Abstract
The bifunctional proline catabolic flavoenzyme, proline utilization A (PutA), catalyzes the oxidation of proline to glutamate via the sequential activities of FAD-dependent proline dehydrogenase (PRODH) and NAD+-dependent Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH) domains. Although structures for some of the domains of PutA are known, a structure for the full-length protein has not previously been solved. Here we report the 2.1 Å resolution crystal structure of PutA from Bradyrhizobium japonicum, along with data from small-angle x-ray scattering, analytical ultracentrifugation, and steady-state and rapid-reaction kinetics. PutA forms a ring-shaped tetramer in solution having a diameter of 150 Å. Within each protomer, the PRODH and P5CDH active sites face each other at a distance of 41 Å and are connected by a large, irregularly shaped cavity. Kinetics measurements show that glutamate production occurs without a lag phase, suggesting that the intermediate, Δ1-pyrroline-5-carboxylate, is preferably transferred to the P5CDH domain rather than released into the bulk medium. The structural and kinetic data imply that the cavity serves both as a microscopic vessel for the hydrolysis of Δ1-pyrroline-5-carboxylate to glutamate semialdehyde and a protected conduit for the transport of glutamate semialdehyde to the P5CDH active site.
Keywords: proline catabolism, substrate channeling
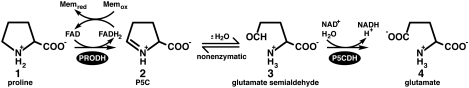
Oxidation of amino acids is a central part of energy metabolism. In eukaryotes and bacteria, proline is oxidized (or catabolized) to glutamate by the conserved enzymes, proline dehydrogenase (PRODH) and Δ1-pyrroline-5-carboxylate (P5C) dehydrogenase (Fig. 1). PRODH catalyzes the FAD-dependent oxidation of proline (1 in Fig. 1) to P5C (2). The intermediate of proline catabolism, P5C, is hydrolyzed nonenzymatically to glutamate γ-semialdehyde (3), which is subsequently oxidized to glutamate (4) by NAD+-dependent P5C dehydrogenase (P5CDH). In humans, proline oxidation is important for cellular redox control, apoptosis and suppression of cancer (1). Inborn errors in PRODH and P5CDH result in types I and II hyperprolinemia disorders (2), and a deficiency in PRODH activity is thought to be a risk factor for schizophrenia (3). Moreover, human PRODH (known as POX) is a p53-induced enzyme that plays a crucial role in apoptosis by serving as a proline-dependent, superoxide generator (1). Other organisms have evolved the ability to harness energy almost exclusively from proline under certain circumstances. Examples include Helicobacter pylori in the human gut (4), procyclic trypanosomes in the midgut of insects (5), and the tsetse fly during flight (6).
Fig. 1.
The reactions catalyzed by PRODH and P5CDH.
Intriguingly, whereas PRODH and P5CDH are separate enzymes in eukaryotes and some bacteria, they are fused into a single polypeptide chain in other bacteria. The fused enzymes, known as proline utilization A, or PutA, are membrane-associated proteins containing 1,000–1,300 amino acid residues, with the PRODH and P5CDH domains located in the N- and C-terminal halves of the polypeptide chain, respectively (7, 8). Some PutAs, such those from Escherichia coli (EcPutA) and Salmonella typhimurium, also have a regulatory function. These “trifunctional” PutAs repress transcription of the put regulon, which contains the genes for PutA and the proline transporter PutP. The DNA-binding domain of trifunctional PutAs is a ribbon-helix-helix domain located in the N-terminal 50 residues (9–11).
PutAs have proven to be challenging structural biology targets. The first structures of PutA domains appeared over two decades after Ratzkin and Roth discovered that PRODH and P5CDH were encoded by a single gene in S. typhimurium (12). These include crystal structures of the PRODH (13–16) and DNA-binding (10, 11) domains of EcPutA, and a solution structure of the DNA-binding domain of Pseudomonas putida PutA (17).
Whereas the domain structures have informed us about the individual functions of PutA, they have not provided insight into how PutAs coordinate multiple functions. Here we report the crystal structure of a full-length bifunctional PutA. The structure and accompanying kinetic data indicate that P5C undergoes hydrolysis to glutamate semialdehyde en route to the P5CDH domain without leaving the confines of the protein.
Results
Tertiary Structure.
Several PutAs were screened for crystallizability, which eventually led us to focus structure determination efforts on the 999-residue PutA from Bradyrhizobium japonicum (BjPutA). The structure of BjPutA was solved using anomalous dispersion diffraction data collected from hexagonal crystals of the Se-Met derivative (Table S1) and was subsequently refined to 2.1 Å resolution using native diffraction data collected from a centered monoclinic crystal (Table 1).
Table 1.
