Abstract
Modification of the cytidine in the first anticodon position of the AUA decoding tRNAIle ( ) of bacteria and archaea is essential for this tRNA to read the isoleucine codon AUA and to differentiate between AUA and the methionine codon AUG. To identify the modified cytidine in archaea, we have purified this tRNA species from Haloarcula marismortui, established its codon reading properties, used liquid chromatography–mass spectrometry (LC-MS) to map RNase A and T1 digestion products onto the tRNA, and used LC-MS/MS to sequence the oligonucleotides in RNase A digests. These analyses revealed that the modification of cytidine in the anticodon of
) of bacteria and archaea is essential for this tRNA to read the isoleucine codon AUA and to differentiate between AUA and the methionine codon AUG. To identify the modified cytidine in archaea, we have purified this tRNA species from Haloarcula marismortui, established its codon reading properties, used liquid chromatography–mass spectrometry (LC-MS) to map RNase A and T1 digestion products onto the tRNA, and used LC-MS/MS to sequence the oligonucleotides in RNase A digests. These analyses revealed that the modification of cytidine in the anticodon of  adds 112 mass units to its molecular mass and makes the glycosidic bond unusually labile during mass spectral analyses. Accurate mass LC-MS and LC-MS/MS analysis of total nucleoside digests of the
adds 112 mass units to its molecular mass and makes the glycosidic bond unusually labile during mass spectral analyses. Accurate mass LC-MS and LC-MS/MS analysis of total nucleoside digests of the 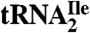 demonstrated the absence in the modified cytidine of the C2-oxo group and its replacement by agmatine (decarboxy-arginine) through a secondary amine linkage. We propose the name agmatidine, abbreviation C+, for this modified cytidine. Agmatidine is also present in Methanococcus maripaludis
demonstrated the absence in the modified cytidine of the C2-oxo group and its replacement by agmatine (decarboxy-arginine) through a secondary amine linkage. We propose the name agmatidine, abbreviation C+, for this modified cytidine. Agmatidine is also present in Methanococcus maripaludis 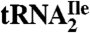 and in Sulfolobus solfataricus total tRNA, indicating its probable occurrence in the AUA decoding tRNAIle of euryarchaea and crenarchaea. The identification of agmatidine shows that bacteria and archaea have developed very similar strategies for reading the isoleucine codon AUA while discriminating against the methionine codon AUG.
and in Sulfolobus solfataricus total tRNA, indicating its probable occurrence in the AUA decoding tRNAIle of euryarchaea and crenarchaea. The identification of agmatidine shows that bacteria and archaea have developed very similar strategies for reading the isoleucine codon AUA while discriminating against the methionine codon AUG.
Keywords: agmatine, decoding, RNA modification, tRNA, wobble pairing
The genetic code table consists of sixteen four-codon boxes. In fourteen of the boxes, all four codons either specify the same amino acid or are split into two sets of two codons, with each set encoding a different amino acid. For example, the UUN box is split into UUU/UUC coding for phenylalanine and UUA/UUG coding for leucine. The wobble hypothesis of Crick proposes how a single phenylalanine tRNA with G in the first anticodon position can base pair with either U or C and a single leucine tRNA with a modified U (or 2-thioU) in the anticodon can base pair with either A or G (1–3). The two remaining boxes, UGN and AUN, are exceptions in that the UGN box is split into UGU/UGC coding for cysteine, UGG coding for tryptophan, and UGA being used as a stop codon, whereas the AUN box is split into AUU/AUC/AUA coding for isoleucine and AUG coding for methionine. The isoleucine codons AUU and AUC can be read by an isoleucine tRNA with G in the anticodon following the wobble pairing rules, but how the AUA codon is read specifically by a tRNAIle without also reading the AUG codon has been a question of much interest over the years.
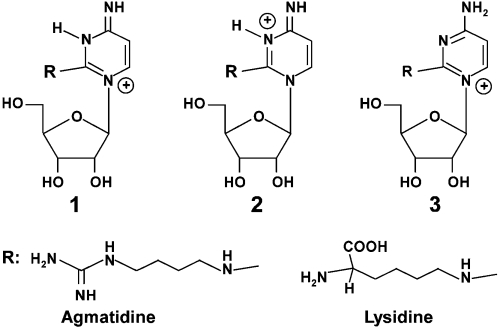
Different organisms have developed different strategies for reading the AUA codon. Bacteria use a tRNAIle with the anticodon LAU (L = lysidine) (4–7). Lysidine is a modified cytidine in which the C2-oxo group of cytidine is replaced by lysine. Exactly how it base pairs with A but not with G is not established. Eukaryotes, on the other hand, contain a tRNAIle with the anticodon IAU (I = inosine), which can read all three isoleucine codons using the wobble pairing rules of Crick. They also contain a tRNAIle with the anticodon ΨAΨ, which is thought to read only the isoleucine codon AUA but not the methionine codon AUG (8). Given these two distinct mechanisms in bacteria and eukaryotes, a question of much interest is how the archaeal tRNAIle accomplishes the task of reading the AUA codon.
In a recent paper, we showed that a tRNA with the anticodon sequence CAU, which was annotated as a methionine tRNA in the archaeon Haloarcula marismortui, was actually aminoacylated with isoleucine in vivo (9). We showed that the cytidine in the anticodon of this tRNA was modified, but not to lysidine as it is in bacteria, and that the same modification was likely present in other haloarchaea and in Methanocaldococcus jannaschii. We also showed that modification of the cytidine was necessary for aminoacylation of the tRNA with isoleucine in vitro. These findings raised the question of the nature of the modification in the archaeal AUA decoding tRNAIle (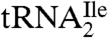 ) and its relationship, if any, to lysidine in Escherichia coli
) and its relationship, if any, to lysidine in Escherichia coli 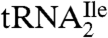 .
.
Here, we describe the purification of two tRNAIle species from H. marismortui. We show that 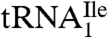 binds to AUC but not to AUA or AUG on the ribosome, whereas
binds to AUC but not to AUA or AUG on the ribosome, whereas 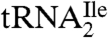 binds to AUA but not to AUC, AUG, or AUU. Mass spectral analysis of nucleosides and oligonucleotides isolated from
binds to AUA but not to AUC, AUG, or AUU. Mass spectral analysis of nucleosides and oligonucleotides isolated from 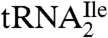 show that the modified cytidine is located in the anticodon wobble position, lacks the C2-oxo group, and has instead agmatine (1-amino 4-guanidino butane) attached to it through a secondary amine linkage. We propose the name agmatidine and the abbreviation C+ for this modified nucleoside. Agmatidine is also present in
show that the modified cytidine is located in the anticodon wobble position, lacks the C2-oxo group, and has instead agmatine (1-amino 4-guanidino butane) attached to it through a secondary amine linkage. We propose the name agmatidine and the abbreviation C+ for this modified nucleoside. Agmatidine is also present in 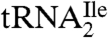 from Methanococcus maripaludis and in total tRNA isolated from Sulfolobus solfataricus, indicating its possible presence in
from Methanococcus maripaludis and in total tRNA isolated from Sulfolobus solfataricus, indicating its possible presence in 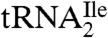 of euryarchaea and crenarchaea.
of euryarchaea and crenarchaea.
