Abstract
Stem cells are essential for tissue generation during the development of multicellular creatures, and for tissue homeostasis in adults. The great therapeutic promise of stem cells makes understanding their regulation a high priority. PUF RNA-binding proteins have a conserved role in promoting self-renewal of germline stem cells. Here we use a genome-wide approach to identify putative target mRNAs for the Caenorhabditis elegans PUF protein known as FBF. We find that putative FBF targets represent ∼7% of all protein-coding genes in C. elegans, implicating FBF as a broad-spectrum gene regulator. These putative FBF targets are enriched for regulators of meiotic entry and other components of the meiotic program as well as regulators of key developmental pathways. We suggest that these targets may be critical for FBF’s role in stem cell maintenance. Comparison of likely FBF target mRNAs with putative PUF target mRNAs from Drosophila and humans reveals 40 shared targets, including several established stem cell regulators. We speculate that shared PUF targets represent part of a broadly used module of stem cell control.
Keywords: germline stem cell, PUF, RIP-Chip, totipotency
Stem cells govern tissue generation during metazoan development, and tissue maintenance during adulthood. A driving force behind stem cell research is the identification of broadly conserved regulatory mechanisms that may facilitate development of therapies to ease human disease. Several major stem cell regulators control gene expression. Best known are the transcription factors Oct4, Sox2, and Nanog, which head a transcriptional network to regulate pluripotency in human embryonic stem (ES) cells (1). In addition, regulators that act posttranscriptionally to govern mRNA translation or stability have also emerged as key stem cell regulators (2). However, their targets within stem cells remain unexplored.
PUF (Pumilio and FBF) proteins are mRNA regulators with a conserved role in stem cell maintenance (2). These sequence-specific RNA-binding proteins are required to maintain Caenorhabditis elegans and Drosophila germline stem cells (GSCs) and planarian totipotent stem cells (neoblasts) (3 –5); they have also been implicated in mammalian stem cell maintenance (6, 7). In addition to their role in stem cell regulation, PUFs can also control embryonic polarity and neuronal functions (e.g., refs. 8 –11). PUF proteins bind regulatory elements, typically in the 3′ untranslated region (3′UTR) of target mRNAs (2); they are best known as repressors, but they can also activate target mRNAs (2, 10, 12, 13).
Putative PUF target mRNAs have been identified on a genomic scale in budding yeast, human HeLa cells, and fly ovaries and embryos (14 –17). In these studies, PUF proteins were immunoprecipitated with bound mRNAs, and those RNAs were then used to probe microarrays, an approach dubbed RIP-Chip. PUF proteins emerge from these studies as broad-spectrum regulators that likely control 7–11% of an organism’s genes. In addition, putative PUF targets often encode functionally related proteins (14 –17). However, these previous studies were not focused on stem cells per se, and therefore PUF target mRNAs in stem cells are largely unknown.
Among the 12 C. elegans PUF proteins, FBF-1 and FBF-2 (fem-3 binding factor) stand out as essential for GSC self-renewal in older larvae and adults (4). FBF-1 and FBF-2, collectively called FBF, are nearly identical and largely redundant (18, 19). GSCs are located where FBF is enriched, and GSC self-renewal fails in fbf-1(0) fbf-2(0) double mutants, which are sterile (20). In addition to its role in germline self-renewal, FBF promotes the oocyte fate (18) and acts in neurons to modulate olfactory adaptation (10). Other C. elegans PUF proteins control progression through meiotic prophase, gametogenesis, and early embryogenesis (21, 22). Therefore, FBF is specialized in the adult germ line for GSC maintenance and germline sex determination.
To date, nine FBF targets have been identified using a candidate approach (Fig. S1); these target mRNAs encode proteins that promote entry into the meiotic cell cycle (e.g., gld-1) (4, 23–25), promote differentiation (e.g., MPK-1, the C. elegans ERK/MAPK; ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase) (26 –28), and promote self-renewal (e.g., FBF-1 and FBF-2 themselves) (19). The FBF repression of differentiation-promoting mRNAs maintains GSCs in an undifferentiated state; its repression of self-renewal-promoting mRNAs may maintain a self-renewal mode that can be overcome to begin the path toward differentiation.
Here we report the identification of FBF target mRNAs on a genome-wide scale. Briefly, FBF-mRNA complexes were purified from adult worm extracts, and microarrays were used to identify FBF-associated mRNAs. We focused on the germline tissue using a tagged FBF under control of a germline promoter. We identified >1000 unique mRNAs bound to FBF, and these mRNAs were enriched for a variety of developmental regulators. Comparison of putative FBF targets with putative PUF targets in other metazoans revealed 40 common targets, including well-established stem cell and differentiation regulators. Given the conserved role of PUF proteins in stem cells, we propose that the shared PUF targets may reveal part of a broadly used regulatory module for stem cell maintenance.
