Abstract
Context: FSH mediates cyclic follicle growth and development and is widely used for controlled ovarian stimulation in women undergoing infertility treatment. The ovarian response of women to FSH is variable, ranging from poor response to ovarian hyperstimulation.
Objective: We investigated whether genetic alterations of the FSH receptor (FSHR) contribute to this variability.
Design and Patients: Our approach was to study women undergoing treatment with in vitro fertilization falling into the edges of the normal distribution of ovarian response to FSH, with respect to age.
Setting: We conducted the study at the Yale Fertility Clinic.
Methods: We extracted RNA from cumulus cells surrounding the oocytes of women undergoing in vitro fertilization and analyzed the FSHR mRNA by RT-PCR and sequencing.
Results: We identified four abnormal FSHR splicing products (three exon deletions and one intron insertion) in the FSHR mRNA in 37% (13 of 35) of women tested. All alterations affected the extracellular ligand-binding portion of the receptor without causing a frameshift. When transfected in HEK293T cells, all four splicing variants showed markedly decreased cAMP activation compared to controls. Untransfected cells showed no response to FSH, whereas all the cell lines showed normal cAMP activation when treated with forskolin, a nonreceptor-mediated cAMP stimulant. None of the normal or mutant forms showed any response to LH or TSH.
Conclusions: Our findings strongly indicate FSHR variants as being an intrinsic genetic cause of some forms of infertility and identify a need for functional characterization of these variants and the investigation of more individualized ovarian stimulation protocols.
FSH receptor splice variants are found in cumulus cells of women undergoing in vitro fertilization and may constitute an intrinsic genetic cause of some forms of infertility.
FSH is a key hormone of mammalian reproduction that is necessary for gonadal development and maturation at puberty and for gamete production during the reproductive phase of life (1). FSH is a member of the glycoprotein hormone family that also includes LH, hCG, and TSH. These hormones share a common α-subunit and differ in their unique β-chains. FSH elicits its effect upon binding to its receptor (FSHR), localized in granulosa and Sertoli cells (2,3,4). FSHR is also expressed in osteoclasts, and its role in bone resorption, osteoporosis, and menopause remains to be determined (5). The FSHR is a G protein-coupled receptor characterized by seven transmembrane helices flanked by intracellular and extracellular domains. The intracellular portion of the FSHR is coupled to a Gs protein and activates the adenyl cyclase enzyme, resulting in elevation of intracellular cAMP and stimulation of the cAMP-dependent protein kinase pathway (6), although it has also been shown to activate additional signal transduction pathways (7).
In humans, inactivating mutations in the FSHR gene cause infertility featuring amenorrhea, hypergonadotropic hypogonadism, ovarian failure, and/or dysgenesis (8,9,10,11,12,13,14,15,16). On the other hand, some cases of ovarian hyperstimulation syndrome have been shown to be due to activating FSHR mutations that cause either constitutive signaling or reduced specificity for FSH and concomitant increased sensitivity to human chorionic gonadotropin (hCG) and/or TSH (17,18,19,20,21). Female mice with targeted disruption of the Fshr gene are infertile (22).
Although inactivating FSHR mutations result in a severe reproductive phenotype, it is plausible that more subtle genetic variations of the receptor can contribute to functional perturbations, subfertility, and/or infertility. FSH is widely used for controlled ovarian stimulation in patients undergoing in vitro fertilization (IVF); however, individual ovarian response to FSH varies significantly, ranging from recruitment of low to high numbers of oocytes (23). Two common FSHR polymorphisms have been associated with altered response to FSH during IVF and different basal FSH levels (24,25). These polymorphisms are two single nucleotide changes in exon 10 of the receptor (SNP Database: rs6165 and rs6166), resulting in two amino acid substitutions (p.307Thr/Ala and p.680Asn/Ser).
In this study, we have hypothesized that additional mutations/polymorphisms in the FSHR gene resulting in altered structure and function of the receptor may influence the number of oocytes produced per cycle. Our approach was to study women falling into the edges of the normal distribution of ovarian response to FSH. This paradigm has been very successful in identifying genes for multifactorial disorders, such as hypertension (26). We specifically recruited patients with a poor or high response during stimulation and examined the FSHR mRNA expressed in their cumulus cells and not only the DNA sequence of the exons. Fertile oocyte donors and women with oocyte numbers within the 25–75% range who were treated for male factor infertility were studied as controls. The ability of all identified variants to initiate a cAMP increase in response to FSH stimulation was studied in vitro in human embryonic kidney (HEK) 293T cells.
Patients and Methods
Patient selection, cumulus cell collection, and RNA extraction
Patients were recruited from the Yale Fertility Center. The procedure was approved by the Yale Institutional Review Board Committee (HIC protocol no. 25431).
All patients’ day 3 FSH levels, before the start of stimulation, were below 10 IU/liter. Treatment was initiated by pituitary suppression with GnRH agonists. Stimulation with gonadotropin was begun only after down-regulation had been achieved (estradiol level <50 pg/ml and absence of follicular cysts >15 mm in size). Stimulation with FSH (150–600 IU/dl, Gonal-F; Serono, Geneva, Switzerland) was monitored by measuring serum estradiol levels and follicle growth. In cases of poor or excessive response, the daily FSH dose was adjusted. hCG (10,000 IU, Pregnyl; Organon, Oss, The Netherlands) was administered when patients had at least two follicles that were 18 mm or greater in diameter. Oocytes were collected 36 h after hCG and rinsed in HEPES-buffered human tubal fluid (Irvine Scientific, Santa Ana, CA). The oocytes were stripped of the surrounding cumulus cells using mechanical manipulation with 135- to 210-μm pipettes in hyaluronidase (Hyase; Irvine Scientific) in separate 30-μl drops for 1–2 min at 37 C. The cumulus cells of each individual patient were collected by aspirating the hyaluronidase solution and wash drops in a 1.5-ml tube and centrifuged at 1000 × g for 10 min.