Agmatidine is in many ways similar to lysidine. In both cases the C2-oxo group of cytidine is replaced by either of two closely related basic amino acids, decarboxy-arginine or lysine. Thus, bacteria and archaea have developed very similar strategies for generating a 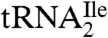 to read the isoleucine codon AUA without also reading the methionine codon AUG. The identification of agmatidine also suggests that agmatine, which is a known neuromodulator (10, 11), and which is essential for polyamine biosynthesis in the archaeon Thermococcus kodakaraensis (12), may also be essential in euryarchaea and in crenarchaea (13) for modification of the cytidine required for decoding the AUA codon by
to read the isoleucine codon AUA without also reading the methionine codon AUG. The identification of agmatidine also suggests that agmatine, which is a known neuromodulator (10, 11), and which is essential for polyamine biosynthesis in the archaeon Thermococcus kodakaraensis (12), may also be essential in euryarchaea and in crenarchaea (13) for modification of the cytidine required for decoding the AUA codon by 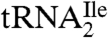 .
.
Results
Purification and Characterization of Isoleucine tRNAs from H. marismortui.
Starting from total tRNA, a two-step procedure, involving hybrid selection of tRNA with biotinylated DNA oligonucleotides bound to streptavidin sepharose followed by native polyacrylamide gel fractionation of the enriched tRNA, was used to isolate pure tRNAIles. The biotinylated oligonucleotides were complementary to nucleotides 54–73 of 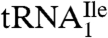 or
or 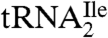 (Fig. 1A) and contained biotin at the 3′-hydroxyl end. The final yields of purified tRNAs from approximately 1,000 A260 units of total tRNA were 6–8 A260 units for
(Fig. 1A) and contained biotin at the 3′-hydroxyl end. The final yields of purified tRNAs from approximately 1,000 A260 units of total tRNA were 6–8 A260 units for 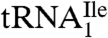 and 0.6–0.8 A260 unit for
and 0.6–0.8 A260 unit for 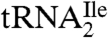 . The significantly lower yield of
. The significantly lower yield of  compared to
compared to 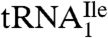 reflects the low abundance of this tRNA and the fact that AUA is a rarely used codon in haloarchaea as it is in E. coli (14).
reflects the low abundance of this tRNA and the fact that AUA is a rarely used codon in haloarchaea as it is in E. coli (14).
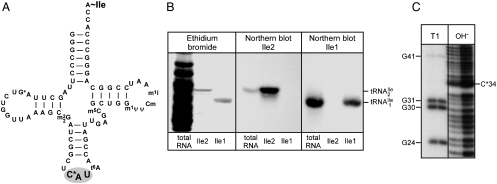
Fig. 1.
(A) Cloverleaf structure of 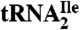 from H. marismortui. Location of modified nucleosides is based on Gupta (46) and LC-MS analysis presented in this work. G+ (archaeosine);
from H. marismortui. Location of modified nucleosides is based on Gupta (46) and LC-MS analysis presented in this work. G+ (archaeosine); 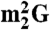 (N2, N2-dimethylguanosine); C* (modified cytidine); t6A (N6-threonylcarbamoyladenosine); m5C (5-methylcytidine); m1ψ (1-methylpseudouridine); ψ (pseudouridine); Cm (2′-O-methylcytidine); m1I (1-methylinosine). (B) Purification of isoleucine tRNAs from H. marismortui. Native PAGE analysis of total tRNA, purified
(N2, N2-dimethylguanosine); C* (modified cytidine); t6A (N6-threonylcarbamoyladenosine); m5C (5-methylcytidine); m1ψ (1-methylpseudouridine); ψ (pseudouridine); Cm (2′-O-methylcytidine); m1I (1-methylinosine). (B) Purification of isoleucine tRNAs from H. marismortui. Native PAGE analysis of total tRNA, purified 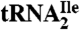 (Ile2) and
(Ile2) and 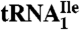 (Ile1) is shown. tRNAs are visualized by ethidium bromide staining or Northern blot analysis using probes specific for
(Ile1) is shown. tRNAs are visualized by ethidium bromide staining or Northern blot analysis using probes specific for 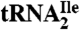 and
and 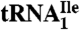 as indicated. (C) Characterization of purified
as indicated. (C) Characterization of purified 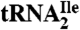 . The homogeneity of
. The homogeneity of 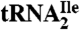 was confirmed by partial RNase T1 digest (lane T1) of 5′-32P-labeled tRNA. 32P-labeled fragments were separated by denaturing PAGE and visualized by autoradiography; lane OH-, partial alkali digest.
was confirmed by partial RNase T1 digest (lane T1) of 5′-32P-labeled tRNA. 32P-labeled fragments were separated by denaturing PAGE and visualized by autoradiography; lane OH-, partial alkali digest.
Fig. 1B shows that the purified tRNAs are essentially homogeneous. 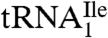 yielded only one band on a native polyacrylamide gel upon staining with ethidium bromide, and this band hybridized to the corresponding complementary oligonucleotide in Northern blots.
yielded only one band on a native polyacrylamide gel upon staining with ethidium bromide, and this band hybridized to the corresponding complementary oligonucleotide in Northern blots. 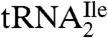 yielded two bands, a strong band and a weaker one, with both bands hybridizing to the oligonucleotide complementary to this tRNA. Based on a comparison of intensities of the hybridization bands and the amount of total tRNA (0.5 A260 unit) and the purified tRNAs (0.005 A260 unit) loaded on the gel, the purified
yielded two bands, a strong band and a weaker one, with both bands hybridizing to the oligonucleotide complementary to this tRNA. Based on a comparison of intensities of the hybridization bands and the amount of total tRNA (0.5 A260 unit) and the purified tRNAs (0.005 A260 unit) loaded on the gel, the purified 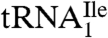 is enriched approximately 57-fold compared to total tRNA, and the purified
is enriched approximately 57-fold compared to total tRNA, and the purified 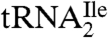 is enriched approximately 500-fold.
is enriched approximately 500-fold.
Additional evidence that the 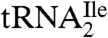 is essentially homogeneous was derived from gel electrophoretic analysis of partial RNase T1 digests of 5′-32P-labeled
is essentially homogeneous was derived from gel electrophoretic analysis of partial RNase T1 digests of 5′-32P-labeled 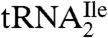 (Fig. 1C). The 32P-labeled bands in the RNase T1 lane are due to cuts at G residues, and these are consistent with the pattern expected for
(Fig. 1C). The 32P-labeled bands in the RNase T1 lane are due to cuts at G residues, and these are consistent with the pattern expected for 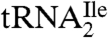 . There is no band due to cleavage by RNase T1 at G26 because G26 is dimethylated in the tRNA and it is known that under conditions of partial RNase T1 digestion there is no cleavage of the phosphodiester bond on the 3′-side of the dimethyl-G residue (15).