Results
Identification of FBF-Associated mRNAs in C. elegans.
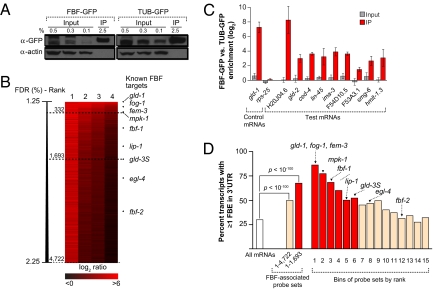
To identify FBF target mRNAs on a genome-wide scale, we purified FBF-mRNA complexes from worm extracts and probed microarrays with the associated mRNAs. For purification, we used an fbf-1-gfp transgene (henceforth, FBF-GFP) that contains the pie-1 promoter and the fbf-1 3′UTR, two regulatory sequences that together ensure appropriate germline expression (25, 26). Abundant FBF-GFP is present in the distal germ line, similar to endogenous FBF protein (4, 19, 25). FBF immunoprecipitations (IPs) were performed from extracts of synchronized adults, where fluorescent FBF-GFP was seen in the germ line but not in the soma. Importantly, FBF-GFP rescued fbf-1(0) fbf-2(0) double mutants to 100% fertility (n = 48). Therefore, FBF-GFP is a functional protein and likely associates with the same germline mRNAs as endogenous FBF. To control for RNAs that nonspecifically coimmunoprecipitate with GFP, TUBULIN-GFP (henceforth, TUB-GFP) driven by the pie-1 promoter was processed in parallel. We used an immobilized anti-GFP antibody to immunoprecipitate the GFP fusions from extracts (Fig. 1A); RNAs in the IPs were then linearly amplified, labeled, and hybridized to C. elegans Affymetrix GeneChips (Materials and Methods and SI Materials and Methods). Four biological replicates were carried out.
Fig. 1.
FBF RIP-Chip. (A) Western blot of FBF-GFP and TUB-GFP IPs, probed with antibodies against GFP or actin (control). (B) Summary of array results for four biological replicates. Columns are array replicates and rows are probe sets. The degree of enrichment (FBF/TUB) is shown on a black-to-red color scale. Probe sets are ordered from Top to Bottom based on their SAM score, with the highest-scoring probe set at the top. Approximate false discovery rates (FDRs) are shown on the left. Dashed lines show selected probe set groups; SAM ranks of validated FBF targets are on the right. (C) qPCR validation. Enrichment of mRNAs present in FBF-GFP compared to TUB-GFP, for input (gray bars) and IPs (red bars), is shown for three biological replicates. Error bars show standard deviation. Data were normalized to eft-3 and analyzed using the ΔΔCT method. SAM rankings for test mRNAs are: H20J04.6, 10; gld-2, 374; ced-4, 448; lin-45, 461; ima-3, 789; F54D10.5, 1004; F53A3.1, 1267; smg-6, 1479; hmit-1.3, 1807. (D) FBE analysis. Percentage of probe sets with ≥1 FBE in the corresponding mRNA 3′UTR is shown for all protein-coding genes (white), for two categories of FBF-associated probe sets (middle), and for 15 bins (4722 FBF-associated probe sets were placed in bins based on their ordered SAM rank, with each bin containing ∼314 probe sets). Bins with the top 1693 probe sets are indicated in red. P values were calculated using the hypergeometric distribution.
To identify mRNAs reproducibly enriched in the FBF-GFP IP (henceforth, FBF IP), we analyzed the array data using significance analysis of microarrays (SAM; Fig. 1B) (29). Briefly, SAM assigns a score to each probe set and estimates their false discovery rates (FDRs). SAM deemed 4722 probe sets as significantly enriched at an FDR of 2.25% or lower (Fig. 1B; see Dataset S1 for the complete list). We designated all 4722 probe sets “FBF-associated” and used downstream analyses to identify the most likely FBF targets (see below).
We used real-time quantitative PCR (qPCR) to validate the enrichment of FBF IP mRNAs with a range of SAM score rankings, from the tenth (H20J04.6) to 1807th-most enriched mRNA (hmit-1.3) (Fig. 1C). Indeed, both the positive control gld-1 as well as all test mRNAs were enriched in the FBF IP compared with the TUB IP, but the negative control rps-25 was not (Fig. 1C, red bars). The FBF IP enrichment was not due to differences in starting samples because the mRNAs were present at similar levels in both FBF and TUB extracts (input) (Fig. 1C, gray bars). We conclude that the array results represent enrichment of specific mRNAs in the FBF IP.