The cumulus cells were lysed in 500 μl of Trizol (Invitrogen, San Diego, CA) using aspiration through a syringe with a fine needle. RNA was extracted according to the manufacturer’s instructions and dissolved in 30 μl of diethylpyrocarbonate-water. RNA was stored at −70 C until used.
RT-PCR
One-step RT-PCR (One-step RT; QIAGEN, Germantown, MD) was performed on 5 μl of RNA according to the manufacturer’s instructions, with the following modification: a mix containing RNA, primers, and diethylpyrocarbonate-water was denatured at 65 C for 5 min and chilled on ice before the addition of the remaining reagents. FSHR primers were: FSHR.1F, 5′GGA GGT TTT TCT CTG CAA ATG CAG3′; and FSHR.2R, 5′CAT ACC CTT CAA AGG CAA GGA CTG3′, flanking the open-reading frame. Actin primers were: actin.F, 5′GAC CTC TAT GCA ACA CAG T3′; and actin.R, 5′TTG CTG ATC CAC ATC TGC T3′. The RT-PCR program was: 50 C 30 min; 95 C 15 min; 10 touch-down cycles: 92 C 30 sec, 65–55 C 20 sec, 72 C 3 min; 20 cycles: 92 C 20 sec, 55 C 15 sec, 72 C 3 min; and 72 C 5 min. Nested PCR (Taq polymerase; QIAGEN) for FSHR produced two overlapping fragments: FSHR.9F, 5′ATG GCC CTG CTC CTG GTC TC3′, with FSHR.10R, 5′CAA ACT CAG TGT ACG TCA TGT C3′; and FSHR.2F, 5′ATC CCA GCC ATT GCT GTG CCT TTG3′, with FSHR.8R, 5′TTA GTT TTG GGC TAA ATG AC3′. The PCR conditions were as above, except that extension was 1 min. The PCR products were analyzed on agarose gels.
Mammalian expression constructs
The full-length human FSHR in pcDNA3.1 (pFSHR-TN) was identified as an expressed sequence tag (American Tissue Culture Collection no. 10089334; IMAGE no. 7001973), and the insert was confirmed by sequencing. We constructed the other common polymorphism (AS allele) using RT-PCR from a patient with primers 9F-8R (see RT-PCR) and cloned into BsmBI and XhoI sites of pFSHR-TN. The splice variants were amplified using RT-PCR from patients as described in RT-PCR, gel-purified (Gel extraction kit; QIAGEN), and cloned into pFSHR-TN using the following enzymes: del ex2, EcoRI-Bsu36I; del ex6, ins ivs8, Bsu36I-BsmBI; and del ex9, Bsu36I-XhoI. All inserts were verified by sequencing. A hemagglutinin (HA) tag was fused in-frame at the 3′ end of all FSHR variants using PCR with primers 9F and 12R: 5′CCC CTC GAG TTA AGC GTA ATC TGG AAC ATC GTA TGG GTA TTC GAA GTT TTG GGC TAA ATG ACT TAG3′ and cloned into BsmBI-XhoI or BsgI-XhoI sites of pFSHR wild-type (WT) or variant constructs, respectively.
Transient transfections
HEK293T cells were cultured at 37 C, 5% CO2 in DMEM (Invitrogen) supplemented with 2 mm l-glutamine, 0.1 mg/ml streptomycin, 0.2 U/ml penicillin, 1 mm sodium pyruvate, and 10% fetal calf serum (Invitrogen). Cells were grown to 70% confluency in 10-cm dishes and transiently transfected using calcium phosphate precipitation. Then, 20 μg of plasmid (QIAGEN) was mixed with 100 μl CaCl2 1 m and water to a final volume of 0.5 ml. The DNA/CaCl2 mixture was added dropwise to 0.5 ml of 2X Hepes buffered saline [1.6% NaCl (wt/vol), 1.2% HEPES (wt/vol), 0.04% Na2HPO4 (wt/vol), pH 7.12]. Twenty-four hours after transfection, the medium was replaced with 10 ml of UltraCHO (BioWhittaker, Inc., Walkersville, MD) supplemented with 0.1 mg/ml streptomycin and 0.2 U/ml penicillin. Recombinant proteins were expressed for 48 h. Then, the cells were stimulated with hormones or lysed.
Western blotting
The cells were washed in cold PBS and lysed at 4 C in lysis buffer [10 mm Tris-HCl (pH 8.0), 2.5 mm MgCl2, 5 mm EGTA (pH 8.0), 0.5% Triton X-100, 1 mm Na3VO4, 50 mm NaF, one tablet of Complete Protease Inhibitor Cocktail (Roche Diagnostics, Indianapolis, IN) per 10 ml of buffer]. Lysates were cleared by centrifugation at 12,000 × g for 30 min at 4 C.