. There is no band due to cleavage by RNase T1 at G26 because G26 is dimethylated in the tRNA and it is known that under conditions of partial RNase T1 digestion there is no cleavage of the phosphodiester bond on the 3′-side of the dimethyl-G residue (15).
The pattern obtained in partial alkali digests shows a pronounced shift in gel electrophoretic mobility of the 5′-32P-labeled oligonucleotide going from nucleotide 33 to 34. We had shown previously that nucleotide 34 contained a modified cytidine (9). The presence of nine bands between G31 and G41 in the alkali digest shows that every phosphodiester bond between G31 and G41 is cleaved by alkali. This means that none of the nucleotides in between contain a substitution in the 2′-hydroxyl group. Therefore, the very pronounced shift in the gel electrophoretic mobility indicates strongly that the modified cytidine has at least one if not more positive charges in the ring, which would retard the electrophoretic migration of the oligonucleotide that contained the modified cytidine (16).
Codon Reading Properties of 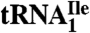 and
and 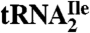 .
.
Purified 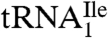 and
and 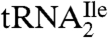 were aminoacylated in vitro with 3H-Ile and the 3H-Ile-tRNAs were used for measuring oligonucleotide-directed binding to H. marismortui ribosomes. The oligonucleotides used as template had the following sequences AUGAUC, AUGAUA, AUGAUG and AUGAUU. Results of ribosome binding experiments show that
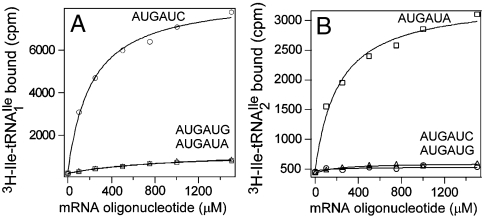
were aminoacylated in vitro with 3H-Ile and the 3H-Ile-tRNAs were used for measuring oligonucleotide-directed binding to H. marismortui ribosomes. The oligonucleotides used as template had the following sequences AUGAUC, AUGAUA, AUGAUG and AUGAUU. Results of ribosome binding experiments show that 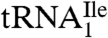 binds to AUC but not to AUA or AUG (Fig. 2A). In contrast,
binds to AUC but not to AUA or AUG (Fig. 2A). In contrast, 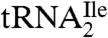 binds to AUA but not to AUC, AUG (Fig. 2B), or to AUU (Table S1). Thus, the modified cytidine in the anticodon wobble position of
binds to AUA but not to AUC, AUG (Fig. 2B), or to AUU (Table S1). Thus, the modified cytidine in the anticodon wobble position of 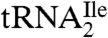 base pairs specifically with A but not with G, C, or U.
base pairs specifically with A but not with G, C, or U.
Fig. 2.
Ribosome binding of H. marismortui 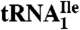 and
and 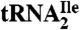 . Template-dependent binding of purified
. Template-dependent binding of purified 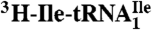 (A) and
(A) and 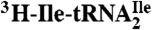 (B) to ribosomes isolated from H. marismortui. Oligonucleotides used were AUGAUG (▵), AUGAUA (□) and AUGAUC (○).
(B) to ribosomes isolated from H. marismortui. Oligonucleotides used were AUGAUG (▵), AUGAUA (□) and AUGAUC (○).
Localization of the Modified Cytidine (C*) in the Anticodon of 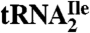 and Determination of Its Mass.
and Determination of Its Mass.
Liquid chromatography–mass spectrometry (LC-MS)/MS RNase mapping and sequencing of nucleic acids has been used extensively for the localization of modified nucleosides in RNAs (17–21). Here, we used separate digestions of 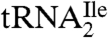 with RNase T1 and RNase A combined with bacterial alkaline phosphatase (BAP) followed by LC-MS/MS to confirm C34 as the site of the unknown modification and to identify the mass of the modified nucleoside. Knowledge of the mass of the modified nucleoside was then used as a search criterion for subsequent LC-MS and LC-MS/MS analysis of the nucleosides present in a total digest of
with RNase T1 and RNase A combined with bacterial alkaline phosphatase (BAP) followed by LC-MS/MS to confirm C34 as the site of the unknown modification and to identify the mass of the modified nucleoside. Knowledge of the mass of the modified nucleoside was then used as a search criterion for subsequent LC-MS and LC-MS/MS analysis of the nucleosides present in a total digest of 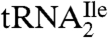 .
.
Digestion of 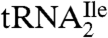 with RNase T1 is predicted to yield a decanucleotide 5′-CUC∗AU[t6A]ACCGp-3′, which would include C34 (designated as C*), the first nucleotide of the anticodon. The predicted molecular mass of this digestion product without the C34 modification is 3327.4 u. Consistent with our previous results (9) showing that C34 is quantitatively modified in
with RNase T1 is predicted to yield a decanucleotide 5′-CUC∗AU[t6A]ACCGp-3′, which would include C34 (designated as C*), the first nucleotide of the anticodon. The predicted molecular mass of this digestion product without the C34 modification is 3327.4 u. Consistent with our previous results (9) showing that C34 is quantitatively modified in 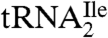 , LC-MS analysis of the RNase T1 digestion mixture failed to identify any m/z values related to the above unmodified sequence. Instead, abundant ions were detected (Fig. S1) at m/z 1718.8 (doubly charged) and m/z 1145.5 (triply charged). These m/z values correspond to an oligonucleotide mass of 3439.6, an increase of 112 over the oligonucleotide with no modification at C34. Collision-induced dissociation tandem mass spectrometry (CID-MS/MS) was attempted on this oligonucleotide, but minimal sequence information could be obtained likely due to the relatively large size of this RNase T1 digestion product and the lability of the glycosidic bond involving the modified C34 (see below).
, LC-MS analysis of the RNase T1 digestion mixture failed to identify any m/z values related to the above unmodified sequence. Instead, abundant ions were detected (Fig. S1) at m/z 1718.8 (doubly charged) and m/z 1145.5 (triply charged). These m/z values correspond to an oligonucleotide mass of 3439.6, an increase of 112 over the oligonucleotide with no modification at C34. Collision-induced dissociation tandem mass spectrometry (CID-MS/MS) was attempted on this oligonucleotide, but minimal sequence information could be obtained likely due to the relatively large size of this RNase T1 digestion product and the lability of the glycosidic bond involving the modified C34 (see below).
