Abstract
Endothelial cells (ECs) respond to changes in mechanical forces, leading to the modulation of signaling networks and cell function; an example is the inhibition of EC proliferation by steady laminar flow. MicroRNAs (miRs) are short noncoding 20–22 nucleotide RNAs that negatively regulate the expression of target genes at the posttranscriptional level. This study demonstrates that miRs are involved in the flow regulation of gene expression in ECs. With the use of microRNA chip array, we found that laminar shear stress (12 dyn/cm2, 12 h) regulated the EC expression of many miRs, including miR-19a. We further showed that stable transfection of miR-19a significantly decreased the expression of a reporter gene controlled by a conserved 3′-untranslated region of the cyclinD1 gene and also the protein level of cyclin D1, leading to an arrest of cell cycle at G1/S transition. Laminar flow suppressed cyclin D1 protein level, and this suppressive effect was diminished when the endogenous miR-19a was inhibited. In conclusion, we demonstrated that miR-19a plays an important role in the flow regulation of cyclin D1 expression. These results revealed a mechanism by which mechanical forces modulate endothelial gene expression.
Keywords: cell cycle, endothelium, flow, gene expression, noncoding RNA
Being constantly exposed to hemodynamic forces, vascular endothelium plays an important role in sensing the alterations in biological, chemical, and physical properties in the flowing blood to maintain physiological homeostasis. Shear stress, the frictional force created by blood flow, exerts a variety of effects on endothelial structure and function. In endothelial cells (ECs), steady laminar flow results in reorganization of cytoskeleton, modulation of molecular signaling, and inhibition of cell proliferation and apoptosis (1, 2). In particular, laminar shear stress tends to maintain ECs in a quiescent and less proliferative state. Such a growth-arresting action of shear stress might contribute to the athero-protective property in the straight portions of the arterial tree (3).
The molecular mechanisms by which ECs respond to changes in mechanical properties of blood flow and undergo structural and functional adaptations remain to be elucidated. Emerging evidence supports the concept that shear stress exerts its physiological effects by modulating the gene expression in ECs. For instance, steady shear stress can regulate the expression of a number of growth-controlling molecules such as p21 and GADD45 (4). The transcriptional regulation of the flow-responsive genes has been extensively investigated. Many transcription factors have been found to mediate the flow-induced cell cycle arrest in ECs, including p53 (4), Krüppel-like factor–2 (5, 6) and STAT3 (7). However, the effects of shear stress on the posttranscriptional control of gene expression are largely unknown.
MicroRNAs (miRs) are a family of highly conserved, small noncoding RNAs that posttranscriptionally repress gene expression via degradation or translational inhibition of their target mRNAs. There is mounting evidence suggesting that miRs are involved in nearly all physiological and pathological processes (8, 9). Recent studies have demonstrated that miRs play critical roles in vascular development and homeostasis (10, 11). The present study was conducted to examine the effects of shear stress on miR expression and, if any, their functional roles in mechanotransduction in ECs.
Results
Differential Expression of miRs in ECs in Response to Laminar Shear Stress.
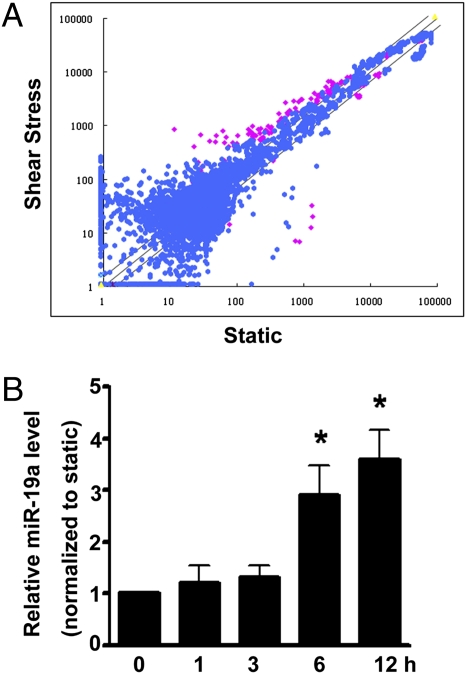
Using the miR microarray approach, we compared the miR expression profiles in human umbilical vein endothelial cells (HUVECs) following 12 h of laminar shear stress at 12 dyn/cm2 with those cultured under static condition. Among 569 individual miRs represented on the microarrays, 35 miRs were found to be significantly up-regulated and 26 were significantly down-regulated in the laminar flow-treated HUVECs in comparison with the static control cells (Fig. 1A and Table S1). Among the miRs that were regulated in HUVECs in response to laminar shear stress, miR-19a was expressed at a high abundance under static condition and its level was greatly increased after 12 h of shear stress exposure. To examine the temporal dynamics of miR-19a expression, HUVECs were exposed to laminar shear stress for various time periods. As detected by quantitative real-time PCR (qRT-PCR), miR-19a was significantly increased as early as 6 h after shear treatment, indicating that miR-19a was an early responsive gene for shear stress in ECs (Fig. 1B).