All previously identified FBF target mRNAs were among the 4722 FBF-associated probe sets, and most had highly ranked SAM scores (Fig. 1B). Four known FBF targets were in the top 332 probe sets, and all but two, fbf-2 and egl-4, were in the top 1693. The low fbf-2 rank is likely due to the FBF IP strain being homozygous for an fbf-2 nonsense mutation; nonsense mRNAs are often degraded by nonsense-mediated mRNA decay (30). Whereas egl-4 is known as an FBF target only in neurons (10), egl-4 mRNA is present in germ lines according to a database of C. elegans mRNA in situ hybridizations (NEXTDB; http://nematode.lab.nig.ac.jp). In addition, two candidate target mRNAs that FBF binds directly in vitro, fog-3 and lin-3 (31, 32), had SAM scores in the top 122 probe sets (Dataset S1). We conclude that all previously identified FBF targets were enriched in the FBF IP, and that most fell among the top 1693 probe sets.
Next, we asked whether FBF-associated mRNAs contained FBF binding elements (FBEs) in their 3′UTRs. The FBE consensus is UGUDHHAU, where D is A, U, or G and H is A, U, or C (33). Indeed, mRNAs containing FBEs were significantly enriched among FBF-associated probe sets: 49% of 4722 FBF-associated probe sets corresponded to an mRNA with ≥1 FBE in the 3′UTR, versus only 30% for all C. elegans genes (P < 10−100; Fig. 1D). To ask whether FBEs were preferentially associated with higher-scoring mRNAs, we divided our list into 15 bins based on their SAM score and determined the percentage of FBE-containing transcripts in each bin (Fig. 1D). The first bin had >85% FBE-containing mRNAs, and this percentage decreased in successive bins. The highly ranked probe sets were also highly enriched for mRNAs with multiple FBEs (Fig. S2). These percentages are likely an underestimate of those mRNAs with FBF binding sites, because PUFs can bind elements in mRNA-coding regions (34), which were excluded in our analysis, and because FBF can bind a distinct element with an unknown consensus (10).
The dramatic enrichment among the highest-scoring probe sets, both for known FBF target mRNAs and for FBE-containing mRNAs, suggested that the highest-scoring probe sets likely represent direct FBF targets. For further analyses, we used a cutoff at gld-3S, a known FBF target and the 1693rd top probe set. These 1693 probe sets collapse into 1350 unique, unambiguous genes, representing ∼7% of all C. elegans protein-coding genes. They contain seven of eight validated germline FBF targets (Fig. 1B) and a high percentage of FBE-containing transcripts (67%) (Fig. 1D). Although these 1350 mRNAs are unverified FBF targets, we refer to them as FBF target mRNAs for simplicity.
One caveat to the RIP-Chip approach is that RNA-binding proteins (RBPs) and mRNAs can reassort in vitro, and purified RBP-RNA complexes may not represent actual in vivo interactions (35). We reasoned that if reassortment had occurred, then the most abundant mRNAs, especially those with FBEs, would appear as FBF targets. To test this prediction, we used a serial analysis of gene expression (SAGE) database that provides the relative abundance of mRNAs in whole young adults (36) (Materials and Methods). Indeed, only 3 of the 20 most abundant FBE-containing mRNAs found in this database were identified in our experiments as FBF targets. Therefore, FBF targets were not enriched for the most abundant mRNAs, suggesting that significant reassortment of FBF-RNA complexes in vitro had not occurred.
Functional Annotation of FBF Targets: GO Analysis.
To explore functional themes among FBF targets, we searched for enriched gene ontology (GO) annotations, focusing on representative terms (Table 1; see Dataset S2 for all enriched GO terms). For biological processes, most enriched GO terms were related to development, cell cycle, or trafficking. Several coincided with FBF’s known role in controlling GSCs (e.g., germ cell development). Several also were related to embryonic development. One attractive idea is that FBF represses diverse developmental regulators to promote totipotency and maintain GSCs. For molecular functions, most enriched GO terms were related to protein-nucleic acid and protein-protein interactions. This enrichment suggests a role for FBF targets in regulation of other genes or complexes and that FBF is a “regulator of regulators,” as previously proposed for human PUM1 (17). For cellular compartments, enriched GO terms were broadly distributed.
Table 1.