To obtain a shorter oligonucleotide more amenable to MS/MS, a combination of RNase A and BAP was used to digest 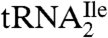 . The sequence of the RNase A and BAP digestion product of
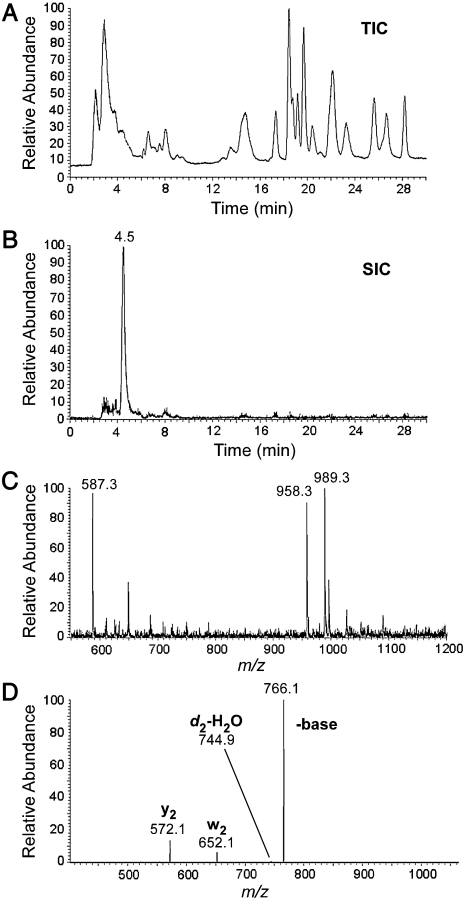
. The sequence of the RNase A and BAP digestion product of 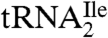 that covers the anticodon is the trinucleoside diphosphate 5′-C*pApU-3′. Based on elution time and m/z values, several oligonucleotides were detected in the total ion chromatograms (TIC) of the RNase A/BAP digest (Fig. 3A). Each of these was sequenced and all except one, m/z 989.3, could be mapped onto the
that covers the anticodon is the trinucleoside diphosphate 5′-C*pApU-3′. Based on elution time and m/z values, several oligonucleotides were detected in the total ion chromatograms (TIC) of the RNase A/BAP digest (Fig. 3A). Each of these was sequenced and all except one, m/z 989.3, could be mapped onto the 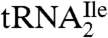 sequence. The selected ion chromatogram (SIC) of the oligonucleotide at m/z 989.3 is shown in Fig. 3B, and the corresponding mass spectrum of the ions eluting at 4.5 min is shown in Fig. 3C. The mass spectrum shows that this oligonucleotide elutes with GpC (m/z 587.3) and ApG+pU (m/z 958.3), which are derived from nucleotides 49–50 and 14–16, respectively, of
sequence. The selected ion chromatogram (SIC) of the oligonucleotide at m/z 989.3 is shown in Fig. 3B, and the corresponding mass spectrum of the ions eluting at 4.5 min is shown in Fig. 3C. The mass spectrum shows that this oligonucleotide elutes with GpC (m/z 587.3) and ApG+pU (m/z 958.3), which are derived from nucleotides 49–50 and 14–16, respectively, of 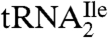 .
.
Fig. 3.
LC-MS/MS analysis of oligonucleotides present in RNase A/BAP digests of H. marismortui 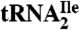 . (A) TIC of RNase A/BAP digestion of H. marismortui
. (A) TIC of RNase A/BAP digestion of H. marismortui 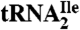 . (B) SIC of the anticodon-derived trinucleoside diphosphate 5′-C*pApU-3′ at m/z 989.3. (C) Mass spectrum of oligonucleotides eluting at 4.5 min. (D) MS/MS analysis of the trinucleoside diphosphate at m/z 989.3 (22).
. (B) SIC of the anticodon-derived trinucleoside diphosphate 5′-C*pApU-3′ at m/z 989.3. (C) Mass spectrum of oligonucleotides eluting at 4.5 min. (D) MS/MS analysis of the trinucleoside diphosphate at m/z 989.3 (22).
Because the difference in mass between the oligonucleotide at m/z 989.3 and the unmodified CpApU is 112 u and because there are no unmodified nucleotide base compositions that match 990.3 u (989.3 + H+) nor any one known modification mass of 112 u, this oligonucleotide is most likely C*pApU. This assignment is confirmed by MS/MS of m/z 989.3 (Fig. 3D). The major fragment ion is the loss of 223 (cytosine + 112), indicating once again the lability of the glycosidic bond and being consistent with the finding above (Fig. 1C) that C* contains at least one positive charge. The fragment ions at m/z 572.1, 652.1, and 744.9 correspond to cleavages of the C*pA phosphodiester bond, yielding y2 and w2 fragment ions, and the ApU phosphodiester bond, yielding a d2-H2O fragment ion, respectively (Fig. S2) (22). Together, these data establish the sequence of the trinucleotide as [C + 112]pApU. Thus, both RNase T1 mapping and RNase A/BAP mapping and sequencing confirm that C34 is modified in 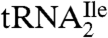 , with a net modification mass of 112 u. A modified nucleoside of mass 355 (cytidine + 112) was, therefore, used as a search criterion during subsequent LC-MS/MS analysis of total nucleosides derived from
, with a net modification mass of 112 u. A modified nucleoside of mass 355 (cytidine + 112) was, therefore, used as a search criterion during subsequent LC-MS/MS analysis of total nucleosides derived from 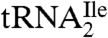 .
.
Modified Nucleosides Present in H. marismortui 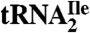 .
.
The purified 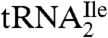 was digested completely to nucleosides and analyzed by LC-UV-MS (Fig. S3). All of the expected modified nucleosides in
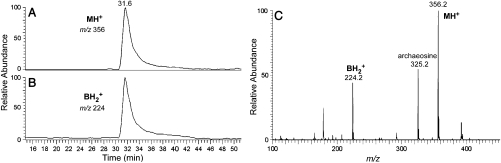
was digested completely to nucleosides and analyzed by LC-UV-MS (Fig. S3). All of the expected modified nucleosides in 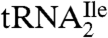 were detected. The presence of 2′-deoxyribo-nucleosides (dG, dA) indicates minor contamination with DNA oligonucleotide used for the purification of the tRNA. SICs for the molecular ion, m/z 356 (MH+), and the base ion, m/z 224 (
were detected. The presence of 2′-deoxyribo-nucleosides (dG, dA) indicates minor contamination with DNA oligonucleotide used for the purification of the tRNA. SICs for the molecular ion, m/z 356 (MH+), and the base ion, m/z 224 (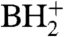 ), were obtained demonstrating that a nucleoside of the expected mass (from the RNase mapping and sequencing data) was present in this sample (Fig. 4A and 4B). The mass difference of 132 u between the molecular ion and the base ion is consistent with location of the unknown modification on the base and not on the ribose of the nucleoside. The mass spectrum from the SIC is shown in Fig. 4C. The unknown modified cytidine C* coelutes with the leading edge of archaeosine (m/z 325.2).