Data Collection and Refinement Statistics*
| Space group | C2 |
| a, b, c, Å | 166.8, 195.8, 108.7 |
| β,° | 121.5 |
| Resolution, Å | 43 - 2.1(2.2 - 2.1) |
| Unique reflections | 172549 |
| Redundancy | 3.8 (3.7) |
| Completeness, % | 99.9 (99.9) |
| Rmerge | 0.103 (0.412) |
| I/σ | 12.1 (2.7) |
| No. of protein atoms | 14647 |
| No. of FAD and NAD+ atoms | 194 |
| No. of water molecules | 795 |
 |
0.200 (0.225) |
| Rfree† | 0.233 (0.281) |
| rmsd bond lengths, Å | 0.005 |
| rmsd bond angles, ° | 0.86 |
| Ramachandran—favored | 1901 |
| Ramachandran—allowed | 39 |
| Ramachandran—outliers | 3 |
| Average B for protein, Å2 | 35 |
| Average B for FAD, Å2 | 24 |
| Average B for NAD+, Å2 | 32 |
| Average B for water, Å2 | 34 |
*Values for the outer resolution shell of data are given in parentheses.
†5% random test set.
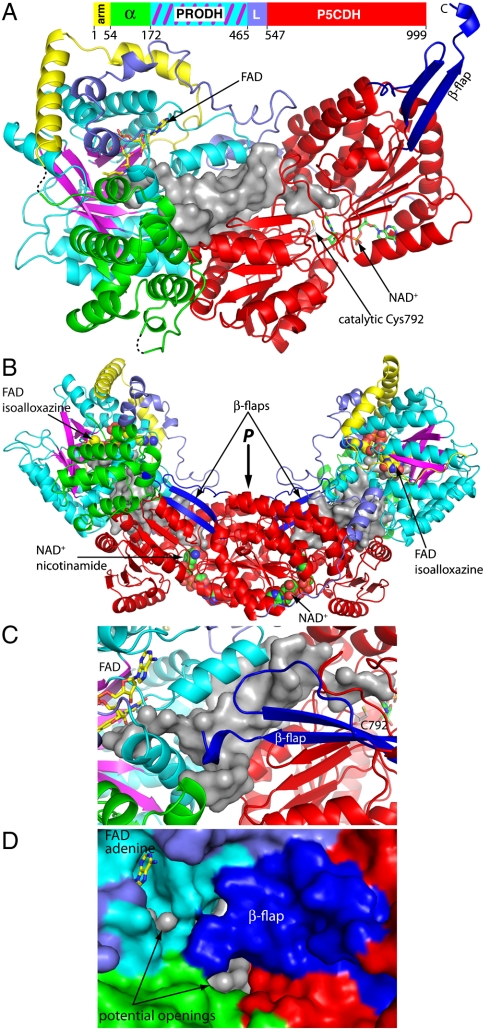
The protomer consists of the two catalytic domains plus three smaller, ancillary domains denoted N-terminal arm, α-helix bundle, and linker (Fig. 2A). The asymmetric unit of the C2 crystal form contains two protomers, which form a U-shaped dimer having dimensions of 150 Å × 70 Å × 80 Å (Fig. 2B). The polypeptide chain begins with a 51-residue arm-like extension that traverses ∼100 Å as it wraps around the PRODH domain (Fig. 2A, Yellow). The arm is followed by the α-helix bundle domain, which consists of six tightly packed α-helices (Fig. 2A, Green) and contacts both catalytic domains. The PRODH and P5CDH domains follow the α-helix bundle and are separated by an 83-residue linker.
Fig. 2.
Structure of BjPutA. (A) Ribbon drawing of the protomer. The domains are color coded as follows: yellow, N-terminal arm; green α-helix bundle domain; cyan/magenta, PRODH domain; slate, linker; and red, P5CDH domain. FAD and NAD+ are drawn as sticks in yellow and green, respectively. The β-flap is colored blue. The silver surface represents the internal cavity that connects the two active sites. (B) The dimer of the asymmetric unit, colored as in A. FAD and NAD+ are drawn as spheres. (C) Close-up view of the dimer showing the region near the β-flap. The coloring scheme of A is used. (D) Surface representation of the β-flap region, shown with the same orientation and coloring scheme as in C.
The PRODH domain resembles that of EcPutA in both the overall fold and details of the active site. This domain has a distorted (βα)8 barrel fold (Fig. 2, Magenta Strands, Cyan Helices), which is considered to be diagnostic of the PRODH family (18). The FAD cofactor is bound noncovalently at the C termini of the strands (Figs. 2A, B). The re face packs tightly against strands 4–6 of the barrel, allowing proline to bind at the si face (Fig. S1). Residues near the isoalloxazine and proline-binding site are highly conserved in PutAs, and many of these residues have similar conformations in BjPutA and EcPutA (Fig. S2A). BjPutA and EcPutA also have identical FAD conformations (Fig. S2A). Finally, there appears to be a sulfate ion bound in the active site. Comparison to the structure of the EcPutA PRODH domain complexed with the proline analog L-tetrahydro-2-furoic acid indicates that the sulfate ion occupies the binding site for the substrate carboxylate group (Fig. S2A).
