Abstract
CTCF sites are abundant in the genomes of diverse species but their function is enigmatic. We used chromosome conformation capture to determine long-range interactions among CTCF/cohesin sites over 2 Mb on human chromosome 11 encompassing the β-globin locus and flanking olfactory receptor genes. Although CTCF occupies these sites in both erythroid K562 cells and fibroblast 293T cells, the long-range interaction frequencies among the sites are highly cell type specific, revealing a more densely clustered organization in the absence of globin gene activity. Both CTCF and cohesins are required for the cell-type-specific chromatin conformation. Furthermore, loss of the organizational loops in K562 cells through reduction of CTCF with shRNA results in acquisition of repressive histone marks in the globin locus and reduces globin gene expression whereas silent flanking olfactory receptor genes are unaffected. These results support a genome-wide role for CTCF/cohesin sites through loop formation that both influences transcription and contributes to cell-type-specific chromatin organization and function.
Keywords: chromatin loops, insulator, CTCF, globin, transcription
Eukaryotic chromatin is dynamically organized to form distinct transcriptionally active or silent domains (1). Chromatin insulators are proposed to play a role in the establishment of such domains within which proper enhancer–gene interactions occur and improper ones are excluded (2). Insulators can function as boundary elements between active and silent chromatin domains and can also interfere with enhancer–gene interaction when placed between them. Evidence also suggests that insulators are involved in higher-order chromatin organization by clustering together with other insulators. Such “insulator bodies” can be visualized at the nuclear periphery of certain cells in Drosophila and are thought to coalesce through the interaction of proteins such as Su(Hw) and Drosophila CTCF (3, 4).
In mammalian cells, insulators are bound by CTCF, a protein with 11 zinc fingers through which it can bind to a range of DNA sequences, to itself, and to nuclear structural proteins such as nucleophosmin and matrix attachment regions (MARs) (2). Although insulators would be expected to reside primarily in intergenic regions where they could provide boundary or enhancer blocking activity, genome scans of CTCF enrichment show that CTCF sites are overrepresented in genes and promoter regions, with only 41–56% at intergenic locations (5). In Drosophila, between about 49% and 64% of insulator sites are located in intergenic regions (4). Thus, although CTCF sites are proposed to be insulators, their in vivo functions may not be limited to insulation. Recently, cohesin has been reported to colocalize at most sites of CTCF enrichment and to play a role in transcriptional insulation, but the significance of this joint occupancy to higher-order chromatin structures is unknown (6).
In the Igf2/H19 locus, CTCF binding to the imprinting control region on the maternal allele is required to loop out the Igf2 gene and prevent interaction between Igf2 and enhancers (7, 8). Similarly, CTCF binding and chromatin organization is necessary for activation of IFNG during T-cell differentiation (9). However, in the mouse β-globin locus, CTCF binding at the flanking HS5 and 3′HS1 sites results in loop formation, but the loop does not appear to be required for normal globin gene regulation in erythroid progenitor cells (10). These functions of CTCF all involve CTCF-mediated long-range chromatin organization, which can be manifest intra- or interchromosomally (11, 12).
In a β-globin transgenic mouse model, we showed that CTCF-binding HS5 has intrinsic, portable enhancer blocking activity (13). When an extra copy of HS5 was placed between the locus control region (LCR) and downstream genes, it formed a new insulator loop with endogenous HS5 that topologically isolated the LCR and nullified its activity. Here we show that CTCF/cohesin sites over a 2-Mb region of human chromosome 11, primarily among silent odorant receptor genes up- and downstream of the β-globin locus, participate in numerous interactions that form loops. CTCF enrichment is maintained in K562 cells and nonerythroid 293T cells; however, the interactions between and among the sites are highly cell type specific and require both CTCF and cohesin. Furthermore, general loss of CTCF site interactions in a human erythroid background results in diminished globin gene transcription coincident with an increase in H3 lysine 9 di-methylation in the locus. The results suggest that CTCF/cohesin site long-range interactions function generally in chromosome organization, occurring not only at sites of insulator function.