Protein concentration was measured using the Bradford assay (Bio-Rad, Hercules, CA). Then, 10 μg of total protein was mixed with 2X sodium dodecyl sulfate (SDS) sample buffer [125 mm Tris-HCl (pH 6.8), 20% glycerol, 4% SDS, 0.1% bromophenol blue, 10% mercaptoethanol], incubated at 55 C for 5 min, and analyzed on an 8% SDS-PAGE gel. Protein was transferred to polyvinylidene difluoride (Bio-Rad) at 50 V overnight. The membrane was blocked in 5% nonfat dry milk in PBST (0.05% Tween-20) and incubated with primary mouse anti-HA antibody (1:1000; Upstate Biotechnology, Lake Placid, NY) and secondary antimouse-horseradish peroxidase antibody (Pierce, Rockford, IL), diluted in 1% milk in PBST. Detection was performed with the ECL Western Blotting Detection System (Amersham Bioscience, Piscataway, NJ) and exposed to film (Kodak, Rochester, NY).
cAMP assays
The HEK293T cells were grown in 24-well plates and transfected with 0.6 μg of pFSHR DNA (QIAGEN) and 1 ng of pCMV-RL (Promega, Madison, WI) as described above. Forty-eight hours after transfection, the cells were preincubated with 0.45 mm 3-isobutyl-1-methylxanthine for 15 min and then stimulated with one of the following for 30 min in the presence of 3-isobutyl-1-methylxanthine: 0–500 ng/ml FSH (GonalF; Serono), 10 μm forskolin (Sigma, St. Louis, MO), 0–675 ng/ml LH (Serono), or 0–600 ng/ml TSH (Sigma). The cells were lysed, and cAMP was measured according to the manufacturer’s instructions (KGE002; R&D Systems, Minneapolis, MN). Luciferase activity of pCMV-RL was measured in 10 μl of the extract (Dual-Luciferase Assay System; Promega) and used to normalize the cAMP across the plate.
Results
Patient recruitment and classification of ovarian response
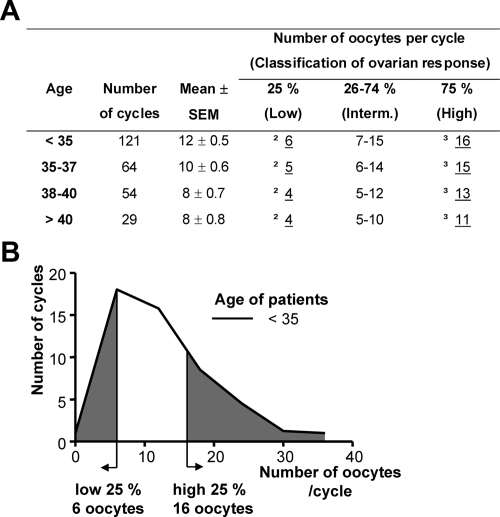
Individual ovarian response to FSH stimulation of women undergoing IVF shows significant variability, ranging from poor to high response (23). To determine the distribution of the response of our patient population and to establish cutoff values for high or low response to FSH, we analyzed all IVF cycles performed at the Yale Fertility Center between 2003 and 2005 (Fig. 1). Data were collected and compared only from patients treated with the protocol described in Patients and Methods. The patients were separated into four age groups: below 35, 35–37, 38–40, and above 40 yr, consistent with the grouping used for outcome analysis by the Society for Assisted Reproductive Technologies (SART). The number of oocytes per cycle defining the 25th (lower) and 75th (higher) percentiles were determined for each age group (Fig. 1). As expected, the mean number of oocytes retrieved per cycle declined with age.
Figure 1.
Oocytes retrieved per cycle after FSH stimulation. A, Cumulative results from the Yale Fertility Center (2003–2005). The number of oocytes that mark the 25% and 75% interval of each age category is underlined. These numbers define low and high responses to FSH in our patient population. B, A graph depicting the data from patients under 35 yr of age. The gray areas represent the lower and higher 25th percentiles of the number of oocytes per cycle.
Next, we assessed whether the ovarian response was a direct consequence of the amount of FSH used to treat the patients. Younger patients required less FSH to achieve ovarian stimulation (Supplemental Table 1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org). Interestingly, for women younger than 40, the amount of FSH received showed an inverse correlation with the number of oocytes retrieved at the end of the cycle. This is likely to be a reflection of the IVF procedure, where the FSH dose of poor responders is raised to achieve an adequate number of eggs and the dose of high responders is lowered to avoid hyperstimulation (see Patients and Methods). These data exclude the possibility of a linear relationship between the ovarian response and the FSH amount received. Therefore, in this patient population, we defined as low responders those patients that produced fewer oocytes than 25% of their respective age group and as high responders those patients that produced more oocytes than 75% of their group. The exact oocyte cutoff number for each age group is shown in Fig. 1.
FSHR splicing variants
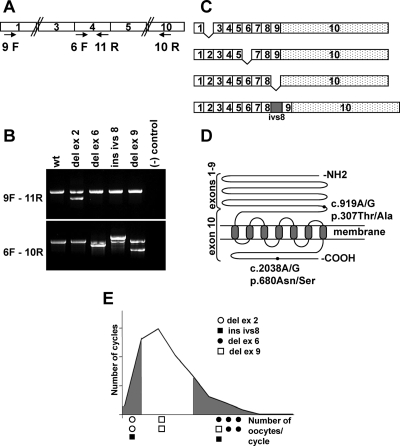
RNA was extracted from cumulus cells from 58 patients undergoing intracytoplasmic sperm injection in 63 cycles (Supplemental Table 2). Five patients had two cycles each. cDNA was synthesized, and RT-PCR for actin was used to assess the quantity and quality of isolated RNA. Eighty-six percent of the samples (54 of 63) were positive for actin and were further analyzed for the FSHR. Amplification of the FSHR cDNA was carried out using nested PCR (Fig. 2A). The receptor was successfully amplified in 35 cases (65%). In the rest of the samples, the amount of FSHR mRNA was below detectable levels. The RT-PCR products were analyzed by agarose gel electrophoresis and sequencing (Figs. 2B and 3). Alternatively spliced products were identified in 35% of the patients (12 of 34) (Table 1). These included exon skipping of exons 2, 6, and 9 and insertion of a 102-bp fragment of intron 8 (Fig. 2C). All patients expressing a splice variant were also expressing the WT form, and none of the patients contained more than one variant. The number of nucleotides on exons 2, 6, and 9 is a multiple of 3, and therefore exon skipping did not cause a frame-shift. All variants affect the extracellular domain of the receptor (Fig. 2D).