The P5CDH domain (Fig. 2A, Red) is similar to other aldehyde dehydrogenases (19, 20). It has two lobes, one that binds NAD+ using a nonclassical Rossmann fold and another that furnishes catalytic Cys792. In addition, there is a β-strand substructure protruding from the Rossmann fold subdomain (β-flap, residues 628–646, 977–989, see Fig. 2A). The β-flap stabilizes the U-shaped dimer by forming an intermolecular β-sheet interaction with the P5CDH domain of the other protomer (Fig. 2C). As with the PRODH active site, there is a sulfate ion bound in the P5CDH active site. Comparison with the structure of the monofunctional P5CDH from Thermus thermophilus shows that the sulfate ion occupies the binding site for the carboxylate group of glutamate semialdehyde (Fig. S2B).
Quaternary Structure.
The oligomeric state of BjPutA in solution was examined by analytical ultracentrifugation as described in SI Text. Data from eighteen parallel sedimentation equilibrium experiments were fit globally to a single-species model, enabling simultaneous estimation of the partial specific volume and molecular weight (Fig. S3). The best-fit values are 0.772 mL/g (0.767–0.777 mL/g) and 442 kDa (428–455 kDa). The partial specific volume is significantly larger than the value of 0.74 mL/g calculated from the sequence, which is consistent with the observation of a large solvent-filled cavity inside the protein (vide infra). The estimated molecular weight is within 3% of the theoretical molecular weight of a tetramer of 430 kDa.
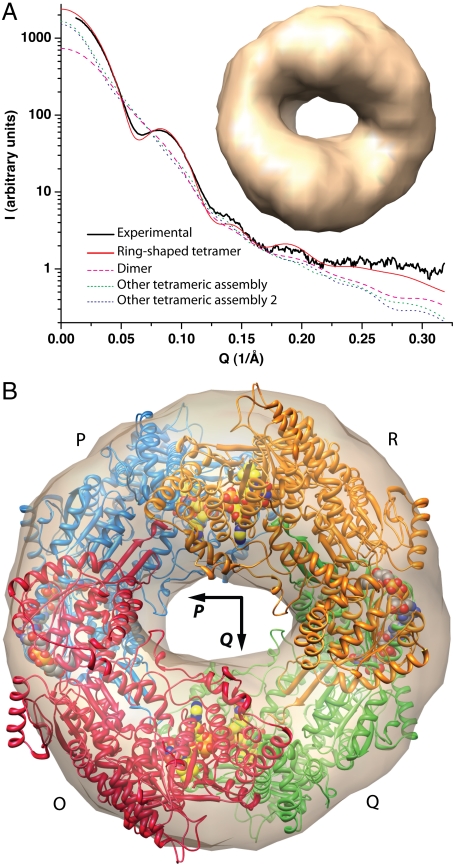
Small-angle x-ray scattering (SAXS) was used to determine the arrangement of protomers within the tetramer as described in SI Text. A typical SAXS profile is shown in Fig. 3A (Black Solid Curve). Guinier analysis suggested a radius of gyration of 56 Å. Calculations of the pair distribution function yielded estimates for the radius of gyration and the maximum particle dimension of 51 and 140 Å, respectively. Shape reconstructions calculated with GASBOR (21) indicated that BjPutA forms a ring-shaped particle in solution (Fig. 3A, inset).
Fig. 3.
SAXS analysis and tetrameric structure of BjPutA. (A) Experimental and simulated scattering profiles. The solid black curve represents a typical experimental scattering curve for BjPutA. The solid red curve represents the profile calculated from the ring-shaped tetramer shown in panel B. The dashed magenta curve was calculated from the dimer of the asymmetric unit. The two dotted curves were calculated from other tetrameric assemblies that are not shaped like a ring. The inset shows an ab initio shape restoration calculated from the experimental SAXS data using GASBOR. (B) Superposition of the SAXS reconstruction and the crystallographic tetramer. The four chains are labeled O (red), P (blue), Q (green), and R (orange). FAD and NAD+ are shown in yellow and gray spheres, respectively.
The C2 lattice was inspected to identify a tetramer of BjPutA having the size and shape of the SAXS reconstruction. Such an assembly could be built by applying the twofold crystallographic symmetry transformation (-x,y,-z - 1) to the U-shaped dimer of the asymmetric unit (Fig. 3B and Fig. S4). Further validation of this particular tetramer as the one formed in solution was obtained from calculations of the scattering curve from atomic models using CRYSOL (22). The curve calculated from the ring-shaped tetramer displays satisfactory agreement with the experimental one (Fig. 3A, compare Red and Black Solid Curves). In contrast, the curves calculated from the U-shaped dimer (Fig. 3A, Dashes), and two other tetrameric assemblies that do not form rings (Fig. 3A, Dots), show poor agreement with the experimental data. Finally, we note that the tetrameric ring is also present in lattices of the hexagonal and orthorhombic crystal forms of BjPutA.