Results and Discussion
Numerous Sites of CTCF Enrichment Reside Among Silent Olfactory Receptor Genes Surrounding the Human β-Globin Locus.
Although several reports have documented sites of CTCF enrichment genome-wide, whether these sites participate generally in chromatin organization through mutual interactions is unknown. To explore this issue, we selected a 2-Mb test region on chromosome 11 surrounding the β-globin locus that is engaged in active transcription in erythroid K562 cells but not in fibroblast 293T cells. In both cell types, the clusters of olfactory receptor genes up- and downstream of the β-globin locus are silent (14).
To accurately predict sites of CTCF interaction, we carried out CTCF chromatin immunoprecipitation (ChIP)-chip analysis for K562 cells and 293T cells using a NimbleGen tiling array of nonrepetitive sequences on human chromosomes 10 and 11. We then analyzed the data using the ACME algorithm (15) with parameters chosen to maximize peak designation to a “training set” of CTCF sites confirmed by ChIP assay (Fig. 1A and Fig. S1). Importantly, the optimized ACME parameters identified peaks at the locus flanking β-globin HS5 and 3′HS1 sites where numerous reports have documented CTCF binding in vivo (10, 13, 16); these sites had not been consistently identified as peaks in earlier genome scans (17, 18). In other respects, such as the degree of overlap in CTCF sites between cell types and the CTCF motif shared by the sites, there was a high level of agreement between this analysis and the earlier studies (Fig. S2).
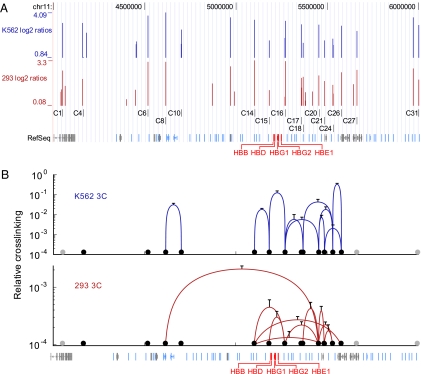
Fig. 1.
CTCF occupancy and chromatin loop formation over 2 Mb of chromosome 11. (A) CTCF localization on chr11:4,000,000–6,000,000 in K562 and 293T cells derived from ChIP-chip experiments. CTCF peaks were predicted using the ACME algorithm optimized to the training set (C1–C31) of sites validated by ChIP. RefSeq genes are depicted at the bottom in red (globin genes), blue (odorant receptor genes), or gray (other genes). (B) Long-range interactions among CTCF/cohesin sites vary between cell types. (Upper) 3C was carried out with K562 cell chromatin. Specific interactions between and among CTCF sites (black circles; gray circles represent CTCF sites not analyzed by 3C) are depicted by blue lines with the height of the curve corresponding to the cross-linking frequency. (Lower) Interactions among CTCF sites in 293T cells are depicted by red curves.Cross-linking is plotted relative to the signal for two fragments in the α-tubulin gene. Note the log scale of the y axis. Error bars represent SD.
We found that the cohesin complex members Rad21, SMC1, and SMC3 were enriched at all of the CTCF training sites, which is consistent with reports that cohesin colocalizes with CTCF at more than 80% of sites genome-wide (19 –21) (Fig. S3). These results validate numerous CTCF/cohesin sites at intervals between 30 and 400 kb flanking the β-globin locus and within the silent olfactory receptor gene clusters up- and downstream on human chromosome 11.
Cell Type–Specific Interactions Among CTCF Sites Are Characteristic Across an Extensive Chromosomal Region.
A crucial step in understanding the chromatin organizational potential of CTCF binding sites is to determine interaction among them. Thus, we carried out chromosome conformation capture (3C) analysis. Primers for each EcoRI fragment with a CTCF enrichment site were designed, and each was used as an anchor primer in tests of proximity in chromatin with every other quantitative PCR (qPCR)-validated CTCF site.
