Abstract
Ferritin is a spherical molecule composed of 24 subunits of two types, ferritin H chain (FHC) and ferritin L chain (FLC). Ferritin stores iron within cells, but it also circulates and binds specifically and saturably to a variety of cell types. For most cell types, this binding can be mediated by ferritin composed only of FHC (HFt) but not by ferritin composed only of FLC (LFt), indicating that binding of ferritin to cells is mediated by FHC but not FLC. By using expression cloning, we identified human transferrin receptor-1 (TfR1) as an important receptor for HFt with little or no binding to LFt. In vitro, HFt can be precipitated by soluble TfR1, showing that this interaction is not dependent on other proteins. Binding of HFt to TfR1 is partially inhibited by diferric transferrin, but it is hindered little, if at all, by HFE. After binding of HFt to TfR1 on the cell surface, HFt enters both endosomes and lysosomes. TfR1 accounts for most, if not all, of the binding of HFt to mitogen-activated T and B cells, circulating reticulocytes, and all cell lines that we have studied. The demonstration that TfR1 can bind HFt as well as Tf raises the possibility that this dual receptor function may coordinate the processing and use of iron by these iron-binding molecules.
Keywords: ferritin, HFE, iron, receptors, endocytosis
Iron is essential for life in most organisms (1 –3). Soluble iron, however, accelerates the generation of reactive oxygen species (ROS) from H2O2, and it is therefore toxic to cells (4). Ferritin allows the storage of iron in high concentration and without access to the substrates that generate ROS (5 –7).
Ferritin is a spherical molecule composed of 24 subunits, a mixture of ferritin H chain (FHC) and ferritin L chain (FLC). FHC is named for its initial isolation from heart, whereas FLC is named for its initial isolation from liver (1). In humans, FHC is also heavier (∼21 kDa) than FLC (∼19 kDa), and the ferritin subunits are sometimes referred to as heavy and light ferritin, respectively (1). The ratio of FHC to FLC within ferritin varies from species to species and from organ to organ, and it also varies in response to inflammation. FHC and FLC are highly homologous, and they can form a ferritin sphere in any proportion; however, only FHC can oxidize Fe2+ to Fe3+, and FHC facilitates the accumulation of iron within ferritin (8). Within ferritin, iron is stored in mineral form as ferrihydrite, and nucleation of ferrihydrite is promoted by FLC (7, 9).
In addition to its role in storing iron within cells, ferritin also circulates. Here, its role is uncertain, but ferritin can bind to a variety of cell types in a saturable manner. These include lymphocytes, placental microvilli, and erythroid precursors (10 –17). On most cell types, binding has been observed for ferritin composed only of FHC (HFt) but not for ferritin composed only of FLC (LFt). Studies of liver cells, however, have suggested a second receptor that binds both HFt and LFt (17). The erythroleukemic cell line K562 binds HFt with an apparent equilibrium association of ∼3 × 10−8 L/mol and an estimated 20,000 receptors per cell (12), whereas MOLT-4 T cells bind HFt with an apparent equilibrium association constant of ∼6.5 × 10−7 L/mol and ∼10,000 receptors sites per cell; subsequently, they endocytose HFt (13). Activated fresh lymphocytes also bind HFt (16), as do erythroid precursors, in which the subsequent endocytosis of HFt permits sufficient uptake of iron to produce hemoglobin (Hb) (18, 19). Although transferrin (Tf) is the usual and dominant source of iron for erythropoiesis, ferritin can provide sufficient iron for this to occur in the absence of Tf (19, 20).
In studies of mouse cells, we recently showed that T cell immunoglobulin and mucin domain protein-2 (TIM-2) serves as an endocytosing receptor for HFt but not LFt. TIM-2 is expressed on activated lymphocytes, but it is also expressed outside the hematopoietic system on bile-duct epithelial cells and kidney tubular epithelial cells (21). Mature rat oligendrocytes also express TIM-2, and they use it as a primary source of intracellular HFt (22). TIM-2 is not, however, expressed in humans (23).
In the studies presented here, we use expression cloning to identify human transferrin receptor-1 (TfR1) as an endocytosing cell-surface receptor for HFt. We show that binding of HFt to TfR1 occurs independently of Tf or the hemochromatosis-associated protein HFE, and instead it is partially blocked by diferric Tf. Binding of HFt to TfR1 results in the uptake of HFt into endosomes and lysosomes. Finally, by using an mAb that blocks the binding of HFt to TfR1, we show that TfR1 accounts for most binding of HFt to cells, including mitogen-activated lymphocytes and circulating reticulocytes.
Results
Molecular Cloning of Receptors for Human HFt Identifies TfR1.
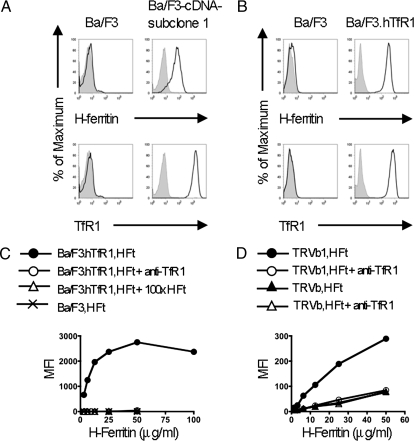
To define human receptors for HFt, we first screened a panel of human cell lines for saturable binding of recombinant human HFt. HFt bound the human B cell line 721.221 but not the mouse pro-B cell line Ba/F3. We, therefore, expressed a cDNA library from 721.221 cells in Ba/F3 cells and screened for binding of HFt; 8 of >100 clones obtained were sequenced in full, and all encoded TfR1. As an example, binding of HFt and anti-TfR mAb to one clone is shown in Fig. 1A.
Fig. 1.