Figure 2.
RT-PCR analysis of FSHR. A, Schematic representation of RT-PCR primers used in nested PCR to amplify the FSHR cDNA. The numbered boxes indicate FSHR exons. B, RT-PCR products from patients carrying FSHR splicing variants. Upper and lower panels show amplification from exons 1–4 and 4–10, respectively. C, Schematic representation of the splicing variants detected in patients. D, Schematic representation of FSHR protein. The NH2 terminus constitutes the hormone binding domain, is extracellular, and is encoded by exons 1–9. The remaining protein is encoded by exon 10. The COOH terminus is intracellular. The positions of the two common polymorphisms are indicated by black circles. c, cDNA; p, protein. The nomenclature is according to the Human Genome Organization (HUGO) guidelines. E, Distribution of the splice variants along the graph of Fig. 1. del, Deletion; ins, insertion; ivs, intervening sequence (intron).
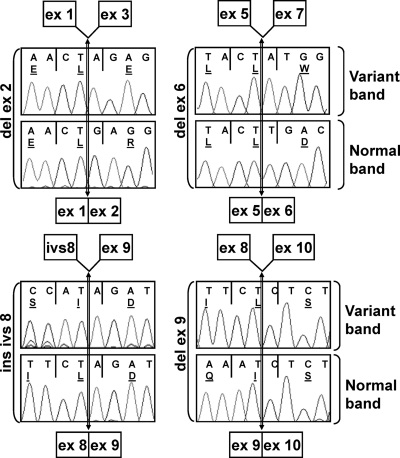
Figure 3.
FSHR mRNA sequencing. The bands shown in Fig. 2B were gel purified and sequenced. The lower panel of each pair shows the sequence of the normal band, and the upper panel shows the variant band. The limits of exons are indicated with a vertical double arrow. A schematic representation of each product is shown above and below each sequence pair.
Table 1.
Description of patients with FSHR splicing variants
| FSHR variant | Patient ID | Age (yr) | No. of oocytes | Response |
|---|---|---|---|---|
| Del ex2 | FR-1A | 33 | 4 | Low |
| FR-2 | 32 | 2 | Low | |
| Del ex6 | FR-1a | 30 | 26 | High |
| FR-1b | 32 | 20 | High | |
| FR-21 | 40 | 12 | High | |
| FR-25 | 25 | 37 | High | |
| FR-57 | 30 | 32 | High | |
| FR-78 | 31 | 34 | High | |
| Ins ivs8 | FR-6 | 31 | 4 | Low |
| FR-64 | 37 | 18 | High | |
| Del ex9 | FR-32 | 29 | 9 | Intermediate |
| FR-35 | 30 | 33 | High | |
| FR-40 | 34 | 8 | Intermediate |
Patient FR-1 was treated twice:
first cycle;
second cycle.
Del ex2 was associated with a low response to FSH (two of five patients), and del ex6 with a high response (five of 19 patients). Finally, ins ivs8 (two patients) and del ex9 (three patients) did not associate with a particular phenotype, and their clinical significance is not clear because they were also observed in good responders (Fig. 2E).
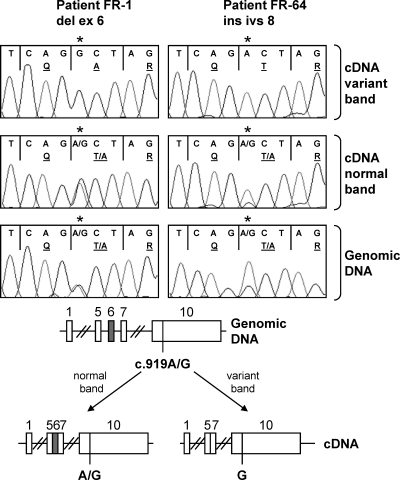
Sequencing of the intron-exon junctions and approximately 100 bp of the flanking introns did not reveal any DNA changes in the patients with the FSHR variants. The introns flanking the affected exons ranged between 2 and 86 kb in length and were not sequenced. However, we took advantage of the fact that some of the patients were heterozygous for the common FSHR polymorphisms lying within exon 10 to follow the origin of the spliced allele (Fig. 4). A PCR amplification of a fragment that encompassed the variant region and the exon 10 polymorphism was performed. The different size bands were sequenced. Interestingly, the variant band was only produced from one of the two alleles of the patient, whereas the WT band was expressed from both alleles. Patient 1 had two cycles, and the variant band originated twice from the same allele carrying the c.919A.
Figure 4.
