Abstract
DNA methylation is a critical epigenetic regulator in mammalian development. Here, we present a whole-genome comparative view of DNA methylation using bisulfite sequencing of three cultured cell types representing progressive stages of differentiation: human embryonic stem cells (hESCs), a fibroblastic differentiated derivative of the hESCs, and neonatal fibroblasts. As a reference, we compared our maps with a methylome map of a fully differentiated adult cell type, mature peripheral blood mononuclear cells (monocytes). We observed many notable common and cell-type-specific features among all cell types. Promoter hypomethylation (both CG and CA) and higher levels of gene body methylation were positively correlated with transcription in all cell types. Exons were more highly methylated than introns, and sharp transitions of methylation occurred at exon–intron boundaries, suggesting a role for differential methylation in transcript splicing. Developmental stage was reflected in both the level of global methylation and extent of non-CpG methylation, with hESC highest, fibroblasts intermediate, and monocytes lowest. Differentiation-associated differential methylation profiles were observed for developmentally regulated genes, including the HOX clusters, other homeobox transcription factors, and pluripotence-associated genes such as POU5F1, TCF3, and KLF4. Our results highlight the value of high-resolution methylation maps, in conjunction with other systems-level analyses, for investigation of previously undetectable developmental regulatory mechanisms.
DNA methylation is an important epigenetic modification that plays critical roles in cellular differentiation, development, and disease. Hypermethylation is strongly associated with heterochromatin and transcriptional silencing (Keshet et al. 1986; Reik et al. 2001). In particular, correct DNA methylation is critical for X-inactivation (Heard et al. 1997), imprinting (Li et al. 1993), and silencing of specific genomic elements, such as transposons (Walsh et al. 1998). Derangements in DNA methylation patterns have been associated with dysregulation of gene expression in cancer cells, particularly down-regulation of genes with tumor suppressor functions by hypermethylation of their promoter regions (Ting et al. 2006).
In mammals, the predominant form of DNA methylation occurs symmetrically on the cytosine residues on both strands of CpG dinucleotides, although there is evidence that cytosine methylation is not limited to those in CpG sequences (Ramsahoye et al. 2000; Haines et al. 2001; Dodge et al. 2002). About 70%–80% of CpG sites in mammalian cells are methylated, but both the CpG sites and their degrees of methylation are unevenly distributed in the genome (Bird et al. 1985). CpG dinucleotides are largely concentrated in small regions termed “CpG islands” (Bird et al. 1985), which are found within the promoters of ∼70% of human genes (Saxonov et al. 2006). CpGs located in promoter-associated CpG islands tend to be unmethylated, but a subset are differentially methylated in specific tissues (Song et al. 2005) or during the course of development (Li 2002), and result in transcriptional repression of the adjacent genes.
Most methods currently used to examine the DNA methylation patterns are biased toward CpG-rich regions of the genome, using methylation-sensitive restriction enzymes (Meissner et al. 2008), affinity enrichment through methyl-cytosine-specific protein domains or antibodies (Cross et al. 1994; Weber et al. 2007), or bisulfite conversion (Frommer et al. 1992). These studies have provided a snapshot of methylation status in a variety of cell types, but have low resolution and limited genomic coverage and are biased toward specific genomic features, such as CpG islands, promoter regions, or subsets of genes (Bibikova et al. 2006; Fouse et al. 2008; Meissner et al. 2008). Recent methodological improvements have revealed subtler methylation patterns that correlate with gene expression or cell type. For example, methylation at CpG sites located at the edges, or “shores,” of promoter-associated CpG islands has been inversely correlated with gene expression (Irizarry et al. 2009). Expressed protein-coding genes in general appear to have low methylation around their promoter region and high methylation over their gene body (Ball et al. 2009). In addition, comparison between human pluripotent stem cells and somatic cells revealed cell-type-specific areas of differential methylation (Deng et al. 2009).
These studies show that as resolution of the genomic methylation profile increases, new, subtler phenomena are revealed, providing new insights into the mechanisms by which cells regulate gene expression. We used next-generation bisulfite sequencing technology and bioinformatics analysis to produce single base pair–resolution whole-genome methylation maps of cells representing progressive stages of cellular differentiation, ranging from pluripotent (undifferentiated hESCs) to differentiated (a fibroblastic derivative of the hESCs and a primary human fibroblast cell line). As a reference standard, we compared the maps of our cultured cell types to a methylome map of terminally differentiated peripheral blood mononuclear cells (monocytes).
The high resolution of our maps and the selection of cell types mapped allowed us to confirm previously reported observations and recognize new subtle differences in methylation that are likely to play important roles in development. Specifically, although mCpGs were the dominant differentially methylated sites in all the cell types, we found a higher level of methylated cytosines in the mCpA context in undifferentiated hESCs compared to differentiated cells (Ramsahoye et al. 2000). mCpA followed the same pattern as mCpG and was found across the genome and gene regions but reduced at promoters. We were also able to identify differences in relative distributions of DNA methylation along the sequences of genes that correlated with their degree of expression in different cell types, and could link DNA methylation with underlying chromatin states and splicing processes. These data, and information about specific genomic regions reported here, provide clues about how multiple levels of regulation of gene expression and complex of DNA modifications are involved in carrying out precisely choreographed developmental programs.