Identification of TfR1 as a receptor for HFt. (A) Example of a clone obtained by transducing mouse Ba/F3 cells with a cDNA library from human 721.221 cells and staining with human HFt (Upper) or anti-TfR1 (Lower). (B) Transduction of Ba/F3 cells with cDNA for TfR1 alone allows expression of TfR1 (Lower) and binding of HFt (Upper). (C) Binding of HFt to Ba/F3 cells expressing TfR1 is saturable (•) and is inhibited either by excess HFt (△) or mAb to TfR1 (○). (D) HFt binds to TRVb1 cells, which express TfR1 (•), and binding is inhibited by mAb to TfR1 (○). Low levels of binding are seen in TRVb cells, which lack TfR1 (▲), and this binding is not blocked by anti-TfR1 (△).
To confirm that binding of HFt was mediated by TfR1, we expressed the cDNA for TfR1 in Ba/F3 cells. These Ba/F3.hTfR1 cells bound HFt (Fig. 1 B and C) but not detectable amounts of LFt (Fig. S1). At concentrations above 50 μg/mL, binding of biotinylated HFt to Ba/F3.hTfR1 was saturable, and at all concentrations it was inhibited by unconjugated HFt, which shows that binding was specific (Fig. 1C). Furthermore, an mAb to TfR1, M-A712, completely blocked the binding of HFt to TfR1, confirming that binding was mediated by TfR1 (Fig. 1C). Additionally, CHO cells lacking an endogenous TfR (TRVb cells) (24) bound only low levels of HFt, but this was notably enhanced in CHO cells expressing human TfR1 (TRVb1 cells) and this enhanced binding was blocked by anti-TfR1 mAb (Fig. 1D). These studies show that TfR1 binds HFt, but they do not exclude the possibility that other proteins might be required for this interaction. We, therefore, examined the interaction between TfR1 and HFt in isolation by coupling soluble his-tagged TfR1 extracellular domain to cobalt beads, using these to assess binding by HFt. In these studies, HFt showed low levels of nonspecific binding to the beads in the absence of TfR1, but the addition of TfR1 markedly enhanced binding of HFt, which confirms that TfR1 binds HFt in the absence of other proteins (Fig. 2A). By similar methods, we have found that horse spleen apoferritin also binds to TfR1 (25).
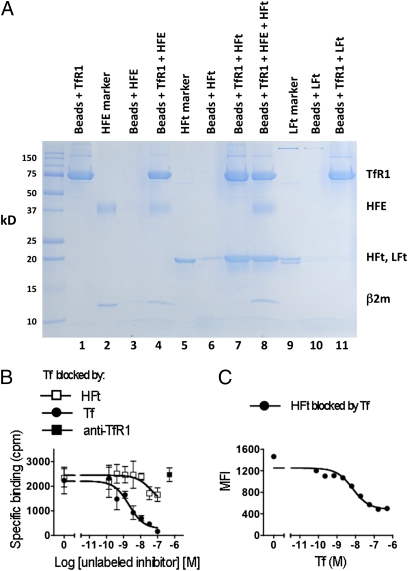
Fig. 2.
Binding of HFt to TfR1 and effects of HFE and Tf. (A) HFt in solution binds to isolated TfR1, and this is not significantly inhibited by HFE. Lanes 1, 3, 4, 6, 7, and 8 show SDS/PAGE of proteins eluted from cobalt beads treated with His-tagged TfR1 only (lane 1), HFE only (lane 3), His-tagged TfR1 followed by HFE (lane 4), HFt only (lane 6), His-tagged TfR1 followed by HFt (lane 7), and His-tagged TfR1 followed by HFE and HFt (HFE:HFt molar ratio = 10:1; lane 8). Lanes 2 and 5 show HFE and HFt, respectively, not exposed to beads but placed directly on the gel. (B) Binding of 125I-Tf to MOLT-4 cells is only partially inhibited by 100× molar excess of HFt (□) and not at all by antibody to TfR1 (■). As a control, binding is inhibited by unlabeled Tf (●). (C) Binding of biotinylated HFt (50 nM) to MOLT-4 cells is inhibited by Tf (●), but inhibition is incomplete, even with a 10-fold molar excess of Tf.
Effects of HFE and Tf on Binding of HFt to TfR1.
The hemochromatosis protein HFE binds tightly to TfR1, reducing binding of transferrin (26). In our pull-down studies, HFE in 10 molar excess had little or no effect on the binding of HFt to soluble TfR1, as shown in Fig. 2A, where HFE reduced recovery of HFt only slightly and in proportion to the recovery of TfR1. Thus, under the conditions of our studies, HFE does not significantly impair binding of HFt to TfR1.
For studies regarding competition between Tf and HFt for TfR1, we returned to studies of intact cells (MOLT-4 T cells), because they have been shown to specifically bind HFt. We confirmed this by flow cytometry, showing that binding of biotinylated HFt to MOLT-4 cells is blocked by unconjugated HFt, but it is not blocked by LFt, indicating that HFt binding is specific and confirming that LFt does not share the binding characteristics of HFt (Fig. S2). We then assessed the effect of HFt on binding by 125I-labeled diferric Tf to MOLT-4 cells. With 125I-Tf at 5 × 10−9 M, binding was blocked as expected by unlabeled diferric Tf, but HFt in 20-fold molar excess blocked binding by diferric Tf less than 50% (Fig. 2B). We also attempted to test the reverse, the blocking of 125I-labeled HFt by diferric Tf, and to use binding by 125I-labeled HFt to perform Scatchard plot analysis of HFt binding; however, these experiments were not possible, because 125I-labeling of HFt by either the Bolton–Hunter method or the use of Iodobeads resulted in loss of specific binding to TfR1. As assessed by flow cytometry, however, binding by 50 nM HFt to MOLT-4 cells was reduced by as little as 6 nM diferric Tf, but even with very high levels of Tf (500 nM), inhibition was only partial, reaching a maximum of ∼70% inhibition (Fig. 2C). The inability of diferric Tf to fully block binding by HFt does not likely reflect a second receptor for HFt on MOLT-4 cells, because mAb to TfR1 completely blocks binding of HFt to these cells (Figs. 3 and 4). Although inhibition by Tf of HFt binding to TfR1 is incomplete, it again shows the specific binding of HFt to TfR1, and it excludes the possibility that this binding is mediated or facilitated by Tf.