The tetramer is a dimer of dimers having 222 symmetry, which can be described by three mutually perpendicular twofold axes, denoted P, Q, and R, intersecting at the centroid of the particle (Fig. 3B and Fig. S4). We adopt the convention of labeling the four chains O, P, Q, and R, such that the P-, Q-, and R-axes transform the O chain into the P, Q, and R chains, respectively.
There are two types of interface in the tetramer, which bury a total of 8360 Å2 of surface area. The larger one is generated by rotation around the P-axis (Fig. 2B). This interface features the swapped β-flap interaction shown in Fig. 2C and is a well-known feature of aldehyde dehydrogenases (23). The other interface is unique to BjPutA and contributes 1760 Å2 of interfacial area. It is generated by rotation around the Q axis and joins two U-shaped dimers to form a ring such that all four β-flaps face the hole of the ring (Fig. 3B). The N-terminal arm, α-helical bundle domain, and linker domain form this interface.
Internal Cavity Implies Substrate Channeling.
Within each protomer, the PRODH and P5CDH active sites face each other at a distance of 41 Å. Between them is an irregularly shaped cavity that connects the si face of the isoalloxazine to the catalytic cysteine of the P5CDH domain (Figs. 2C, 4). The cavity volume was estimated with VOIDOO (24) to be 1400 Å3, which is equivalent to about 170 water molecules. All domains except the arm contribute to the cavity (Fig. 2C). The β-flap evidently serves as a lid that covers the cavity of the P-related protomer (Figs. 2C, 2D); analysis with the program MOLE (25) indicates potential openings (3–5 Å wide) to the bulk medium in this region (Fig. 2D). Because the cavity connects the active sites of a protomer, the structure implies the potential for substrate channeling within, rather than between, protomers.
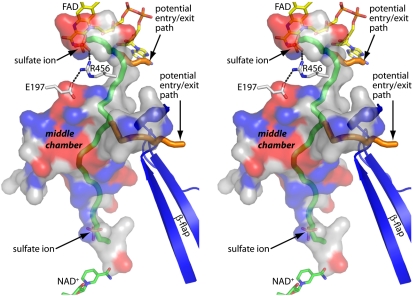
Fig. 4.
Stereographic view of the cavity. The cavity is represented as a semitransparent surface and is colored to indicate the locations of positively (blue) and negatively (red) charged residues lining the cavity. The tubes represent pathways identified by the program MOLE. The green tube guides the eye from the PRODH active site (Top) to the P5CDH active site (Bottom). The orange tubes represent possible pathways leading to the bulk medium.
The cavity has three sections, corresponding to the two active sites and a large, intervening chamber (Fig. 4 and Fig. S5). The middle chamber is 24 Å by 14 Å in its two largest dimensions, and 3–7 Å in the third dimension. It has a volume of about 1325 Å3, which represents over 90% of the total volume of the entire cavity system. Thus, the middle chamber is large enough to accommodate P5C (molecular volume 102 Å3) and glutamate semialdehyde (120 Å3). The active sites connect to the middle chamber via constrictions that have widths approximately corresponding to the diameter of a water molecule.
The connection between the PRODH active site and the middle chamber is interesting because of its circuitous path (Fig. 4 and Fig. S5). It runs down the ribityl chain of the FAD and then passes over a potential opening to the bulk medium and under the side chain of Arg456 before connecting to the middle chamber. A more direct path into the middle chamber appears to be blocked by the ion-pairing residues Arg456 and Glu197. Both residues are identically conserved in PutAs, and Arg456 plays an essential role in substrate binding by forming a critical ionic interaction with the carboxylate of the substrate.
Finally, the walls of the cavity are highly ionic, and thus the cavity itself is very hydrophilic. The cavity walls are lined with twelve arginine, three lysine, ten glutamate, and seven aspartate residues, which are represented by the blue (Arg, Lys) and red (Glu, Asp) patches in Fig. 4 and Fig. S5.
Kinetic Evidence for Substrate Channeling.
