Abstract
Nonsense-mediated decay (NMD) degrades mRNA containing premature translation termination codons. In yeast, NMD substrates are decapped and digested exonucleolytically from the 5′ end. Despite the requirement for translation in recognition, degradation of nonsense-containing mRNA is considered to occur in ribosome-free cytoplasmic P bodies. We show decapped nonsense-containing mRNA associate with polyribosomes, indicating that recognition and degradation are tightly coupled and that polyribosomes are major sites for degradation of aberrant mRNAs.
Cells use intricate mechanisms to ensure fidelity of gene expression and to safeguard against the accumulation of aberrant mRNAs and proteins. In eukaryotes, mRNAs harboring a premature translation termination codon (PTC, or nonsense codon) within their protein-coding regions are efficiently detected and rapidly degraded1–4. This surveillance pathway, termed nonsense-mediated mRNA decay (NMD), prevents the accumulation of truncated polypeptides that would potentially be toxic to the cell. NMD targets a heterogeneous class of mRNAs, including those that have acquired a PTC as a consequence of gene mutation or errors during transcription. Moreover, inaccurate or alternative pre-mRNA splicing generates a major subset of PTC-containing mRNAs, and unproductively rearranged immunoglobulin and T-cell receptor genes represent an important physiological class of NMD substrates1. NMD also modulates the levels of ~1–10% of cellular mRNA lacking a PTC, thereby serving an additional role in regulating gene expression1,2.
Three phylogenetically conserved proteins, up-frameshift protein 1 (UPF1), UPF2 and UPF3, constitute the core of the NMD machinery and are required for both the recognition of nonsense-containing mRNA and their targeting for rapid degradation1,2. In the budding yeast Saccharomyces cerevisiae, destruction of NMD substrates is initiated by removal of the 5′ cap (decapping) followed by 5′→3′ exonucleolytic digestion5. In mammals, NMD triggers mRNA decapping and 5′ digestion but also targets substrates for simultaneous degradation at the 3′ end catalyzed by the cytoplasmic exosome6. Moreover, NMD targets in human cells7 can be cleaved endonucleolytically at a position proximal to the PTC, similar to the predominant pathway for degradation observed in Drosophila melanogaster cells1.
Recognition of premature termination by the NMD pathway requires mRNA translation3. Consistent with this, both the NMD machinery and its substrates are found associated with polyribosomes8,9. Degradation of nonsense-containing mRNA, in contrast, is hypothesized to occur after dissociation of the mRNA from ribosomes and in cytoplasmic foci known as P bodies10,11. In support of this, UPF1 inhibits translation of PTC-containing mRNA and also functions as a repressor of normal mRNA translation10–12. Moreover, the decapping holoenzyme (DCP1 and DCP2), 5′→3′ exoribonuclease (XRN1), UPF proteins and NMD substrates localize to P bodies, proposed sites for mRNA storage and decay10. Physical interactions observed between UPF1 and the decapping enzyme (DCP2) are hypothesized to mediate targeting of NMD substrates to P bodies3,11. Based on these and additional observations, UPF1 is proposed to translationally repress and target the aberrant mRNA to P bodies, where UPF2 and UPF3 activate decapping of the mRNA10.
We have recently shown that decapping and 5′→3′ degradation of normal mRNA in yeast occurs co-translationally and that dissociation of mRNA from ribosomes is not a prerequisite for its destruction13. Based on these findings, we hypothesized that decapping of nonsense-containing mRNA mediated by the NMD pathway also occurs while the mRNA is associated with polyribosomes. Consistent with a view that NMD substrates are degraded co-translationally, NMD can be uncoupled from the visible aggregation of P bodies in yeast, Drosophila and mammalian cells14–17.