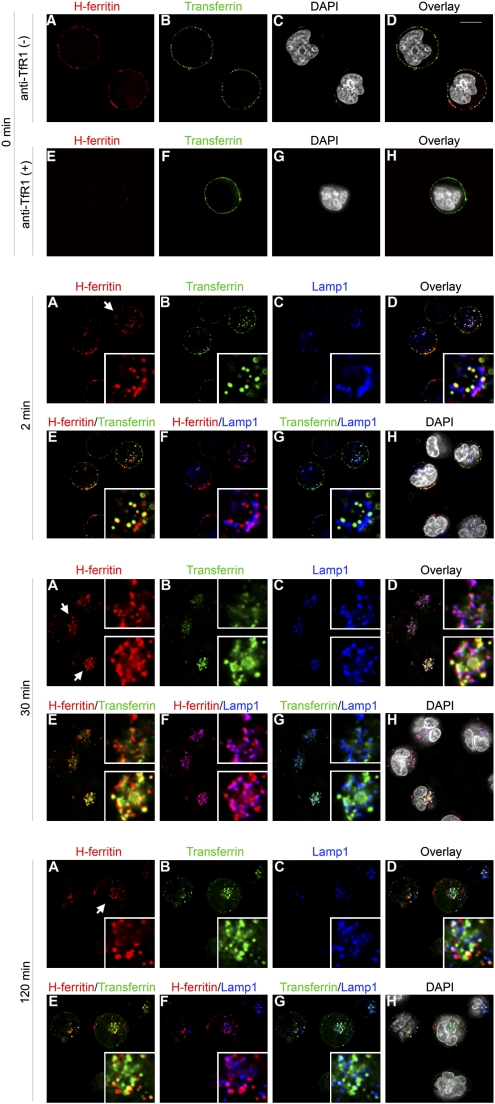
Fig. 3.
Binding of HFt to TfR1 results in endocytosis of HFt and trafficking of HFt to lysosomes in MOLT-4 T cells. MOLT-4 cells were incubated for 60 min on ice with HFt-Alexa 568 (50 μg/mL) and Tf-Alexa 488 (50 μg/mL). During this period, both HFt and Tf bound to the cell surface (top row), but anti-TfR1 (50 μg/mL) blocked the binding of HFt but not Tf (second row). Cells were subsequently incubated at 37 °C for 2–120 min, followed by staining with the lysosomal marker Lamp1. The examples presented include enlargements of one or two selected cells, boxed to the right side of each frame, and designated by the white arrow in frame A for each time point. At 2 min, HFt appeared in early endosomes, as shown by colocalization with Tf (2 min; E), but even at this early time point, HFt was also found in lysosomes, as shown by colocalization with Lamp1 (2 min; F). At 30 min, the majority of HFt was found primarily in lysosomes, and only little was seen in early endosomes (30 min; cell at top right); however, in some cells, HFt was still in early endosomes as well as lysosomes (30 min; cell at lower right). At 120 min, some HFt was still seen in endosomes or lysosomes, but HFt was also seen in vesicles that lacked Lamp1 or Tf, presumably endosomes from which Tf has been sorted (120 min; E and F). As expected, and in contrast to HFt, Tf did not sort into lysosomes (2 min to 120 min; G). (Scale bar: 10 μm.)
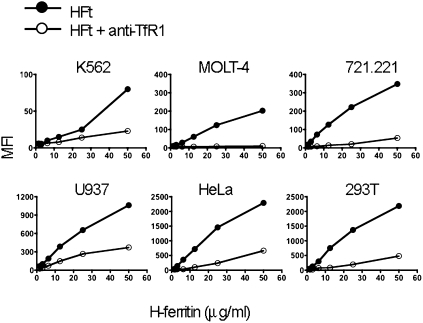
Fig. 4.
Anti-TfR1 mAb blocks binding by HFt to six human cell lines: K562 (erythroleukemia), MOLT-4 (T cell), 721.221 (B cell), U937 (monocyte/macrophage), HeLa (ovarian cancer), and 293T cells (kidney). The y axis shows mean fluorescence intensity (MFI) of staining with biotinylated HFt/streptavidin.
Binding of HFt to TfR1 Permits Entry of HFt into Endosomes and Lysosomes.
Because diferric Tf incompletely blocks binding by HFt, we were able to use fluorescence microscopy to examine the simultaneous binding and uptake of diferric Tf and HFt by MOLT-4 cells. Consistent with results obtained by flow cytometry, binding of HFt to MOLT-4 cells on ice was blocked by anti-TfR1, whereas binding of Tf was not (Fig. 3, top two panels; Fig. S3 shows magnification of Fig. 3). On warming of the cells to 37 °C, HFt and Tf were both rapidly internalized. At early time points, much of the HFt colocalized with Tf, which is known to be restricted to endosomes (Fig. 3). Unlike Tf, however, HFt was also transferred to lysosomes, which was assessed by colocalization with Lamp1 (Fig. 3). We conclude that HFt can dissociate from TfR1 in endosomes, allowing for the translocation of HFt to lysosomes.
TfR1 Accounts for Most of the Binding of HFt to Cell Lines, Activated Lymphocytes, and Reticulocytes.