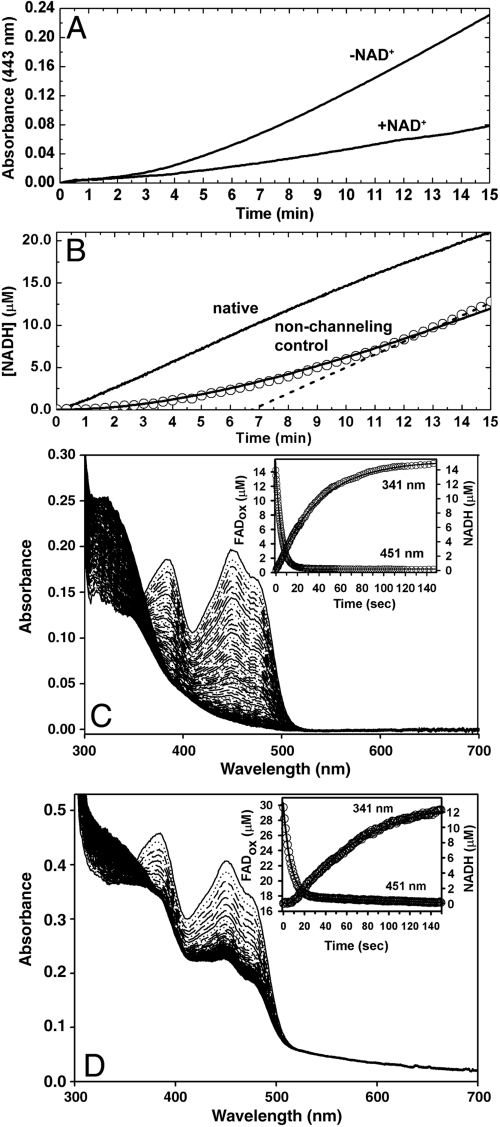
Kinetic measurements were performed as described in SI Text. The results of a P5C trapping assay are shown in Fig. 5A. The trapping experiment uses an excess of o-aminobenzaldehyde (o-AB), which forms a yellow complex with P5C. In the absence of NAD+, formation of the o-AB-P5C complex is evident, indicating that P5C is released into bulk solvent. In contrast, with NAD+ present (0.2 mM), formation of the o-AB-P5C complex is diminished, suggesting that some of the P5C generated in the PRODH active site is converted to glutamate without leaving the enzyme. Under the conditions of this assay, the apparent fraction of P5C that is channeled is 0.7.
Fig. 5.
Kinetic data. (A) P5C trapping experiments performed in the presence and absence of NAD+. (B) NADH formation by native BjPutA (upper solid curve) and a nonchanneling control consisting of an equimolar mixture of monofunctional variants R456M and C792A (circles). The solid curve overlaying the data for the nonchanneling control was calculated from Eq. S2 of SI Text. The dashed line represents the extrapolation used to estimate the transient time for the nonchanneling control. For clarity, only a subset of the experimental data points for the nonchanneling control is displayed. (C) Rapid-reaction kinetic data for native BjPutA acquired under single turnover conditions. PutA (14.26 µM), NAD+ (0.1 mM), and proline (40 mM) were rapidly mixed (concentrations after mixing) and monitored by stopped-flow multiwavelength absorption. The spectra shown were recorded at 0.0025–250 sec after mixing. Inset: Plots of FAD reduction and NADH formation monitored at 451 nm and 341 nm, respectively. The observed rate constants for reduction of FAD and NAD+ are 0.2 s-1 and 0.025 s-1, respectively, estimated from single exponential fits of the data as described in SI Text. (D) Rapid-reaction kinetic data for the nonchanneling control acquired under single turnover conditions. BjPutA variants C792A and R456M (14.9 µM each), NAD+ (0.1 mM), and proline (40 mM) were rapidly mixed (concentrations after mixing) and monitored by stopped-flow multiwavelength absorption. The spectra shown were recorded at 0.0025–300 sec after mixing. Inset: Plots of FAD reduction and NADH formation monitored at 451 nm and 341 nm, respectively. The observed rate constants for reduction of FAD and NAD+ are 0.144 s-1 and 0.016 s-1, respectively, estimated from double exponential fits of the data as described in SI Text.
The time-dependence of NADH production from proline was measured to see whether a lag phase occurs. The progress curve for native BjPutA is approximately linear, and a lag phase is not apparent (Fig. 5B). This result contrasts with the expectations for noninteracting PRODH and P5CDH enzymes. The progress curve for two noninteracting enzymes is predicted to exhibit a lag phase with transient time, τ, equal to the ratio of Km to Vmax for the second enzyme (26). The value of τ estimated from the kinetic constants in Table S2 is ∼8 min. The absence of such a lag phase in the experimental progress curve suggests that channeling occurs in BjPutA.
The NADH production assay was repeated using a nonchanneling control containing equal concentrations of the two BjPutA monofunctional mutant enzymes R456M and C792A (Fig. 5B, circles). PRODH activity is disabled in R456M, but the P5CDH activity is identical to that of the native enzyme (Table S2). Conversely, the C792A mutation abolishes P5CDH activity but does not affect the PRODH activity. If channeling occurs within the protomer, as suggested by the structure, the progress curve for an equimolar mixture of R456M and C792A is expected to exhibit a lag phase. As predicted, the progress curve for the nonchanneling control exhibits an obvious lag phase. Furthermore, the data agree well with the theoretical progress curve for noninteracting PRODH and P5CDH enzymes calculated from Eq. S2 of SI Text (Fig. 5B). The transient time extrapolated from the experimental progress curve is about 7 min (Fig. 5B, dashed line), which is close to the theoretical value based on Km/Vmax of 8.6 min. These results validate the use of the monofunctional mutants as a nonchanneling control. Moreover, the substantial difference between the progress curves for the native enzyme and the nonchanneling control provides additional evidence of substrate channeling in BjPutA.