), were obtained demonstrating that a nucleoside of the expected mass (from the RNase mapping and sequencing data) was present in this sample (Fig. 4A and 4B). The mass difference of 132 u between the molecular ion and the base ion is consistent with location of the unknown modification on the base and not on the ribose of the nucleoside. The mass spectrum from the SIC is shown in Fig. 4C. The unknown modified cytidine C* coelutes with the leading edge of archaeosine (m/z 325.2).
Fig. 4.
Mass spectral analysis of the modified cytidine C∗. Purified H. marismortui 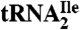 was digested to nucleosides and analyzed by LC-UV-MS (for complete analysis see Fig. S3). Selected ion chromatograms of (A) the molecular ion at m/z 356 (MH+) and (B) the base ion at m/z 224 (
was digested to nucleosides and analyzed by LC-UV-MS (for complete analysis see Fig. S3). Selected ion chromatograms of (A) the molecular ion at m/z 356 (MH+) and (B) the base ion at m/z 224 (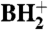 ). (C) Mass spectrum of the modified cytidine C*; C* coelutes with the leading edge of archaeosine.
). (C) Mass spectrum of the modified cytidine C*; C* coelutes with the leading edge of archaeosine.
Identification of C* as Agmatidine.
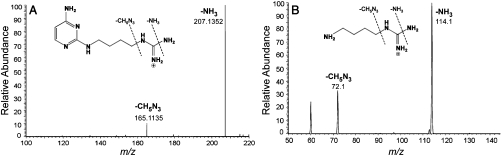
Accurate mass analysis of the protonated modified cytidine yielded an elemental composition of C14H26N7O4 (measured 356.2038, expected 356.2040, error 0.6 ppm). Accurate mass analysis of the protonated modified base yielded an elemental composition of C9H18N7 (measured 224.1617, expected 224.1618, error 0.4 ppm). The difference in elemental composition of the nucleoside and base is consistent with the absence of the C2-oxo group in this modified cytidine. Accurate mass analysis of the CID-MS/MS fragments of the base ion (Fig. 5A) yielded elemental compositions consistent with the loss of NH3 (m/z 207.1352) and CH5N3 (m/z 165.1135) from the modified cytidine.
Fig. 5.
Identification of the modified cytidine C*. (A) Accurate mass analysis of CID-MS/MS fragments of the base ion of C* (measured m/z 224.1617; expected m/z 224.1618). m/z 207.1352 and m/z 165.1135 are consistent with the loss of NH3 and CH5N3, respectively, from the base ion of C*. (B) MS fragmentation pattern of an agmatine standard. m/z 114.1 and m/z 72.1 are consistent with the loss of NH3 and CH5N3, respectively, from agmatine.
The absence of the C2-oxo group, the relatively high nitrogen content, and the fragmentation pattern suggested that the modification was attachment of agmatine to the C2 of cytidine. To confirm that loss of ammonia and a guanidino group are consistent with the presence of agmatine (23), a solution of agmatine was electrosprayed and subjected to MS/MS (Fig. 5B). The major fragment ions were indeed loss of NH3 and CH5N3. Thus, taken together, these data support the structure of C* as shown in Fig. 6. Similar to lysidine, at neutral pH, agmatidine has the potential of forming several tautomeric structures, three of which are shown in Fig. 6.
Fig. 6.
Comparison of agmatidine present in the anticodon wobble position of the archaeal AUA-decoding 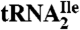 to lysidine present in the corresponding bacterial tRNA. Possible tautomeric structures of protonated agmatidine and lysidine are shown. Tautomers 1 and 2 could base pair with A.
to lysidine present in the corresponding bacterial tRNA. Possible tautomeric structures of protonated agmatidine and lysidine are shown. Tautomers 1 and 2 could base pair with A.
The mass spectral data by themselves do not rule out an alternate structure for agmatidine in which the C2 oxygen of cytidine is replaced by hydrogen and agmatine is attached to the C5 or C6. However, this is most unlikely for several reasons: (i) such a modified nucleoside would make 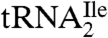 base pair with AUG instead of AUA (Fig. 2); (ii) the
base pair with AUG instead of AUA (Fig. 2); (ii) the 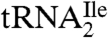 would be aminoacylated with methionine instead of isoleucine (8, 9); (iii) it would not explain the block in reverse transcriptase reaction by agmatidine (9); and (iv) it would require, in addition to a reductase to reduce the C2 oxygen, the biochemically difficult linkage of C5 or C6 of cytidine to the amino group of agmatine.
would be aminoacylated with methionine instead of isoleucine (8, 9); (iii) it would not explain the block in reverse transcriptase reaction by agmatidine (9); and (iv) it would require, in addition to a reductase to reduce the C2 oxygen, the biochemically difficult linkage of C5 or C6 of cytidine to the amino group of agmatine.
Agmatidine Is also Present in 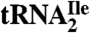 from M. maripaludis and in Total tRNA from S. solfataricus.
from M. maripaludis and in Total tRNA from S. solfataricus.
The LC/MS analysis of nucleosides from digests of partially purified 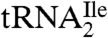 from M. maripaludis and total tRNA from S. solfataricus (Fig. S4) shows that agmatidine is also present in tRNAs from these archaeal organisms. Identification of agmatidine was confirmed by the SICs and mass spectral data. Thus, agmatidine is present in the AUA decoding
from M. maripaludis and total tRNA from S. solfataricus (Fig. S4) shows that agmatidine is also present in tRNAs from these archaeal organisms. Identification of agmatidine was confirmed by the SICs and mass spectral data. Thus, agmatidine is present in the AUA decoding 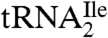 of euryarchaea and crenarchaea.
of euryarchaea and crenarchaea.
Discussion
A clear conclusion of the work described in here is that archaea and bacteria use very similar strategies to read the isoleucine codon AUA without also reading the methionine codon AUG. Agmatidine (abbreviation C+), identified here to be in the anticodon wobble position of the AUA decoding archaeal 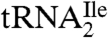 , is in many respects similar to lysidine found in the corresponding tRNA of bacteria. In both cases, the C2-oxo group of cytidine is replaced by a long side chain derived from either lysine, an amino acid, or by agmatine, a decarboxylated amino acid. The only difference is that in lysidine, the linkage involves the side chain amino group of lysine, whereas in agmatidine, it involves the former α-amino group of arginine and not the guanidino side chain. It is striking that archaea and bacteria use basically similar, although different, types of base modification to generate a cytidine derivative that can base pair only with A.