We next examined the role of TfR1 in binding of HFt to different cell lines, using the capacity of the anti-TfR1 mAb M-A712 to specifically block binding of HFt to TfR1. The cell lines examined included MOLT-4 T cells and K562 myeloid/erythroid leukemia cells, both of which have been previously shown to bind HFt (12, 13). All of the lines examined expressed TfR1 (Fig. S4), and all bound HFt (Fig. 4). Binding of HFt to all of the cell lines was substantially blocked by anti-TfR1; in MOLT-4 cells, it was blocked almost completely (Fig. 4). K562 cells have previously been used to show up-regulation of transferrin receptors after iron depletion by desferrioxamine (27). We confirmed that TfR1 was up-regulated on K562 cells after treatment with desferrioxamine, and we found that this was accompanied by increased binding of HFt (Fig. S5). We conclude that prior descriptions of HFt binding by MOLT-4 and K562, as well as binding by the other cell lines tested, can be attributed largely, if not entirely, to their expression of TfR1.
Prior studies have shown saturable binding of HFt to activated lymphocytes (16, 17). In our studies, fresh, unactivated T cells expressed only low levels of TfR1 and did not bind detectable levels of HFt (Fig. 5A Left); however, after 60 h of stimulation with the mitogen phytohemagglutinin (PHA), levels of TfR1 were increased, binding of HFt was detected, and most HFt binding was blocked by mAb to TfR1 (Fig. 5B Left). Thus, binding of HFt to mitogen-activated T cells reflects up-regulation of TfR1.
Fig. 5.
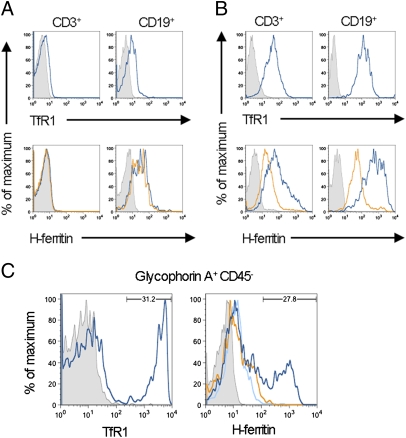
HFt binds to TfR1 on activated lymphocytes and reticulocytes. (A Upper) Fresh, unstimulated peripheral blood T cells (CD3+; Left) express little if any TfR1 (blue line), whereas fresh B cells express low levels (CD19+; Right). In these and all stains with anti-TfR1, the control is staining with an isotype-matched mAb (shaded curve). (Lower) HFt does not bind to fresh T cells (CD3+; Left). The blue line represents staining by biotinylated HFt plus SA-APC. The gold line indicates staining by HFt in the presence of anti-TfR1. The shaded curve is SA-APC only. Binding of HFt to fresh B cells can be detected (CD19+, blue line; Right), but this is not blocked by anti-TfR1 (gold line). (B Upper) After exposure to PHA, both T cells (CD3+; Left) and B cells (CD19+; Right) up-regulate TfR1 (blue line). (Lower) After activation, both B and T cells bind HFt (blue line), and much, but not all, of this binding is blocked by anti-TfR1 (gold line). (C) In a normal donor, glycophorin A+CD45- reticulocytes were coisolated with PBMC. About 30% of these expressed high levels of TfR1 (Left). A similar percentage of cells bound HFt at high levels (blue line; Right), and binding was blocked by anti-TfR1 (gold line; Right) or 100-fold excess of unlabeled HFt (light blue line; Right).
Similarly, fresh B cells expressed only low levels of TfR1 (Fig. 5A Right). They also bound low levels of HFt, but this binding was not blocked by anti-TfR1 (Fig. 5A Lower Right). After stimulation of peripheral blood mononuclear cells with PHA, B cells expressed increased TfR1 in the same manner as T cells, presumably because of the release of cytokines by activated T cells; these B cells bound HFt, and most binding was inhibited by anti-TfR1 (Fig. 5B Lower Right). Thus, activated B cells, as well as T cells, bind HFt by up-regulating TfR1.
Blood cells from one volunteer included a population of non-T, non-B cells that expressed high levels of TfR1 and that bound high levels of HFt. These cells lacked CD45 but expressed glycophorin A, indicating that they were erythroid cells; staining of these cells with new methylene blue after sorting confirmed that they were reticulocytes that had copurified with PBMC. About 30% of these glycophorin A+/CD45- cells expressed high levels of TfR1, and a similar percent bound HFt in a manner that was blocked by anti-TfR1 (Fig. 5C). Prior studies have shown that erythroid precursors bind and take up ferritin (11, 28) and that this can provide sufficient iron for the production of Hb (18, 19). Our findings show that the binding of HFt to reticulocytes is specifically and uniquely mediated by TfR1.
Discussion
Our studies show that TfR1 serves as a receptor for HFt. Binding occurs in the absence of Tf or HFE and is significantly inhibited by diferric Tf, although not completely. TfR1 accounts for most of the binding of HFt to all cell lines that we have studied, as well as to mitogen-stimulated blood lymphocytes and fresh reticulocytes.
After binding to TfR1, HFt enters not only endosomes but also lysosomes. Other studies have shown that cytoplasmic ferritin similarly reaches lysosomes and that lysosomes may facilitate the release of iron from ferritin (29, 30). De Domenico et al. (31) recently showed that lysosomal degradation of cytosolic ferritin can be by induced by autophagy. Alternatively, cytosolic ferritin can be ubiquitinated and subjected to proteasomal degradation (3, 31, 32). It will be of interest to define the fate of HFt after it reaches lysosomes after endocytosis through TfR1, as well as the fate of iron contained within the HFt.