Rapid-reaction kinetic measurements employing single turnover conditions were also performed (anaerobic, no external electron acceptor for the reduced flavin). BjPutA, proline, and NAD+ were rapidly mixed, and the absorbance spectra of the FAD cofactor and NADH product were recorded using a stopped-flow instrument. For the native enzyme, NADH production occurs without a lag phase (Fig. 5C and Fig. S6). In contrast, single turnover experiments with the nonchanneling control enzymes show a lag in the appearance of NADH of approximately 10 s (Fig. 5D and Fig. S6). These results are also consistent with substrate channeling in the native enzyme.
Discussion
The BjPutA structure reveals, for the first time, the architecture of a PutA protein. The most significant result is that the two active sites are separated by 41 Å and oriented so that the exit path of the PRODH active site faces the substrate entrance tunnel of the P5CDH active site. The cavity connecting the two active sites strongly suggests the possibility of substrate channeling within the protomer, which is supported by kinetic measurements.
Although substrate channeling is not unique to PutA, some aspects of channeling in PutA are different from other bifunctional enzymes. In particular, the irregular shape of the cavity is rather different from the tunnels that channel indole and ammonia in tryptophan synthase and carbamoyl phosphate synthetase (27, 28). The PutA cavity better resembles that of dimethylglycine oxidase (29, 30).
The demonstration of substrate channeling in BjPutA is consistent with kinetic data reported by Surber and Maloy a decade ago for the trifunctional PutA from S. typhimurium (31), the only other study of channeling in PutAs. As with BjPutA, initial production of NADH from proline occurred without a lag phase for S. typhimurium PutA. Surber and Maloy also used 14C-labeled proline to show that exogenous P5C does not efficiently compete for P5CDH activity in the presence of proline, clear evidence of channeling. The fact that channeling has been demonstrated in two PutAs suggests that it may be a conserved aspect of PutAs. However, there are likely to be differences in the details of the channeling mechanisms of trifunctional and bifunctional PutAs.
Significantly, our results imply that the P5C produced by PutA is hydrolyzed to glutamate semialdehyde within the confines of the protein rather than in the bulk medium. It is tempting to suggest that the hydrolysis reaction occurs in the middle chamber, which is large enough to accommodate several molecules of P5C and/or glutamate semialdehyde. The middle chamber could facilitate the hydrolysis reaction by shifting the equilibrium between P5C and glutamate semialdehyde to favor the latter species. The equilibrium has a strong pH-dependence, with the reverse reaction (toward P5C) favored above pH 6.5 (32). This transition point corresponds to the pKa of the pyrrolinium moiety of P5C, indicating that protonation of the pyrrolinium favors hydrolysis. The highly ionic character of the walls of the middle chamber could create an electrostatic field inside the cavity that effectively increases the pKa of the pyrrolinium moiety, thus favoring the formation of glutamate semialdehyde.
The structure of the cavity suggests that protein dynamics might play a role in channeling. For example, the conserved Arg456-Glu197 ion pair blocks a direct route from the active site to the middle chamber (Fig. 4 and Fig. S5). If P5C formation and concomitant reduction of the FAD induce rupture of the ion pair, the PRODH cavity would coalesce with the middle chamber, opening a direct route. Thus, the ion pair could potentially act as a gate that opens and closes in response to product release and substrate binding, respectively. We note that there are several examples of gates and constrictions blocking channeling pathways in other bifunctional enzymes (33–36).
The discovery that BjPutA forms a tetramer was unexpected. In the only other biophysical study of the oligomeric state of a PutA, Brown and Wood concluded that EcPutA is an elongated dimer in solution (37), which contrasts with the donut shape of BjPutA. An important difference between the two proteins is that EcPutA functions as a transcriptional repressor of the put regulon, in addition to being a bifunctional enzyme. We previously showed that the repressor domain has a ribbon-helix-helix fold (10). It thus appears that bifunctional and trifunctional PutAs differ in quaternary structure, which was unanticipated. This observation raises the question of how the oligomeric states of PutAs are related to function. The relationship seems clear for trifunctional PutAs; ribbon-helix-helix domains must dimerize to bind DNA. In particular, an intermolecular β-sheet of the dimer inserts into the major groove of DNA so that side chains of the sheet make specific contacts with DNA bases. The significance of the U-shaped dimer of BjPutA is also clear. The β-flap of one protomer covers the substrate channel of the other protomer of the dimer. On the other hand, the role of the tetramer for bifunctional PutAs is, as yet, unknown.
Finally, our work raises the question of the biological relevance of substrate channeling in proline catabolism. Channeling potentially affords the advantages of decreased transit time between enzymes, protection of labile intermediates, and isolation of intermediates from competing enzymatic reactions, among others (26–28). Channeling in PutA may be particularly beneficial for bacteria that have adapted to use proline as a fuel source. One example is H. pylori, which utilizes proline as a preferred energy source in the gut environment, where proline levels in the gastric juice of infected patients are 10-fold higher compared to uninfected individuals (4, 38). Also, channeling segregates the P5C produced by proline catabolism from the proline biosynthetic pathway, which also uses P5C as an intermediate. In the absence of channeling, a futile cycle of interconversion between proline and P5C could occur. Channeling might also be relevant in humans, because P5C is involved in a number of diverse biological functions, including apoptosis, and oxidative stress (1). Although human PRODH (POX) and P5CDH are distinct (i.e., not fused) mitochondrial enzymes, the fact that the two enzymes are fused in PutA, along with the clear demonstration of channeling in PutA, suggests that the human enzymes may interact and engage in substrate channeling to protect P5C from competing biological pathways.