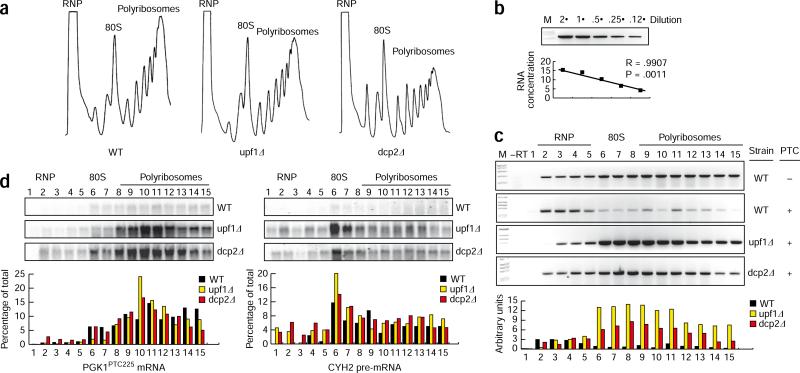
A model for NMD featuring repression of mRNA translation would predict that in cells lacking mRNA decapping activity, substrate mRNA should be stabilized but should not be found bound to ribosomes. The association of an efficient substrate for NMD in yeast, GFPPTC67 mRNA (Supplementary Table 1, Supplementary Data and Supplementary Fig. 1), with a translating messenger ribonucleoprotein (mRNP) was monitored by sucrose density gradient sedimentation. Quantitative RT-PCR (qRT-PCR) was used to assess mRNA abundance in gradient fractions representing the nontranslating cellular RNA (RNP), monoribosomes (80S) and polyribosomes (Fig. 1a,b). In wild-type (WT) cells, the vast majority of GFPPTC67 mRNA was detected in light fractions (such as RNP), with very little mRNA co-sedimenting with polyribosomes (Fig. 1c). For cells lacking UPF1, the NMD factor implicated in translational repression10–12, increased levels of GFPPTC67 mRNA were detected and the bulk of mRNA co-sedimented with polyribosomes, consistent with its stabilization and continued translation in the absence of NMD. Similarly, GFP mRNA lacking a PTC was stable and polysome associated (Fig. 1c). Notably, GFPPTC67 mRNA observed in the absence of UPF1 in heavy gradient fractions represents bona fide polyribosomes and not an association with some other dense particle (see below).
Figure 1.
NMD substrates are bound by polyribosomes when mRNA decapping is inhibited. (a) Polyribosome analysis of WT, upf1Δ and dcp2Δ cells. RNPs, 80S ribosomes and polyribosomes are indicated. (b) RT-PCR analysis for GFPPTC67 mRNA is quantitative for the analysis shown in c. (c) Distribution of GFP mRNA (lacking a premature termination codon, – PTC) and GFPPTC67 mRNA (harboring a premature termination codon, + PTC) in sucrose-gradient fractions. Arbitrary levels of GFPPTC67 mRNA are compared for WT, upf1Δ and dcp2Δ cells. (d) Northern blot analysis to monitor the sedimentation in sucrose gradients of CYH2 pre-mRNA and PGK1PTC225 reporter mRNA in WT, upf1Δ and dcp2Δ cells. The abundance of mRNA in each fraction in terms of the percentage of the total is shown.
The diminished levels of GFPPTC67 mRNA levels associated with polyribosomes for WT cells versus cells lacking UPF1 is not inconsistent with inhibition of mRNA translation and degradation of the mRNA once it is ribosome-free. However, the observation is also compatible with destruction of the mRNA while associated with the translating mRNP, where PTC recognition occurs. To discriminate between these possibilities and determine whether GFPPTC67 mRNA is removed from polyribosomes by the NMD machinery before its decay, we performed polyribosome analysis on cells defective in mRNA decapping (dcp2Δ). In these cells, NMD substrates are recognized by the NMD machinery and are observed to be translationally inhibited but are not destabilized due to the absence of cellular decapping activity12. In dcp2Δ cells, GFPPTC67 mRNA levels were elevated, with a majority of the mRNA co-sedimenting with polyribosomes (dcp2Δ; Fig. 1c). Unexpectedly, the sedimentation pattern of GFPPTC67 mRNA was indistinguishable between decapping-defective cells (dcp2Δ) and those inhibited for NMD (upf1Δ), and, notably, was not enhanced in the RNP in dcp2Δ cells (Fig. 1c). These data suggest that in the absence of decapping, NMD substrates are not dissociated from polyribosomes by UPF1 but rather remain bound by ribosomes. Consistent with qRT-PCR analysis of GFPPTC67 mRNA, additional NMD substrates (such as PGK1PTC225 reporter mRNA and the endogenous CYH2 pre-mRNA) were found by northern blotting to be associated with polyribosomes in dcp2Δ cells, and their sedimentation patterns corresponded to the length of the coding region (Fig. 1d). Collectively, these data indicate that UPF1 does not promote a ribosome-free mRNP and support a view that NMD substrates are degraded while still associated with polyribosomes.