Konijn and coworkers (18, 19) have shown that erythroid precursors can internalize and use iron from HFt in the production of Hb, and this process is blocked by inhibitors of endocytosis (18, 19). Our studies show that the binding of HFt to fresh reticulocytes is dependent on TfR1 and that TfR1 mediates endocytosis of HFt as well as diferric Tf. It is, thus, highly likely that binding of HFt to TfR1 on erythroid precursors is required for its use in supplying iron for erythropoiesis.
Anti-TfR1 blocks binding of HFt but not diferric Tf to TfR1, suggesting that the binding sites for HFt and Tf may not be identical. Nonetheless, diferric Tf inhibits binding of HFt to TfR1, indicating either that the binding sites for HFt and Tf overlap or that Tf alters TfR1 in a way that reduces binding by HFt. Reciprocally, high concentrations of HFt partially block binding by diferric Tf to TfR1. This likely explains the observation by Tsunoo and Sussman (33) over 25 years ago that human placental ferritin or horse splenic ferritin inhibit binding of Tf to placental extracts. We did not observe a significant effect of HFE on binding by HFt to TfR1, unlike its known effect on binding by Tf (26, 34). Importantly, neither HFE nor Tf is required for binding by HFt to TfR1; binding of HFt to TfR1 occurs in the absence of either. HFt consists of 24 identical subunits, and TfR1 consists of two identical subunits, so the interactions between these proteins on the cell surface may be complex. We were not able to perform Scatchard analysis of binding by 125I-HFt, because iodination reduced binding of HFt to TfR1; however, other approaches will be of interest to examine this interaction.
The selective binding by TfR1 of HFt, not LFt, is consistent with prior observations that cell lines and hematopoietic cells selectively bind HFt (12 –17). In this regard, it may be noteworthy that the proinflammatory cytokines TNFα and IL-1α selectively induce transcription of FHC but not FLC through induction of the transcription factor NFkB, thereby increasing the content of the FHC in ferritin (5). Although an increase in circulating FHC in inflammation has not yet been directly shown, the induction of FHC by inflammatory cytokines suggests the possibility that FHC may serve as a secondary signal in inflammation. Studies to test this can now focus in particular on TfR1.
Because TfR1 seems to bind HFt but not LFt, our findings do not explain prior evidence that human liver cells can bind LFt as well as HFt (17), and this receptor remains to be identified. In mice, Li et al. (35) recently identified Scara5 as a receptor for LFt in the developing kidney and showed that this accounted for iron uptake in the absence of TfR1 or TfR2. However, Scara5 is not expressed in liver (36), and human orthologs have not yet been described.
In mice, we previously identified Tim-2 as a receptor for HFt, but Tim-2 is not expressed in humans (21). Interestingly, we have so far found no evidence that murine HFt or human HFt binds to murine TfR1, suggesting that these mechanisms for recognition of HFt have diverged between humans and mice. The extracellular domains of mouse and human TfR1 protein are 76% identical at the protein level (86% conserved), but areas of nonhomology are found throughout these domains. We have not yet studied binding by HFt to the more distantly related (but still 46% identical) TfR2.
Of particular relevance to our studies, Li et al. (37) have recently shown that the human chemokine receptor CXRC4 binds cytosolic FHC in a manner that is dependent on the serine phosphorylation of FHC at position 178. In cells overexpressing CXCR4, its interaction with cytosolic FHC is increased by the internalization of CXCR4 after binding by its ligand CXCL12, and FHC inhibits activation of ERK1/2 by CXCL12 in these cells (37). This may relate to prior observations by Pham et al. (38) that showed that FHC inhibits JNK activation by inhibiting ROS. It will be important to determine if HFt imported through the TfR1 can reach the cell compartments where it could contribute to these effects.
The shared use of TfR1 to bind and internalize both Tf and HFt suggests that this pathway may serve to coordinate the uptake and use of iron by these two ligands. Tf binds ferric iron (Fe3+), but in the endosomes of erythrocyte precursors, this is reduced to ferrous iron (Fe2+) by ferroreductases, notably Steap3 (39, 40). Thus, ferrous iron would be available for uptake by HFt in endosomes. We have not observed this in preliminary studies, but further studies are in progress. Alternatively, or perhaps additionally, Tf and HFt may serve as redundant mechanisms for the uptake of iron by cells.
Materials and Methods
Cell Culture.
MOLT-4, K562, 721.221, U937, HeLa, and 293T cells were obtained from the American Type Culture Collection, and Ba/F3 cells were provided by Lewis Lanier. These cell lines were grown in RPMI-1640 medium (Mediatech, Manassas,VA), or for 293T cells, DMEM (Cellgro). All media were supplemented with 10% FBS (Atlantic Biologics) and penicillin-streptomycin-glutamine (Gibco). Growth of Ba/F3 cells was sustained by adding 1 ng/mL IL-3 (BD Biosciences). TRVb and TRVb1 cells (24), provided by Timothy E. McGraw, Weill Cornell Medical College, New York, NY, were maintained in Ham’s nutrient F12 medium (Invitrogen) supplemented with 5% FBS and penicillin-streptomycin-glutamine. To test the effects of iron chelation, K562 cells were incubated for 18 h with desferrioxamine (Sigma) in concentrations ranging from 2 to 50 μM.
Expression Cloning of the Human HFt Receptor.
The retroviral cDNA library from 721.221 human B cells was generated by using methods previously described (41). The library was transfected into Phoenix cells by FuGENE 6 (Roche Applied Science). The virus-containing supernatant was collected 48 h later and used to infect Ba/F3 cells with N-[1-(2,3-Dioleoyloxy)propyl]-N,N,Ntrimethylammonium methylsulfate (DOTAP) liposomal transfection reagent (10 μg/mL; Roche Applied Science). Two days later, Ba/F3 cells were stained with biotinylated human HFt plus streptavidin APC (SA-APC; Caltag) and selected by cell sorting. This enrichment was repeated five times, after which >90% of the cells stained with HFt. The genomic DNA was isolated (Qiagen), and PCR was carried out by using an advantage-GC genomic PCR kit (Clontech) with the primers 5′-GGT GGA CCA TCC TCT AGA CT-3′ (sense) and 5′-TTT ATT TTA TCG TCG ATC GAC C-3′ (antisense).