Of all possible (78) combinations, 10 interactions between CTCF sites were confirmed in K562 cells, including the known interaction between β-globin HS5 and 3′HS1 (Fig. 1B, Upper). Six of the 10 interactions occurred between adjacent sites, with 3 clusters of 4 sites. The average distance between interacting sites was about 70 kb with the largest interval being 185 kb. No interactions were detected for the sites most distant from the globin locus; however, these sites might interact with sites more 5′ or 3′ that were not analyzed. To confirm the specificity of the contacts, a primer for each interacting site was paired with one from the non-CTCF fragment adjacent to each of its partner CTCF sites (Fig. S4). In each case, reduced interaction frequencies (18% or less compared to partner CTCF frequencies) between CTCF sites and non-CTCF sites were observed.
ChIP experiments confirmed that the CTCF sites occupied in K562 cells were also occupied in 293T cells (Fig. S3). To test whether the interactions between CTCF sites are also conserved, 3C was carried out on 293T cells. Fig. 1B, Lower, shows that 10 interactions were confirmed of which 4 were conserved with K562 cells, including the interaction between β-globin HS5 and 3′HS1, and 6 were specific for 293T cells. The average interval between these sites was 190 kb, which is considerably greater than in K562 cells. Five interactions were over even greater distances, up to nearly 1 Mb. There were four clusters where multiple CTCF sites came together, including three with four contacts and one with six contacts, whereas only four interactions were with nearest neighbors. Thus, clustered and longer-range interactions of CTCF/cohesin sites predominated in 293T cells, where the β-globin locus is inactive, perhaps reflecting a more condensed chromatin milieu. This clustering is reminiscent of the insulator bodies described in Drosophila cells and might facilitate recruitment of the locus to a nuclear territory inhospitable to transcription (22).
These data show that CTCF sites have the capacity to interact with each other over a wide genomic area in both paired and clustered arrangements. Most of these sites would not be expected to be functional insulators as they reside among silent odorant receptor genes where they do not mark boundaries between regions of differing gene activity (see below). It is important to note that the interactions detected by 3C are a composite of those occurring in a large population of cells; in individual cells, a subpopulation of the interactions may occur, as well as others that we did not query (23, 24). We suggest that CTCF site interaction is a common phenomenon in the genome that exists in both active and repressed gene neighborhoods.
Of the interactions that we observed, 60% were erythroid cell type specific. A chromosome conformation capture-on-chip (4C) study revealed that the β-globin locus interacts over very long distances with a different set of loci on mouse chromosome 7 in an erythroid versus a nonerythroid background (24); this study was unable to resolve looping interactions within 10 Mb of the globin locus itself, and the factors that participate in the interactions are unknown. The association of CTCF with boundaries of active and repressive genomic regions was also found to be cell type specific (25). How the specificity of CTCF-dependent chromatin loops is established is not known but might involve cell-type-specific proteins that support more frequent or more strongly stabilized higher-order chromatin interactions or might relate to lineage-specific recruitment of cohesin (26).
CTCF Is Required for Long-Range Interaction Among Its Binding Sites and for Enrichment of Cohesin at These Sites.
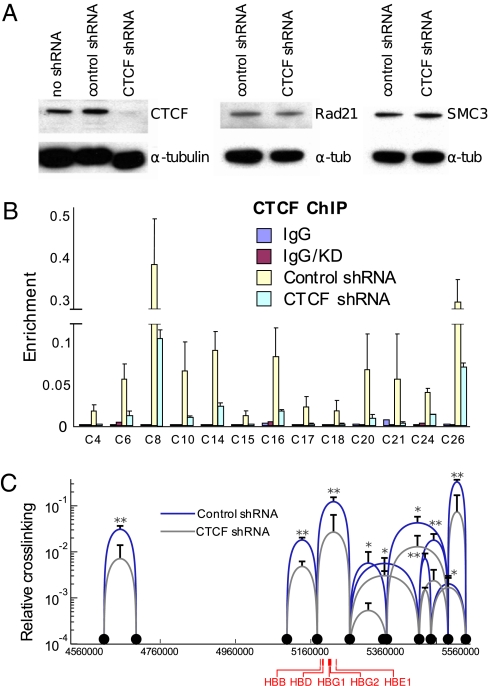
To ascertain the dependence of the long-range interactions on CTCF protein, lentivirus-mediated shRNA knock-down of CTCF was conducted in K562 cells. Western blots showed that CTCF protein was very significantly reduced compared to nontransduced cells or cells transduced with a control shRNA whereas the levels of Rad 21 and SMC3 were unaffected (Fig. 2A). ChIP indicated that enrichment of CTCF was reduced fivefold on average compared to K562 cells infected with control virus (Fig. 2B). Long-range interaction frequencies observed in 3C experiments in CTCF knock-down cells (Fig. 2C, gray curves) were significantly reduced (5–10-fold, although at 2 sites the reduction was only 2-fold) compared to those in control cells (blue curves), confirming the importance of CTCF protein to the interactions.
