Abstract
MicroRNAs (miRs) can regulate many cellular functions, but their roles in regulating responses of vascular endothelial cells (ECs) to mechanical stimuli remain unexplored. We hypothesize that the physiological responses of ECs are regulated by not only mRNA and protein signaling networks, but also expression of the corresponding miRs. EC growth arrest induced by pulsatile shear (PS) flow is an important feature for flow regulation of ECs. miR profiling showed that 21 miRs are differentially expressed (8 up- and 13 downregulated) in response to 24-h PS as compared to static condition (ST). The mRNA expression profile indicates EC growth arrest under 24-h PS. Analysis of differentially expressed miRs yielded 68 predicted mRNA targets that overlapped with results of microarray mRNA profiling. Functional analysis of miR profile indicates that the cell cycle network is highly regulated. The upregulation of miR-23b and miR-27b was found to correlate with the PS-induced EC growth arrest. Inhibition of miR-23b using antagomir-23b oligonucleotide (AM23b) reversed the PS-induced E2F1 reduction and retinoblastoma (Rb) hypophosphorylation and attenuated the PS-induced G1/G0 arrest. Antagomir AM27b regulated E2F1 expression, but did not affect Rb and growth arrest. Our findings indicate that PS suppresses EC proliferation through the regulation of miR-23b and provide insights into the role of miRs in mechanotransduction.
Keywords: cell cycle, shear, bioinformatics, gene regulation, mechanotransduction
Hemodynamic forces, e.g., stretch and shear stress, act constantly on the vascular endothelial cells (ECs) to modulate EC signaling, gene expression, and physiological functions (1). Atherosclerotic lesions in the arterial tree are found mainly at branch points, where blood flow is disturbed with a limited forwarding direction, but are generally spared at the straight parts of the arterial tree, where the flow is laminar with a large forwarding direction (2). Exposure of ECs to 24 h of steady laminar shear flow at 12 dyn/cm2 (approximating the hemodynamic force in straight parts of arteries) leads to antiproliferative (3) and antiinflammatory (4) responses. In contrast, ECs exposed to disturbed flow, mimicking the hemodynamic force at branch points, exhibit opposite responses (5, 6). The laminar shear-induced EC growth arrest involves the expression of CDK inhibitors (e.g., p21cip, p27kip), tumor suppressor p53, and retinoblastoma (Rb) hypophosphorylation (3, 7). Whereas there is considerable knowledge on mechanotransduction at protein and mRNA levels, there is little information on the role of microRNAs (miRs) in this process.
miRs are small noncoding RNAs (∼21–25 nucleotides) that regulate gene expression by binding to target mRNAs to cause their degradation or translational repression (8). It is estimated that miRs regulate ∼30% of human protein-coding genes. More than 800 miRs have been identified in the human genome and registered in the Sanger miRBase. These small RNAs provide a powerful mechanism for posttranscriptional control of gene expression at the mRNA level. There is growing evidence that miRs are involved in many biological processes in health and disease, including cell proliferation and differentiation, oncogenesis, and angiogenesis (9–12), as well as cardiovascular homeostasis (13, 14). In ECs, knockdown of dicer (a key molecule in biogenesis of miRs) altered the expression of genes affecting EC biology and reduced EC proliferation and angiogenesis in vitro (15). When both Dicer and Drosha (another important molecule in miR biogenesis) were knocked down in ECs, capillary sprouting and tube-forming activity were significantly reduced (16). Thus, the global reduction of miRs through the knockdown of Dicer and/or Drosha significantly affects EC functions in vitro and in vivo, suggesting the important roles of miRs in regulating vascular functions (16–19).
We investigated the role of miRs in regulating EC gene expression and functions in response to pulsatile shear (PS) flow with a significant forwarding direction. The results indicate that miR-23b plays an important regulatory role in the inhibition of EC proliferation by PS.
Results
Long-Term Pulsatile Laminar Flow Keeps Vascular Endothelium in a Quiescent State.
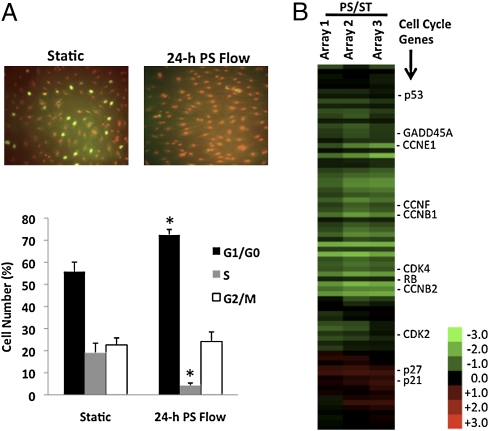
In agreement with previous studies on steady and pulsatile laminar shear stresses (3, 7, 20), we found that human umbilical vein endothelial cells (HUVECs) subjected to 24-h PS had a significant reduction in proliferation rate in comparison with static condition (ST). Twenty-four-hour PS led to a substantial reduction of BrdU-positive cells (Fig. 1A, Top). Flow cytometry (Fig. 1A, Bottom) showed that ECs subjected to 24-h PS had a significant increase in percentage of cells in G0/G1 phase (72.43 ± 2.46%) compared to ST (55.8 ± 4.32%). PS also significantly reduced percentage of cells in S phase (4.18 ± 1.12%) vs. ST (18.99 ± 4.35%), but there is no significant difference for cells in G2/M phase between PS (24.13 ± 4.31%) and ST (22.57 ± 3.2%). Thus, 24-h PS causes growth arrest by preventing ECs from entering the S phase.