Fig. 1.
miR-19a is rapidly induced by shear stress in ECs. (A) Scatter plot of miR expression profiles in ECs in response to laminar shear stress (12 dyn/cm2 for 12 h, y axis) and control ECs kept under static condition (x axis). The miRs differentially expressed with statistical significance are marked in red. (B) qRT-PCR shows that miR-19a was induced in ECs after being exposed to laminar shear stress for various time durations. U6 snRNA level was used for normalization. Data are presented as the mean ± SEM (n = 3). *P < 0.05 compared with static control).
Cyclin D1 Is a Target of miR-19a in ECs.
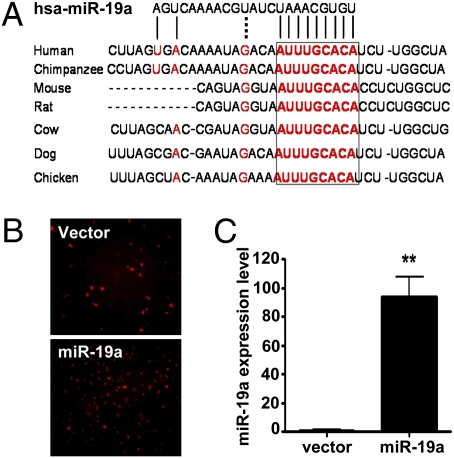
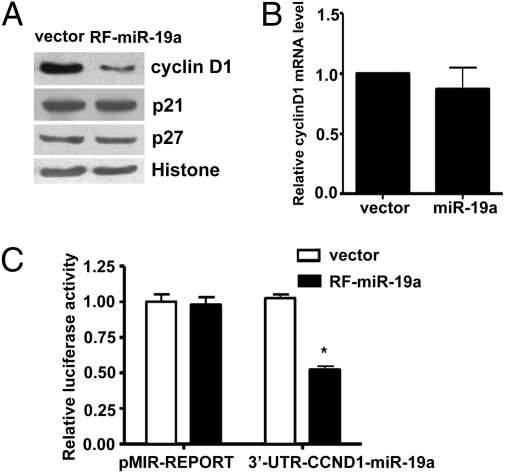
The base-pairing of miRs with the 3′-UTR of target mRNAs leads to translational repression and degradation of mRNA. The 5′-sequences of miRs, particularly those at nucleotide positions 2–7 relative to their 5′-end, are important for binding to the mRNA target and are known as “seed” sequences. Sequence alignment of human miR-19a with 3′-UTR of the human cyclin D1 gene (CCND1) identified a miR-19a binding site (nucleotides 1,778–1,785 in human CCND1) that is well conserved among different species (Fig. 2A). To screen the targets regulated by the miR-mediated RNA silencing pathway, we generated the EC lines (EA.hy.926) stably overexpressing miR-19a (Fig. 2B). A significantly increased expression of miR-19a in the transfected cells was confirmed by qRT-PCR (Fig. 2C). This level was about 8-fold higher than that in flow-stimulated ECs. Western blot analysis revealed that the protein level of cyclin D1, but not p21 or p27, was markedly reduced in the cells overexpressing miR-19a compared with the cell lines transfected with control vector (Fig. 3A). qRT-PCR analysis was conducted to determine the mechanism by which miR-19a inhibited cyclin D1 abundance. No significant difference in cyclin D1 mRNA levels was observed between the EA.hy.926 cells with stable miR-19a expression and the control cells (Fig. 3B), indicating that miR-19a regulates cyclin D1 gene expression at the posttranscriptional rather than the transcriptional level. To further examine whether miR-19a direct targets CCND1, the luciferase reporter containing the CCND1 3′-UTR fragment with the miR-19a binding site (3′-UTR-CCND1-miR-19a) was transfected into EA.hy.926 cells overexpressing miR-19a (RF-miR-19a) or control cell line. Luciferase assays showed that the cyclin D1 reporter, but not the MIR-REPORT control, was suppressed in RF-miR-19a. There was no suppression for either reporter in the control cell line (Fig. 3C). Taken together, these results suggested that miR-19a repressed cyclin D1 through a specific 3′-UTR binding site.