Haplotype analysis of the variant band. An RT-PCR from exons 4–10 (primers 6F-10R) including the common c.919A/G polymorphism on exon 10 was performed from patients FR-1 and FR-64 who are heterozygous for this SNP. The normal and variant bands were isolated and sequenced. The location of the c.919A/G SNP is indicated with a star above the sequence. The top panel shows the sequence of exon 10 of the variant band. The middle panel shows the normal band. The bottom panel shows a genomic PCR of exon 10 to demonstrate that the patient was heterozygous. The variant band originated exclusively from one of the two alleles. The normal band originated from both. The graph depicts the preferential production of the variant band from one of the two FSHR alleles.
cAMP production upon FSH, LH, or TSH stimulation
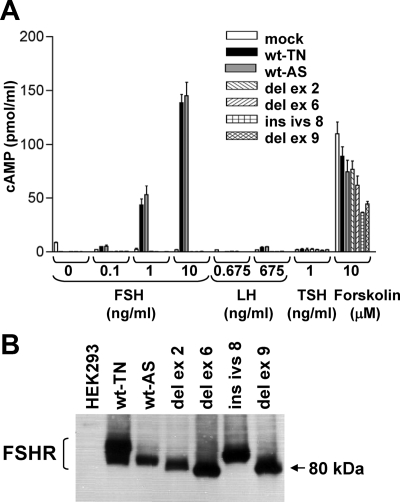
To assess the functional capacity of the FSHR splicing variants identified, we cloned the WT and the variants in the mammalian expression vector pcDNA3 and performed transient transfections in HEK293T cells. We cloned both common WT polymorphisms, named TN (for p.307Thr and p.680Asn) and AS (for p.307Ala and p.680Ser). The transfected cells were stimulated with increasing amounts of FSH, and the amount of cAMP produced was quantified (Fig. 5A). The two WT proteins showed a dose-response to increasing amounts of FSH used to stimulate the cells. In contrast, none of the splice variants were able to up-regulate cAMP production upon FSH stimulation. To verify that expression of the FSHR constructs did not impair the basic ability of the cells to increase cAMP, we treated transfected cells with forskolin and showed a marked elevation of cAMP production in all constructs (Fig. 5A). In all cases, transfection efficiency was measured using cotransfection with a luciferase-expressing plasmid.
Figure 5.
A, cAMP increase upon FSH stimulation. HEK293T cells were transiently transfected with expression plasmids carrying FSHR variants. cAMP was measured after FSH and LH stimulation. Only the two WT constructs (TN, AS) showed cAMP increase. Forskolin activation was used as a positive control. None of the constructs show stimulation with LH or TSH. B, Western blot analysis of transient transfections with the WT and variants of FSHR. The receptor was tagged with HA in the carboxyl terminus, and the detection was done with an anti-HA antibody.
Because all variants eliminate a portion of the extracellular, ligand-binding domain resulting in a possible conformation change, they may have acquired the ability to interact with similar hormones. We performed similar transient transfection experiments to test LH and TSH. None of the WT or variants were stimulated by LH or TSH (Fig. 5A). Adequate expression of all variants was confirmed with Western blot analysis (Fig. 5B).
Discussion
In this study, we have directly analyzed the FSHR mRNA extracted from cumulus cells in women undergoing fertility treatment and showed that, in this selected female infertility population, a remarkably high proportion of patients exhibit splicing variants of the FSHR. In particular, we identified splicing variants associated with low (del ex2) or high (del ex6) response to controlled ovarian stimulation with FSH. The identification of alternatively spliced products in 35% of patients has significant clinical implications in demonstrating a possible genetic cause to female infertility in some women and improves our understanding of the genetic basis of infertility.
Our approach based on the analysis of FSHR mRNA extracted from cumulus cells differs from previous studies because it enabled us to evaluate the posttranscriptional processing of FSHR mRNA, as well as the genomic sequence of FSHR. In contrast, all previous studies of FSHR in women undergoing IVF were performed on genomic DNA and/or have limited their investigations to the two common alleles. The majority of these studies have concentrated somewhat inconclusively on correlating the common polymorphisms to FSH amount received by the patients, stimulation outcome, and pregnancy rates (24,27,28). The use of cumulus cells is ideal because they are accessible, express FSHR, are routinely removed from the oocytes before intracytoplasmic sperm injection incurring no distress to the patients’ treatment, and allow us to gain information from a patient’s actual treatment cycle.
Genetic alterations of gonadotropin receptors have previously been reported. Point mutations in the FSHR have been shown to cause a gain or loss of function in specific cases (29). Gain-of-function point mutations have been found in the extracellular and the transmembrane domains of FSHR. Only one activating point mutation was found in a man, and it was shown to have constitutive activity (30). In all other cases found in women, the receptor lost its specificity, acquired affinity for hCG and/or TSH, and resulted in ovarian hyperstimulation syndrome during pregnancy (17,18,19,20,21). Loss-of-function mutations interfere with proper sexual development and/or ovarian function and result in infertility (11,12,14,15). These mutations have been found in any region of the receptor and can either suppress hormone binding or disrupt signal transduction (31). In the case of gain-of-function mutations, heterozygosity is sufficient to cause phenotypic variations and show a dominant inheritance (29). However, loss-of-function mutations are autosomal recessive.
Because all variants in the current study coexisted with the WT receptor, it is possible that their presence modulates the activity of the WT receptor in a dominant manner. It has been previously described that splice variants of the LH receptor interfere with trafficking of the full-length LH receptor and its expression on the cell surface and consequently with its ability to bind the hormone (32). Moreover, amino acid substitution of a cysteine residue of the FSHR carboxyl-terminal segment severely impaired its plasma membrane expression (33). Another possibility is that the variant proteins interfere with the internalization of the WT receptor or the transcriptional regulation of the gene.
Although point mutations on the FSHR can cause severe perturbations on sexual development and/or function, the two common FSHR polymorphisms have been associated with different basal FSH levels in women of reproductive age (34). Because the secretion of FSH is in a negative feedback loop with the action of FSHR, the basal levels of FSH are often indicative of the function of FSHR and the protein cascade downstream of the receptor. In this study, we identified a FSHR alternative splicing variant (del ex2) that is associated with a decreased pool of follicle growth.