Fig. 2.
Reduction of CTCF results in loss of long-range interactions between and among its binding sites. (A) Western blot indicating reduction of CTCF protein by shRNA without alteration of Rad21 or SMC3 levels. (B) K562 cells were transduced with control or CTCF shRNA. ChIP was performed with an antibody to CTCF. Error bars represent SEM. (C) 3C was carried out using chromatin from K562 cells transduced with control or CTCF shRNA. Interactions between and among CTCF sites are indicated by blue curves, and reduction of these interactions after knock-down of CTCF is indicated by gray curves. Error bars represent SD. **P < 0.01, *P < 0.05.
ChIP was next carried out on CTCF knock-down K562 cells with antibodies against cohesin. Although the expression levels of Rad21 and SMC3 were unaffected by CTCF reduction (Fig. 2A), ChIP showed that enrichment of these proteins at CTCF sites decreased on average fivefold, similar to the CTCF reduction, compared to control K562 cells (Fig. S5). Thus, the presence of CTCF is required for cohesin binding to the sites that we studied, consistent with earlier work indicating that CTCF is required to recruit cohesin complex members to shared sites (19 –21). The direct interaction of CTCF with cohesin subunit SCC3 may underlie such recruitment (27).
Cohesin Can Influence Stable CTCF Binding, and Its Reduction Results in Loss of Long-Range Interactions Among CTCF Sites.
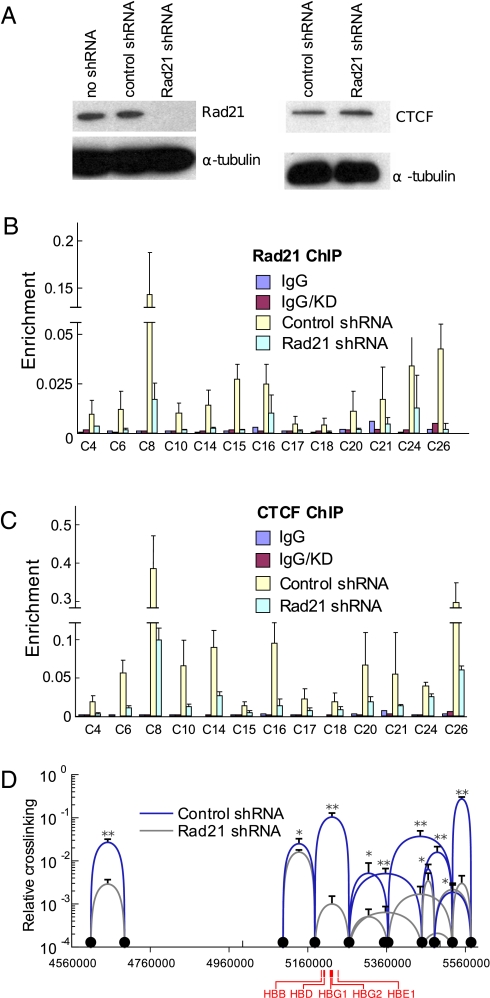
Some studies reported that knock-down of Rad21 had no effect on CTCF enrichment, whereas others reported that Rad21 knock-down reduced CTCF enrichment about 50% at some sites (19, 20). To shed light on this issue, Rad21 was knocked down with lentivirus shRNA in K562 cells. Western blotting showed that the expression of Rad21 was nearly abolished whereas the protein level of CTCF was not affected (Fig. 3A). ChIP experiments showed that the enrichment of Rad21 at CTCF sites was significantly reduced (average of fivefold) compared to that in control K562 cells (Fig. 3B). Significantly, enrichment of CTCF was also reduced (average of threefold) in K562 cells subjected to Rad21 knock-down compared to K562 cells (Fig. 3C), which suggests that Rad21 contributes to stable CTCF binding at sites surrounding the β-globin locus.