Results
To investigate the dynamics of DNA methylation at different states of differentiation, we sequenced bisulfite-converted and unconverted DNA from three cultured cell types that represent progressive stages of differentiation: pluripotent undifferentiated hESCs, fibroblast-like cells differentiated from hESCs (hESC-Fibro), and primary neonatal foreskin fibroblasts (Fibro). For an adult cell reference standard, we compared base-level maps of these cultured cell methylomes with a base-level methylation map of fully differentiated adult cells, a preparation of peripheral blood mononuclear cells (monocytes) (Y Li, J Zhu, G Tian, N Li, Q Li, M Ye, H Zheng, J Yu, H Wu, J Sun, et al., unpubl.). As assessed by hierarchical clustering of global gene expression data, the hESC-derived fibroblasts were similar to differentiated primary cell lines (Supplemental Fig. 1A; Muller et al. 2008). Differential gene expression analysis of the same data suggested that the hESC-derived fibroblasts are a developmental intermediate between hESC and neonatal fibroblasts (Supplemental Fig. 1B).
In total, an average of 542 million aligned reads was generated for each cell type with read lengths up to 75 bp, of which about 400 million could be uniquely mapped to the Hg18 reference genome. The data were filtered, and only the nonredundant, uniquely mapped reads were used for subsequent analysis, resulting in a median coverage of 9 reads per base. Out of the 583 million cytosines in the haploid genome, >60% of all Cs and >70% of Cs in CpG dinucleotides, were covered by at least 3 three reads and, to ensure accuracy, only the ≥3× covered sites were used for determining the methylation status. The bisulfite conversion rates estimated from the bisulfite-PCR followed by sequencing of selective regions of known methylation states showed nearly complete (98.5%–99.3%) conversion efficiency. The details of the sequencing methods and mapping summaries are provided in the Supplemental material (Supplemental Fig. 2; Supplemental Table 1).
Because we analyzed populations of cultured cells, the methylation status of a particular C could range continuously from 0% to 100%. Therefore, the methylation status for each C was expressed as a frequency (β), and each C was classified into one of five categories: methylated (M: >80%), intermediate between partially methylated and methylated (M_P: 60%–80%), partially methylated (P: 40%–60%), intermediate between unmethylated and partially methylated (U_P: 20%–40%), or unmethylated (U: <20%). To validate our data processing and methylation calling strategy, we compared the bisulfite-seq results with data from an independent array-based analysis (Illumina Infinium HumanMethylation27 BeadChip microarray) generated from the same cell preparations. The HumanMethylation27 array interrogates the methylation status of 27,318 CpG sites from 14,495 promoter regions, representing 0.1% of total CpG sites in the genome and 0.01% of the total number of Cs covered by bisulfite sequencing. The array methylation status of the CpG sites was subdivided into the same five categories used to classify the sequencing data. Agreement between the array and sequencing data was high, with 79% of the common CpGs showing concordant methylation status (Supplemental Table 2). Discordant calls were mainly derived from low-coverage (≤3 reads per C) sites in the bisulfite sequencing data. These results allowed us to place high confidence in calls where the coverage was ≥3 reads.
Genome-wide DNA methylation profiles reveal significant non-CpG methylation in hESCs
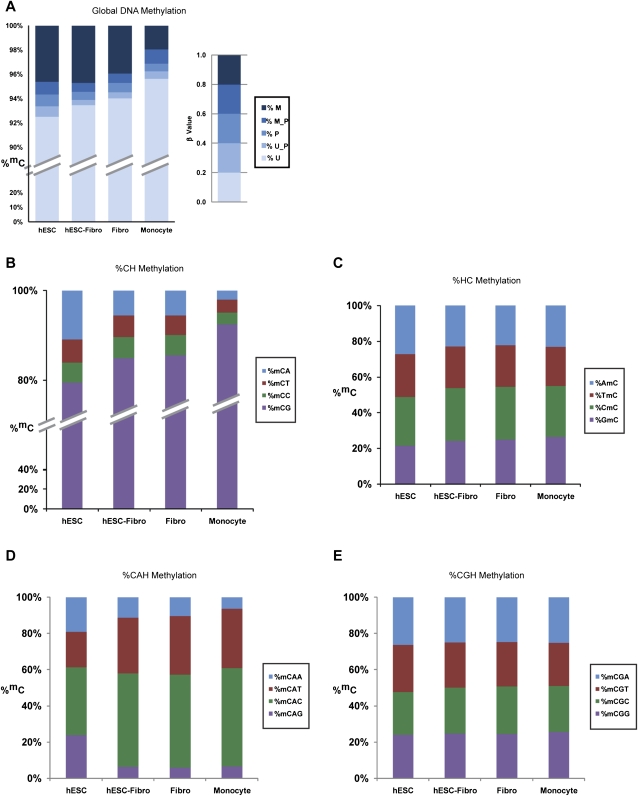
Consistent with results from previous work (Rollins et al. 2006), cytosine residues were mostly unmethylated, with 92%–95% of Cs unmethylated in the four cell types surveyed. We found that the degree of global DNA methylation was inversely correlated with differentiation status. The highest level of methylation was seen in the undifferentiated hESCs (Fig. 1A), suggesting that a global reduction in DNA methylation occurs during differentiation.
Figure 1.
Distribution of DNA methylation levels and the corresponding sequence context. (A) The percentage of methylated Cs was determined by taking the ratio of the number of methylated Cs over the total number of covered Cs. Methylation levels were grouped into five categories: unmethylated (U), intermediate between unmethylated and partially methylated (U_P), partially methylated (P), intermediate between partially methylated and methylated (M_P), and methylated (M). Levels of methylation were found to be highest in undifferentiated hESCs at ∼6% with a reduction in the differentiated cells. The fully differentiated peripheral blood mononuclear cells (monocytes) had the lowest methylation levels at ∼3%. Note: the monocyte data were provided as a base-level methylome map for comparison purposes only (Jun Wang, Beijing Genome Institute). (B) DNA methylation in various combinations of sequence contexts (CH, HC, CHH; H = any four nucleotides) throughout the genome was examined. In the CH sequence context, CpG methylation was the predominant form, but a significant fraction of methylated cytosines were found at CpA sites, particularly in hESCs (where CpA methylation represented >10% of methylcytosines). Levels of CpA methylation were lower in differentiated cells (with the lowest levels in monocytes, 2% of methylcytosines found at CpA sites). (C) In the HC sequence context, the position immediately 5′ of the methylcytosine did not appear to influence the methylation rate, as levels of methylation of the four categories of HC were equally distributed. (D,E) In the CHH sequence context, the predominant methylation type was CGH, followed by CAH. The position immediately 3′ to the dinucleotide had a weak effect on the methylation and was largely dependent on the identity of the second base of the dinucleotide.