Fig. 2.
miR-19a represses cyclin D1 expression through a conserved 3′-UTR binding site. (A) Sequence alignment of the miR-19a base-pairing sites in the 3′-UTR of cyclinD1 mRNAs showed that the regions complementary to the 9 nt of miR-19a are highly conserved among human, mouse, rat, cow, dog, and chicken. The “seed” sequences of miR-19a complementary to cyclin D1 are shown in red and boxed. (B) Endothelial cell stable lines were generated by transfecting EA.hy.926 cells with pEF1-RF-miR-19 or pEF-RF and by the puromycin selection. Photomicrographs show ubiquitous expression of red fluorescence protein in both transfected cells. (C) Overexpression of miR-19a was confirmed in the RF-miR-19a stable cell line with the use of qRT-PCR. U6 was used as the internal control. Data are presented as mean ± SEM (n = 3). **P < 0.01.
Fig. 3.
Cyclin D1 is a target of miR-19a in ECs. (A) Western blot analysis showed that protein level of cyclinD1 was decreased in the miR-19a–transfected cell line. (B) Cyclin D1 mRNA was not different between miR-19a–transfected cells and control cells as detected by qRT-PCR. (C) Luciferase activity of the reporter of the cyclin D1 3′-UTR containing miR-19a site (3′-UTR-CCND1-miR19a), but not the pMIR-REPORT control, was decreased in the miR19a-expressing cells. Data are presented as mean ± SEM of three experiments. *P < 0.05 compared with that in the control cell line.
miR-19a Attenuates G1/S Transition of Cell Cycle in ECs.
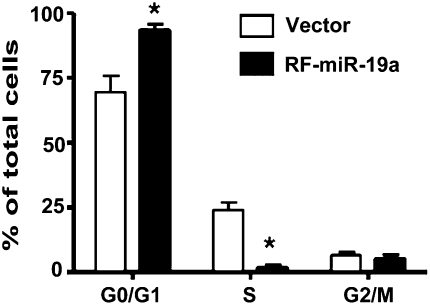
Cyclin D1 is an important regulator of G1 to S phase progression in many cell types. It forms active complexes with cyclin-dependent kinase 4 and 6 (CDK4 and CDK6) to promote cell cycle progression by phosphorylating and inactivating the retinoblastoma protein (Rb) (12). To examine the function of miR-19a in EC proliferation, cell lines overexpressing miR-19a were synchronized by serum starvation (0.1% FBS) for 24 h, and then stimulated with 10% FBS for 18 h. Cell cycle analysis showed that the miR-19a-overexpressing cells had a lower proportion of cells in S phase and a higher proportion in G0/G1 phase, as compared with control cells (Fig. 4). These results demonstrated that the overexpression of miR-19a inhibited the proliferation of ECs to control cell cycle progression. In combination with the results shown in Fig. 3, our findings indicate that miR-19a might inhibit EC proliferation by negatively regulating cyclin D1.
Fig. 4.
Overexpression of miR-19a arrests cell cycle at the G1 phase in ECs. Flow-cytometric analysis indicated that the population of cells in G0/G1 phase was increased and that in S phase decreased in miR-19a–transfected cells. Data are presented as mean ± SEM of three repeated experiments. *P < 0.05 compared with control cells.
Blockade of miR-19a Attenuates Suppressive Effects of Laminar Flow on Cyclin D1 Expression in ECs.