Fig. 3.
Reduction of cohesin components diminishes long-range interactions among CTCF sites. (A) Western blots showing reduction of Rad21 by shRNA and lack of effect of this reduction on CTCF protein. (B) K562 cells were transduced with control or Rad21 shRNA. ChIP was performed with an antibody to Rad21. Error bars represent SEM. (C) ChIP was carried out as described in B with antibodies to CTCF. (D) 3C was carried out using chromatin from K562 cells transduced with control or Rad21 shRNA. Interactions between and among CTCF sites are indicated by blue curves, and reduction of interactions after knock-down of Rad21 is indicated by gray curves. Error bars represent SEM. **P < 0.01, *P < 0.05.
Because reduction of Rad21 leads to lower CTCF enrichment at the shared sites, the interaction frequencies between CTCF sites would be expected to be reduced because of the requirement for CTCF. To examine this prediction, 3C was carried out on Rad21 knock-down cells. As shown in Fig. 3D, the interaction frequencies of CTCF sites in K562 cells with reduced Rad21 (gray curves) were significantly lower (average reduction of threefold) than those in control cells (blue curves): CTCF site interactions were also reduced in SMC3 knock-down cells (Fig. S6). No changes in cell cycle distribution, growth rate, or apoptosis were observed for cells treated with cohesin (or CTCF) shRNAs (Fig. S7). Thus, our data suggest the importance of both cohesin and CTCF to long-range interactions between and among CTCF sites. In contrast, two CTCF/cohesin site looping interactions at the IFNG cytokine locus were reduced after Rad21 protein reduction even though CTCF remained bound (9).
CTCF Reduction Alters the Epigenetic Environment Within the β-Globin Locus and Negatively Affects γ-Globin Expression.
Recent studies in which one or a few specific CTCF sites were lost due to deletion or mutation have revealed both positive and negative effects on transcription of local genes (8, 21, 28, 29). However, deletion of the mouse β-globin HS5 and 3′HS1 CTCF sites individually or in combination did not affect β-globin gene expression, suggesting that, although these sites interact in vivo, they are not required to function as insulators at their endogenous location in the mouse genome (30 –32). In other studies, partial knock-down of CTCF protein or mutation of the 3′HS1 CTCF site were found to have no effect on β-globin transcription in mouse erythroid cells before the onset of active globin gene transcription (10).
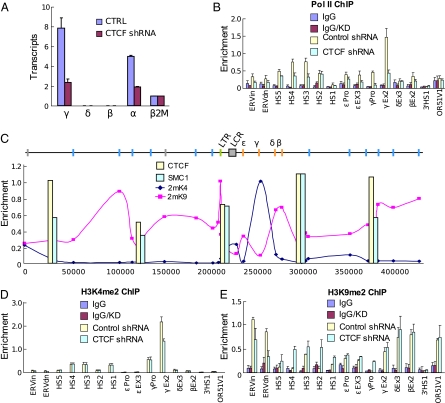
CTCF knock-down in K562 cells reduced γ-globin transcription by 50% (Fig. 4A). Transcription of α-globin was also reduced, suggesting that CTCF function is required in this unlinked locus. RNA pol II enrichment was reduced at γ-globin (Fig. 4B) and at HS3 and HS4 of the LCR, sites known to recruit pol II (33). To ask if reduction of CTCF looping interactions changed the epigenetic landscape within the β-globin locus, we determined occupancy of histone H3K4 di-methylation (H3K4me2), a mark of active transcription and of histone H3 K9 di-methylation (H3K9me2), a repressive mark. There is an inverse pattern of these marks over silent odorant receptor genes flanking the locus (lines, Fig. 4C), which is punctuated by sites of CTCF and Rad21 binding (bars, Fig. 4C). Between the HS5 and 3′HS1 CTCF sites flanking the globin locus, the two modifications vary as a function of gene activity: H3K4me2 marks the LCR and γ-globin gene whereas H3K9me2 is low at these positions but high at the silent δ- and β-globin genes. Upon reduction of CTCF, H3K4me2 was reduced about twofold at γ-globin, and other changes throughout the locus were not observed (Fig. 4D). In contrast, there was an increase in H3K9me2 (twofold to over fivefold at HS1-4) over the LCR, and this mark was detected at γ-globin (Fig. 4E), in parallel with reduced transcription and reduced pol II enrichment.
