300RPM. 1ml aliquots were plated onto LB-agar plates containing 300ng/ml norfloxacin, 7.5µg/ml ampicillin, 15µg/ml kanamycin, 8µg/ml tetracycline and 25µg/ml chloramphenicol, respectively, and grown for 24 hours at 37°C. Approximately 100 ampicillin-treated colonies from each primary drug selection were purified by streaking them onto LB-agar plates containing the same selective antibiotic. Double the volume of untreated control cells were plated for primary resistance selection as compared to the ampicillin-treated cells and these untreated colonies were also purified as described above. Plates were placed at 37°C for 24 hours; these strains were then transferred via replica plating onto LB-agar plates containing norfloxacin, ampicillin, kanamycin, tetracycline or chloramphenicol. Cross-resistance for each primary antibiotic selection following the 5-day ampicillin treatment or the no drug treatment were determined after 24 hours of growth at 37°C by counting the colonies which displayed growth on the various drug-containing replicated plates.
The MIC of 44 of the above isolates and the MG1655 control strain were determined for ampicillin and norfloxacin, respectively. Overnight cultures of each strain were diluted 1:10000 into 100µl of LB plus varying concentration of antibiotic (four replicates per strain per drug concentration) in 96-well plates. Plates were incubated at 37°C and 300RPM for 24 hours after which time, the OD600 was measured using a SPECTRAFluor Plus (Tecan). The median OD600 was calculated for each drug concentration and the MIC was determined as the concentration that inhibited 90% of growth based on OD600.
Wildtype S. aureus were grown for 5 days in the presence of 35ng/ml ampicillin or no drug (untreated) as described above. E. coli clinical isolate NCDC C771 was grown for 5 days in the presence of 1mg/ml ampicillin or no drug (untreated) as described above. These long-term treated cultures were diluted 1:1000 into 25ml LB in 250ml flasks and grown for 3 hours at 37°C and 300RPM. For S. aureus, 1ml aliquots were plated onto LB-agar plates containing 3µg/ml norfloxacin, 7.5µg/ml ampicillin, 15µg/ml kanamycin, 8µg/ml tetracycline and 25µg/ml chloramphenicol, respectively, and grown for 24 hours at 37°C. For NCDC C771, 1ml aliquots were plated onto LB-agar plates containing 400ng/ml norfloxacin, 8.5µg/ml ampicillin, 20µg/ml kanamycin, and 15µg/ml chloramphenicol. Tetracycline cross-resistance was not quantified for NCDC C771 as this strain is resistant to tetracycline (MIC >35µg/ml). Approximately 100 ampicillin-treated colonies from each primary drug selection were purified by streaking them onto LB-agar plates containing the same selective antibiotic. An equal volume of untreated S. aureus or the E. coli clinical isolate cells were plated for primary resistance selection as compared to the ampicillin-treated cells, and these untreated colonies were also purified as described above. The remainder of the cell growth and cross-resistance determination was performed as described above for wildtype E. coli.
Sequencing of ampicillin-treated, norfloxacin-resistant or kanamycin-resistant mutants
Six ampicillin-treated, norfloxacin-resistant isolates and six ampicillin-treated, kanamycin-resistant isolates from the cross-resistance experiment described above, as well as the untreated MG1655 control strain, were grown to a cell density of approximately 109 cfu/ml. Genomic DNA was extracted from each sample using a Qiagen genomic DNA extraction kit according to the manufacturer’s instructions. Primers from IDT (Table S1) were utilized to PCR amplify, using Phusion DNA polymerase (Finnzyme), the regions surrounding gyrA, gyrB, tolC, acrA, marRA, ampC, rpsL, icdA, iscR, sdhB, arcA, and cpxR. These samples were sequenced by Agencourt Bioscience Corporation using primers from IDT (Table S1). Sequences were analyzed using Clone Manager 7 (Scientific & Educational Software) and Sequence Scanner v1.0 (Applied Biosystems).