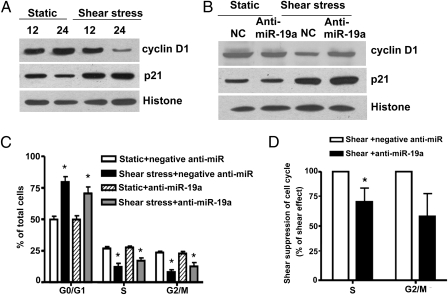
To examine the effect of laminar flow on cyclinD1 gene expression, we applied a laminar shear stress at a physiological level of 12 dyn/cm2 to confluent HUVECs for 12 and 24 h and compared with static control. The protein level of cyclinD1 was determined following shear stress by Western blotting. As shown in Fig. 5A, laminar flow at 12 dyn/cm2 for 24 h (but not 12 h) decreased the expression level of cyclinD1 protein compared with static control. To study the effect of loss-of-function of miR-19a in laminar shear-treated ECs, miR-19a inhibitor (anti–miR-19a) was introduced into HUVECs. The inhibition of the endogenous miR-19a with anti–miR-19a attenuated the suppressive effects of 24-h laminar flow on cyclinD1 protein level, thus confirming that the shear inhibition of expression of cyclinD1 protein is under miR-19a control (Fig. 5B). Next, we asked whether endogenous miR-19a was required for the shear-induced EC growth arrest by using miR-19a inhibitor. Although the introduction of miR-19a inhibitor was not able to totally abrogate the shear-induced growth arrest of ECs, it did result in a decrease in the proportion of EC entering proliferating cycle (Fig. 5C). In the presence of anti–miR-19a, the suppressive effect of shear stress on S and G2/M progression was reduced to 72.3% and 58.76% respectively. (Fig. 5D). These results suggested that the laminar flow–induced miR-19a contributed, at least partially, to the antiproliferative effect of shear stress.
Fig. 5.
miR-19a mediates the suppressive effect of laminar flow on cyclin D1 expression in HUVECs. (A) Effects of laminar flow on the expression of cyclin D1 and p21, another cell cycle–related genes. Confluent HUVECs were exposed to 12 dyn/cm2 shear stress or static control for 12 h and 24 h. Results shown are representative of three independent experiments. (B) Pretreatment with anti–miR-19a prevented the inhibitory effect of laminar flow on cyclin D1 expression. HUVECs were pretreated with anti–miR-19a or control oligonucleotides (negative anti-miR) for 48 h followed by laminar flow for 24 h, and cyclinD1 expression was examined with Western blotting. (C) miR-19a inhibitor partially attenuated the laminar flow-induced cell cycle arrest. HUVECs were transfected with anti–miR-19a or anti-miR negative control for 48 h, followed by 12 dyn/cm2 shear stress or static control for 24 h, and cell cycle distribution of ECs was analyzed with flow cytometry. *P < 0.05 compared with static plus anti-miR negative control. (D) Shear stress–induced suppression of ECs entry S and G2/M phases were reduced by anti–miR-19a. *P < 0.05 compared with anti-miR negative control. Results shown are representative of three independent experiments.
Discussion
The present study demonstrated that expressions of miRs in ECs are regulated by laminar shear stress. Specifically, we have shown that miR-19a is an early responsive gene that mediates the antiproliferative effect of laminar shear stress by directly targeting cyclin D1, a key regulator of cell cycle progression.
Endothelial cells are subjected to the shear stress resulting from blood flow and are able to sense and convert mechanical stimuli into intracellular signals, leading to rapid adaptations in their cellular functions and structures. In the straight parts of the arterial tree, the laminar shear stress suppresses EC turnover and the maintenance of junction integrity keeps lipoprotein permeability at low levels. In contrast, vascular areas with flow disturbances, such as those located near branch points and bifurcations, often have increased EC turnover and accumulation of lipoproteins (1). Such in vivo observations have been confirmed by the studies of the direct effects of defined mechanical forces applied to EC monolayers in culture (13, 14). It is known that ECs undergo rapid and differential responses to shear stress depending on the flow magnitudes and directions. Recently, cDNA microarray studies from several groups have led to the general conclusion that hundreds of genes are regulated by laminar shear stress and that more genes are suppressed than induced under a sustained flow (24 h), including those with products involved in proliferation, inflammation, and oxidative stress. The miR-mediated gene silencing mechanism described in the present study adds to our knowledge as to how mechanical forces fine-tune the endothelial gene expression at the posttranscriptional level. Besides the previously identified flow-dependent transcriptional regulations, this miR machinery may particularly explain how ECs can restrict the expression of preexisting transcripts to adapt to alterations of flow.