The methylation levels of CpG dinucleotides had a bimodal distribution, consistent with previous reports (Supplemental Fig. 3; Meissner et al. 2008). However, in contrast to data generated by methods biased toward regions of high CpG density, which indicated 40% methylated and 20% unmethylated CpGs, our whole-genome approach revealed that 55% of CpGs were methylated (β ≥ 80%) and 20% were unmethylated (β ≤ 20%). The remainder was partially methylated. The higher percentage of methylated CpGs detected in our study likely reflects the existence of a high level of methylated CpGs within low CpG density regions of the genome that were not detected in the earlier studies.
We recorded the relative prevalence of DNA methylation in different sequence contexts throughout the genome by systematically evaluating for base preference at the −1, +1, and +2 positions relative to cytosines (HC, CH, and CHH). All of the cell types had a strong preference for cytosine methylation in the CpG context (>80%). Of the four cell types studied, the hESCs had the highest level of non-CpG methylation, with 20% of the methylcytosines in non-CpG dinucleotides (Fig. 1B). Of the non-CpG methylation types, mCpAs were the most common, and the prevalence of mCpAs showed the same pattern as the CpG methylation, decreasing as the degree of differentiation increased. The validity of CpA methylation calls was confirmed by bisulfite conversion followed by PCR and sequencing of selected regions (Supplemental Fig. 4). In contrast, mCpC and mCpT methylations were rare and did not vary significantly among the four cell types. The identity of the bases at the −1 and +2 positions had little impact on DNA methylation level (Fig. 1C–E), with a weak preference (C > T > A > G) at the +2 position for methylated CpA dinucleotides as the only observable bias (Fig. 1D). Although there was a global decrease in non-CpG (primarily CpA) methylation level with differentiation, we found that the CpA methylation profile was ∼93% conserved among the three differentiated cell types, the hESC-derived fibroblasts (hESC-Fibro), neonatal fibroblasts, and monocytes, suggesting that the CpA methylation pattern was nonrandom. The undifferentiated hESCs had much more CpA methylation than the other cell types, but the hESC profile included the same mCpA sites as the differentiated cells.
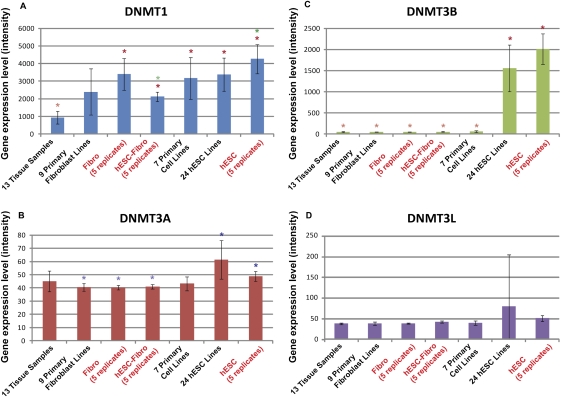
In mammals, DNMT3s are responsible for de novo methylation of CpGs, but the mechanism of cytosine methylation in a non-CpG context is not clear. The nonsymmetric nature of mCpA sites would seem to preclude the re-establishment of their methylation during replication by CpG-methylating DNMTs, but one possible mechanism is nonspecific DNMT3 activity (Ramsahoye et al. 2000). Our gene expression analysis showed the cell type that had the highest level of non-CpG methylation, hESCs, also had the highest expression levels of DNMT3A and DNMT3B, with DNMT3B showing particularly high expression in hESCs (Fig. 2). To evaluate whether the methylation of CpA could have resulted from nearby CpG methylation activity, we determined the density of mCpG surrounding mCpAs. The results showed that, although the density of CpGs is generally higher surrounding CpAs, the density of mCpG surrounding mCpA was no different from the density of mCpG found in any random selected CpA site. Taken together, these data suggest that CpA methylation may be linked to DNMT3b activity, but does not appear to be the result of nonspecific methylation resulting from nearby CpG methylation.
Figure 2.
DNMT1, DNMT3A, DNMT3B, and DNMT3L gene expression in the hESC, hESC-Fibro, and Fibro cell types in the context of a large collection of tissue, primary cell, and hESC cell samples. Gene expression was extracted from microarray data (Muller et al. 2008). Gene expression levels measured as quantiles-normalized signal intensity are indicated on the y-axis. Error bars, SD. Data from five biological replicates of the cell lines used for bisulfite sequencing (hESC, hESC-Fibro, and Fibro) have red labels. Significant differences in expression between cell types are indicated by sets of colored asterisks; the cell types with higher expression are marked with darker asterisks, and the cell types with lower expression are marked with lighter asterisks of the same color (e.g., in A, DNMT1 expression is significantly lower in the Tissue group compared to all of the other groups with the exception of the Primary Fibroblast group).