Fig. 1.
Twenty-four-hour PS reduces cell proliferation and regulates cell cycle-related gene expression. (A) HUVECs were subjected to 24-h PS, with BrdU added during the last 4 h. Cells were fixed and stained (Upper) with anti-BrdU and 7-AAD (for DNA staining). Flow cytometry (Lower) showed a significant increase of cell number in G0/G1 phase and a significant decrease in S phase. The number of cells in G2 + M was not significantly changed. (B) Hierarchical clustering of differentially expressed genes in cell cycle under PS vs. ST. Biological repeats are shown at the Upper. Right column lists the selected gene symbols. Color of each band reflects fold change of mRNA in PS vs. ST. Red bands represent increase, and green bands decrease. Exemplar mRNAs representing cell cycle components are shown in this heat map.
Expression Profiling Analysis of mRNA in ECs Under PS and ST.
We investigated genomewide mRNA expression profile in relation to EC growth arrest under PS in three independent biological experiments. The results were analyzed using the GeneSpring GX software (Agilent). Gene expression ratios from each experiment were calculated by using the average normalized intensities (Materials and Methods). Determination of differential gene expressions between PS and ST with a cutoff at changes ≥1.75-fold and P-value <0.01 yielded 1,227 genes, of which 569 (46%) were up- and 658 (54%) were downregulated by PS.
The up- and downregulated gene lists were submitted to DAVID online tool (http://david.abcc.ncifcrf.gov/home.jsp) (21, 22) for gene ontology (GO) annotation (23) and pathway enrichment analysis. GO terms are grouped into three categories: biological process, cellular component, and molecular function. Table 1 lists the GO terms (top) representing biological process that have the highest enriched score and most significant P-value. The top five GO terms under biological process are all cell cycle related, including cell cycle, mitotic cell cycle, cell division, DNA replication, and cell cycle check point. The GO terms representing cellular compartment and molecular function are listed in Table S1. Functional classification of the differentially expressed mRNA transcripts by KEGG pathway database (24, 25) (Table 1, lower) also shows that the downregulated genes are highly associated with cell cycle, p53 signaling, and DNA synthesis pathways, whereas the upregulated genes are mostly associated with metabolic pathways. Unsupervised clustering of the expression profile for cell cycle genes in ECs under PS vs. ST is shown in Fig. 1B. Most cell cycle-related genes are downregulated (e.g., CDK2, CDK4, most cyclins, and E2F1), whereas growth arrest genes are unchanged (e.g., p53, Rb) or upregulated (e.g., p21cip, p27kip) in PS vs. ST.
Table 1.
Gene ontology and KEGG pathway analysis of up- and down-regulated genes in ECs under PS vs. ST
| Gene ontology analysis | |||
| GO* | Biological process | Enriched score | P value |
| GO:0007049 | Cell cycle | 42.17 | 2.20E-56 |
| GO:0000278 | Mitotic cell cycle | 42.17 | 6.20E-43 |
| GO:0051301 | Cell division | 42.17 | 1.10E-41 |
| GO:0006260 | DNA Replication | 12.14 | 1.10E-10 |
| GO:0000075 | Cell cycle check point | 11.42 | 1.00E-08 |
| KEGG pathway analysis | |||
| Pathway | Count | P value | |
| Up | Glutathione metabolism | 9 | 9.30E-03 |
| Up | Arachidonic acid metabolism | 10 | 2.50E-02 |
| Up | Biosynthesis of steroids | 6 | 3.50E-02 |
| Up | Glycerolipid metabolism | 9 | 3.80E-02 |
| Down | Cell cycle | 47 | 7.10E-22 |
| Down | p53 signaling pathway | 20 | 1.10E-06 |
| Down | Pyrimidine metabolism | 20 | 6.60E-05 |
| Down | Purine metabolism | 27 | 9.40E-05 |
| Down | DNA polymerase | 9 | 4.30E-04 |
*Only the top five GO terms representing biological process are listed in the table.
These results indicate that 24-h PS prevents ECs from entering the S phase by downregulating key proteins in the cell cycle network and upregulating CDK inhibitors.
miR Expression Profiles in ECs Under PS and ST.