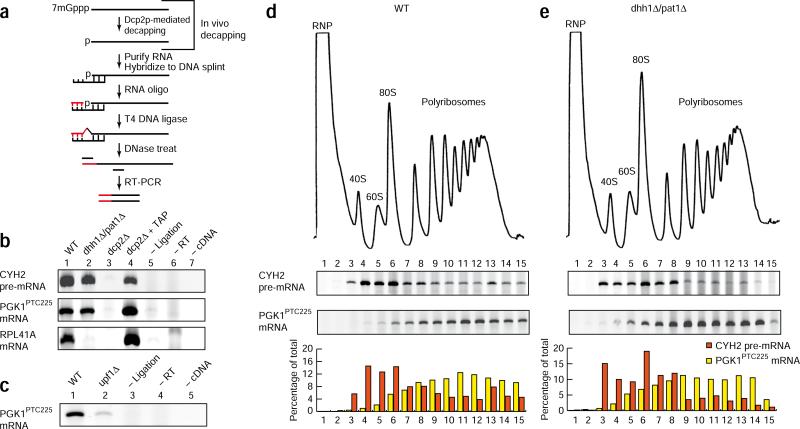
To evaluate whether bulk levels of decapped nonsense-containing mRNA can be found associated with polyribosomes, we performed quantitative primer extension analysis on sucrose-gradient fractions from xrn1Δ cells, where the products of decapping are enriched13. We detected the majority (~80%) of decapped PGK1PTC225 reporter mRNA on polyribosomes (Supplementary Fig. 2). To examine whether the decapping of NMD substrates occurs coincidentally with the mRNA's association with ribosomes, we used splinted ligation followed by RT-PCR to detect products of the decapping reaction that are short-lived in WT cells13. In this procedure, an RNA adaptor is ligated to the 5′ end of a decapped mRNA (assisted by a DNA splint) and the intermediate used as a template for cDNA synthesis and amplification of a DNA product by PCR (Fig. 2a). Decapped products of ‘normal’ (for example, RPL41a mRNA) and nonsense-containing mRNA (endogenous CYH2 pre-mRNA and PGK1PTC225 mRNA) were detected in WT cells but not in dcp2Δ cells, where mRNA decapping is inhibited, showing that product formation is dependent on decapping in vivo (Fig. 2b, lane 1 versus lane 3). Consistent with this, removal of the mRNA 5′ cap in vitro by tobacco acid pyrophosphatase resulted in detection of decapped mRNA from dcp2Δ cells (Fig. 2b, lane 4). Notably, decapping of PGK1PTC225 mRNA required NMD, as products were greatly reduced in cells lacking UPF1 (Fig. 2c). Splinted ligation analysis of RNA recovered from sucrose-gradient fractionation of WT cells established that the bulk of decapped CYH2 pre-mRNA and PGK1PTC225 mRNA co-sedimented with polyribosomes (Fig. 2d). Critically, the sedimentation pattern of decapped mRNA correlated with the open reading frame length of these two mRNAs, illustrating that sedimentation was a result of polyribosome association. Specifically, the product of unspliced CYH2 pre-mRNA (encoding an 18-residue polypeptide) associated predominantly with 80S and light polyribosome regions, whereas PGK1PTC225 mRNA (encoding a 224-residue polypeptide) was detected deep in the gradient and with high-density polyribosomes (Fig. 2d). Given the transient nature of uncapped mRNA in WT cells, we interpret the association of these decay intermediates with polyribosomes as a consequence of their co-translational generation.
Figure 2.
The association of decapped NMD substrates with polyribosomes. (a) The splinted ligation RT-PCR assay for detecting decapped mRNA. (b) Detection of decapped CYH2 pre-mRNA, PGK1PTC225 mRNA and RPL41A mRNA in WT, dhh1Δ/pat1Δ and dcp2Δ cells. Addition of tobacco alkaline phosphatase before splinted ligation RT-PCR (dcp2Δ + TAP), without splinted ligation step (– Ligation), without reverse transcription step (– RT) and without cDNA template added to the PCR reaction (– cDNA). (c) Detection of decapped PGK1PTC225 mRNA in WT and upf1Δ cells. (d,e) Splinted ligation RT-PCR analysis on RNA recovered from sucrose-gradient fractionation of wild-type (d) or dhh1Δ/pat1Δ (e) cells. RNPs, 40S and 60S ribosomal subunits, 80S ribosomes and polyribosomes are indicated. The abundance of mRNA in each fraction in terms of the percentage of the total is shown.