Supplementary Material
Acknowledgments
We thank D. Dwyer, B. Davies and K. Allison for helpful discussions during the course of this work. We thank Q. Beg for help running the anaerobic chamber. This work was supported by the National Institutes of Health through the NIH Director’s Pioneer Award Program, grant number DP1 OD003644, and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alekshun MN, Levy SB. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson DI. Persistence of antibiotic resistant bacteria. Curr Opin Microbiol. 2003;6:452–456. doi: 10.1016/j.mib.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Ariza RR, Li Z, Ringstad N, Demple B. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J Bacteriol. 1995;177:1655–1661. doi: 10.1128/jb.177.7.1655-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- Becnel Boyd L, Maynard MJ, Morgan-Linnell SK, Horton LB, Sucgang R, Hamill RJ, Jimenez JR, Versalovic J, Steffen D, Zechiedrich L. Relationships among ciprofloxacin, gatifloxacin, levofloxacin, and norfloxacin MICs for fluoroquinolone-resistant Escherichia coli clinical isolates. Antimicrob Agents Chemother. 2009;53:229–234. doi: 10.1128/AAC.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumaghim JL, Li Y, Henle E, Linn S. Effects of hydrogen peroxide upon nicotinamide nucleotide metabolism in Escherichia coli: changes in enzyme levels and nicotinamide nucleotide pools and studies of the oxidation of NAD(P)H by Fe(III) J Biol Chem. 2003;278:42495–42504. doi: 10.1074/jbc.M306251200. [DOI] [PubMed] [Google Scholar]
- Bryskier A. Antimicrobial Agents: Antibacterials and Antifungals. Washington, D.C.: ASM Press; 2005. [Google Scholar]
- Carlsson J, Carpenter VS. The recA+ gene product is more important than catalase and superoxide dismutase in protecting Escherichia coli against hydrogen peroxide toxicity. J Bacteriol. 1980;142:319–321. doi: 10.1128/jb.142.1.319-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I, O'Neill AJ, Miller K. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist Updat. 2003;6:137–145. doi: 10.1016/s1368-7646(03)00041-4. [DOI] [PubMed] [Google Scholar]
- Cirz RT, Chin JK, Andes DR, de Crecy-Lagard V, Craig WA, Romesberg FE. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SP, McMurry LM, Hooper DC, Wolfson JS, Levy SB. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989;33:1318–1325. doi: 10.1128/aac.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- de Lencastre H, Figueiredo AM, Tomasz A. Genetic control of population structure in heterogeneous strains of methicillin resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1993;12 Suppl 1:S13–S18. doi: 10.1007/BF02389872. [DOI] [PubMed] [Google Scholar]
- Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Kohanski MA, Collins JJ. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol. 2009;12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridovich I. The biology of oxygen radicals. Science. 1978;201:875–880. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd edn. Washington, D.C.: ASM Press; 2006. [Google Scholar]
- George AM, Levy SB. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983;155:531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, Taddei F. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science. 2001;291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- Girgis H, Hottes A, Tavazoie S. Genetic architecture of intrinsic antibiotic susceptibility. PLoS one. 2009;4 doi: 10.1371/journal.pone.0005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein F. The potential clinical impact of low-level antibiotic resistance in Staphylococcus aureus. J Antimicrob Chemother. 2007;59:1–4. doi: 10.1093/jac/dkl429. [DOI] [PubMed] [Google Scholar]