Fig. 4.
Effect of CTCF shRNA on K562 cells. (A) RT–PCR analysis of globin gene transcription in K562 cells after transduction with control or CTCF shRNA. Results of three RNA preparations are shown ±SEM. (B) ChIP performed on chromatin from K562 cells after treatment with control or CTCF shRNA using an antibody against RNA pol II. Error bars represent SEM. Amplicons include ERVin and ERVdn, within and downstream (unique amplicon) of ERV9-LTR; HS5 and 3′HS1, globin locus-flanking CTCF sites; HS1–HS4, locus control region DNase I hypersensitive sites; globin gene promoters and exons as indicated; and OR51V1, odorant receptor gene in the globin locus 3′ flank. (C) ChIP was performed using K562 cell chromatin as described in B with antibodies to H3K4me2, H3K9me2, CTCF, and SMC1. H3 methylation is plotted with the highest value for each modification set to 1 in the line graphs. CTCF or cohesin ChIP enrichment is represented by superimposed bars. Diagram at the top: orange lines, globin genes; gray box, LCR; blue lines, odorant receptor genes; gray lines, other silent genes; green lines, ERV9-LTR. Note the depiction of chromosome 11 in the 5′ to 3′ direction, which is reversed compared to the Genome browser view in Figs. 1–3. Panel C is drawn to scale but the numbers below the graph are arbitrary. (D) ChIP was performed as described in B with antibodies to H3K4me2. (E) ChIP was performed as described in B using an antibody to H3K9me2.
Thus, global loss of CTCF site interactions negatively affects active globin gene transcription coincident with an increase in H3K9me2 in the locus. We consider it likely that this effect is directly due to the loss of CTCF occupancy. This conclusion is supported by Western blot analysis, indicating that CTCF reduction does not affect the protein levels of the important globin gene activators GATA-1 and NF-E2 (Fig. S8). Inactive genes up- and downstream of the globin locus were unaffected by CTCF reduction (Fig. S8) in agreement with earlier work (10).
The differing effect of CTCF reduction on human globin gene transcription compared to loss of either or both of the mouse HS5 and 3′HS1 sites (10, 30–32) has several possible explanations. First, in our study, CTCF site interactions are lost globally, perhaps allowing more generalized spread of negative histone modifications into the globin locus. Second, it is conceivable that fetal γ-globin transcription is more sensitive to CTCF site loss than adult β-globin transcription. Finally, solitary ERV9-LTR insertions flank human HS5 and 3′HS1, and adjacent, unique ChIP amplicons (ERVdn and OR51V1 in Fig. 4E) are highly enriched for H3K9me2. Possibly, CTCF insulator function in the human globin locus excludes H3K9me2 from encroaching on the LCR and globin genes, similar to the role proposed for the HS4 insulator (HS5 ortholog) lying between the chicken globin locus and a 16-kb heterochromatic region (34). The mouse locus lacks these retroviral insertions and may not require the insulator activity (35, 36).
Because CTCF sites have an intrinsic loop-forming capability even at ectopic locations (13), we favor the idea that loops are central to CTCF site function even though in these experiments we cannot separate CTCF occupancy, per se, from CTCF site loop formation. Collectively, the results suggest that loss of chromosomal CTCF organizational structure through disruption of loops exerts a negative effect on active globin gene transcription at an endogenous locus. Interestingly, this effect seems to be dominant over LCR activity because the silent genes surrounding the globin locus remain silent; this outcome could also relate to the absence of odorant receptor gene-activating factors in erythroid cells. Considering the broad and diverse distribution of CTCF sites revealed in genome scans, we suggest that the insulation function of CTCF/cohesin sites may be secondary to their broader organizational role in the genome. The intrinsic insulation/looping capacity of the sites might then be manifest at particular positions where the chromatin surrounding an active locus is not conducive to gene transcription.