Cellular Expression of Human TfR1.
TfR1 cDNA was obtained from the ATCC and confirmed by sequencing. The cDNA was amplified by PCR, cloned into the pMX-puromycin expression vector (Lewis Lanier, University of California San Francisco, CA), and transfected into Phoenix packaging cells using Fugene 6 (Roche). Two days later, the viral supernatant was harvested and used to infect Ba/F3 cells with the use of DOTAP (10 μg/mL; Roche). Clones were selected and maintained in RPMI-1640 plus 1 μg/mL puromycin (Sigma).
Preparation and Purification of Soluble Proteins.
Human HFt and LFt were produced in Escherichia coli and purified as described (42). Horse spleen apoferritin was from Calbiochem. HFE and His-tagged soluble TfR1 were produced as previously described (43). Diferric Tf was from Sigma.
Isolation and Culture of Human PBMC.
Peripheral blood mononuclear cells were isolated from blood of normal human donors by Ficoll-hypaque gradient centrifugation (Amersham Biosciences) using a protocol approved by the University of California San Francisco and San Francisco Veterans Affairs Medical Center Committees on Human Research. Cells were cultured for 60 h in RPMI 1640 with 10% FBS in the presence of 10 μg/mL PHA. GlycophorinA+/CD45− cells were isolated by flow cytometry using a FACSAria flow cytometer.
Flow Cytometry.
Monoclonal antibodies were from BD Bioscience, unless otherwise indicated, and they included: mouse anti-human CD3 (clone UCHT1, FITC), mouse anti-human CD19 (clone HIB19, FITC), mouse anti-human CD45 (clone H130, PerCP-Cy5.5; eBioscience), mouse anti-human glycophorin A (clone GA-R2, FITC), mouse anti-human TfR1 (clone M-A712, phycoerythrin, or biotinylated). Human HFt and LFt were biotinylated by a using EZ-Link sulfo-NHS-LC-biotin kit (Pierce) according to the manufacture’s directions. Fresh PBMC or PHA-stimulated PBMC was stained with biotinylated HFt (20 μg/mL) with or without the presence of anti-TfR1 (50 μg/mL), and binding was detected with streptavidin coupled to allophycocyanin (APC) fluor. Expression of the human TfR1 was assessed by using PE-conjugated anti-TfR1 or as indicated, biotinylated anti-TfR1. Binding to cell lines of biotinylated HFt or anti-TfR1 was detected by streptavidin-APC (SA-APC). Cells were analyzed on a FACSCalibur flow cytometer (BD Biosciences). Data were displayed and analyzed by using FlowJo software.
Pulldown Assay.
Soluble His-tagged TfR1 (100 μg, 0.715 nmol) was allowed to bind to 50 μL of cobalt chelate resin (Profound Pull-Down Poly-His Protein:Protein Interaction Kit; Pierce) following the procedure of the manufacturer. TfR1-conjugated beads or unconjugated beads were exposed to 90 μg HFt (0.176 nmol) for 90 min at 4 °C, with or without 78.5 μg HFE (1.76 nmol). The beads were washed, and proteins were eluted in 100 μL 290 mM imidazole, which was followed by analysis with SDS/PAGE.
Assays for Cross-Blocking by Tf, HFt, and LFt.
125I-labeled human diferric Tf, labeled by a modification of the Hunter and Greenwood methods, was purchased from PerkinElmer Life Sciences. Before binding of labeled Tf, 5 × 105 MOLT-4 cells were incubated for 30 min on ice with 100 μL saline or with cold HFt or Tf (Sigma) at concentrations ranging from 0.27 to 200 nM. 125I-labeled Tf was added in equal volume to a final concentration of 5 nM, and cells were incubated on ice for 90 min. Cells were then washed three times in PBS and suspended in PBS for detection of bound 125I. The capacity of Tf or LFt to inhibit binding by HFt was assessed by flow cytometry: 5 × 105 MOLT-4 cells were incubated for 30–90 min on ice with biotinylated HFt (50 nM) together with unlabeled Tf at concentrations from 0.07 to 500 nM, or with unlabeled LFt at concentrations from 5 to 500 nM. Cells were washed, and HFt was detected with SA-APC.
Deconvolution Fluorescence Microscopy of Endocytosis.
HFt was labeled with Alexa 568 (Invitrogen) according to the manufacture’s instructions. Internalization of HFt by MOLT-4 T cells was performed as previously described for mouse cells (21). Briefly, MOLT-4 cells were incubated for 30 min at 37 °C in PBS supplemented with 10 mM Tris-HCl, 10 mM Hepes, 5 mM glucose, and 1 mg/mL BSA at pH 7.4. Cells were pelleted and resuspended in cold buffer with or without anti-TfR1 mAb (50 μg/mL), and then they were incubated on ice for 30 min, which was followed by the addition of HFt-Alexa 568 (50 μg/mL) and Tf-Alexa 488 (50 μg/mL; Invitrogen); further incubation was on ice for 90 min. Cells were then washed with cold PBS and resuspended in buffer at 37 °C to allow endocytosis. At defined time points, internalization was stopped by adding cold PBS/0.02% sodium azide. Cells were pelleted and resuspended in 4% paraformaldehyde, and they were plated on poly-L-lysine–treated coverslips (BD Biosciences) for 60 min. Cells were permeabilized with 0.5% Triton X-100 in PBS for 5 min and then blocked with 0.5% BSA and 0.1% Triton X-100 in PBS for 30 min at room temperature. To identify lysosomes, cells were incubated with an anti-Lamp1 mAb (1:200, clone H4A3; Invitrogen) overnight at 4 °C. Cells were then washed three times with PBS and incubated with goat anti-mouse IgG1 conjugated with Alexa 647 (1:500; Invitrogen). Images were collected by an API DeltaVision DV3 Restoration microscope using a MicroMax 5 MHz cooled CCD camera (Roper Scientific); deconvolution was performed using API SoftWoRx software.