GO terms enriched among FBF targets
| Category | Term | Count | P value |
| Biological process | GO:0009790∼embryonic development | 361 | 3.75 × 10−30 |
| GO:0000003∼reproduction | 229 | 2.47 × 10−10 | |
| GO:0030154∼cell differentiation | 65 | 4.80 × 10−8 | |
| GO:0007281∼germ cell development | 16 | 2.91 × 10−5 | |
| GO:0048513∼organ development | 89 | 3.21 × 10−5 | |
| GO:0051301∼cell division | 79 | 6.96 × 10−22 | |
| GO:0007049∼cell cycle | 79 | 2.53 × 10−21 | |
| GO:0000279∼M phase | 45 | 3.47 × 10−14 | |
| GO:0007126∼meiosis | 28 | 6.43 × 10−12 | |
| GO:0007067∼mitosis | 21 | 4.28 × 10−5 | |
| GO:0016043∼cellular component organization and biogenesis | 146 | 6.08 × 10−17 | |
| GO:0051641∼cellular localization | 71 | 2.07 × 10−11 | |
| GO:0006996∼organelle organization and biogenesis | 73 | 1.40 × 10−8 | |
| GO:0000226∼microtubule cytoskeleton organization and biogenesis | 26 | 1.09 × 10−6 | |
| GO:0016192∼vesicle-mediated transport | 29 | 9.28 × 10−6 | |
| GO:0046903∼secretion | 22 | 3.94 × 10−5 | |
| GO:0006259∼DNA metabolic process | 58 | 1.19 × 10−7 | |
| GO:0006974∼response to DNA damage stimulus | 31 | 9.73 × 10−7 | |
| GO:0006281∼DNA repair | 30 | 1.06 × 10−6 | |
| Cellular compartment | GO:0005622∼intracellular | 334 | 1.39 × 10−10 |
| GO:0005634∼nucleus | 159 | 1.86 × 10−6 | |
| GO:0043231∼intracellular membrane-bound organelle | 195 | 2.97 × 10−6 | |
| GO:0005694∼chromosome | 28 | 1.01 × 10−5 | |
| GO:0015630∼microtubule cytoskeleton | 23 | 6.81 × 10−5 | |
| Molecular function | GO:0005515∼protein binding | 329 | 4.29 × 10−31 |
| GO:0003676∼nucleic acid binding | 184 | 1.30 × 10−5 |
Functional Annotation of FBF Targets: Directed Analyses.
To identify FBF targets with specific roles in germline development, we sought targets with functional annotations related to meiosis and/or development. First, we searched WormBase (http://www.wormbase.org) for genes with GO annotations and/or RNAi phenotypes relevant to the meiotic cell cycle. Among 247 “meiosis” genes represented on the array, 84 (34%) were found among the FBF targets (Dataset S3); functions embraced multiple stages of the meiotic cell cycle, including meiotic entry (e.g., gld-2) (24), meiotic progression (e.g., prom-1) (37), chromosomal synapsis (e.g., him-3) (38), homologous recombination (e.g., rad-51) (39), and chromosome segregation (e.g., klp-16) (40). Thus, FBF appears to exert a multifaceted control of the meiotic program.
For developmental regulators, we focused first on Ras/MAPK-, Notch-, Wnt-, and Hedgehog-related signaling pathways. Each had at least four major components among FBF targets (Dataset S4). Most striking was the Ras/MAPK pathway (27, 28). All 58 known components of the C. elegans MAP kinase signaling pathway were represented on the array (28, 41, 42), and 15 (26%) were FBF targets, including three core components: lin-45/RAF, ksr-2, and the previously identified FBF target mpk-1/MAPK (26, 28).
Another key developmental process is programmed cell death. Of 28 core components of the programmed cell death pathway represented on the arrays (43), 9 (32%) were FBF targets (Dataset S4). These include egl-1 and ced-4, two genes required for cell death (44, 45).
We also found other critical developmental regulators among FBF targets, including regulators of sperm and oocyte fates (e.g., rnp-8) (46), small RNAs (e.g., prg-1/PIWI) (47), aging (e.g., daf-16/FOXO) (48), and the mitotic cell cycle (e.g., cdk-1) (49), which are all included in Dataset S4 and Dataset S5. Coupled with the broader gene ontology analyses (see above), these more directed searches suggest that FBF controls a broad network of key cellular and developmental regulators.
Shared PUF Targets from Diverse Organisms.
We considered the idea that common PUF target mRNAs might reflect ancient PUF functions. First, we demonstrated that FBF targets were enriched for conserved genes (Fig. 2A, upper pie charts). Whereas only ∼32% of all C. elegans protein-coding genes were conserved in the human genome, ∼56% of FBF targets were conserved (P < 10−50; Fig. 2A, upper pie charts). We then compared the lists of putative targets for nematode FBF, Drosophila Pumilio, and both human PUM1 and PUM2 (15 –17). Numerous mRNAs were shared (Dataset S6), and the overlap between FBF and human PUM target datasets was significant. Fig. 2A (lower pie charts) compares FBF and one dataset of human PUM1 targets (16): among all C. elegans genes conserved in humans, ∼16% were orthologous to a PUM1 target, but among all FBF targets conserved in humans, ∼23% were orthologous to a PUM1 target. A similar enrichment was consistent for all three human PUM datasets. Others have reported a significant overlap between Drosophila and human PUF targets (P < 0.04) (17). Our data extend this result to nematode FBF and human PUMs, and suggest that certain PUF target mRNAs have been broadly conserved.
Fig. 2.