Materials and Methods
Cell Culture.
K562 cells and 293T cells were cultured in RPMI1640 and DMEM, respectively, supplemented with 10% FBS. Cells were maintained at 37 °C in a 5% CO2 incubator.
ChIP-chip and Data Analysis.
ChIP DNA was amplified as recommended (Nimblegen), and DNA samples were hybridized to a human genome tiling array consisting of 50 mers positioned every 100 bp along nonrepeating sequences of chromosomes 10 and 11. Raw data were collected by Nimblegen. On the basis of the inspection of individual data tracks at a false discovery rate (FDR) of <0.2, 31 primer combinations were chosen to represent CTCF peaks and negative positions. CTCF ChIP and real-time qPCR (Fig. S1) results validated 16 positive CTCF sites whereas 15 sites were negative. Next, for each cell type, biweight-scaled log2 ratios from three biological replicates were averaged to reduce biological variation. The averaged log2 ratios were used as input to the ACME tiling array algorithm. To call significant peaks, ACME uses a χ2 test on a 2 × 2 contingency table by using the number of probes above and below a threshold both inside and outside a moving window. The final ACME parameters (threshold of 0.984, p value 7.58e-14 for K562; threshold of 0.9890, p value 1.67e-9 for 293T; window size 1000 bp) were chosen by an iterative process to obtain the best fit to sites of positive CTCF binding as determined by the qPCR data.
Chromatin Immunoprecipitation.
ChIP assays were carried out as described (37). Briefly, ∼50 million cells were cross-linked with 1% formaldehyde, and nuclei were sonicated and digested with 200 U/mL of MNase to an average chromatin fragment size of 200–500 bp. Precleared chromatin was incubated with antibodies overnight at 4 °C and immunoprecipitated with protein A/G agarose beads. Immunoprecipitated material was extensively purified after reversal of the cross-links and was dissolved in Tris-EDTA for ChIP-chip or real-time PCR. The ratio of input DNA to immunoprecipitated DNA isolated from the same number of cells was maintained at 1/25.
Quantitative Real-Time PCR.
Real-time PCR using SYBR green chemistry was performed to quantify enriched DNA from ChIP and 3C and cDNA from reverse transcription on an ABI Prism 7900HT (PE Applied Biosystems). TaqMan chemistry was used to obtain the data in Fig. 4 B–E. The threshold was set to cross a point at which PCR amplification was linear, and the number of cycles (Ct) required to reach the threshold was recorded. The analyses were performed in duplicate for at least three experimental samples. Primer sequences for ChIP, 3C, and RT–PCR appear in Table S1.
Chromatin Conformation Capture.
The 3C assay was performed as described (13). Briefly, formaldehyde-fixed nuclei were digested with EcoRI to generate conveniently sized fragments, followed by ligation with T4 DNA ligase at 16 °C for 4 h. Cross-links were reversed, and DNA was extensively purified before PCR amplification. Specific ligation between two fragments was confirmed by sequencing the PCR products. Primer efficiency and ligation efficiency were determined as described (38). Quantification of ligated products was performed by real-time qPCR on three or more chromatin samples. To compare results from different cell types or cells with different lentiviruses, the results were normalized to the ligation frequency of two fragments in the α-tubulin gene.
shRNA Lentivirus Knock-Down.
Control, CTCF-, Rad21-, and SMC3-directed lentiviral shRNAs plasmids were purchased from Open Biosystems. Plasmids were transduced into 293FT cells with Virapower packaging mix (Invitrogen). Virus was harvested from the media on day 3 by centrifugation at 3000 × g for 15 min at 4 °C. K562 cells were incubated with viral supernatant in the presence of 10 μg/mL of polybrene. After 1 day of incubation, the medium was changed to RPMI 1640. Puromycin was added 2 days after transduction, and cells were cultured for another 2 days before collection.