Materials and Methods
Protein Preparation, Crystallization, and Structure Determination.
Preparation of native BjPutA and the growth of three crystal forms have been described (39). See SI Text for detailed information regarding the preparation and crystallization of selenomethionyl BjPutA, phasing by anomalous dispersion using the hexagonal crystal form (Table S1), and molecular replacement calculations for the orthorhombic and C2 crystal forms. The structure was refined to 2.1 Å resolution in space group C2 (Table 1). The final model includes one BjPutA dimer consisting of 1957 of the expected 1998 residues, two FAD cofactors, and two NAD+ cofactors.
Supplementary Material
Acknowledgments.
We thank Dr. Jay Nix of ALS beamline 4.2.2 and Dr. Stephan L. Ginell of the APS SBC beamlines for assistance with data collection and processing. This research was supported by National Institutes of Health grants GM065546, GM061068, and P20 RR-017675. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract DE-AC02-05CH11231. Results shown in this report are derived, in part, from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3HAZ).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906101107/DCSupplemental.
References
- 1.Phang JM, Donald SP, Pandhare J, Liu Y. The metabolism of proline, a stress substrate, modulates carcinogenic pathways. Amino Acids. 2008;35(4):681–690. doi: 10.1007/s00726-008-0063-4. [DOI] [PubMed] [Google Scholar]
- 2.Phang JM, Hu CA, Valle D. Disorders of proline and hydroxyproline metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. Metabolic and Molecular Basis of Inherited Disease. New York: McGraw Hill; 2001. pp. 1821–1838. [Google Scholar]
- 3.Willis A, Bender HU, Steel G, Valle D. PRODH variants and risk for schizophrenia. Amino Acids. 2008;35(4):673–679. doi: 10.1007/s00726-008-0111-0. [DOI] [PubMed] [Google Scholar]
- 4.Nagata K, et al. L-Serine, D- and L-proline and alanine as respiratory substrates of Helicobacter pylori: correlation between in vitro and in vivo amino acid levels. Microbiology+ 2003;149(Pt 8):2023–2030. doi: 10.1099/mic.0.26203-0. [DOI] [PubMed] [Google Scholar]
- 5.Bringaud F, Riviere L, Coustou V. Energy metabolism of trypanosomatids: Adaptation to available carbon sources. Mol Biochem Parasitol. 2006;149(1):1–9. doi: 10.1016/j.molbiopara.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 6.Saktor B. Biochemical adaptations for flight in the insect. Biochem Soc Symp. 1976;41:111–131. [PubMed] [Google Scholar]
- 7.Menzel R, Roth J. Purification of the putA gene product. J Biol Chem. 1981;256(18):9755–9761. [PubMed] [Google Scholar]
- 8.Ling M, Allen SW, Wood JM. Sequence analysis identifies the proline dehydrogenase and pyrroline-5-carboxylate dehydrogenase domains of the multifunctional Escherichia coli PutA protein. J Mol Biol. 1994;243:950–956. doi: 10.1006/jmbi.1994.1696. [DOI] [PubMed] [Google Scholar]
- 9.Gu D, et al. Identification and characterization of the DNA-binding domain of the multifunctional PutA flavoenzyme. J Biol Chem. 2004;279(30):31171–31176. doi: 10.1074/jbc.M403701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larson JD, et al. Crystal structures of the DNA-binding domain of Escherichia coli proline utilization A flavoprotein and analysis of the role of Lys9 in DNA recognition. Protein Sci. 2006;15:1–12. doi: 10.1110/ps.062425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y, et al. Structural basis of the transcriptional regulation of the proline utilization regulon by multifunctional PutA. J Mol Biol. 2008;381(1):174–188. doi: 10.1016/j.jmb.2008.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ratzkin B, Roth J. Cluster of genes controlling proline degradation in Salmonella typhimurium. J Bacteriol. 1978;133(2):744–754. doi: 10.1128/jb.133.2.744-754.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee YH, Nadaraia S, Gu D, Becker DF, Tanner JJ. Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein. Nat Struct Biol. 2003;10(2):109–114. doi: 10.1038/nsb885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M, et al. Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors. Biochemistry. 2004;43(39):12539–12548. doi: 10.1021/bi048737e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W, et al. Redox-induced changes in flavin structure and roles of flavin N(5) and the ribityl 2′-OH group in regulating PutA--membrane binding. Biochemistry. 2007;46(2):483–491. doi: 10.1021/bi061935g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostrander EL, Larson JD, Schuermann JP, Tanner JJ. A conserved active site tyrosine residue of proline dehydrogenase helps enforce the preference for proline over hydroxyproline as the substrate. Biochemistry. 2009;48(5):951–959. doi: 10.1021/bi802094k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halouska S, Zhou Y, Becker DF, Powers R. Solution structure of the Pseudomonas putida protein PpPutA45 and its DNA complex. Proteins. 2009;75(1):12–27. doi: 10.1002/prot.22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanner JJ. Structural biology of proline catabolism. Amino Acids. 2008;35(4):719–730. doi: 10.1007/s00726-008-0062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inagaki E, et al. Crystal structure of Thermus thermophilus Delta1-pyrroline-5-carboxylate dehydrogenase. J Mol Biol. 2006;362(3):490–501. doi: 10.1016/j.jmb.2006.07.048. [DOI] [PubMed] [Google Scholar]