Conserved PUF targets and model. (A) Top pie charts show the proportion of all C. elegans genes (Left) or all FBF targets (Right) with human orthologs; bottom pie charts show subsets of conserved genes orthologous to a human PUM1 target from one data set (16); the FBF targets are significantly enriched compared with all C. elegans human orthologs (asterisk). P value was calculated using the hypergeometric distribution. (B) FBF is a broad-spectrum regulator that controls stem cell totipotency. FBF is required for germline stem cells and likely controls ∼7% of the genome. Its best-known role is as a repressor, which is shown here, but we note that FBF can also activate mRNAs (see text). We suggest that FBF modulates the expression of many developmentally critical genes and that this action may provide a molecular solution for totipotency.
At least 40 PUF targets are shared among C. elegans, Drosophila, and humans (summary in Table 2 and complete list in Table S1). These shared PUF targets include cell-signaling components, endocytic regulators, and cyclins. The signaling pathways found among the 40 common PUF targets (Ras/MAPK, PI3K/Akt, NF-κB, and Notch) are all critical for stem cell regulation (50). We suggest that the 40 common PUF targets represent part of a broadly conserved regulatory module for stem cell control (Discussion). Not included among the list of 40 are the fbf and pum mRNAs themselves. Yet FBF controls fbf-1 and fbf-2 (19), and the pumilio and pum mRNAs IP with fly and human PUF proteins, respectively (15 –17). This omission reflects the stringency of the program used to identify orthologs, and shows that 40 is likely an underestimate of common PUF targets.
Table 2.
Shared PUF target mRNAs in humans, C. elegans, and Drosophila
| Human | C. elegans | Drosophila | Protein description |
| ARAF*† | lin-45 | phl‡ | MAP kinase kinase kinase/RAF (MAPK pathway) |
| ECT2§ | ect-2 | pbl‡ | RhoGEF |
| RKTG*§ | Y67A10A.8 | CG7530‡¶ | Haemolysin-III-related protein/RAF inhibitor |
| PDK1§ | pdk-1 | Pk61C‡ | 3-phosphoinositide-dependent protein kinase (PI3/Akt pathway) |
| FOXO3*§ | daf-16 | foxo‡ | Forkhead/HNF3 transcription factor (PI3/Akt pathway) |
| NFKBIA*†§ | C33A11.1 | cact‡ | NF-κB transcription factor inhibitor (NF-κB pathway) |
| CSL*§ | lag-1 | Su(H)‡ | CSL transcription factor (Notch pathway) |
| LMBR1*§ | R05D3.2 | CG5807‡ | Lipocalin transmembrane receptor family |
| PARD3§/B* | par-3 | baz‡ | Atypical PKC-interacting protein |
| CCNT2* | cit-1.1/1.2 | CycT‡ | Cyclin T |
| CCNB1§/2*§ | cyb-1/2.1/2.2 | CycB‡ | Cyclin B |
| ITSN2* | itsn-1 | Dap160‡ | Intersectin |
| SMAP1* | W09D10.1 | CG8243‡ | ARF GTPase-activating protein |
| AP1S1*† | aps-1 | AP-1σ‡ | Clathrin adapter complex, σ1 subunit |
| RAB5B*†§ | rab-5 | Rab5‡ | Rab GTPase |
| SLC25A40*† | C16C10.1 | Tyler‡ | Mitochondrial solute carrier |
| SLC37A3§ | T10C6.6 | CG10069‡ | Major facilitator superfamily solute carrier |
| LARP5§ | larp-5 | CG11505‡ | Metazoan-specific La protein |
| RBM7* | Y37D8A.21 | CG11454‡ | RRM-containing RNA-binding protein |
| HNRNPA3† | H28G03.1 | Hrb98DE‡ | hnRNA-binding protein |
| TIS11*§ | oma-1/2 | Tis11‡ | CCCH zinc finger protein |
| ZCCHC11§ | pup-2 | CG11418‡ | Nucleotidyltransferase proteins |
| EIF4E*†§ | ife-5 | eIF-4E‡ | Translation initiation factor 4E |
| RBM25*†§ | W04D2.6 | CG4119‡ | snRNP complex protein |
| MBTD1* | lin-61 | Sfmbt‡ | Polycomb group protein |
| HMG20A§ | W02D9.3 | CG9418¶ | High-mobility group DNA-binding protein |
| EPC2† | epc-1 | E(Pc)‡ | Enhancer of Polycomb family |
| CYP3A4† | cyp-13B1 | Cyp6a19‡¶ | Cytochrome P450 |
| OSBPL9*§/10*§ | obr-4 | CG1513‡ | Oxysterol-binding protein |
| ZDHHC6* | M18.8 | CG5196‡ | DHHC zinc finger, putative palmitoyltransferase |
| MAPRE3* | ebp-2 | Eb1‡ | Microtubule-binding protein |
| CHPF* | mig-22 | CG4351‡ | Chondroitin N-acetylgalactosaminyltransferase |
| WIPI2* | atg-18 | CG8678‡ | Autophagy protein |
Discussion
FBF Is a Broad-Spectrum Gene Regulator.