Western Blotting.
Cells were lysed with protein loading buffer. Lysates were denatured at 70° C for 10 min, briefly sonicated, and centrifuged to remove debris. Samples were separated by SDS–PAGE, and immunoblotting was performed as described (37). Immunoblots were developed with the ECL Plus detection system (Bio-Rad).
Flow Cytometry.
Cells were fixed on day 4 after lentivirus transduction and then were stained with annexin-V or propidium iodide and analyzed on a FACSCalibur flow cytometer (BD Sciences).
RNA and Reverse Transcription.
RNA was isolated with TRIzol reagent (Invitrogen). One microgram of RNA was treated with RNase-free DNaseI before reverse transcription with SuperScript III kit (Invitrogen). cDNA was diluted 10-fold, and 5 μl was used as template for real-time PCR.
Supplementary Material
Acknowledgments
We thank Elissa Lei for helpful comments and Qihui Gong for assistance with ChIP experiments. This research was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912087107/DCSupplemental.
References
- 1.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 2.Wallace JA, Felsenfeld G. We gather together: Insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capelson M, Corces VG. Boundary elements and nuclear organization. Biol Cell. 2004;96:617–629. doi: 10.1016/j.biolcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Göndör A, Ohlsson R. Chromatin insulators and cohesins. EMBO Rep. 2008;9:327–329. doi: 10.1038/embor.2008.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 8.Kurukuti S, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadjur S, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Splinter E, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling JQ, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 12.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 13.Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc Natl Acad Sci USA. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulger M, et al. Conservation of sequence and structure flanking the mouse and human β-globin loci: The β-globin genes are embedded within an array of odorant receptor genes. Proc Natl Acad Sci USA. 1999;96:5129–5134. doi: 10.1073/pnas.96.9.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scacheri PC, Crawford GE, Davis S. Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol. 2006;411:270–282. doi: 10.1016/S0076-6879(06)11014-9. [DOI] [PubMed] [Google Scholar]
- 16.Bulger M, et al. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol Cell Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Parelho V, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 21.Stedman W, et al. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terranova R, et al. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell. 2008;15:668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 23.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 25.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubio ED, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gombert WM, Krumm A. Targeted deletion of multiple CTCF-binding elements in the human C-MYC gene reveals a requirement for CTCF in C-MYC expression. PLoS One. 2009;4:e6109. doi: 10.1371/journal.pone.0006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bender MA, et al. Description and targeted deletion of 5′ hypersensitive site 5 and 6 of the mouse β-globin locus control region. Blood. 1998;92:4394–4403. [PubMed] [Google Scholar]
- 31.Bender MA, et al. Flanking HS-62.5 and 3′ HS1, and regions upstream of the LCR, are not required for beta-globin transcription. Blood. 2006;108:1395–1401. doi: 10.1182/blood-2006-04-014431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farrell CM, et al. A large upstream region is not necessary for gene expression or hypersensitive site formation at the mouse β-globin locus. Proc Natl Acad Sci USA. 2000;97:14554–14559. doi: 10.1073/pnas.97.26.14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson KD, Christensen HM, Zhao B, Bresnick EH. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- 34.Prioleau MN, Nony P, Simpson M, Felsenfeld G. An insulator element and condensed chromatin region separate the chicken beta-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrell CM, West AG, Felsenfeld G. Conserved CTCF insulator elements flank the mouse and human β-globin loci. Mol Cell Biol. 2002;22:3820–3831. doi: 10.1128/MCB.22.11.3820-3831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long Q, Bengra C, Li C, Kutlar F, Tuan D. A long terminal repeat of the human endogenous retrovirus ERV-9 is located in the 5′ boundary area of the human beta-globin locus control region. Genomics. 1998;54:542–555. doi: 10.1006/geno.1998.5608. [DOI] [PubMed] [Google Scholar]
- 37.Song S-H, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range β-globin locus control region function. Mol Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.