- 20.Moore SA, et al. Sheep liver cytosolic aldehyde dehydrogenase: The structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure. 1998;6(12):1541–1551. doi: 10.1016/s0969-2126(98)00152-x. [DOI] [PubMed] [Google Scholar]
- 21.Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from x-ray solution scattering. Biophys J. 2001;80(6):2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svergun D, Barberato C, Koch MHJ. CRYSOL—A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28(6):768–773. [Google Scholar]
- 23.Liu ZJ, et al. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat Struct Biol. 1997;4(4):317–326. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- 24.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D. 1994;50(Pt 2):178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 25.Petrek M, Kosinova P, Koca J, Otyepka M. MOLE: A Voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure. 2007;15(11):1357–1363. doi: 10.1016/j.str.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Anderson KS. Fundamental mechanisms of substrate channeling. Methods Enzymol. 1999;308:111–145. doi: 10.1016/s0076-6879(99)08008-8. [DOI] [PubMed] [Google Scholar]
- 27.Miles EW, Rhee S, Davies DR. The molecular basis of substrate channeling. J Biol Chem. 1999;274(18):12193–12196. doi: 10.1074/jbc.274.18.12193. [DOI] [PubMed] [Google Scholar]
- 28.Huang X, Holden HM, Raushel FM. Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu Rev Biochem. 2001;70:149–180. doi: 10.1146/annurev.biochem.70.1.149. [DOI] [PubMed] [Google Scholar]
- 29.Leys D, Basran J, Scrutton NS. Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase. EMBO J. 2003;22(16):4038–4048. doi: 10.1093/emboj/cdg395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tralau T, et al. An internal reaction chamber in dimethylglycine oxidase provides efficient protection from exposure to toxic formaldehyde. J Biol Chem. 2009;284(26):17826–17834. doi: 10.1074/jbc.M109.006262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Surber MW, Maloy S. The PutA protein of Salmonella typhimurium catalyzes the two steps of proline degradation via a leaky channel. Arch Biochem Biophys. 1998;354(2):281–287. doi: 10.1006/abbi.1998.0697. [DOI] [PubMed] [Google Scholar]
- 32.Bearne SL, Wolfenden R. Glutamate gamma-semialdehyde as a natural transition state analogue inhibitor of Escherichia coli glucosamine-6-phosphate synthase. Biochemistry. 1995;34(36):11515–11520. doi: 10.1021/bi00036a026. [DOI] [PubMed] [Google Scholar]
- 33.Chaudhuri BN, et al. Crystal structure of imidazole glycerol phosphate synthase: a tunnel through a (beta/alpha)8 barrel joins two active sites. Structure. 2001;9(10):987–997. [PubMed] [Google Scholar]
- 34.van den Heuvel RH, et al. The active conformation of glutamate synthase and its binding to ferredoxin. J Mol Biol. 2003;330(1):113–128. doi: 10.1016/s0022-2836(03)00522-9. [DOI] [PubMed] [Google Scholar]
- 35.Manjasetty BA, Powlowski J, Vrielink A. Crystal structure of a bifunctional aldolase-dehydrogenase: Sequestering a reactive and volatile intermediate. Proc Natl Acad Sci USA. 2003;100(12):6992–6997. doi: 10.1073/pnas.1236794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaRonde-LeBlanc N, Resto M, Gerratana B. Regulation of active site coupling in glutamine-dependent NAD(+) synthetase. Nat Struct Mol Biol. 2009;16(4):421–429. doi: 10.1038/nsmb.1567. [DOI] [PubMed] [Google Scholar]
- 37.Brown ED, Wood JM. Redesigned purification yields a fully functional PutA protein dimer from Escherichia coli. J Biol Chem. 1992;267(18):13086–13092. [PubMed] [Google Scholar]
- 38.Nakajima K, et al. Possible involvement of put A gene in Helicobacter pylori colonization in the stomach and motility. Biomed Res. 2008;29(1):9–18. doi: 10.2220/biomedres.29.9. [DOI] [PubMed] [Google Scholar]
- 39.Schuermann JP, White TA, Srivastava D, Karr DB, Tanner JJ. Three crystal forms of the bifunctional enzyme proline utilization A (PutA) from Bradyrhizobium japonicum. Acta Crystallogr F. 2008;64(Pt 10):949–953. doi: 10.1107/S174430910802842X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.