A major finding in this study is that, among the many shear-regulated miRs, miR-19 directly targets cyclin D1 expression. This was validated by the reporter assay and Western blotting analysis. Importantly, we found that the shear induction of miR-19a was necessary for the suppression of the cyclin D1 protein level in ECs by laminar shear stress. This result provides a potential explanation for the previous observation that laminar shear stress increased the cyclin D1 transcript but caused arrest of cell cycle progression (15). As an endogenous gene in the genome, miR-19a is a member of miR-17–92 cluster of seven miRs (miR-17–5p, miR-17–3p, miR-18, miR-19a, miR-20, miR-19b-1, and miR-92-1). The genomic region encoding the miR-17–92 miR cluster is often amplified in lymphoma and other cancers (16, 17). Of these clustered miRs, the levels of the miR-17–5p/miR-20a are inversely correlated with cyclin D1 abundance in human breast tumors and cell lines, whereas miR-17/20 suppressed breast cancer cell proliferation by negatively regulating cyclin D1 translation via a conserved 3′ UTR-binding site (18). Intriguingly, miR-19 was also previously predicted to target phosphatase and tensin homolog deleted on chromosome 10 (PTEN) by a high-throughput approach performed in HeLa cells (19). It was reported that miR-19a inhibition led to growth arrest and survival in cancer cells (20, 21). These reported data indicate that miR19a may function as either a progrowth and prosurvival gene in cancer cells or an antigrowth gene in ECs. It is likely that the same miR performs context-specific functions, depending on the types of stimulation and tissues. Given that the gain-of-function data were obtained by using the EC stable lines other than primary cultured ECs, it is possible that the selection process of stable cells may also modify the cell characteristics. To address this concern, we used a loss-of-function approach to specifically inhibit the function of miR-19a in HUVEC with the miR-19a antagomir. The results from these experiments indicated that miR-19a contributes to the cyclin D suppression in HUVECs.
Blocking endogenous miR-19a abrogated the cyclin D1 suppression and partially reversed the cell cycle arrest by laminar shear stress in ECs. However, anti–miR-19a had little effect on the cell cycle progression under static conditions. These results suggest that the induced miR-19a may play a more important role in a flow-mediated context. Although miR-19a induction plays a critical role in the shear regulation of cyclin D1, the overall suppression of endothelial proliferation may require multiple miRs to act in a coordinated manner. The notion is in accordance with the finding that 24 h of pulsatile shear stress induced miR-23b, which mediates the dephosphorylation of Rb protein by flow (22). The shear-induced miR-19a and miR-23b may have distinct yet interrelated functions because cyclin D1 and Rb are known to orchestrate cell cycle progression. Cyclin D1 forms a complex with and functions as a regulatory subunit of cyclin-dependent kinases (CDK) 4 or 6. The activity of cyclin D/CDK4/6 complex is required for G1/S transition (23). The flow-induced miR-19a targets cyclin D1 to negatively regulate the cell cycle. Rb is switched between the hypophosphorylated (growth-suppressive) and phosphorylated (inactive) states (24). The flow-induced miR-23b keeps Rb hypophosphorylated and also functions to block the G1/S transition. Because complexes of cyclins/CDKs can phosphorylate Rb and inhibit its activity (25), it is plausible that the miR-19a–induced decrease in cyclin D1 may facilitate the effect of miR-23b on Rb activity. Interestingly, we found that miR-23b was only minimally increased at 12 h after the exposure to steady shear stress (Table S1), whereas miR-19a was up-regulated in the ECs as an early response at 12 h but not after 24 h of shear treatment. Such temporal-specific regulations of the miR lead to our postulation that the rapidly induced miR-19a brakes the G1/S progression via preventing the translation of the preexisting cyclin D1 transcripts; this growth-braking effect is reinforced by miR-23b, which inhibits the phosphorylation of Rb protein and restrains E2F transcription factors in their inactive form, thus leading to the continued transcriptional repression of multiple growth-promoting genes. In addition, by using bioinformatics tools, we have predicted that many of the shear-regulated miRs potentially target the genes involved in cell proliferation, survival, and inflammation. The biological functions of these miRs in mechanotransduction, individually or in combination, remain to be further examined.