, is in many respects similar to lysidine found in the corresponding tRNA of bacteria. In both cases, the C2-oxo group of cytidine is replaced by a long side chain derived from either lysine, an amino acid, or by agmatine, a decarboxylated amino acid. The only difference is that in lysidine, the linkage involves the side chain amino group of lysine, whereas in agmatidine, it involves the former α-amino group of arginine and not the guanidino side chain. It is striking that archaea and bacteria use basically similar, although different, types of base modification to generate a cytidine derivative that can base pair only with A.
The archaeal translation apparatus is known to be a mosaic of bacterial and eukaryotic features (24–26). Archaeal mRNAs are bacterial-like in lacking the 5′-terminal cap and 3′-terminal PolyA sequences and in using Shine–Dalgarno-like sequences and leaderless mRNAs for translation initiation. However, the archaeal translation initiation factors, of which six have been identified, are a subset of eukaryotic initiation factors (27, 28). In addition, translation elongation and termination in archaea and eukaryotes are similar based on patterns of antibiotic sensitivity of the archaeal translation system (29) and on the biochemical and genomic analysis of translation elongation and termination factors, for example, the presence of diphthamide, a modified histidine, in the elongation factor a/eEF2 (30), the presence of hypusine, a critical modified lysine, in the elongation factor a/eIF5A (31), and the presence of a single Class I transcription termination factor a/eRF1, which reads all three stop codons instead of two termination factors (RF1 and RF2) in bacteria. In view of these substantial similarities between archaea and eukaryotes, our finding that archaea use a strategy for translation of the AUA codon that is similar to that used by bacteria but different from that used by eukaryotes is quite surprising.
As for lysidine, the identification of agmatidine in the AUA specific archaeal 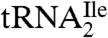 raises the question of how agmatidine base pairs specifically with A on the ribosome. Agmatidine has the potential for forming several tautomeric structures (Fig. 6), and it is not known which tautomeric form is present in the tRNA and whether this form undergoes changes in the context of the ribosome. Detailed structural studies will be necessary to understand the mechanism of specific recognition of the AUA codon by the corresponding archaeal and bacterial tRNAIles.
raises the question of how agmatidine base pairs specifically with A on the ribosome. Agmatidine has the potential for forming several tautomeric structures (Fig. 6), and it is not known which tautomeric form is present in the tRNA and whether this form undergoes changes in the context of the ribosome. Detailed structural studies will be necessary to understand the mechanism of specific recognition of the AUA codon by the corresponding archaeal and bacterial tRNAIles.
The identification of agmatidine in 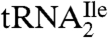 from H. marismortui and M. maripaludis and in total tRNA from S. solfataricus suggests that agmatidine is present in the AUA decoding
from H. marismortui and M. maripaludis and in total tRNA from S. solfataricus suggests that agmatidine is present in the AUA decoding 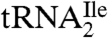 of euryarchaea and crenarchaea. This conclusion is supported by the finding that all sixty-six of the euryarchaeal and crenarchaeal genomes sequenced have only one tRNA with the anticodon GAU annotated as an isoleucine tRNA. This tRNA can read the isoleucine codons AUU and AUC but not AUA. Interestingly, all sixty-six of these archaeal genomes also contain at least three tRNAs with the anticodon CAU, which are annotated as methionine tRNA (KEGG database; http://www.genome.jp/kegg/). As shown by us here and elsewhere for the H. marismortui
of euryarchaea and crenarchaea. This conclusion is supported by the finding that all sixty-six of the euryarchaeal and crenarchaeal genomes sequenced have only one tRNA with the anticodon GAU annotated as an isoleucine tRNA. This tRNA can read the isoleucine codons AUU and AUC but not AUA. Interestingly, all sixty-six of these archaeal genomes also contain at least three tRNAs with the anticodon CAU, which are annotated as methionine tRNA (KEGG database; http://www.genome.jp/kegg/). As shown by us here and elsewhere for the H. marismortui 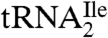 (9) and for the M. maripaludis
(9) and for the M. maripaludis 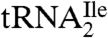 , one of these putative methionine tRNAs is actually the AUA decoding isoleucine tRNA. Therefore, it is likely that in the rest of euryarchaea and crenarchaea also, the anticodon wobble nucleoside C of one of the putative methionine tRNAs is modified to agmatidine to generate a tRNA, which is aminoacylated with isoleucine and which reads the isoleucine codon AUA. It is interesting to note that in contrast to H. marismortui, in which AUA is a very rare codon (3.6/1000 codons), in S. solfataricus it is the most frequent codon (49.4/1000 codons) (http://www.kazusa.or.jp/codon/). Thus, the use of agmatidine in
, one of these putative methionine tRNAs is actually the AUA decoding isoleucine tRNA. Therefore, it is likely that in the rest of euryarchaea and crenarchaea also, the anticodon wobble nucleoside C of one of the putative methionine tRNAs is modified to agmatidine to generate a tRNA, which is aminoacylated with isoleucine and which reads the isoleucine codon AUA. It is interesting to note that in contrast to H. marismortui, in which AUA is a very rare codon (3.6/1000 codons), in S. solfataricus it is the most frequent codon (49.4/1000 codons) (http://www.kazusa.or.jp/codon/). Thus, the use of agmatidine in 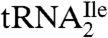 does not depend on the frequency of utilization of the AUA codon in mRNA.
does not depend on the frequency of utilization of the AUA codon in mRNA.
In contrast to euryarchaea and crenarchaea, in the only available genome sequences of a nanoarchaeum and a korarchaeum, two tRNA genes are annotated as coding for isoleucine tRNAs (32–34). One of them would have the anticodon GAU, whereas the other one would have the anticodon UAU, which is likely modified to ΨAΨ as in eukaryotic 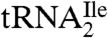 (8). This latter tRNA would have the potential for reading the AUA codon although how it would be prevented from also reading the methionine codon AUG is unknown. The genome sequences could suggest that nanoarchaea and korarchaea use a eukaryote-like strategy to read the isoleucine codon AUA. However, before reaching this conclusion, it is important to sequence additional nanoarchaeal and korarchaeal genomes to determine whether other isolates of nanoarchaea and korarchaea also have a
(8). This latter tRNA would have the potential for reading the AUA codon although how it would be prevented from also reading the methionine codon AUG is unknown. The genome sequences could suggest that nanoarchaea and korarchaea use a eukaryote-like strategy to read the isoleucine codon AUA. However, before reaching this conclusion, it is important to sequence additional nanoarchaeal and korarchaeal genomes to determine whether other isolates of nanoarchaea and korarchaea also have a 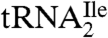 with the anticodon UAU (possibly ΨAΨ) or have C*AU (C∗ = agmatidine) as in euryarchaea and crenarchaea.
with the anticodon UAU (possibly ΨAΨ) or have C*AU (C∗ = agmatidine) as in euryarchaea and crenarchaea.