Supplementary Material
Acknowledgments
We thank Kenneth Scalapino, M.D., for his expert advice and help with flow cytometry, and Eric Huang, M.D. for assistance with microscopy. This work was supported by National Institutes of Health Grants R01 AI061164-01A1 (to W.E.S.) and R37DK42412 (to F.M.T.) and by the Veterans Administration.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913192107/DCSupplemental.
References
- 1.Theil EC. Ferritin: Structure, gene regulation, and cellular function in animals, plants, and microorganisms. Annu Rev Biochem. 1987;56:289–315. doi: 10.1146/annurev.bi.56.070187.001445. [DOI] [PubMed] [Google Scholar]
- 2.Ganz T. Iron homeostasis: Fitting the puzzle pieces together. Cell Metab. 2008;7:288–290. doi: 10.1016/j.cmet.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 3.MacKenzie EL, Iwasaki K, Tsuji Y. Intracellular iron transport and storage: From molecular mechanisms to health implications. Antioxid Redox Signal. 2008;10:997–1030. doi: 10.1089/ars.2007.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halliwell B, Gutteridge JM. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torti FM, Torti SV. Regulation of ferritin genes and protein. Blood. 2002;99:3505–3516. doi: 10.1182/blood.v99.10.3505. [DOI] [PubMed] [Google Scholar]
- 6.Arosio P, Levi S. Ferritin, iron homeostasis, and oxidative damage. Free Radic Biol Med. 2002;33:457–463. doi: 10.1016/s0891-5849(02)00842-0. [DOI] [PubMed] [Google Scholar]
- 7.Harrison PM, Arosio P. The ferritins: Molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- 8.Levi S, et al. Mechanism of ferritin iron uptake: Activity of the H-chain and deletion mapping of the ferro-oxidase site. A study of iron uptake and ferro-oxidase activity of human liver, recombinant H-chain ferritins, and of two H-chain deletion mutants. J Biol Chem. 1988;263:18086–18092. [PubMed] [Google Scholar]
- 9.Ford GC, et al. Ferritin: Design and formation of an iron-storage molecule. Philos Trans R Soc Lond B Biol Sci. 1984;304:551–565. doi: 10.1098/rstb.1984.0046. [DOI] [PubMed] [Google Scholar]
- 10.Liao QK, Kong PA, Gao J, Li FY, Qian ZM. Expression of ferritin receptor in placental microvilli membrane in pregnant women with different iron status at mid-term gestation. Eur J Clin Nutr. 2001;55:651–656. doi: 10.1038/sj.ejcn.1601195. [DOI] [PubMed] [Google Scholar]
- 11.Meyron-Holtz EG, Fibach E, Gelvan D, Konijn AM. Binding and uptake of exogenous isoferritins by cultured human erythroid precursor cells. Br J Haematol. 1994;86:635–641. doi: 10.1111/j.1365-2141.1994.tb04797.x. [DOI] [PubMed] [Google Scholar]
- 12.Fargion S, et al. Characteristics and expression of binding sites specific for ferritin H-chain on human cell lines. Blood. 1988;71:753–757. [PubMed] [Google Scholar]
- 13.Moss D, Powell LW, Arosio P, Halliday JW. Characterization of the ferritin receptors of human T lymphoid (MOLT-4) cells. J Lab Clin Med. 1992;119:273–279. [PubMed] [Google Scholar]
- 14.Bretscher MS, Thomson JN. Distribution of ferritin receptors and coated pits on giant HeLa cells. EMBO J. 1983;2:599–603. doi: 10.1002/j.1460-2075.1983.tb01469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takami M, et al. Human placental ferritin receptor. Biochim Biophys Acta. 1986;884:31–38. doi: 10.1016/0304-4165(86)90223-0. [DOI] [PubMed] [Google Scholar]
- 16.Fargion S, et al. Specific binding sites for H-ferritin on human lymphocytes: Modulation during cellular proliferation and potential implication in cell growth control. Blood. 1991;78:1056–1061. [PubMed] [Google Scholar]
- 17.Moss D, et al. Functional roles of the ferritin receptors of human liver, hepatoma, lymphoid and erythroid cells. J Inorg Biochem. 1992;47:219–227. doi: 10.1016/0162-0134(92)84067-w. [DOI] [PubMed] [Google Scholar]
- 18.Gelvan D, Fibach E, Meyron-Holtz EG, Konijn AM. Ferritin uptake by human erythroid precursors is a regulated iron uptake pathway. Blood. 1996;88:3200–3207. [PubMed] [Google Scholar]
- 19.Leimberg MJ, Prus E, Konijn AM, Fibach E. Macrophages function as a ferritin iron source for cultured human erythroid precursors. J Cell Biochem. 2008;103:1211–1218. doi: 10.1002/jcb.21499. [DOI] [PubMed] [Google Scholar]
- 20.Leimberg JM, Konijn AM, Fibach E. Developing human erythroid cells grown in transferrin-free medium utilize iron originating from extracellular ferritin. Am J Hematol. 2003;73:211–212. doi: 10.1002/ajh.10355. [DOI] [PubMed] [Google Scholar]
- 21.Chen TT, et al. TIM-2 is expressed on B cells and in liver and kidney and is a receptor for H-ferritin endocytosis. J Exp Med. 2005;202:955–965. doi: 10.1084/jem.20042433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Todorich B, Zhang X, Slagle-Webb B, Seaman WE, Connor JR. Tim-2 is the receptor for H-ferritin on oligodendrocytes. J Neurochem. 2008;107:1495–1505. doi: 10.1111/j.1471-4159.2008.05678.x. [DOI] [PubMed] [Google Scholar]