In contrast to del ex2, which was found in two patients with only two and four oocytes, del ex6 observed in five separate patients was related to a higher response to ovarian stimulation. The consistent observation of this deletion with the same phenotype even in an older woman provides strong evidence that specific mutations in the FSHR are correlative to a functional phenotype. It is intriguing that the del ex6 splice variant was found in patients with a high response yet fails to show a cAMP increase in vitro. However, the cAMP up-regulation is the main but not the only signaling cascade downstream of FSHR (7). Further analysis of this variant in granulosa cells to test the activation of downstream targets is under way.
Exon skipping of exons 2 and 5 has been described for the mouse Fshr (35), and omission of exons 4, 5, and 9 has been observed in cattle (36). In rat and sheep, alternative splicing of the receptor results in soluble protein or modification of the intracellular carboxyl terminus (37,38). However, the physiology of the mouse and human ovaries differs significantly, including the fact that only one dominant follicle develops in human ovaries in a given cycle per month, as opposed to eight to 12 in the mouse. It is intriguing to postulate that differential regulation of the FSHR splicing and/or expression may play an important role in these interspecies differences.
In humans, skipping of exons 6 and 9 and insertion of intron 8 have been described in infertile males (39). Skipping of exon 9 has also been found in normal human testis (40). In light of these previous observations in males, our current findings are the first characterization of FSHR variants in relation with a measurable phenotype in women. The cause of alternative splicing in the patients we studied is elusive. No mutations were found in intron-exon junctions. Using single nucleotide polymorphisms (SNPs) in the coding region of FSHR as markers, we found that in patients that underwent several ovarian stimulation cycles the splice variant was always produced from the same allele. This is an indication that the alternative splicing observed is not due to random splicing regulation, but rather a specific event.
In our studies, we found that the presence of variant FSHRs is associated with a deviation from the normal ovarian response to FSH. The question remains as to whether this response is a cause of the patients’ infertility problems or is exacerbated by the use of controlled ovarian hyperstimulation regimens. It is also possible that the alternative splicing of the FSHR was mediated by the action of the hCG the patients receive to achieve oocyte maturation. A more complete assessment of female infertility patients is needed; however, the high incidence observed in our select group of patients indicates that this factor should now be considered.
The study of the genetics of reproductive disorders requires a different approach than other diseases. Due to the very nature of reproductive disorders, linkage analysis in large families across several generations is not applicable. A careful examination of the symptoms, physiological and morphological appreciation of tissue biopsies, and pharmacological response may provide more hints of the signal cascade contributing to the disorder. Here, we undertook a candidate gene approach and described FSHR variants seen in women with a normal menstrual cycle that show an abnormal response to FSH stimulation. These variants may play a role well beyond a woman’s reproductive years and contribute to her global health during menopause.
Supplementary Material
Acknowledgments
We thank Dr. Rosa Maduro for help with the collection of cumulus cells.
Footnotes
This work was funded by a Serono Special Grant (to D.S. and M.D.L.) and RO3 Grant HD055274-01 (to M.D.L.) from the National Institutes of Health/National Institute of Child Health and Human Development.
Present address for M.N.T.: Gray Institute for Radiation Oncology and Biology, University of Oxford, Oxford, United Kingdom.
Disclosure Summary: T.G., M.N.T., D.Z., and E.S. have nothing to disclose. M.D.L. and D.S. received grant support (2003–2008) from Serono.
First Published Online January 8, 2010
Abbreviations: del ex2, Deletion of exon 2; FSHR, FSH receptor; HA, hemagglutinin; hCG, human chorionic gonadotropin; HEK, human embryonic kidney; IVF, in vitro fertilization; SDS, sodium dodecyl sulfate; SNP, single nucleotide polymorphism; WT, wild-type.
References
- Huhtaniemi IT, Themmen AP 2005 Mutations in human gonadotropin and gonadotropin-receptor genes. Endocrine 26:207–217 [DOI] [PubMed] [Google Scholar]
- Camp TA, Rahal JO, Mayo KE 1991 Cellular localization and hormonal regulation of follicle-stimulating hormone and luteinizing hormone receptor messenger RNAs in the rat ovary. Mol Endocrinol 5:1405–1417 [DOI] [PubMed] [Google Scholar]
- Sprengel R, Braun T, Nikolics K, Segaloff DL, Seeburg PH 1990 The testicular receptor for follicle stimulating hormone: structure and functional expression of cloned cDNA. Mol Endocrinol 4:525–530 [DOI] [PubMed] [Google Scholar]