In summary, we demonstrated that laminar shear stress modulates miR expression and that miR-19a plays an important role in the flow regulation of cyclin D1 expression and endothelial proliferation. These results revealed a mechanism by which mechanical forces modulate gene and protein expressions and cell proliferation in endothelial cells.
Materials and Methods
Cell Culture.
HUVECs were cultured as previously described (26). For all experiments, HUVECs within passage 3 were used. EA.hy.926 cells, an endothelial cell line, were grown in high-glucose DMEM (Invitrogen) supplemented with 10% FBS and penicillin–streptomycin at 37°C in a humidified atmosphere of 5% CO2.
Flow Experiments.
Flow experiments were performed as previously described (27). A parallel-plate flow system was used to impose a laminar shear stress of 12 dyn/cm2 by perfusing the culture media over a confluent monolayer of ECs seeded on a glass slide on the bottom of the flow chamber. The flow system was kept at 37°C and ventilated with 95% humidified air containing 5% CO2.
Microarray for miRs.
Confluent HUVECs were exposed to laminar shear stress or static control for 12 h. RNA samples were isolated, size fractioned, and labeled with Cy3 or Cy5. Paired labeled samples were hybridized to dual-channel microarray using the μParaflo microfluidics chips (miR human 10.0 miR array chips; LC Sciences), which were based on Sanger miRBase release 10.0. Raw data were normalized by the LOWESS method on the background-subtracted data. A Student t test was performed to analyze the statistical significance of the signal differences between the two groups.
qRT-PCR.
qRT-PCR was performed to detect miR expression using the TaqMan miR assay kits (Applied Biosystems) according to the manufacturer’s protocol. Levels of miRs were normalized using rRNA U6 as a reference. Dicer mRNA expression were examined using specific primers (forward: 5′-GTACGACTACCACAAGTACTTC; reverse: 5′-ATAGTACACCTGCCAGACTGT). The mRNA level was normalized by housekeeping gene GADPH.
Western Blotting.
Western blotting was performed as previously described (28) using primary antibodies against cyclinD1, p21, p27, or histone (Santa Cruz) and an HRP-conjugated secondary antibody, followed by enhanced chemiluminescence detection.
Flow Cytometry.
Adherent cells were harvested, fixed, and stained with propidium iodide (50 μg/mL with RNase A for 30 min). Cell cycle distribution was analyzed with a FACScan (Becton-Dickinson).
Generation of Stable Cell Lines Expressing Primary miR.
Primary miR-19a including ≈100 bp of upstream and downstream flanking regions of its precursor was PCR-amplified from the human genomic DNA and subcloned into the pEF1-rPuro-RF vector to make the pEF1-RF-miR-19a–expressing plasmid. To generate miR-19a–expressing stable cell lines, EA.hy.926 cells were transfected with pEF1-RF-miR-19a or control plasmid pEF-RF that expresses only the red fluorescence protein. Cells were grown in the presence of 2.5μg/mL puromycin to select the stably transfected clones. Single clones were selected to generate monoclonal cell lines.
miR-Reporter and Luciferase Assays.
To generate reporter vectors bearing miR-19a binding site, sense, and antisense strands of the oligonucleotides of the CCND1-3′-UTR containing the miR-19a–binding site were synthesized, annealed, and subcloned into pMIR-REPORT vector (Ambion). The CCND1-3′-UTR-miR19a or control plasmid was transfected into the EC stable lines using Lipofectamine 2000 (Invitrogen). pRSV-β-Gal was cotransfected for normalization of transfection efficiency. Luciferase and β-galactosidase activities were measured as described previously (28).
Inhibitor of miR.
The miR-19a inhibitor, anti–miR-19a, and anti-miR negative control oligonucleotide were obtained from Ambion and transfected into HUVECs at a final concentration of 40 nM using Lipofectamine 2000. After 48 h, the cells were exposed to the laminar shear stress or kept under static conditions.