The very similar structures of agmatidine and lysidine suggest a simple pathway for biosynthesis of agmatidine, similar to that of lysidine, involving activation of the C2 of cytidine by adenylation followed by nucleophilic attack on the activated C2 by the primary amino group of agmatine (35, 36). Agmatine is derived by decarboxylation of arginine and occurs naturally as one of the intermediates in polyamine biosynthesis. In the thermophilic euryarchaeon T. kodakaraensis, agmatine is essential for polyamine biosynthesis (12). Agmatine is also an important neuronal modulator, which binds to various receptors such as the α2-adrenergic receptor, NMDA receptors, etc. (10, 11). It has also been suggested to be a neurotransmitter, although that remains to be proven (37). In view of the critical role of agmatidine in archaeal AUA decoding 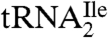 function, the potential involvement of agmatine in agmatidine biosynthesis suggests that agmatine may also be essential in euryarchaea and crenarchaea for the reading of the genetic code.
function, the potential involvement of agmatine in agmatidine biosynthesis suggests that agmatine may also be essential in euryarchaea and crenarchaea for the reading of the genetic code.
Although agmatidine and lysidine are very similar modified nucleosides, both derived from cytidine, in spite of extensive bioinformatics searches, no homolog of TilS, the enzyme responsible for biosynthesis of lysidine in bacteria, has been detected in archaea (38). This raises the interesting question of how similar the enzyme responsible for biosynthesis of agmatidine is to TilS and whether the two enzymes are products of convergent evolution. Clearly, an analysis of the evolutionary history of these enzymes would be of much interest.
Materials and Methods
Purification of 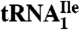 and
and 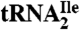 from H. marismortui.
from H. marismortui.
H. marismortui ATCC 43,049 was kindly provided by Peter Moore (Yale University). General manipulations of H. marismortui were performed according to standard procedures (39). Cells were grown at 37 ºC, harvested in late-log phase, and crude RNA was isolated by acid guanidinium thiocyanate-phenol-chloroform extraction (40). Ribosomal RNA was removed by precipitation with 1 M NaCl yielding total tRNA. The yield of total tRNA was approximately 1,000 A260 units from a 15 L-culture. Purification of tRNAs was carried out essentially as described by Suzuki and Suzuki (41). Details are given in SI Text.
Binding of 3H-Ile-tRNAIle to Ribosomes.
H. marismortui ribosomes were isolated essentially as described (42) with slight modifications. Pelleted cells from a 2 L-culture were suspended in 10 mM Tris-HCl pH 7.5, 100 mM Mg(OAc)2 and 3.4 M KCl (buffer 1) and lysed by French Press (Constant Cell Disruption System), and the lysate was cleared of cell debris by centrifugation first at 27,000 × g for 15 min and then at 65,000 × g for 15 min. Ribosomes were then pelleted at 255,000 × g for 4 hr and suspended in buffer 1. The ribosome suspension was divided into aliquots, quick frozen, and stored at -80 ºC. Ribosome binding was adapted from Bayley and Griffiths (43) with modifications. Details are given in SI Text.
Digestion of tRNA to Nucleosides.
Purified H. marismortui 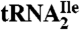 (0.2 A260 unit), purified M. maripaludis
(0.2 A260 unit), purified M. maripaludis 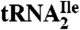 (0.6 A260 unit), H. marismortui total tRNA (0.6 A260 unit) and S. solfataricus total tRNA (1.0 A260 unit) were digested to nucleosides using nuclease P1 (Sigma), snake venom phosphodiesterase I (Worthington Biochemical Corporation) and antarctic phosphatase (New England Biolabs) as described by Crain (44).
(0.6 A260 unit), H. marismortui total tRNA (0.6 A260 unit) and S. solfataricus total tRNA (1.0 A260 unit) were digested to nucleosides using nuclease P1 (Sigma), snake venom phosphodiesterase I (Worthington Biochemical Corporation) and antarctic phosphatase (New England Biolabs) as described by Crain (44).
Digestion of 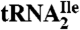 with Nucleases and LC-MS and LC-MS/MS.
with Nucleases and LC-MS and LC-MS/MS.
LC-MS to map RNase A and T1 digestion products onto the tRNA, LC-MS/MS to sequence the oligonucleotides in RNase A digests, and accurate mass LC-MS and LC-MS/MS analysis of total nucleoside digests of the 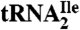 were carried out as described (45) with modifications. Details are given in SI Text.
were carried out as described (45) with modifications. Details are given in SI Text.
Supplementary Material
Acknowledgments.
We thank Annmarie McInnis for her useful cheerfulness and care in the preparation of this manuscript. This work was supported by grants from the National Institutes of Health [GM17151 (to U.L.R.), GM22854 (to D.Söll), RR19900 (to P.A.L.)], National Science Foundation [CHE0602413 and CHE0910751 (to P.A.L.)], the University of Cincinnati (to P.A.L.), and the U.S. Department of Energy [DE-FG36-08GO88055 (to P.B.)].
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914869107/DCSupplemental.
References
- 1.Crick FH. Codon-anticodon pairing: The wobble hypothesis. J Mol Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 2.Agris PF. Decoding the genome: A modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Söll D, RajBhandary UL. Studies on polynucleotides. LXXVI. Specificity of transfer RNA for codon recognition as studied by amino acid incorporation. J Mol Biol. 1967;29:113–124. doi: 10.1016/0022-2836(67)90184-2. [DOI] [PubMed] [Google Scholar]
- 4.Harada F, Nishimura S. Purification and characterization of AUA specific isoleucine transfer ribonucleic acid from Escherichia coli B. Biochemistry. 1974;13:300–307. doi: 10.1021/bi00699a011. [DOI] [PubMed] [Google Scholar]
- 5.Muramatsu T, et al. A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli. J Biol Chem. 1988;263:9261–9267. doi: 10.1351/pac198961030573. [DOI] [PubMed] [Google Scholar]
- 6.Muramatsu T, et al. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988;336:179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- 7.Grosjean H, Björk GR. Enzymatic conversion of cytidine to lysidine in anticodon of bacterial tRNAIle—an alternative way of RNA editing. Trends Biochem Sci. 2004;29:165–168. doi: 10.1016/j.tibs.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Senger B, Auxilien S, Englisch U, Cramer F, Fasiolo F. The modified wobble base inosine in yeast tRNAIle is a positive determinant for aminoacylation by isoleucyl-tRNA synthetase. Biochemistry. 1997;36:8269–8275. doi: 10.1021/bi970206l. [DOI] [PubMed] [Google Scholar]
- 9.Köhrer C, et al. Identification and characterization of a tRNA decoding the rare AUA codon in Haloarcula marismortui. RNA. 2008;14:117–126. doi: 10.1261/rna.795508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halaris A, Plietz J. Agmatine: Metabolic pathway and spectrum of activity in brain. CNS Drugs. 2007;21:885–900. doi: 10.2165/00023210-200721110-00002. [DOI] [PubMed] [Google Scholar]