This work identifies 1,350 likely direct targets of C. elegans FBF, an RNA-binding protein with a conserved role in stem cell maintenance (see the Introduction). We reason that most are bona fide FBF target mRNAs, because they include previously validated FBF targets and are strongly enriched for 3′UTRs with consensus FBEs. Moreover, the 1350 are not enriched for the most abundant FBE-containing mRNAs.
The FBF target mRNAs represent ∼7% of the C. elegans protein-coding genes, and PUF proteins in humans and Drosophila likely control a similar proportion (7–11%) of their respective transcriptomes (15 –17). Such broad-spectrum gene regulators are a common mechanism of stem cell control. For example, the transcription factors Oct4, Sox2, and Nanog control ES pluripotency and individually occupy the promoters of 3–9% of all protein-coding genes (1, 51). The Polycomb group (PcG) chromatin factors are also important for ES pluripotency, and PcG subunit SUZ12 localizes to ∼8% of all genes in ES cells (52 –55). An appealing idea is that broad-spectrum DNA and RNA regulators complement each other to ensure that stem cells are maintained in an undifferentiated state. However, this idea awaits the genomic analysis of DNA and RNA regulators in the same type of stem cells.
Implications of FBF Targets for a GSC Program.
FBF target mRNAs are enriched for regulators of developmental and cellular pathways, including the meiotic cell cycle, intercellular signaling, programmed cell death, and intracellular trafficking. We suggest that these functions require tight control in totipotent or pluripotent stem cells (Fig. 2B). Interestingly, these broad functions only partially overlap with those of putative targets for key ES cell transcriptional regulators. For example, putative SUZ12 targets, like FBF targets, are enriched for developmental regulators, but, unlike FBF targets, they are not enriched for trafficking proteins (55). PUF targets in humans and Drosophila are enriched for genes related to trafficking as well (15 –17). Therefore, PUF targets may include stem cell factors previously overlooked in analyses of transcriptional regulators.
Control of meiosis-promoting genes is likely to be key for GSC maintenance. Previous work showed that FBF represses two key regulators of entry into the meiotic cell cycle (gld-1 and gld-3) (4, 56, 57). Here we find that FBF also likely controls an additional key regulator of meiotic entry (gld-2) plus numerous components of the machinery driving meiosis-specific events (e.g., homologous recombination). These additional targets confirm the role of FBF as a key regulator of meiotic entry and extend its role to the meiotic program more broadly.
How are the 1350 mRNAs controlled? FBF can repress (e.g., gld-1) or activate (e.g., egl-4) target mRNA expression (e.g., refs. 4, 10). Therefore, conclusions about how FBF controls specific mRNAs must be anchored in genetic and biochemical analyses of their expression. Nonetheless, the simple view that FBF lowers expression is true for most germline mRNAs analyzed to date and is likely to be the primary role for FBF in stem cell control (20). A key task for the future is to find which FBF targets among the 1350 affect stem cell maintenance and how the modes of FBF activation or repression feed into stem cell control.
Common PUF Targets May Reflect Conserved Stem Cell Control.
C. elegans, Drosophila, and human putative PUF targets include 40 common mRNAs (Table 2). Previous studies reported an overlap between PUF targets in Drosophila and humans, but downplayed its significance, probably because the overlap was not dramatic (16, 17). Here we argue that these shared targets are likely important, because the overlap is significant and consistent between independent datasets.
The first indication that conserved PUF targets may be important for stem cell control emerged from studies of ERK/MAPK homologs in C. elegans and human ES cells (26). Our analysis reveals additional conserved PUF target mRNAs that encode proteins affecting stem cells, including components of Ras/MAP kinase, NF-κB, Notch, and PI3/Akt signaling pathways. MAPK signaling inhibits self-renewal of mouse ES cells and promotes differentiation (58), and NF-κB signaling seems to have a similar function (59). Notch signaling controls cell-fate choices in human ES cells (60) and promotes survival of neural stem cells (61, 62). PI3/Akt signaling has been linked to both ES cell and hematopoietic stem cell controls (63, 64). Indeed, the PUF control of Ras/MAPK signaling to maintain cells in an undifferentiated state is particularly well conserved: Yeast PUF5, also known as Mtp5, represses the Ras/MAPK signaling component Ste7/MAPKK to inhibit filamentous differentiation (65). We propose that PUF proteins and their shared target pathways represent a broadly conserved gene regulatory module that is central to maintenance of cells in an undifferentiated state.
Perspectives.
Understanding stem cell controls is integral to the development of stem-cell-based therapies. Here we identify likely mRNA targets of the conserved stem cell regulator FBF, some of which are shared with PUF proteins in other organisms. Because PUF proteins function posttranscriptionally, FBF targets could contain key stem cell genes overlooked by analyses of transcriptional regulators. Therefore, this work may serve as a database for potential regulators that are broadly important for stem cell control.