The decapping of mRNA represents the critical, rate-determining step in the degradation of both normal and NMD substrates, and we have shown that normal mRNA decapping occurs co-translationally13. To ensure that the accumulation of decapped PGK1PTC225 mRNA on polyribosomes was due to recognition and targeting by the NMD pathway and was not a consequence of shunting of the mRNA into the deadenylation-dependent pathway of decay used for the metabolism of normal mRNA, we performed splinted ligation analysis on cells in which decapping of normal but not PTC-containing mRNA is defective. Deletion of the activators of mRNA decapping, DHH1 and PAT1, leads to marked mRNA stabilization and accumulation of deadenylated, capped intermediates for normal but not nonsense-containing mRNA18 (Supplementary Figs. 3 and 4). Consistent with this, analysis of mRNA decapping by splinted ligation RT-PCR failed to detect decapped products for RPL41a mRNA in dhh1Δ/pat1Δ cells as compared to WT cells (Fig. 2b). In contrast, the products of decapping were readily detected for two nonsense-containing mRNAs in dhh1Δ/pat1Δ cells (CYH2 pre-RNA and PGK1PTC225; Fig. 2b). Moreover, destabilization of PTC- containing mRNA in dhh1Δ/pat1Δ cells required NMD as demonstrated by its dependence on UPF1 (Supplementary Fig. 4d). These data establish that decapping of nonsense-containing mRNA in dhh1Δ/pat1Δ cells occurs as a consequence of targeting by NMD. We therefore evaluated the sedimentation pattern of decapped CYH2 pre-mRNA and PGK1PTC225 mRNA by sucrose-gradient analysis using splinted ligation RT-PCR in dhh1Δ/pat1Δ cells. Notably, decapped products of nonsense-containing mRNA from these cells sedimented in a similar manner to that observed for WT (Fig. 2d versus e). We conclude that polyribosome association of the decapped products of NMD substrates observed in both WT and dhh1Δ/pat1Δ mutant cells are the result of decapping triggered by NMD.
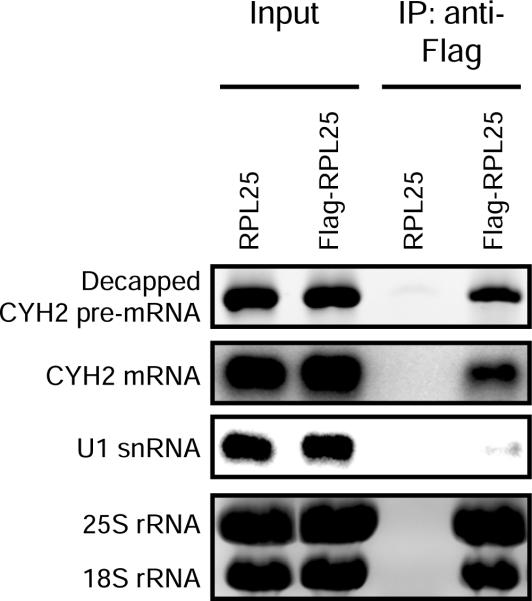
To further demonstrate the association of decapped intermediates of nonsense-containing mRNA with ribosomes, we purified ribosomes from cell extracts before assaying for decapped RNA products19. Immunoprecipitation of affinity-tagged ribosomal protein RPL25 co-purified 25S and 18S ribosomal RNA and ribosome-bound mRNA (such as CYH2 mRNA) but not nontranslated RNA (such as U1 snRNA) as detected by northern blotting (Fig. 3). Critically, splinted ligation RT-PCR analysis showed that decapped products of CYH2 pre-mRNA, an endogenous NMD substrate, also purified with ribosomes (Fig. 3). Notably, a similar product was not amplified from cells lacking epitope-tagged RPL25 protein. The co-purification of decapped nonsense-containing mRNA with ribosomes supports a mechanism in which the first step in the degradation of NMD substrates occurs co-translationally.
Figure 3.