Genomic features associated with DNA methylation
The methylation profiles across each chromosome were examined. Using a 100-kb sliding window, we calculated the average methylcytosine intensities of total mC, mCpG, and mCpA separately across the genome. Overall, the baseline level of constitutive methylation varied among chromosomes, with higher levels of methylation on chromosomes 16, 17, 19, and 22, which correlates with the higher gene density on these chromosomes (Supplemental Fig. 5).
When CpG methylation was examined along the length of each chromosome, we noted a uniform baseline level of methylation, with sporadic hypomethylated regions (Fig. 3A; Supplemental Fig. 6). Using ±3 standard deviations (SDs) as a cutoff, 286 regions were identified in hESCs as hypomethylated mCpG regions, primarily in clusters of promoters and CpG islands. The four most significantly hypomethylated regions in hESCs corresponded to the HOXA, HOXC, HOXB, and HOXD loci. Looking closely at the HOX clusters in the four cell types, we saw a progressive increase in methylation of these genes with extent of differentiation (Fig. 3B). In the differentiated hESC (hESC-Fibro), 103 out of these 286 hypomethylated regions had higher methylation levels than the hESCs (>5 SDs), while only 71 of the 286 hESC-associated hypomethylated regions remained hypomethylated in hESC-Fibro cells.
Figure 3.
(A) Methylation profile of chromosome 7 in hESC sample. (Dark blue bars) The positions of RefSeq genes; (green bars) the positions of CpG islands; (light blue trace) CpG methylation. The region surrounding the HOXA locus is expanded to show the level of hypomethylation. (B) The differential methylation profiles in relation to differentiation within the clusters of four HOX loci. Overall DNA methylation intensity of these clusters was the lowest in hESC and highest in monocytes and Fibro cells. (C) Percent total cytosine methylation for genomic repeat elements. Error bars, SD. The types of repeat elements shown are Alu, ERV, LINE, LTR, microsatellite, and SINE. Comparison of methylation levels of hESC, hESC-Fibro, and Fibro cell lines shows lower methylation in more differentiated cells for all of these types of repeat elements except microsatellites.
We also determined the methylation level of known repeat elements on both strands. Some repeats, such as Alus, were slightly hypermethylated relative to the average methylation level of ∼8.5% (in hESCs), while others, such as LINE elements, were relatively hypomethylated. Interestingly, several types of repeat elements, including LTRs, showed lower levels of methylation with differentiation, beyond the overall drop in average methylation seen in the differentiated cell types (Fig. 3C).
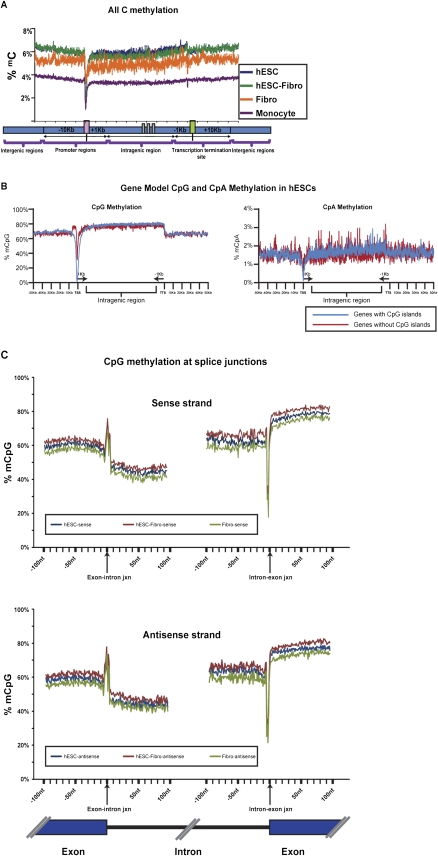
High-resolution methylation mapping allowed us to closely examine the relationship of DNA methylation to transcription. We mapped the DNA methylation data for each gene to a “gene model,” which contained annotated genomic features in the neighborhood of transcribed genes, including promoters/transcription start sites (TSSs), gene body, transcription termination sites (TTSs), and intergenic regions. Promoters were defined as −10 kb to +1 kb of the TSS, TTS regions were defined as −1 kb to +10 kb of the TTS, gene body regions were defined as +1 kb from the TSS to −1 kb from the TTS, and intergenic regions consisted of regions not included in the above three categories. The density of DNA methylation in each region was calculated as the percent of methylcytosine over total covered Cs. By scaling and normalizing the profiles for all 17,578 RefSeq genes, a clear pattern emerged, which was similar for all four cell types (Fig. 4A). There was high methylation throughout most of the promoter region with a sharp dip starting ∼1 kb upstream of the TSS with a nadir immediately upstream of the TSS, followed by a more gradual increase in methylation over the first part of the transcribed region. Methylation remained high throughout the rest of the intragenic region, and then showed a small but sharp step down at the TTS to a level that was maintained in the intergenic regions.
Figure 4.
(A) Average distribution of DNA methylation mapped onto a gene model. Overall methylation levels at the TSS (transcription start site) region were lower in hESCs compared to the differentiated cell types. (B) The CpG and CpA methylation distribution surrounding genes with and without CpG islands (CpI), shown for hESCs. CpG and CpA methylation levels were lower at the TSS region in both genes with and genes without CpG islands at the promoter. However, the level of methylation was lower for genes with promoters containing CpG islands. Promoters without CpG islands showed a peak of CpG methylation ∼1.5–5.0 kb upstream of the TSS. Data for all cell types are shown in Supplemental Figure 7. (C) CpG methylation across splice junctions. The percent of mCpG across a 100-bp window spanning the exon/intron junctions was mapped. Both sense (upper panel) and antisense (lower panel) strands showed a sharp spike in CpG methylation at the exon/intron junction, followed by a steep decrease in methylation that gradually increases with proximity to the next exon. Another sharp spike, of decreased methylation in this case, is followed by a steep rise in methylation as the next exon begins.