Materials and Methods
Nematode Strains.
Strains JK4091, fbf-1(ok91) fbf-2(q738); axIs1459[unc-119(+) Ppie-1::gfp::fbf-1+3′UTR] (26), and AZ244, unc-119(ed3); ruIs57[unc-119(+) Ppie-1::gfp::tubulin] (66), were maintained at 20 °C.
Immunoprecipitations.
IPs were done as described (15), with minor modifications to accommodate C. elegans and our specific reagents (SI Materials and Methods).
Western Analysis.
Western blots were probed with either mouse primary antibodies against GFP (JL-8; Clontech; 1:1,000 dilution) or actin (C4; MP Biomedicals; 1:40,000 dilution) followed by donkey HRP-conjugated anti-mouse secondary antibodies (Jackson ImmunoResearch; 1:40,000 dilution).
Microarrays and Statistical Analyses.
Microarrays were carried out by the University of Wisconsin–Madison Gene Expression Center. IP RNA was linearly amplified, labeled, and used to probe C. elegans Affymetrix GeneChips following standard procedures (SI Materials and Methods). Array data were extracted, normalized, and analyzed by standard procedures (SI Materials and Methods).
Real-Time Quantitative PCR.
qPCR was carried out by standard procedures (SI Materials and Methods).
Bioinformatics.
C. elegans 3′UTR sequences and ortholog assignments were obtained from BioMart (http://www.biomart.org). SAGE data were obtained from the Genome Sciences Centre C. elegans Gene Expression Consortium (http://elegans.bcgsc.bc.ca). Gene ontology (GO) analysis was carried out using the DAVID Bioinformatics Database (http://david.abcc.ncifcrf.gov) (67, 68). See SI Materials and Methods for database release information.
Supplementary Material
Acknowledgments
We thank André Gerber for helpful comments and discussions about RIP protocols and experimental setup and Audrey Gasch and Michael Newton for questions regarding data analysis. We thank the Kimble laboratory for helpful comments and insights during the course of this work and Anne Helsley-Marchbanks for assistance with manuscript preparation. National Institutes of Health Grant GM069454 supported this work. J.K. is an investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data have been deposited in the Gene Expression Omnibus database, http://www.ncbi.nlm.nih.gov/geo, accession #GSE19922.
This article contains supporting information online at www.pnas.org/cgi/content/full/1000495107/DCSupplemental.
References
- 1.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 3.Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 4.Crittenden SL, et al. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans . Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 5.Salvetti A, et al. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development. 2005;132:1863–1874. doi: 10.1242/dev.01785. [DOI] [PubMed] [Google Scholar]
- 6.Moore FL, et al. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc Natl Acad Sci USA. 2003;100:538–543. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu EY, Chang R, Salmon NA, Reijo Pera RA. A gene trap mutation of a murine homolog of the Drosophila stem cell factor Pumilio results in smaller testes but does not affect litter size or fertility. Mol Reprod Dev. 2007;74:912–921. doi: 10.1002/mrd.20687. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann R, Nüsslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- 9.Schweers BA, Walters KJ, Stern M. The Drosophila melanogaster translational repressor pumilio regulates neuronal excitability. Genetics. 2002;161:1177–1185. doi: 10.1093/genetics/161.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaye JA, Rose NC, Goldsworthy B, Goga A, L’Etoile ND. A 3′UTR Pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron. 2009;61:57–70. doi: 10.1016/j.neuron.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubnau J, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 12.Archer SK, Luu VD, de Queiroz RA, Brems S, Clayton C. Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle. PLoS Pathog. 2009;5:e1000565. doi: 10.1371/journal.ppat.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suh N, et al. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics. 2009;181:1249–1260. doi: 10.1534/genetics.108.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster . Proc Natl Acad Sci USA. 2006;103:4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galgano A, et al. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris AR, Mukherjee N, Keene JD. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol Cell Biol. 2008;28:4093–4103. doi: 10.1128/MCB.00155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, et al. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 19.Lamont LB, Crittenden SL, Bernstein D, Wickens M, Kimble J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev Cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 20.Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans . Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- 21.Lublin AL, Evans TC. The RNA-binding proteins PUF-5, PUF-6, and PUF-7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis. Dev Biol. 2007;303:635–649. doi: 10.1016/j.ydbio.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Noble SL, Allen BL, Goh LK, Nordick K, Evans TC. Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J Cell Biol. 2008;182:559–572. doi: 10.1083/jcb.200802128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis R, Barton MK, Kimble J, Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans . Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadyk LC, Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans . Development. 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- 25.Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee M-H, et al. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007;3:e233. doi: 10.1371/journal.pgen.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee M-H, et al. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics. 2007;177:2039–2062. doi: 10.1534/genetics.107.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sundaram MV. RTK/Ras/MAPK signaling. WormBook. February 11, 2006 10.1895/wormbook.1.80.1. [Google Scholar]