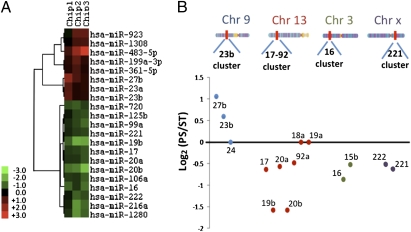
To examine the involvement of miRs in PS-regulated gene expression and growth arrest, we analyzed miR profiles in ECs under PS and ST with LC Sciences μParaflo microfluidic miRNA microarray, which contains quadruplicate probes for 856 human miRs with the Sanger miRbase sequence database (release 12.0). After filtering out the signals below the threshold level for detection and applying the criteria of fold change ≥1.25 and P < 0.01, the data of three independent biological experiments revealed that 21 miRs were differentially regulated in PS vs. ST (Fig. 2A), with 8 up- and 13 downregulated. PS caused the upregulation of miR-23b cluster (miR-23b and miR-27b), and the downregulations of miR-17–92 cluster (miR-17, -19b, -20a, -20b, -92a), miR-16 cluster (miR-15b and -16), and miR-221 cluster (miR-221 and -222) (Fig. 2B). We selected 9 miRs on the basis of functional interest for validation with RT-PCR, which showed that 7 of them (miR-23b, -27b, -923, -16, -106, -26b, and let-7) matched the microarray data, but 2 (miR-19b and -221) did not (Table S2).
Fig. 2.
miRNA expression profile under PS vs. ST. Expression levels of 856 human miRs (Sanger miRBase 12.0) in ECs under PS and ST were measured with miR microarray (n = 3). (A) Unsupervised hierarchical clustering analysis of miR expression profile. Heat map shows PS vs. ST miR log2 ratios with a threshold of 1.25-fold and P < 0.05. Red bands represent upregulated miRs and green bands, downregulated miRs. (B) Differential expression of miR clusters. miRs are considered to belong to the same cluster if they are located in close proximity to each other in the genome. PS upregulates the miR-23b cluster in chromosome 9 and downregulates miR-17–92, -16, and -221 clusters in chromosomes 13, 3, and X, respectively. The images of chromosomes are copied from http://genomics.energy.gov.
We searched for potential mRNA targets of the 21 differentially expressed miRs by using the algorithms TargetScan and PicTar (26–29). By taking the consensus targets selected by both, we identified 1,073 putative targets. Among these, 68 genes (5.5% of the putative targets) were differentially expressed in ECs at the mRNA level under PS vs. ST (Fig. S1). The overlapping GO terms (representing biological process) between the predicted targets and the differentially expressed genes are listed in Table S3, and cell cycle-related processes are among the top list.
Correlation of miR Levels with Functional Gene Sets.
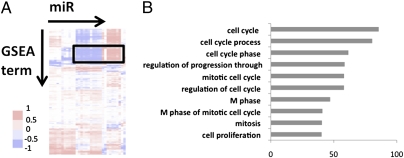
To investigate the functional roles of the PS-regulated miRs, we calculated the correlation coefficients between PS-induced miRs and differentially expressed genes (1,227 mRNAs) and ranked and used them to perform the gene set enrichment analysis (GSEA) (30, 31). A total of 394 functional gene sets (FGSs) were identified from the curated gene sets in the c.2 collection representing the functional pathways in the GSEA database. The associations of miRs with these FGSs were generated, hierarchically clustered, and plotted as a matrix on the basis of the statistical significance (P-value) of the enrichment score obtained from GSEA analysis. We identified the potential associations between miRs and the FGSs (Fig. 3A); the strongest associations are shown in the boxed region. We extracted protein-coding genes from these FGSs and performed GO analysis. Among the top 10 GO terms (Fig. 3B), cell cycle-related ones are again highly enriched and strongly associated with the miR expression profile. The miR-23b cluster (miR-23b and -27b) is negatively associated with these functional categories. Integration of miR and mRNA expression profiles, together with computational bioinformatics approaches, revealed the potential roles of miR-23b and -27b in regulating the PS-induced EC growth arrest.
Fig. 3.
Association matrix of miR levels with functional gene sets under PS vs. ST. (A) Columns represent 394 functional gene sets, and rows represent flow-regulated miRs. Functional categories are positively (red bands), negatively (blue bands), or not (white bands) associated with miR expression profiles under PS vs. ST. The boxed region shows high associations between functional categories and miRs. The genes extracted from the functional gene sets in the box were used to perform the gene ontology analysis. (B) The −log (P values) of top 10 GO terms are plotted.
Blockade of miR-23b, but Not miR-27b, Attenuated the PS-Induced Growth Arrest.