- Kelton CA, Cheng SV, Nugent NP, Schweickhardt RL, Rosenthal JL, Overton SA, Wands GD, Kuzeja JB, Luchette CA, Chappel SC 1992 The cloning of the human follicle stimulating hormone receptor and its expression in COS-7, CHO, and Y-1 cells. Mol Cell Endocrinol 89:141–151 [DOI] [PubMed] [Google Scholar]
- Sun L, Peng Y, Sharrow AC, Iqbal J, Zhang Z, Papachristou DJ, Zaidi S, Zhu LL, Yaroslavskiy BB, Zhou H, Zallone A, Sairam MR, Kumar TR, Bo W, Braun J, Cardoso-Landa L, Schaffler MB, Moonga BS, Blair HC, Zaidi M 2006 FSH directly regulates bone mass. Cell 125:247–260 [DOI] [PubMed] [Google Scholar]
- Means AR, Dedman JR, Tash JS, Tindall DJ, van Sickle M, Welsh MJ 1980 Regulation of the testis Sertoli cell by follicle stimulating hormone. Annu Rev Physiol 42:59–70 [DOI] [PubMed] [Google Scholar]
- Ulloa-Aguirre A, Zariñán T, Pasapera AM, Casas-González P, Dias JA 2007 Multiple facets of follicle-stimulating hormone receptor function. Endocrine 32:251–263 [DOI] [PubMed] [Google Scholar]
- Layman LC, Lee EJ, Peak DB, Namnoum AB, Vu KV, van Lingen BL, Gray MR, McDonough PG, Reindollar RH, Jameson JL 1997 Delayed puberty and hypogonadism caused by mutations in the follicle-stimulating hormone β-subunit gene. N Engl J Med 337:607–611 [DOI] [PubMed] [Google Scholar]
- Matthews CH, Borgato S, Beck-Peccoz P, Adams M, Tone Y, Gambino G, Casagrande S, Tedeschini G, Benedetti A, Chatterjee VK 1993 Primary amenorrhoea and infertility due to a mutation in the β-subunit of follicle-stimulating hormone. Nat Genet 5:83–86 [DOI] [PubMed] [Google Scholar]
- Phillip M, Arbelle JE, Segev Y, Parvari R 1998 Male hypogonadism due to a mutation in the gene for the β-subunit of follicle-stimulating hormone. N Engl J Med 338:1729–1732 [DOI] [PubMed] [Google Scholar]
- Aittomäki K, Lucena JL, Pakarinen P, Sistonen P, Tapanainen J, Gromoll J, Kaskikari R, Sankila EM, Lehväslaiho H, Engel AR, Nieschlag E, Huhtaniemi I, de la Chapelle A 1995 Mutation in the follicle-stimulating hormone receptor gene causes hereditary hypergonadotropic ovarian failure. Cell 82:959–968 [DOI] [PubMed] [Google Scholar]
- Beau I, Touraine P, Meduri G, Gougeon A, Desroches A, Matuchansky C, Milgrom E, Kuttenn F, Misrahi M 1998 A novel phenotype related to partial loss of function mutations of the follicle stimulating hormone receptor. J Clin Invest 102:1352–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty E, Pakarinen P, Tiitinen A, Kiilavuori A, Huhtaniemi I, Forrest S, Aittomäki K 2002 A novel mutation in the FSH receptor inhibiting signal transduction and causing primary ovarian failure. J Clin Endocrinol Metab 87:1151–1155 [DOI] [PubMed] [Google Scholar]
- Meduri G, Touraine P, Beau I, Lahuna O, Desroches A, Vacher-Lavenu MC, Kuttenn F, Misrahi M 2003 Delayed puberty and primary amenorrhea associated with a novel mutation of the human follicle-stimulating hormone receptor: clinical, histological, and molecular studies. J Clin Endocrinol Metab 88:3491–3498 [DOI] [PubMed] [Google Scholar]
- Touraine P, Beau I, Gougeon A, Meduri G, Desroches A, Pichard C, Detoeuf M, Paniel B, Prieur M, Zorn JR, Milgrom E, Kuttenn F, Misrahi M 1999 New natural inactivating mutations of the follicle-stimulating hormone receptor: correlations between receptor function and phenotype. Mol Endocrinol 13:1844–1854 [DOI] [PubMed] [Google Scholar]
- Allen LA, Achermann JC, Pakarinen P, Kotlar TJ, Huhtaniemi IT, Jameson JL, Cheetham TD, Ball SG 2003 A novel loss of function mutation in exon 10 of the FSH receptor gene causing hypergonadotrophic hypogonadism: clinical and molecular characteristics. Hum Reprod 18:251–256 [DOI] [PubMed] [Google Scholar]
- Smits G, Olatunbosun O, Delbaere A, Pierson R, Vassart G, Costagliola S 2003 Ovarian hyperstimulation syndrome due to a mutation in the follicle-stimulating hormone receptor. N Engl J Med 349:760–766 [DOI] [PubMed] [Google Scholar]
- Montanelli L, Delbaere A, Di Carlo C, Nappi C, Smits G, Vassart G, Costagliola S 2004 A mutation in the follicle-stimulating hormone receptor as a cause of familial spontaneous ovarian hyperstimulation syndrome. J Clin Endocrinol Metab 89:1255–1258 [PubMed] [Google Scholar]
- Vasseur C, Rodien P, Beau I, Desroches A, Gérard C, de Poncheville L, Chaplot S, Savagner F, Croué A, Mathieu E, Lahlou N, Descamps P, Misrahi M 2003 A chorionic gonadotropin-sensitive mutation in the follicle-stimulating hormone receptor as a cause of familial gestational spontaneous ovarian hyperstimulation syndrome. N Engl J Med 349:753–759 [DOI] [PubMed] [Google Scholar]
- De Leener A, Caltabiano G, Erkan S, Idil M, Vassart G, Pardo L, Costagliola S 2008 Identification of the first germline mutation in the extracellular domain of the follitropin receptor responsible for spontaneous ovarian hyperstimulation syndrome. Hum Mutat 29:91–98 [DOI] [PubMed] [Google Scholar]