Decapped products of nonsense-containing mRNA co-purify with ribosomes. Immunoprecipitation of ribosomes (mediated by affinity-tagged large ribosomal protein, RPL25) co-purifies decapped CYH2 pre-mRNA as detected by splinted ligation and RT-PCR analysis. Co-purification of CYH2 mRNA and ribosomal RNA (rRNA) were detected by 1.4% agarose gel followed by northern blot and ethidium bromide staining, respectively. U1 snRNA did not co-purify with ribosomes as monitored by northern blot.
In summary, we have presented evidence that decapping of nonsense-containing mRNA by NMD occurs while the substrate is bound by ribosomes. Our findings are consistent with the association of the NMD factors UPF1, UPF2 and UPF3 with polyribosomes8,9, as well as DCP2 and XRN1, the catalytic activities for decapping and 5′→ 3′ exonucleolytic RNA digestion9,13,20. Moreover, the rapid degradation of NMD substrates upon removal of cycloheximide, an inhibitor of translation elongation, also supports a co-translational decay mechanism4. Our data indicate that dissociation of nonsense-containing mRNA from ribosomes does not occur before their decay, which is inconsistent with their sequestration away from polyribosomes followed by targeting and destruction in P bodies. In agreement with this, the rapid decay of NMD substrates can be uncoupled from detectable P-body aggregation in several organisms, including yeast14–17. Notably, endonucleolytic cleavage of NMD substrates proximal to the PTC observed in Drosophila and human cells suggests that the prematurely terminating ribosome helps in delineating the site of the nonsense codon and that these substrates are also degraded while ribosome associated.
Although our observations indicate that recognition and decay of nonsense-containing mRNA by NMD are tightly coupled, they raise several mechanistic questions. Of particular interest are recruitment of the decapping complex and how the mRNA is rapidly decapped, particularly in light of its association with the translation machinery. It has been previously observed that translation of PTC-containing mRNAs is inhibited and that UPF1 functions as a general translational repressor10–12. In the context of co-translational decapping, it will be important to determine whether UPF1 acts to inhibit translation elongation or to facilitate rapid rearrangement of translational initiation factors at the mRNA 5′ end, thereby allowing the decapping enzyme accessibility to the cap. Our finding that NMD substrates associate with polyribosomes in dcp2Δ cells supports a mechanism of translation inhibition downstream of initiation; however, UPF1 interacts with eIF3, a critical factor in 80S ribosomal-subunit joining11. To reconcile these observations, it will be necessary to determine whether ribosomes associated with NMD substrates are impaired in elongation in response to the action of UPF1 (and/or UPF2 and UPF3) or whether the kinetics of 5′-end remodeling and decapping are much faster than the rate of ribosomal run-off once translational repression ensues.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank members of the Baker and Coller laboratories for critical evaluation of the manuscript. W.H. is supported by the American Heart Association. C.P. is supported by an undergraduate stipend provided by the US National Science Foundation. Funding was provided by the US National Institutes of Health (GM080465).
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Isken O, Maquat LE. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 2.Behm-Ansmant I, et al. FEBS Lett. 2007;581:2845–2853. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 3.Amrani N, Sachs MS, Jacobson A. Nat. Rev. Mol. Cell Biol. 2006;7:415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- 4.Baker KE, Parker R. Curr. Opin. Cell Biol. 2004;16:293–299. doi: 10.1016/j.ceb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Muhlrad D, Parker R. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 6.Lejeune F, Li X, Maquat LE. Mol. Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 7.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. Nat. Struct. Mol. Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 8.Atkin AL, et al. J. Biol. Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 9.Mangus DA, Jacobson A. Methods. 1999;17:28–37. doi: 10.1006/meth.1998.0704. [DOI] [PubMed] [Google Scholar]
- 10.Sheth U, Parker R. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isken O, et al. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muhlrad D, Parker R. Mol. Biol. Cell. 1999;10:3971–3978. doi: 10.1091/mbc.10.11.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stalder L, Muhlemann O. RNA. 2009;15:1265–1273. doi: 10.1261/rna.1672509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kshirsagar M, Parker R. Genetics. 2004;166:729–739. doi: 10.1093/genetics/166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Decker CJ, Teixeira D, Parker R. J. Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coller J, Parker R. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inada T, et al. RNA. 2002;8:948–958. doi: 10.1017/s1355838202026018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. Proc. Natl. Acad. Sci. USA. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.