- Grassi GG. Drug-inactivating enzymes of bacteria grown in subminimal inhibitory concentrations of antibiotics. Rev Infect Dis. 1979;1:852–857. doi: 10.1093/clinids/1.5.852. [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci U S A. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegreness M, Shoresh N, Damian D, Hartl D, Kishony R. Accelerated evolution of resistance in multidrug environments. Proc Natl Acad Sci U S A. 2008;105:13977–13981. doi: 10.1073/pnas.0805965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol. 2006;59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- Imlay JA, Linn S. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J Bacteriol. 1986;166:519–527. doi: 10.1128/jb.166.2.519-527.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA, Linn S. Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Bacteriol. 1987169;169:2967–2976. doi: 10.1128/jb.169.7.2967-2976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore DM. Bacterial resistance: origins, epidemiology, and impact. Clin Infect Dis. 2003;36:S11–S23. doi: 10.1086/344654. [DOI] [PubMed] [Google Scholar]
- Lopez E, Elez M, Matic I, Blazquez J. Antibiotic-mediated recombination: ciprofloxacin stimulates SOS-independent recombination of divergent sequences in Escherichia coli. Mol Microbiol. 2007;64:83–93. doi: 10.1111/j.1365-2958.2007.05642.x. [DOI] [PubMed] [Google Scholar]
- Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, Hearst JE. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie GJ, Rosenberg SM. Adaptive mutations, mutator DNA polymerases and genetic change strategies of pathogens. Curr Opin Microbiol. 2001;4:586–594. doi: 10.1016/s1369-5274(00)00255-1. [DOI] [PubMed] [Google Scholar]
- Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science. 2004;305:1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- Miller JH. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol. 1996;50:625–643. doi: 10.1146/annurev.micro.50.1.625. [DOI] [PubMed] [Google Scholar]
- Miller K, O'Neill AJ, Chopra I. Response of Escherichia coli hypermutators to selection pressure with antimicrobial agents from different classes. J Antimicrob Chemother. 2002;49:925–934. doi: 10.1093/jac/dkf044. [DOI] [PubMed] [Google Scholar]
- Mouneimne H, Robert J, Jarlier V, Cambau E. Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:62–66. doi: 10.1128/aac.43.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris JR, Ribbons DW. Methods in Microbiology. Orlando, FL: Academic Press Inc; 1969. [Google Scholar]
- Novogrodsky A, Ravid A, Rubin AL, Stenzel KH. Hydroxyl radical scavengers inhibit lymphocyte mitogenesis. Proc Natl Acad Sci U S A. 1982;79:1171–1174. doi: 10.1073/pnas.79.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, El Amin N, Davies T, Appelbaum PC, Edlund C. gyrA mutations associated with quinolone resistance in Bacteroides fragilis group strains. Antimicrob Agents Chemother. 2001;45:1977–1981. doi: 10.1128/AAC.45.7.1977-1981.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Capilla T, Baquero MR, Gomez-Gomez JM, Ionel A, Martin S, Blazquez J. SOS-independent induction of dinB transcription by beta-lactam-mediated inhibition of cell wall synthesis in Escherichia coli. J Bacteriol. 2005;187:1515–1518. doi: 10.1128/JB.187.4.1515-1518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repine JE, Fox RB, Berger EM. Hydrogen peroxide kills Staphylococcus aureus by reacting with staphylococcal iron to form hydroxyl radical. J Biol Chem. 1981;256:7094–7096. [PubMed] [Google Scholar]
- Rosche WA, Foster PL. Determining mutation rates in bacterial populations. Methods. 2000;20:4–17. doi: 10.1006/meth.1999.0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Ma WT, Sandri GH. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica. 1992;85:173–179. doi: 10.1007/BF00120324. [DOI] [PubMed] [Google Scholar]
- Setsukinai K, Urano Y, Kakinuma K, Majima HJ, Nagano T. Development of novel fluorescence probes that can reliably detect reactive oxygen species and distinguish specific species. J Biol Chem. 2003;278:3170–3175. doi: 10.1074/jbc.M209264200. [DOI] [PubMed] [Google Scholar]
- Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill FX, Guesnier F, Guibert V, Timinouni M, Demartin M, Polomack L, Grimont PA. Multidrug resistance in Salmonella enterica serotype Typhimurium from humans in France (1993 to 2003) J Clin Microbiol. 2006;44:700–708. doi: 10.1128/JCM.44.3.700-708.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser ER, Kolter R. Escherichia coli evolution during stationary phase. Res Microbiol. 2004;155:328–336. doi: 10.1016/j.resmic.2004.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.