The abundance of miR-23b and miR-27b in HUVECs and their upregulation by 24-h PS (Fig. 2) indicate that the miR-23b cluster may play significant roles in EC growth arrest. Therefore, we tested whether their antagomirs can reverse the growth arrest effect of PS.
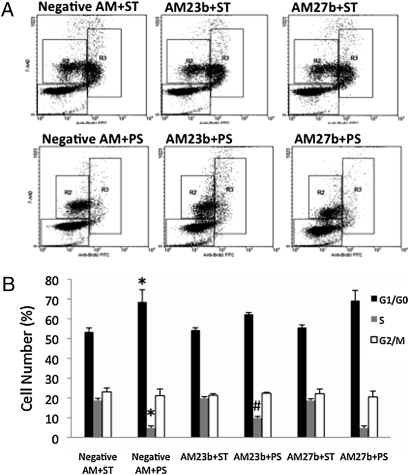
HUVECs were transfected with antagomir-23b (AM23b), antagomir-27b (AM27b), or anti-miR negative control (Neg. AM) and subjected to 24-h PS/ST with BrdU labeling during the last 4 h. Transfection with Neg. AM did not affect the growth arrest induced by PS (Figs. 1 and 4). AM23b partially reversed the PS-induced cell cycle arrest; under 24-h PS, the cell number in S phase was markedly higher at 9.87 ± 0.79% in AM23b + PS than 4.88 ± 0.96% in Neg. AM + PS (Fig. 4). AM27b, however, did not block the growth arrest caused by 24-h PS. Thus, the antiproliferative response of HUVECs under 24-h PS is partially regulated by mechanisms mediated by miR-23b, but not miR-27b.
Fig. 4.
Antagomir against miR-23b attenuates the flow-induced EC growth arrest. (A) Flow cytometry for cells transfected with anti-miR negative control (Right), AM23b (Center), and AM27b (Left) in ST (Upper) vs. 24-h PS (Lower). R1, R2, and R3 region gates denote cells in G1/G0, G2 + M, and S phases, respectively. (B) The bar graph summarized the percentage of cells in G1/G0, G2/M, and S phases under PS and ST with antagomir transfection. *,P < 0.05 in comparison with the corresponding data for negative AM transfection under static condition. #, P < 0.05 in comparison with the corresponding data for negative AM transfection under PS condition.
Roles of miR-23b and mir-27b in Modulating the Signaling Molecules that Mediate PS-Induced Growth Arrest.
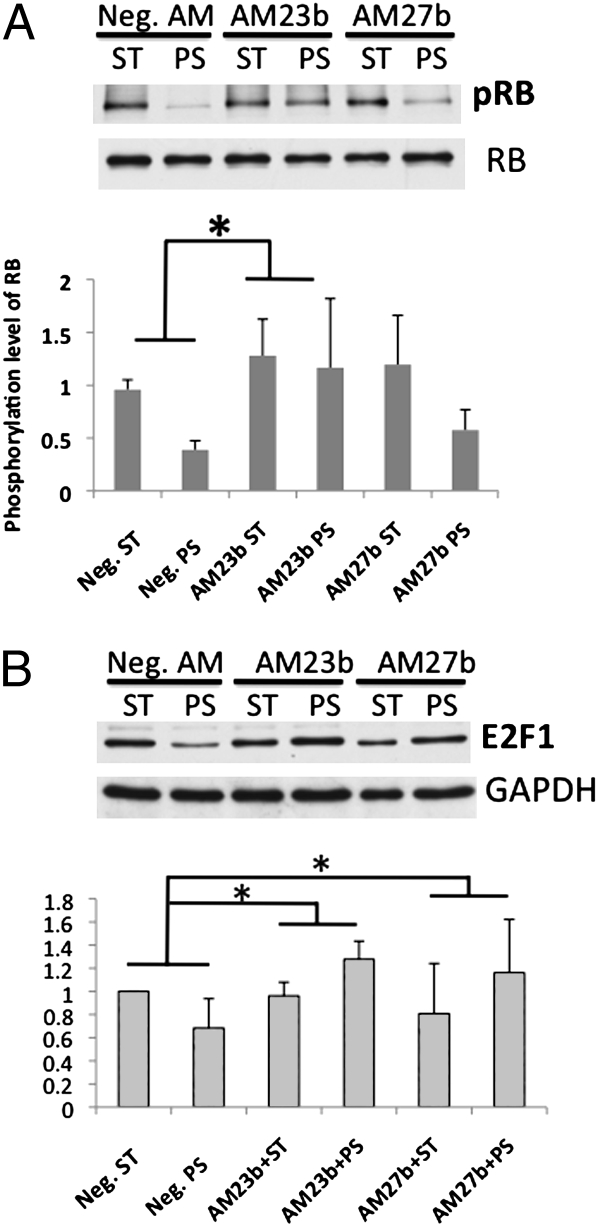
To investigate the signaling molecules that mediate the effects of miR-23b cluster on the 24-h PS suppression of EC growth, we examined the actions of AM23b and AM27b on several cell cycle proteins regulating G1-to-S transition under PS. Our previous study showed that steady laminar shear causes dephosphorylation of Rb protein (3). Rb in its hypophosphorylation state binds to E2F family proteins and suppresses cell proliferation by inhibiting the cell cycle progression through G1 into S phase and preventing DNA synthesis (32, 33). Twenty-four-hour PS decreased Rb phosphorylation without affecting the total Rb expression (Fig. 5A), and this Rb hypophosphorylation would inhibit EC proliferation. The PS-induced Rb hypophosphorylation was prevented by AM23b, but AM27b had no significant effect. Both AM23b and AM27b reversed the reduction of E2F1 expression caused by 24-h PS (Fig. 5B), but did not affect the expressions of p53 and p27kip (CDKN1B) (Fig. S2). These results suggest that miR-27b plays a role in downregulating EC expression of E2F1, but not sufficiently to modulate the PS-induced EC growth arrest without the accompanied Rb hypophosphorylation. In contrast, the PS-induced miR-23b expression can cause growth arrest through downregulation of E2F family proteins and Rb hypophosphorylation.