To dissect subtle methylation features that might be involved in regulation of gene expression, we plotted CpG and CpA methylation separately for genes with CpG-island-containing promoters (CPI promoters) and non-CPI-containing promoters (non-CPI promoters) (Fig. 4B). The CpG traces were very similar to the total C traces, with the exception of the TSS for genes with non-CPI promoters: a small but sharp rise in CpG methylation occurred just before the start of the dip at the TSS in all cell lines. This small peak was located between 1.5 and 5 kb upstream of the non-CPI promoters, and the intensity of this peak did not appear to correlate with transcription (data not shown). The CpA traces were not as distinct, but they still showed a clear dip in methylation at the TSS and a stepdown at the TTS (Fig. 4B).
In agreement with earlier reports on chromosome 21 (Zhang et al. 2009), we observed a strong inverse correlation in all cell types between CpG density and overall methylation level around the promoter regions. Specifically, the CPI promoters had very low levels of CpG methylation at the TSS (∼1.2%–2% in hESCs), whereas the non-CPI promoters showed markedly higher levels of CpG methylation (35%–40% in hESCs) (Fig. 4B; Supplemental Fig. 7). CPI promoters have been associated with constitutively expressed genes, while non-CPI promoters have been associated with developmentally regulated genes. This pattern suggests that the dramatically higher levels of DNA methylation at the TSS of non-CPI promoters may make these genes more susceptible to regulation by DNA methylation. The difference in methylation of CpG and non-CpG island promoters was most marked in hESCs, suggesting that this cell type may be poised to change the activation state of developmentally regulated genes.
We examined the methylation profiles of introns, exons, and across splice junctions, and observed that exons were methylated at markedly higher levels (10.5% in hESCs) compared to introns, which were methylated at rates close to the genomic average. Interestingly, we observed a striking change in methylation across the splice junctions on both the sense and antisense strands at both ends of the intron. There was a sharp spike in DNA methylation at the 5′ splice site and a sharp dip at the 3′ splice site of the intron/exon boundary (Fig. 4C). This steep change in methylation level is probably influenced by the donor/acceptor sequence context around the splice junctions. A downward gradient was seen going across exons from 5′ to 3′, while an upward gradient of DNA methylation was seen traveling from 5′ to 3′ across introns (Fig. 4C). While this is the first report on the splice junction methylation spikes, recent reports show that the intron–exon boundaries also appear to be marked by gradients in chromatin features, including nucleosomes (Schwartz et al. 2009) and the H3K36me3 histone mark (Kolasinska-Zwierz et al. 2009). Taken together, our data suggest that coupling of transcription and splicing may be regulated by DNA methylation as well as by other epigenetic marks.
DNA methylation and gene expression
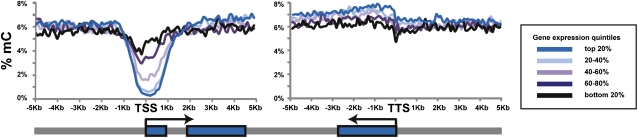
DNA methylation is linked to gene silencing and is considered to be an important mechanism in the regulation of gene expression. To explore the relationship between gene expression and methylation, we used genome-wide gene expression data from three biological replicates of the same hESC, hESC-Fibro, and Fibro cell lines, generated with the Human WG-6 Gene Expression BeadChip (Illumina). The 25,159 unique genes were divided into five categories ranked by expression level, and the average DNA methylation values for each category were mapped onto the gene model. We found that DNA methylation around the TSS was negatively correlated with gene expression, whereas methylation levels in the gene body and TTS regions were positively correlated with gene expression (Fig. 5). For example, in the hESCs, the region ±1 kb of the TSS was only 0.42% methylated in the most highly expressed gene category and 4.8% methylated in the least expressed gene category.
Figure 5.
Correlation of methylation profile with expression level in hESCs. The expression levels of genes in hESCs (from microarray analysis) were divided into five categories. The 20% most highly expressed genes exhibited the lowest methylation levels with the nadir of the hypomethylated “valley” centered at ±1 kb from their TSS. As the gene expression decreased, the valley became more shallow. Interestingly, the levels of methylation found in the gene bodies of the most highly expressed genes were slightly higher than in genes expressed at lower levels.
Correlation between DNA methylation and histone modifications
Histone modification is another major epigenetic modification that is associated with DNA methylation and contributes to transcriptional regulation (Meissner et al. 2008). The H3K4me3 mark is associated with active transcriptional initiation, while H3K27me3 is associated with inactive promoters. In murine ESCs, developmentally regulated genes have been reported to be enriched for bivalent marks that include both H3K4me3 and H3K27me3 (Bernstein et al. 2006). To explore the relationship between DNA methylation and the chromatin state, H3K4me3 and H3K27me3 data were obtained from the same hESC line used in our study (Mikkelsen et al. 2007). Using a stringent cutoff (false discovery rate = 0.01), 15,517 H3K4me3 sites and 6560 H3K27me3 sites were identified. Among these sites, 11,630 regions were marked by H3K4me3 alone, 3887 by both H3K4me3 and H3K27me3 (bivalent), and 3094 regions were marked by only H3K27me3. Examination of DNA methylation in these regions revealed a strong anti-correlation between H3K4me3 binding and DNA methylation, but no correlation between H3K27me3 and DNA methylation (Supplemental Fig. 8).