- 23.Kuchroo VK, Umetsu DT, DeKruyff RH, Freeman GJ. The TIM gene family: Emerging roles in immunity and disease. Nat Rev Immunol. 2003;3:454–462. doi: 10.1038/nri1111. [DOI] [PubMed] [Google Scholar]
- 24.McGraw TE, Greenfield L, Maxfield FR. Functional expression of the human transferrin receptor cDNA in Chinese hamster ovary cells deficient in endogenous transferrin receptor. J Cell Biol. 1987;105:207–214. doi: 10.1083/jcb.105.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lebrón JA. Pasadena, CA: California Institute of Technology; 1999. Biochemical and biophysical characterization of the hemochromatosis protein HFE and its interaction with the transferrin receptor. PhD thesis. [Google Scholar]
- 26.Giannetti AM, Björkman PJ. HFE and transferrin directly compete for transferrin receptor in solution and at the cell surface. J Biol Chem. 2004;279:25866–25875. doi: 10.1074/jbc.M401467200. [DOI] [PubMed] [Google Scholar]
- 27.Mattia E, Rao K, Shapiro DS, Sussman HH, Klausner RD. Biosynthetic regulation of the human transferrin receptor by desferrioxamine in K562 cells. J Biol Chem. 1984;259:2689–2692. [PubMed] [Google Scholar]
- 28.Meyron-Holtz EG, et al. Regulation of intracellular iron metabolism in human erythroid precursors by internalized extracellular ferritin. Blood. 1999;94:3205–3211. [PubMed] [Google Scholar]
- 29.Vaisman B, Fibach E, Konijn AM. Utilization of intracellular ferritin iron for hemoglobin synthesis in developing human erythroid precursors. Blood. 1997;90:831–838. [PubMed] [Google Scholar]
- 30.Kidane TZ, Sauble E, Linder MC. Release of iron from ferritin requires lysosomal activity. Am J Physiol Cell Physiol. 2006;291:C445–C455. doi: 10.1152/ajpcell.00505.2005. [DOI] [PubMed] [Google Scholar]
- 31.De Domenico I, Ward DM, Kaplan J. Specific iron chelators determine the route of ferritin degradation. Blood. 2009;12:4546–4551. doi: 10.1182/blood-2009-05-224188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwok JC, Richardson DR. Examination of the mechanism(s) involved in doxorubicin-mediated iron accumulation in ferritin: Studies using metabolic inhibitors, protein synthesis inhibitors, and lysosomotropic agents. Mol Pharmacol. 2004;65:181–195. doi: 10.1124/mol.65.1.181. [DOI] [PubMed] [Google Scholar]
- 33.Tsunoo H, Sussman HH. Characterization of transferrin binding and specificity of the placental transferrin receptor. Arch Biochem Biophys. 1983;225:42–54. doi: 10.1016/0003-9861(83)90005-x. [DOI] [PubMed] [Google Scholar]
- 34.Feder JN, et al. The hemochromatosis gene product complexes with the transferrin receptor and lowers its affinity for ligand binding. Proc Natl Acad Sci USA. 1998;95:1472–1477. doi: 10.1073/pnas.95.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li JY, et al. Scara5 is a ferritin receptor mediating non-transferrin iron delivery. Dev Cell. 2009;16:35–46. doi: 10.1016/j.devcel.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Y, Oliver P, Davies KE, Platt N. Identification and characterization of murine SCARA5, a novel class A scavenger receptor that is expressed by populations of epithelial cells. J Biol Chem. 2006;281:11834–11845. doi: 10.1074/jbc.M507599200. [DOI] [PubMed] [Google Scholar]
- 37.Li R, Luo C, Mines M, Zhang J, Fan GH. Chemokine CXCL12 induces binding of ferritin heavy chain to the chemokine receptor CXCR4, alters CXCR4 signaling, and induces phosphorylation and nuclear translocation of ferritin heavy chain. J Biol Chem. 2006;281:37616–37627. doi: 10.1074/jbc.M607266200. [DOI] [PubMed] [Google Scholar]
- 38.Pham CG, et al. Ferritin heavy chain upregulation by NF-kappaB inhibits TNFalpha-induced apoptosis by suppressing reactive oxygen species. Cell. 2004;119:529–542. doi: 10.1016/j.cell.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 39.Ohgami RS, et al. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet. 2005;37:1264–1269. doi: 10.1038/ng1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sendamarai AK, Ohgami RS, Fleming MD, Lawrence CM. Structure of the membrane proximal oxidoreductase domain of human Steap3, the dominant ferrireductase of the erythroid transferrin cycle. Proc Natl Acad Sci USA. 2008;105:7410–7415. doi: 10.1073/pnas.0801318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arase H, Saito T, Phillips JH, Lanier LL. Cutting edge: The mouse NK cell-associated antigen recognized by DX5 monoclonal antibody is CD49b (alpha 2 integrin, very late antigen-2) J Immunol. 2001;167:1141–1144. doi: 10.4049/jimmunol.167.3.1141. [DOI] [PubMed] [Google Scholar]
- 42.Rucker P, Torti FM, Torti SV. Recombinant ferritin: Modulation of subunit stoichiometry in bacterial expression systems. Protein Eng. 1997;10:967–973. doi: 10.1093/protein/10.8.967. [DOI] [PubMed] [Google Scholar]
- 43.Lebrón JA, et al. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.