- 11.Rawls SM, Gerber K, Ding Z, Roth C, Raffa RB. Agmatine: Identification and inhibition of methamphetamine, kappa opioid, and cannabinoid withdrawal in planarians. Synapse. 2008;62:927–934. doi: 10.1002/syn.20571. [DOI] [PubMed] [Google Scholar]
- 12.Fukuda W, Morimoto N, Imanaka T, Fujiwara S. Agmatine is essential for the cell growth of Thermococcus kodakaraensis. FEMS Microbiol Lett. 2008;287:113–120. doi: 10.1111/j.1574-6968.2008.01303.x. [DOI] [PubMed] [Google Scholar]
- 13.Schleper C, Jurgens G, Jonuscheit M. Genomic studies of uncultivated archaea. Nat Rev Microbiol. 2005;3:479–488. doi: 10.1038/nrmicro1159. [DOI] [PubMed] [Google Scholar]
- 14.Goo YA, et al. Low-pass sequencing for microbial comparative genomics. BMC Genomics. 2004;5:3. doi: 10.1186/1471-2164-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.RajBhandary UL, Chang SH. Studies on polynucleotides. LXXXII. Yeast phenylalanine transfer ribonucleic acid: Partial digestion with ribonuclease T-1 and derivation of the total primary structure. J Biol Chem. 243:598–608. [PubMed] [Google Scholar]
- 16.Köhrer C, RajBhandary UL. The many applications of acid urea polyacrylamide gel electrophoresis to studies of tRNAs and aminoacyl-tRNA synthetases. Methods. 2008;44:129–138. doi: 10.1016/j.ymeth.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA. A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Res. 1993;21:4577–4585. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCloskey JA, et al. Post-transcriptional modification in archaeal tRNAs: Identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 2001;29:4699–4706. doi: 10.1093/nar/29.22.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hossain M, Limbach PA. Mass spectrometry-based detection of transfer RNAs by their signature endonuclease digestion products. RNA. 2007;13:295–303. doi: 10.1261/rna.272507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartmer R, et al. RNase T1 mediated base-specific cleavage and MALDI-TOF MS for high-throughput comparative sequence analysis. Nucleic Acids Res. 2003;31:e47. doi: 10.1093/nar/gng047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng Z, Limbach PA. RNase mapping of intact nucleic acids by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry (ESI-FTICRMS) and 18O labeling. Int J Mass Spectrom. 2004;234:37–44. [Google Scholar]
- 22.McLuckey SA, Van Berkel GJ, Glish GL. Tandem mass spectrometry of small, multiply charged oligonucleotides. J Am Soc Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 23.Feistner GF. Liquid chromatography-electrospray tandem mass spectrometry of danyslated polyamines and basic amino acids. J Mass Spectrom. 1995;30:1546–1552. [Google Scholar]
- 24.Londei P. Translation in Archaea. In: Cavicchioli R, editor. Archaea. Washington, D.C.: ASM Press; 2007. pp. 175–197. [Google Scholar]
- 25.Srinivasan G, Krebs MP, RajBhandary UL. Translation initiation with GUC codon in the archaeon Halobacterium salinarum: Implications for translation of leaderless mRNA and strict correlation between translation initiation and presence of mRNA. Mol Microbiol. 2006;59:1013–1024. doi: 10.1111/j.1365-2958.2005.04992.x. [DOI] [PubMed] [Google Scholar]
- 26.Dennis PP. Ancient ciphers: Translation in Archaea. Cell. 1997;89:1007–1010. doi: 10.1016/s0092-8674(00)80288-3. [DOI] [PubMed] [Google Scholar]
- 27.Kyrpides NC, Woese CR. Archaeal translation initiation revisited: The initiation factor 2 and eukaryotic initiation factor 2B alpha-beta-delta subunit families. Proc Natl Acad Sci USA. 1998;95:3726–3730. doi: 10.1073/pnas.95.7.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyrpides NC, Woese CR. Universally conserved translation initiation factors. Proc Natl Acad Sci USA. 1998;95:224–228. doi: 10.1073/pnas.95.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hummel H, Böck A. 23S ribosomal RNA mutations in halobacteria conferring resistance to the anti-80S ribosome targeted antibiotic anisomycin. Nucleic Acids Res. 1987;15:2431–2443. doi: 10.1093/nar/15.6.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kessel M, Klink F. Archaebacterial elongation factor is ADP-ribosylated by diphtheria toxin. Nature. 1980;287:250–251. doi: 10.1038/287250a0. [DOI] [PubMed] [Google Scholar]
- 31.Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translational elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waters E, et al. The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci USA. 2003;100:12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Randau L, Pearson M, Söll D. The complete set of tRNA species in Nanoarchaeum equitans. FEBS Lett. 2005b;579:2945–2947. doi: 10.1016/j.febslet.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 34.Elkins JG, et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci USA. 2008;105:8102–8107. doi: 10.1073/pnas.0801980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soma A, et al. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol Cell. 2003;12:689–698. doi: 10.1016/s1097-2765(03)00346-0. [DOI] [PubMed] [Google Scholar]
- 36.Ikeuchi Y, et al. Molecular mechanism of lysidine synthesis that determines tRNA identity and codon recognition. Mol Cell. 2005;19:235–246. doi: 10.1016/j.molcel.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Regunathan S. Agmatine: Biological role and therapeutic potentials in morphine analgesia and dependence. AAPS J. 2006;8:E479–484. doi: 10.1208/aapsj080356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grosjean H, Marck C, Gaspin C, Decataur WA, de Crecy-Lagard V. RNomics and Modomics in the halophilic archaea Haloferax volcanii: Identification of RNA modification genes. BMC Genomics. 2008;9:470. doi: 10.1186/1471-2164-9-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DasSarma S, Fleischmann EM. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1995. Archaea—A Laboratory Manual. Halophiles. [Google Scholar]
- 40.Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat Protoc. 2006;1:581–585. doi: 10.1038/nprot.2006.83. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki T, Suzuki T. Chaplet column chromatography: Isolation of a large set of individual RNAs in a single step. Methods Enzymol. 2007;425:231–239. doi: 10.1016/S0076-6879(07)25010-4. [DOI] [PubMed] [Google Scholar]
- 42.Shevack A, Gewitz HS, Hennemann B, Yonath A, Wittmann HG. Characterization and crystallization of ribosomal particles from Halobacterium marismortui. FEBS Lett. 1985;184:68–71. [Google Scholar]
- 43.Bayley ST, Griffiths E. A cell-free amino acid incorporating system from an extremely halophilic bacterium. Biochemistry. 1968;7:2249–2256. doi: 10.1021/bi00846a030. [DOI] [PubMed] [Google Scholar]
- 44.Crain PF. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 45.Pomerantz SC, McCloskey JA. Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol. 1990;193:796–824. doi: 10.1016/0076-6879(90)93452-q. [DOI] [PubMed] [Google Scholar]
- 46.Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem. 1984;259:9461–9671. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.