Differentially methylated regions associated with differentiation
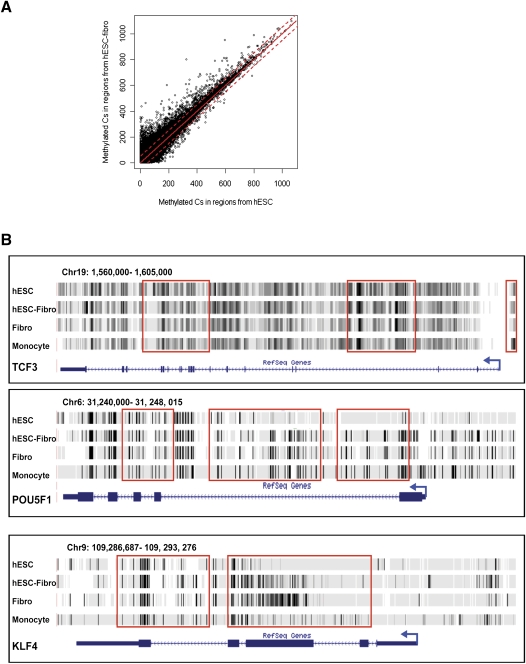
To gain insights into the role of DNA methylation in cellular differentiation, we identified differentially methylated regions (DMRs) by comparing the hESCs and hESC-Fibro cells, and asked if the genes associated with the DMRs showed cell-type-specific expression. While the overall DNA methylation patterns between hESCs and hESC-Fibro cells showed high similarity (correlation coefficient = 0.879) (Fig. 6A), 4772 nonoverlapping DMRs were identified (Supplemental Table 3). While the overall level of methylation decreased with differentiation, more than fourfold more DMRs showed increased rather than decreased methylation in the transition from hESC to differentiated cells (hESC-Fibro), suggesting that differentiation is associated with a more restricted pattern of gene expression. Among the 3899 regions that showed increased methylation with differentiation and the 873 regions with reduced methylation, more than 50% were associated with gene regions (promoter, gene body, or TTS). Genes associated with pluripotency, development, and imprinting were differentially methylated; for example, in hESCs compared with hESC-Fibro cells, POU5F1 (also known as OCT4) and KLF4 had significantly lower TSS methylation, and TCF3 had higher gene body methylation, consistent with their higher expression and functional importance in hESCs (Fig. 6B). In contrast, all four HOX gene clusters were extensively hypermethylated in hESC-Fibro cells. Gene Ontology annotations for all DMR-associated genes showed significant enrichments for genes in the developmental processes, transcriptional regulation, and cell–cell communication categories. With differentiation from hESC to hESC-Fibro, we saw increased promoter and TTS methylation of transcription factors, particularly homeobox transcription factors such as ALX1 and CDX1 (Supplemental Fig. 9), and increased gene body methylation of cell adhesion molecules and genes associated with G-protein signaling (Supplemental Table 4).
Figure 6.
Differentially methylated regions (DMRs) in hESCs and hESC-Fibros. (A) Scatterplot of methylation level in hESCs (x-axis) versus in hESC-Fibro (y-axis). The red line indicates the cutoff of 5 SDs. The distribution is very similar in the two cell types, with a correlation coefficient of 0.879. (B) Examples of DMRs found in the pluripotence-associated transcription factors TCF3, POU5F1, and KLF4.
Discussion
Whole-genome bisulfite sequencing offers unprecedented breadth and depth of genomic coverage at single-base-pair resolution. The use of this approach, along with the range of differentiation states represented in this study, have allowed us to address questions raised by earlier studies that used less powerful analysis methods on more restricted sets of cellular phenotypes. These include general questions about the relationships between DNA methylation and local sequence features, gene expression level, and histone modifications, as well as detailed questions about the effects of cellular differentiation on DNA methylation globally and at specific nucleotides.
We observed many properties of DNA methylation that were common to both undifferentiated and differentiated cells. For example, when the DNA methylation level of individual RefSeq genes was mapped to a gene model, the same pattern emerged for all cell types, consisting of high DNA methylation in the intergenic and distal promoter regions, with a deep valley at the TSS, high methylation in the gene body region, and a sharp shallow stepdown at the TTS. Consistent with many previous reports, promoters associated with CpG islands had 10-fold lower levels of CpG methylation at the TSS than those without CpG islands.
Our data confirm previous reports that the extent of TSS hypomethylation is anticorrelated with gene expression, while the methylation level upstream of the TSS and in the gene body are directly correlated with gene expression. It has been shown that relatively higher methylation over gene bodies is a general phenomenon both in Arabidopsis (Cokus et al. 2008; Lister et al. 2008) and in mammals (Ball et al. 2009), while selective examples indicated that hypomethylated gene bodies are associated with repressed gene activity (Shann et al. 2008). A high level of gene body methylation may improve transcription efficiency of actively transcribed genes by interfering with nonproductive transcription initiation within transcribed regions (Ball et al. 2009).
Our analysis also indicated that the level of DNA methylation is closely associated with local chromatin conformation. We observed a strong anticorrelation between the level of DNA methylation and binding of H3K4me3, a mark of active transcription. These results confirm a previous more limited study that showed that sequences occupied by H3K4me3 tended to be less methylated than other regions (Meissner et al. 2008). We also observed that exons had a distinctively higher level of methylation than introns. This pattern is likely to be influenced by the higher GC content of exons compared to their surrounding introns, which correlates with nucleosome occupancy and level of the histone modifications (Kolasinska-Zwierz et al. 2009; Schwartz et al. 2009).