Fig. 5.
Effects of miR-23b and 27b on cell cycle proteins. HUVECs were transfected with Neg. AM, AM23b, and AM27b. Twenty-four-hour posttransfection, cells were subjected to PS or ST for 24 h. Western blot analysis of cell cycle regulatory proteins, including (A) phosphorylation levels of Rb protein and (B) E2F1 expressions on total cell extracts, were determined with antibodies against phospho-Rb (BD Biosciences), Rb, and E2F1 (Santa Cruz). Blots shown here are representative of three independent experiments with similar results. *, P < 0.05 in comparison with the corresponding data for neg. AM transfection under ST.
Discussion
The differential effects of flow patterns on ECs form the fluid mechanical basis of the predilection of regions of disturbed flow to atherosclerosis and the atheroprotection in regions of laminar flow with a significant forwarding direction. There have been many studies on the effects of different flow patterns on EC gene expression at the mRNA level (34–36). Following the recent discovery that miRs provide a powerful mechanism for posttranscriptional control of gene expression and that they are involved in many biological processes, it is logical and timely to investigate the role of miRs in the laminar shear-induced antiproliferative action on ECs.
Besides the steady laminar and disturbed flows that are commonly studied in the literature, there is evidence that the pulsatility imposed on the laminar flow due to heart beats may also contribute to the regulation of vascular cell functions (37, 38). In the present study the effects of PS with a significant forwarding direction (12 ± 4 dynes/cm2) in modulating miR expression was studied, and the miR modulation was related to mRNA expression by using bioinformatics analyses. The results led to the discovery that miR-23b plays a significant role in mediating the PS-induced suppression of several cell cycle genes and the consequent growth arrest.
We found that application of 24-h PS to HUVECs causes (a) growth arrest and (b) downregulation of cell cycle genes (mRNA microarray); thus, PS and steady laminar have the same antiproliferation effects. The findings on PS-regulated gene expression correspond well with the cell cycle network shown in the red boxes of Fig. S3. Determination of the miR expression profile of ECs show that PS vs. ST causes differential regulation of miRs, with the upregulation of miR-23b cluster (miR-23b and miR-27b) and downregulations of miR-17–92, miR-16, and miR-221 clusters in ECs. The differential expressions of these miR clusters in response to PS provide an example of the potential roles of miRs in mechanotransduction.
Searching of potential mRNA targets of the 21 differentially expressed miRs by using the algorithms TargetScan and PicTar (26–29) led to the identification of 1,073 putative targets that were in common with both algorithms, but only 68 of these genes (5.5%) were differentially expressed in ECs at the mRNA level. This finding indicates that the computational prediction can generate many false positives and that it needs validation with experimental approaches. It is also possible that miR regulation is through the inhibition of translation, which would not be reflected in miR target prediction. Analysis shows that the overlapping GO terms (representing biological process) between the predicted targets and the differentially expressed genes are highly enriched with cell cycle-related terms.
By ranking the correlation coefficients between the PS-induced miRs and the differentially expressed 1,227 mRNAs and using the GSEA, we found that 12 of the FGSs that represent cell cycle had the strongest association (Fig. 3A). miR-23b and miR-27b were negatively associated with these FGSs. Integration of miR and mRNA expression profiles, together with computational approaches, revealed the potential roles of miR-23b and -27b in regulating the PS-induced EC growth arrest.