- 29.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 31.Thompson BE, et al. Dose-dependent control of proliferation and sperm specification by FOG-1/CPEB. Development. 2005;132:3471–3481. doi: 10.1242/dev.01921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson BE, Lamont LB, Kimble J. Germ-line induction of the Caenorhabditis elegans vulva. Proc Natl Acad Sci USA. 2006;103:620–625. doi: 10.1073/pnas.0510264103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muraro NI, et al. Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J Neurosci. 2008;28:2099–2109. doi: 10.1523/JNEUROSCI.5092-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKay RM, McKay JP, Avery L, Graff JM. C. elegans: A model for exploring the genetics of fat storage. Dev Cell. 2003;4:131–142. doi: 10.1016/s1534-5807(02)00411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jantsch V, et al. Caenorhabditis elegans prom-1 is required for meiotic prophase progression and homologous chromosome pairing. Mol Biol Cell. 2007;18:4911–4920. doi: 10.1091/mbc.E07-03-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zetka MC, Kawasaki I, Strome S, Müller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takanami T, et al. Characterization of a Caenorhabditis elegans recA-like gene Ce-rdh-1 involved in meiotic recombination. DNA Res. 1998;5:373–377. doi: 10.1093/dnares/5.6.373. [DOI] [PubMed] [Google Scholar]
- 40.Colaiácovo MP, et al. A targeted RNAi screen for genes involved in chromosome morphogenesis and nuclear organization in the Caenorhabditis elegans germline. Genetics. 2002;162:113–128. doi: 10.1093/genetics/162.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Canevascini S, Marti M, Fröhli E, Hajnal A. The Caenorhabditis elegans homologue of the proto-oncogene ect-2 positively regulates RAS signalling during vulval development. EMBO Rep. 2005;6:1169–1175. doi: 10.1038/sj.embor.7400574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kritikou EA, et al. C. elegans GLA-3 is a novel component of the MAP kinase MPK-1 signaling pathway required for germ cell survival. Genes Dev. 2006;20:2279–2292. doi: 10.1101/gad.384506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conradt B, Xue D. Programmed cell death. WormBook. October 6, 2005 doi: 10.1895/wormbook.1.32.1. 10.1895/wormbook.1.32.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans . Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 45.Conradt B, Horvitz HR. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 46.Kim KW, et al. Antagonism between GLD-2 binding partners controls gamete sex. Dev Cell. 2009;16:723–733. doi: 10.1016/j.devcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 48.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 49.van den Heuvel S. Cell-cycle regulation. WormBook. September 21, 2005 doi: 10.1895/wormbook.1.28.1. 10.1895/wormbook.1.28.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dreesen O, Brivanlou AH. Signaling pathways in cancer and embryonic stem cells. Stem Cell Rev. 2007;3:7–17. doi: 10.1007/s12015-007-0004-8. [DOI] [PubMed] [Google Scholar]
- 51.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The Polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 54.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 55.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eckmann CR, Crittenden SL, Suh N, Kimble J. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans . Genetics. 2004;168:147–160. doi: 10.1534/genetics.104.029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hansen D, Wilson-Berry L, Dang T, Schedl T. Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development. 2004;131:93–104. doi: 10.1242/dev.00916. [DOI] [PubMed] [Google Scholar]
- 58.Burdon T, Stracey C, Chambers I, Nichols J, Smith A. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev Biol. 1999;210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- 59.Torres J, Watt FM. Nanog maintains pluripotency of mouse embryonic stem cells by inhibiting NFκB and cooperating with Stat3. Nat Cell Biol. 2008;10:194–201. doi: 10.1038/ncb1680. [DOI] [PubMed] [Google Scholar]
- 60.Yu X, et al. Notch signaling activation in human embryonic stem cells is required for embryonic, but not trophoblastic, lineage commitment. Cell Stem Cell. 2008;2:461–471. doi: 10.1016/j.stem.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Androutsellis-Theotokis A, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 62.Woo SM, et al. Notch signaling is required for maintaining stem-cell features of neuroprogenitor cells derived from human embryonic stem cells. BMC Neurosci. 2009;10:97. doi: 10.1186/1471-2202-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paling NR, Wheadon H, Bone HK, Welham MJ. Regulation of embryonic stem cell self-renewal by phosphoinositide 3-kinase-dependent signaling. J Biol Chem. 2004;279:48063–48070. doi: 10.1074/jbc.M406467200. [DOI] [PubMed] [Google Scholar]
- 64.Miyamoto K, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 65.Prinz S, Aldridge C, Ramsey SA, Taylor RJ, Galitski T. Control of signaling in a MAP-kinase pathway by an RNA-binding protein. PLoS One. 2007;2:e249. doi: 10.1371/journal.pone.0000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans . Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 68.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.