A close examination of the methylation profile spanning exon–intron boundaries revealed unexpected observations that may shed light on the control of transcript splicing. There was a very distinctive pattern of methylation across exon–intron boundaries. The profile consisted of a methylation spike followed by a sharp drop in methylation at the 5′ exon–intron boundary, a slow rise in methylation across the following intron, and a downward deflection followed by a sharp transition to a higher methylation level at the 3′ intron–exon boundary. This methylation profile occurred on both the sense and antisense strands, suggesting that the sharp transition may serve as a signal for regulation of mRNA splicing. This observation, in light of reports that splicing is influenced by chromatin structure (Sims et al. 2007; Loomis et al. 2009), supports the idea that chromatin modification and DNA methylation work in concert to regulate the generation of specific splice variants. The differential methylation patterns may modulate the chromatin conformation to facilitate the recognition of exon–intron boundaries, and the binding of specific histones may, in turn, affect DNA methylation. The specific roles of chromatin and methylation in differential splicing will have to be determined by experimental studies, but there are indications that the cross-talk between epigenetic signals may be mediated through the actions of RNA polymerase; during transcription, RNA polymerase II is found to associate with splicing machinery (Custodio et al. 2007) as well as chromatin modifiers (Batsche et al. 2006).
Our finding of global non-CpG methylation adds another level of complexity to current models of epigenetic control. Although non-CpG methylation has demonstrated functional significance in plants, it has not been well characterized in mammals. Earlier reports indicated that mouse ESCs have detectable methylation at CpA and CpT dinucleotides (Ramsahoye et al. 2000), and CpA and CpT methylations have been detected at specific loci during development (Haines et al. 2001). Comprehensive analyses of non-CpG methylation from our study highlighted two additional important points. First, we demonstrated that the mCpA is the predominant form of non-CpG methylation. Secondly, the CpA methylation profile is similar to the CpG methylation profile in transcribed regions, showing hypomethylation in both CPI promoters and non-CPI promoters, and consistent methylation across the gene body. The mechanism of non-CpG methylation is unclear. Previous studies demonstrated that DNMT3B and DNMT1 cooperatively maintain virtually all methylation in human cancer cells (Rhee et al. 2002), and some evidence suggested that DNMT3A (in mouse) or DNMT2 (in Drosophila) may be responsible for the establishment of non-CpG methylation (Lyko et al. 2000; Ramsahoye et al. 2000). In our studies, the expression level of DNMT3B had the best correlation with the level of non-CpG DNA methylation, but no causal relationship has been established.
The breadth of unbiased coverage offered by whole-genome bisulfite sequencing has allowed us to evaluate subtle changes in methylation profile that occur with differentiation. hESCs exhibited the most complex DNA methylation pattern, including the highest global methylation level and the greatest frequency of non-CpG methylation. This complex pattern may represent an epigenetically primed state in hESCs that is followed during the early phases of differentiation by an increase in methylation of a subset of genes in the context of a general reduction of global methylation. Detailed analysis of CpG methylation profiles identified regions of hypomethylated CpGs in the hESCs, which were found preferentially in promoter/CpG island areas, and progressively disappeared with differentiation. Detailed examination of HOX gene clusters, which were the four most significant hypomethylated regions, showed an increase in methylation with differentiation of hESCs to fibroblasts (hESC Fibro). We also observed that repetitive elements tended to be progressively demethylated with differentiation; we speculate that reduction in methylation of repetitive elements may help increase accessibility of regulatory elements such as transcription factor binding sites involved in differentiation (Bourque et al. 2008).
Cell-type-specific differentially methylated regions (DMRs) are associated with differences in gene expression (Dindot et al. 2009; Irizarry et al. 2009). By comparing undifferentiated and differentiated cells, we found that dynamic DNA methylation is closely associated with changes in gene expression during differentiation. We identified DMRs that were methylated or demethylated during differentiation by comparing hESCs with their fibroblast-like derivatives (hESC-Fibro), and these DMRs were preferentially located in promoter, gene body, and TTS regions. Even though the global level of methylation decreased with differentiation, almost 80% of DMRs showed increased methylation with differentiation. Many key pluripotency and differentiation-associated genes were found in DMRs.
Overall, our results underscore the advantages of using an unbiased whole-genome approach to methylome mapping, combined with genome-wide gene expression and chromatin binding studies, to develop integrated models of gene regulation in complex processes such as cellular differentiation. Our findings point to many potential areas for future investigation, including exploration of the functional significance of non-CpG methylation in the establishment and maintenance of the pluripotent state, and a possible role for DNA methylation in cell-type-specific gene splicing. In conjunction with other systems-level data such as global mRNA and microRNA expression profiling, comprehensive DNA methylation maps will enable us to better understand the mechanisms that guide specific pathways of differentiation during human development.
Methods
Cell culture and DNA preparation
The WA09 hESC line (Thomson et al. 1998) was cultured feeder-free on Matrigel (Becton-Dickinson) in StemPro medium (Lifetech, Inc.), and passaged with Accutase (Lifetech, Inc.). They were harvested at passage 41. hESC-derived fibroblasts (hESC-Fibro) were derived as a stable proliferating population from spontaneously differentiating passage 40 WA09 hESC cells (Gonzalez et al. 2008); after their differentiation they were expanded in two batches, to passage 11 and passage 13 before harvesting. The neonatal fibroblast line (Fibro) cell line was obtained from GlobalStem, Inc (newborn human foreskin fibroblasts, untreated) and was harvested for analysis at passage 13. All cell lines were cultured at 37°C, ambient oxygen, and 5% CO2. The hESC-Fibro and Fibro cell lines were cultured in DMEM + 10%FBS and passaged with trypsin. The monocyte mapping data were provided prior to publication for the purpose of comparison with the other cell lines (J Wang, Beijing Genome Institute, unpubl.), and were prepared from a single individual using established methods (Wang et al. 2008; J Wang, pers. comm.). Genomic DNA was purified using the DNeasy Blood & Tissue Kit (QIAGEN) and quantified using the Picogreen reagent (Lifetech, Inc.).