Both miR-23b and miR-27b are highly conserved and lie within 0.5 kilobases (kb) of each other on human chromosome 9, and both are upregulated under PS vs. ST (Fig. 2). We showed that AM23b reversed the EC growth arrest under PS; this is in concert with the finding that miR-23b overexpression decreases hepatocelluar carcinoma cell proliferation (39). Although both AM23b and AM27b can regulate E2F1 expression, only AM23b significantly reduced the PS-induced Rb hypophosphorylation and cell cycle arrest. It is known that increases of Rb protein inhibits cell cycle progression and that Rb activities depend on its phosphorylation status (40). The hypophosphorylated Rb binds to E2F family proteins in a cell cycle-dependent manner to modulate proliferation (32, 33). Blocking Rb–E2F interaction with its endogenous genes has been reported to prevent the Rb-mediated growth arrest (41), and this probably involves the recruitment of other repressors (e.g., HDAC) to the Rb–E2F complex (42). It seems that the decrease of E2F1 per se is not sufficient to cause growth arrest, but the hypophosphorylation of Rb with E2F1 binding plays an important role in regulating EC growth. Both AM23b and AM27b did not have any effect on the PS-induced expressions of p53 and p27kip; hence these molecules may not mediate the effects of miR-23b on growth arrest.
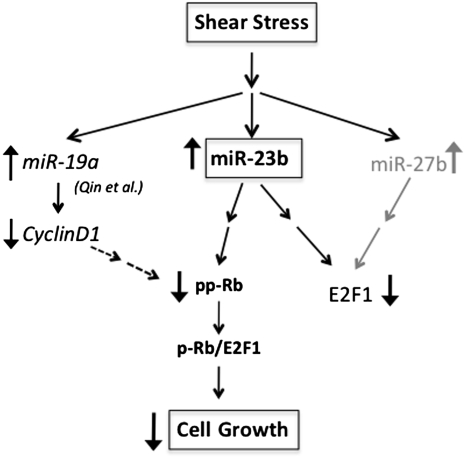
The overall effects of miR-23b in mediating the PS-induced growth arrest of ECs is summarized in Fig. 6. In this figure, we propose a unique mechanism by which PS causes upregulation of miR-23b, which in turn decreases Rb phosphorylation, leading to growth arrest. Because the regulatory mechanisms through miRs generally involve a network, there must be other miRs that also mediate this PS-induced effect. It is most likely that miR-23b regulates EC proliferation through more complicated mechanisms than that shown in Fig. 6, including the interplays among different miRs and mRNAs. The incomplete blockade of the PS actions by AM23b is in keeping with this line of thinking. Rb phosphorylation is also modulated by cyclins, CDKs, p53, and p27. Our mRNA microarray results demonstrated that CDKs 1 and 4 and cyclins A, B, and E are downregulated (Fig. S3), which may contribute to Rb hypophosphorylation. Although we found that miR-23b did not significantly modulate p53, p27, CyclinD1, and CDK4, the effects of miR-23b on other cyclins and CDKs remain to be determined. The GO analysis of predicted miR targets and mRNA data indicates that the regulation of kinase activities is an important feature for EC gene signature in response to shear (Table S3); hence the signaling pathways may also involve kinase/phosphatase cascades. Moreover, the mechanism by which PS regulates miR-23b expression remains unclear. Recent study indicated that c-Myc, a known stimulator for cell proliferation, suppresses miR-23b expression in prostate cancer cells (43). Further investigations will be needed to determined the role of myc in the PS regulation of miRs.
Fig. 6.
Schematic representation of the role of miR-23b in flow regulation of EC growth. PS increases the expression of miR-23b (bold), which alters Rb into a hypophosphorylated state to complex with E2F family proteins and suppress EC growth. The upregulation of miR-27b (gray letters) also decreased the expression of E2F1, which is not sufficient for cell growth arrest without modulating Rb status. Qin et al. (44) show that miR-19 (italics) regulates cyclin D1 expression to cause EC growth arrest in response to steady laminar flow, and this effect may also be mediated via Rb hypophosphorylation.
The miR regulation of gene and protein expressions not only involves a spatial network, but also temporal dynamics. In Qin et al. (44), miR profiles were deteremined in HUVECs under steady laminar flow at 12 and 24 h. The results indicate that 12-h shearing caused a significant expression of miR-19a, but not after 24-h shearing. In contrast, miR-23b was induced under 24-h shearing, which is the duration in this study, but not at 12 h. Whereas miR-23b is shown in this study to regulate EC growth through the modulation of E2F expression and Rb phosphorylation, Qin et al. (44) found that the regulation of EC growth by miR-19a is mediated mainly through the modulation of cyclin D1 at 24 h. The downregulation of cyclin D1 may also contribute to the Rb dephosphorylation and lead to the cell growth arrest. However, preliminary tests showed that combined uses of AM23b and AM19a did not have any synergistic affect on EC growth arrest. There is a need for further investigations on the interactions between different miRs on EC functions. These two studies together show the participation of two miRs in regulating the PS-induced growth arrest with different temporal dynamics and different signaling pathways. These findings exemplify the complexity of the miR-mediated regulation of cellular functions.