- De Leener A, Montanelli L, Van Durme J, Chae H, Smits G, Vassart G, Costagliola S 2006 Presence and absence of follicle-stimulating hormone receptor mutations provide some insights into spontaneous ovarian hyperstimulation syndrome physiopathology. J Clin Endocrinol Metab 91:555–562 [DOI] [PubMed] [Google Scholar]
- Dierich A, Sairam MR, Monaco L, Fimia GM, Gansmuller A, LeMeur M, Sassone-Corsi P 1998 Impairing follicle-stimulating hormone (FSH) signaling in vivo: targeted disruption of the FSH receptor leads to aberrant gametogenesis and hormonal imbalance. Proc Natl Acad Sci USA 95:13612–13617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser BC, Diedrich K, Devroey P 2008 Predictors of ovarian response: progress towards individualized treatment in ovulation induction and ovarian stimulation. Hum Reprod Update 14:1–14 [DOI] [PubMed] [Google Scholar]
- Perez Mayorga M, Gromoll J, Behre HM, Gassner C, Nieschlag E, Simoni M 2000 Ovarian response to follicle-stimulating hormone (FSH) stimulation depends on the FSH receptor genotype. J Clin Endocrinol Metab 85:3365–3369 [DOI] [PubMed] [Google Scholar]
- Sudo S, Kudo M, Wada S, Sato O, Hsueh AJ, Fujimoto S 2002 Genetic and functional analyses of polymorphisms in the human FSH receptor gene. Mol Hum Reprod 8:893–899 [DOI] [PubMed] [Google Scholar]
- Lifton RP, Gharavi AG, Geller DS 2001 Molecular mechanisms of human hypertension. Cell 104:545–556 [DOI] [PubMed] [Google Scholar]
- Laven JS, Mulders AG, Suryandari DA, Gromoll J, Nieschlag E, Fauser BC, Simoni M 2003 Follicle-stimulating hormone receptor polymorphisms in women with normogonadotropic anovulatory infertility. Fertil Steril 80:986–992 [DOI] [PubMed] [Google Scholar]
- Jun JK, Yoon JS, Ku SY, Choi YM, Hwang KR, Park SY, Lee GH, Lee WD, Kim SH, Kim JG, Moon SY 2006 Follicle-stimulating hormone receptor gene polymorphism and ovarian responses to controlled ovarian hyperstimulation for IVF-ET. J Hum Genet 51:665–670 [DOI] [PubMed] [Google Scholar]
- Meduri G, Bachelot A, Cocca MP, Vasseur C, Rodien P, Kuttenn F, Touraine P, Misrahi M 2008 Molecular pathology of the FSH receptor: new insights into FSH physiology. Mol Cell Endocrinol 282:130–142 [DOI] [PubMed] [Google Scholar]
- Gromoll J, Simoni M, Nieschlag E 1996 An activating mutation of the follicle-stimulating hormone receptor autonomously sustains spermatogenesis in a hypophysectomized man. J Clin Endocrinol Metab 81:1367–1370 [DOI] [PubMed] [Google Scholar]
- Rannikko A, Pakarinen P, Manna PR, Beau I, Misrahi M, Aittomäki K, Huhtaniemi I 2002 Functional characterization of the human FSH receptor with an inactivating Ala189Val mutation. Mol Hum Reprod 8:311–317 [DOI] [PubMed] [Google Scholar]
- Apaja PM, Tuusa JT, Pietilä EM, Rajaniemi HJ, Petäjä-Repo UE 2006 Luteinizing hormone receptor ectodomain splice variant misroutes the full-length receptor into a subcompartment of the endoplasmic reticulum. Mol Biol Cell 17:2243–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe A, Zariñán T, Pérez-Solis MA, Gutiérrez-Sagal R, Jardón-Valadez E, Piñeiro A, Dias JA, Ulloa-Aguirre A 2008 Functional and structural roles of conserved cysteine residues in the carboxyl-terminal domain of the follicle-stimulating hormone receptor in human embryonic kidney 293 cells. Biol Reprod 78:869–882 [DOI] [PubMed] [Google Scholar]
- de Koning CH, Benjamins T, Harms P, Homburg R, van Montfrans JM, Gromoll J, Simoni M, Lambalk CB 2006 The distribution of FSH receptor isoforms is related to basal FSH levels in subfertile women with normal menstrual cycles. Hum Reprod 21:443–446 [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy PJ, Marsh P, Dudley K 1994 Follicle-stimulating hormone receptor mRNA in the mouse ovary during post-natal development in the normal mouse and in the adult hypogonadal (hpg) mouse: structure of alternate transcripts. Mol Cell Endocrinol 101:197–201 [DOI] [PubMed] [Google Scholar]
- Rajapaksha WR, Robertson L, O'Shaughnessy PJ 1996 Expression of follicle-stimulating hormone-receptor mRNA alternate transcripts in bovine granulosa cells during luteinization in vivo and in vitro. Mol Cell Endocrinol 120:25–30 [DOI] [PubMed] [Google Scholar]
- Kraaij R, Verhoef-Post M, Grootegoed JA, Themmen AP 1998 Alternative splicing of follicle-stimulating hormone receptor pre-mRNA: cloning and characterization of two alternatively spliced mRNA transcripts. J Endocrinol 158:127–136 [DOI] [PubMed] [Google Scholar]
- Sairam MR, Jiang LG, Yarney TA, Khan H 1996 Follitropin signal transduction: alternative splicing of the FSH receptor gene produces a dominant negative form of receptor which inhibits hormone action. Biochem Biophys Res Commun 226:717–722 [DOI] [PubMed] [Google Scholar]
- Song GJ, Park YS, Lee YS, Lee CC, Kang IS 2002 Alternatively spliced variants of the follicle-stimulating hormone receptor gene in the testis of infertile men. Fertil Steril 77:499–504 [DOI] [PubMed] [Google Scholar]
- Gromoll J, Gudermann T, Nieschlag E 1992 Molecular cloning of a truncated isoform of the human follicle stimulating hormone receptor. Biochem Biophys Res Commun 188:1077–1083 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.