Supplementary Material
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (30890040, 30670848, 30821001, 30700292 and 30971160), the Major National Basic Research Program of China (2006CB503906), and the National Institutes of Health (United States Public Health Service) (HL080518 and HL085159).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914882107/DCSupplemental.
References
- 1.Chien S, Li S, Shyy YJ. Effects of mechanical forces on signal transduction and gene expression in endothelial cells. Hypertension. 1998;31:162–169. doi: 10.1161/01.hyp.31.1.162. [DOI] [PubMed] [Google Scholar]
- 2.Li YS, Haga JH, Chien S. Molecular basis of the effects of shear stress on vascular endothelial cells. J Biomech. 2005;38:1949–1971. doi: 10.1016/j.jbiomech.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 3.Chien S. Mechanotransduction and endothelial cell homeostasis: The wisdom of the cell. Am J Physiol Heart Circ Physiol. 2007;292:H1209–H1224. doi: 10.1152/ajpheart.01047.2006. [DOI] [PubMed] [Google Scholar]
- 4.Lin K, et al. Molecular mechanism of endothelial growth arrest by laminar shear stress. Proc Natl Acad Sci USA. 2000;97:9385–9389. doi: 10.1073/pnas.170282597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekker RJ, et al. KLF2 provokes a gene expression pattern that establishes functional quiescent differentiation of the endothelium. Blood. 2006;107:4354–4363. doi: 10.1182/blood-2005-08-3465. [DOI] [PubMed] [Google Scholar]
- 6.Wang N, et al. Shear stress regulation of Krüppel-like factor 2 expression is flow pattern-specific. Biochem Biophys Res Commun. 2006;341:1244–1251. doi: 10.1016/j.bbrc.2006.01.089. [DOI] [PubMed] [Google Scholar]
- 7.Ni CW, Hsieh HJ, Chao YJ, Wang DL. Shear flow attenuates serum-induced STAT3 activation in endothelial cells. J Biol Chem. 2003;278:19702–19708. doi: 10.1074/jbc.M300893200. [DOI] [PubMed] [Google Scholar]
- 8.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Suárez Y, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci USA. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonauer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 12.Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: Polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25:1620–1628. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- 13.Olgac U, Kurtcuoglu V, Poulikakos D. Computational modeling of coupled blood-wall mass transport of LDL: Effects of local wall shear stress. Am J Physiol Heart Circ Physiol. 2008;294:H909–H919. doi: 10.1152/ajpheart.01082.2007. [DOI] [PubMed] [Google Scholar]
- 14.Hwang J, et al. Pulsatile versus oscillatory shear stress regulates NADPH oxidase subunit expression: Implication for native LDL oxidation. Circ Res. 2003;93:1225–1232. doi: 10.1161/01.RES.0000104087.29395.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, et al. Improved significance test for DNA microarray data: Temporal effects of shear stress on endothelial genes. Physiol Genomics. 2002;12:1–11. doi: 10.1152/physiolgenomics.00024.2002. [DOI] [PubMed] [Google Scholar]
- 16.Fontana L, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Z, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182:509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 20.Takakura S, et al. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008;99:1147–1154. doi: 10.1111/j.1349-7006.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang KC, et al. Role of microRNA-23b in flow-regulation of Rb phosphorylation and endothelial cell growth. Proc Natl Acad Sci USA. 2009;107:3234–3239. doi: 10.1073/pnas.0914825107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherr CJ. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 24.Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 25.Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang N, et al. Adenovirus-mediated overexpression of dominant-negative mutant of c-Jun prevents intercellular adhesion molecule-1 induction by LDL: A critical role for activator protein-1 in endothelial activation. Arterioscler Thromb Vasc Biol. 2001;21:1414–1420. doi: 10.1161/hq0901.095549. [DOI] [PubMed] [Google Scholar]
- 27.Qin X, et al. Laminar shear stress up-regulates the expression of stearoyl-CoA desaturase-1 in vascular endothelial cells. Cardiovasc Res. 2007;74:506–514. doi: 10.1016/j.cardiores.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin X, et al. Peroxisome proliferator-activated receptor-delta induces insulin-induced gene-1 and suppresses hepatic lipogenesis in obese diabetic mice. Hepatology. 2008;48:432–441. doi: 10.1002/hep.22334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.