In conclusion, our findings indicate that the 24-h PS-induced EC growth arrest is partially mediated through the regulation of miR-23b. Twenty-four-hour PS significantly increases the miR-23b level, which decreases the expression level of E2F1 and causes Rb hypophosphorylation. These results provide unique insights into the role of miRs in mechanotransduction. It is suggested that miRs may act as a master switch to modulate various posttranscriptional processes in human ECs. Our findings have demonstrated that the PS-induced miR signature and the corresponding mechanisms are important for the understanding of the mechanisms of regulation of cardiovascular homeostasis in health and disease.
Materials and Methods
Cell Culture and Antagomir Transfection.
HUVECs were obtained from Cell Application and used before passage 7. Antagomirs against miR-23b and -27b, as well as anti-miR negative control molecule, were purchased from Ambion. The oligonucleotides were individually transfected with siPort NeoFx reagent (Ambion) at the final concentration of 50 nM, 24-h posttransfection.
Shear Experiments.
An in vitro circulating flow system was used to impose PS stress to cultured HUVECs. Before the shear experiment, HUVECs were seeded on a collagen I (BD Biosciences) -coated glass slide, which was assembled into the flow chamber and exposed to PS at 12 ± 4 dyn/cm2 at 1 Hz for 24 h, and the cells obtained from concurrent unsheared sample were used as the ST.
miR and mRNA Microarray Analyses.
Total RNA was isolated from ECs with mirVana kit (Ambion) after PS or ST. Five micrograms of total RNA of each sample was sent to LC Sciences for miR profiling. The samples were enriched for small RNAs, labeled with fluorescent dyes, and hybridized to μParaFlo microfluidic chips, which contain 856 mature human microRNA probes (miRBase version 12.0). The raw data were normalized to the control probes on individual chips. Significantly regulated miRs were selected by using a cutoff of 1.25-fold change and P < 0.01.
The cDNA microarray experiments were performed by the Genomic Core Facility in University of California, San Diego (Biomedical Genomics Laboratory, Biogem); 10 μg of total RNA was hybridized on Agilent 44K oligonucleotide microarrays (Agilent). Three biological repeats were performed for all array studies.
miR Real-Time PCR.
PCR was performed with miR-specific primers from the TaqMan miR assays (Applied Biosystems) in the 7900HT real-time PCR machine (Applied Biosystems) according to manufacturer’s protocol. Three biological replicates were used for analysis and all reactions were run in triplicates. The relative expression levels of miRs in ECs were determined with ΔΔCT method and compared with internal controls.
BrdU Incorporation Assay and Flow Cytometry.
HUVECs were pulse labeled with 10 mM BrdU during the last 4 h of shearing and static incubation. After labeling, the HUVECs were assayed using Cell Cycle Analysis kit (BD Bioscience) according to the manufacturer’s protocol. The cells were subjected to flow cytometric analysis with FACScan (BD Bioscience).
Statistical Analysis.
Data are expressed as mean ± SEM and compared among separate experiments. For comparisons between two groups, statistical analyses were performed using the two-sample independent groups t test. Comparison of multiple mean values was made by one-way ANOVA, and statistical significance among multiple groups was determined by post hoc analysis (for pairwise comparisons of means between PS and ST. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Research Grants HL064382, HL085159, and HL080518 (to S.C.); HL087375 and DK074868 (to S.S.); and HL89940 (to J.S.) and Taiwan National Science Council TMS-094-1A-004 (fellowship to K.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914825107/DCSupplemental.
References
- 1.Chien S. Mechanotransduction and endothelial cell homeostasis: The wisdom of the cell. Am J Physiol Heart Circ Physiol. 2007;292:H1209–H1224. doi: 10.1152/ajpheart.01047.2006. [DOI] [PubMed] [Google Scholar]
- 2.Chien S. Mechanical and chemical regulation of endothelial cell polarity. Circ Res. 2006;98:863–865. doi: 10.1161/01.RES.0000219686.29872.e2. [DOI] [PubMed] [Google Scholar]
- 3.Lin K, et al. Molecular mechanism of endothelial growth arrest by laminar shear stress. Proc Natl Acad Sci USA. 2000;97:9385–9389. doi: 10.1073/pnas.170282597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang N, et al. Shear stress regulation of Krüppel-like factor 2 expression is flow pattern-specific. Biochem Biophys Res Commun. 2006;341:1244–1251. doi: 10.1016/j.bbrc.2006.01.089. [DOI] [PubMed] [Google Scholar]
- 5.Matharu NM, Rainger GE, Vohra R, Nash GB. Effects of disturbed flow on endothelial cell function: Pathogenic implications of modified leukocyte recruitment. Biorheology. 2006;43:31–44. [PubMed] [Google Scholar]
- 6.Hsiai TK, et al. Monocyte recruitment to endothelial cells in response to oscillatory shear stress. FASEB J. 2003;17:1648–1657. doi: 10.1096/fj.02-1064com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akimoto S, Mitsumata M, Sasaguri T, Yoshida Y. Laminar shear stress inhibits vascular endothelial cell proliferation by inducing cyclin-dependent kinase inhibitor p21(Sdi1/Cip1/Waf1) Circ Res. 2000;86:185–190. doi: 10.1161/01.res.86.2.185. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 11.Bonauer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 12.Suárez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104:442–454. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parmacek MS. MicroRNA-modulated targeting of vascular smooth muscle cells. J Clin Invest. 2009;119:2526–2528. doi: 10.1172/JCI40503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carè A, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 15.Suárez Y, Fernández-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 16.Kuehbacher A, Urbich C, Dimmeler S. Targeting microRNA expression to regulate angiogenesis. Trends Pharmacol Sci. 2008;29:12–15. doi: 10.1016/j.tips.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Hua Z, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poliseno L, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 19.Shilo S, Roy S, Khanna S, Sen CK. Evidence for the involvement of miRNA in redox regulated angiogenic response of human microvascular endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28:471–477. doi: 10.1161/ATVBAHA.107.160655. [DOI] [PubMed] [Google Scholar]
- 20.Levesque MJ, Nerem RM, Sprague EA. Vascular endothelial cell proliferation in culture and the influence of flow. Biomaterials. 1990;11:702–707. doi: 10.1016/0142-9612(90)90031-k. [DOI] [PubMed] [Google Scholar]
- 21.Dennis G, Jr, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 22.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Ashburner M, et al. The Gene Ontology Consortium. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, et al. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34(Database issue):D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 27.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Wang X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 2006;34:1646–1652. doi: 10.1093/nar/gkl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Creighton CJ, Nagaraja AK, Hanash SM, Matzuk MM, Gunaratne PH. A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions. RNA. 2008;14:2290–2296. doi: 10.1261/rna.1188208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 32.Sun A, Bagella L, Tutton S, Romano G, Giordano A. From G0 to S phase: A view of the roles played by the retinoblastoma (Rb) family members in the Rb-E2F pathway. J Cell Biochem. 2007;102:1400–1404. doi: 10.1002/jcb.21609. [DOI] [PubMed] [Google Scholar]
- 33.Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- 34.McCormick SM, et al. DNA microarray reveals changes in gene expression of shear stressed human umbilical vein endothelial cells. Proc Natl Acad Sci USA. 2001;98:8955–8960. doi: 10.1073/pnas.171259298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Cardeña G, Comander J, Anderson KR, Blackman BR, Gimbrone MA., Jr Biomechanical activation of vascular endothelium as a determinant of its functional phenotype. Proc Natl Acad Sci USA. 2001;98:4478–4485. doi: 10.1073/pnas.071052598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen BP, et al. DNA microarray analysis of gene expression in endothelial cells in response to 24-h shear stress. Physiol Genomics. 2001;7:55–63. doi: 10.1152/physiolgenomics.2001.7.1.55. [DOI] [PubMed] [Google Scholar]
- 37.Davies PF, Dewey CF, Bussolari SR, Gordon EJ, Gimbrone MA., Jr Influence of hemodynamic forces on vascular endothelial function. In vitro studies of shear stress and pinocytosis in bovine aortic cells. J Clin Invest. 1984;73:1121–1129. doi: 10.1172/JCI111298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M, Scott DE, Shandas R, Stenmark KR, Tan W. High pulsatility flow induces adhesion molecule and cytokine mRNA expression in distal pulmonary artery endothelial cells. Ann Biomed Eng. 2009;37:1082–1092. doi: 10.1007/s10439-009-9684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salvi A, et al. MicroRNA-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells. FEBS J. 2009;276:2966–2982. doi: 10.1111/j.1742-4658.2009.07014.x. [DOI] [PubMed] [Google Scholar]
- 40.Poznic M. Retinoblastoma protein: A central processing unit. J Biosci. 2009;34:305–312. doi: 10.1007/s12038-009-0034-2. [DOI] [PubMed] [Google Scholar]
- 41.He Y, Armanious MK, Thomas MJ, Cress WD. Identification of E2F-3B, an alternative form of E2F-3 lacking a conserved N-terminal region. Oncogene. 2000;19:3422–3433. doi: 10.1038/sj.onc.1203682. [DOI] [PubMed] [Google Scholar]
- 42.Brehm A, et al. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 43.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin X, et al. MicroRNA-19a mediates the suppressive effect of laminar flow on cyclin D1 expression in human umbilical vein endothelial cells. Proc Natl Acad Sci. 2010;107:3240–3244. doi: 10.1073/pnas.0914882107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.