Genome-wide bisulfite sequencing
For the whole-genome mapping of DNA methylation at single-nucleotide resolution, genomic DNA was extracted from the individual cell lines and fragmented via nebulization to sizes between 300 bp and 500 bp. The fragmented DNA was end-polished and ligated with Illumina methylated PE adaptors followed by two consecutive bisulfite treatments using the EpiTect Bisulfite Kit (QIAGEN) to ensure maximal conversion rate. To estimate the conversion efficiency, the C → T conversion rate found in the non-CpG context was used first to estimate the conversion rate to be ∼98%–99%. The bisulfite-treated DNA was enriched by 10 cycles of PCR with primers complementary to the adaptor sequences by uracil-insensitive Taq polymerase (Pfu Turbo Cx Hotstart DNA Polymerase; Stratagene), during which the uracil residues produced by bisulfite conversion of unmethylated cytosines were amplified as thymines. Sizes between 250 bp and 300 bp (including the adaptors) were selected, and two sets of strands corresponding to the original plus and minus strands of the genome were produced, which were subsequently sequenced with the Illumina Genome Analyzer using the PE sequencing method at 75-bp read length (Supplemental Fig. 2).
Sequence alignment and identification of methylcytosines
Sequenced reads generated were first processed by Illumina standard sequencing pipeline for base calling and quality filtering. Reads that passed filtering were extracted and aligned to the reference genome hg18. The monocyte reads were mapped by Jun Wang and colleagues at the Beijing Genome Institute, who graciously allowed us to use their mapping data in advance of publication (Y Li, J Zhu, G Tian, N Li, Q Li, M Ye, H Zheng, J Yu, H Wu, J Sun, et al., in prep.) for comparison with the other three cell types (see Acknowledgments), using the SOAP2 alignment program (Li et al. 2009). The hESC, hESC-Fibro, and Fibro reads were mapped using two different methods that yielded essentially the same results—SOAP2 and a method employing brute-force exhaustive matching of the reads (I Rigoutsos, in prep.). To accommodate the conversion of unmethylated cytosines to thymines by bisulfite conversion, we used a “reduced,” three-letter alphabet comprising A, G, and {C or T} to represent the genome. Up to two mismatched positions were allowed during the alignment. Methylation levels of a C within an aligned read were gauged by the ratio of reads that contained a methylated C at that location versus all the reads that covered the location, and resulted in an assignment to one of five levels. The number of Cs and Ts, corresponding to methylated and unmethylated Cs in the original sequence, for each cytosine position in the reference genome was used to calculate its methylation status (Supplemental Fig. 2). Because individual cells in the analyzed populations could have different methylation status at specific sites, we called the methylation status as a continuous β-value, where β = number of methylated reads/(number of methylated reads + number of unmethylated reads). The β-value was then assigned to discrete bins: 0.8 < β ≤ 1.0 methylated (M), 0.6 < β ≤ 0.8 intermediate between partially methylated and methylated (M_P), 0.4 < β ≤ 0.6 partially methylated (P), 0.2 < β ≤ 0.4 intermediate between unmethylated and partially methylated (U_P), and 0 ≤ β ≤ 0.2 unmethylated (U). To facilitate interpretation of our results, the β-values were expressed as percentages in the text and figures. Methylation status was confirmed for selected regions using PCR amplification of bisulfite-treated and untreated DNA, followed by capillary DNA sequencing (Supplemental Fig. 4).
Methylcytosine mapping to gene coding regions
Of the 32,127 targets, the longest transcripts for each gene symbol in the RefSeq database were selected, leaving 19,296 genes. Selecting genes with length ≥2 kb (17,578 genes), 8880 genes were on the + strand and 8698 genes were on the − strand. Cs with β-values >60% were considered to be methylated. Surrounding the TSS and TTS, the genomic regions were divided into 100-bp bins. For each bin, the ratio of methylated Cs to total covered Cs in the bins was calculated. For intragenic regions, each gene was divided into 1000 equal segments. For each segment, the ratio of methylated Cs to total covered Cs was calculated.
Identification of differentially methylated regions
Differentially methylated regions (DMRs) in a comparison of hESCs and hESC-Fibro cells were identified using a 2-kb sliding window with a 1-kb step size. The numbers of Cs with 60% or more methylated calls were counted in the sliding window. For a valid comparison of the differentially methylated regions, the number of methylated Cs in each window from both cell lines was required to be at least five. There were about 1.8 million such windows. The difference in the number of methylated Cs between the cell types for each window was calculated, and the histogram of the differences approximated a normal distribution. The mean and SD of the differences were calculated. Regions with a >5 SD difference in global methylation level from the mean (which corresponds to a P-value < 10−6 for a normal distribution) were identified as DMRs. We annotated each DMR by mapping the position of its center relative to features of proximal genes. TSS DMRs were centered [−10 kb, +1 kb] to a TSS, TTS DMRs were centered [−1 kb, +10 kb] to a TTS, intragenic DMRs were centered in the region >[+1 kb] to a TSS, and <[−1 kb] to the corresponding TTS. All other DMRs were annotated as intergenic. Gene Ontology analysis was performed on six sets of differentially methylated genes (Supplemental Table 4). The Bonferroni correction was applied for multiple testing of pathway, biological process, and molecular functions in the Gene Ontology analysis. The human NCBI gene list was used as the reference genome. The P-value for significance was set at 0.05.
Gene expression microarrays
Gene expression analysis was performed on three biological replicates for the hESC, hESC-Fibro, and Fibro cells using the Human WG-6 version 3 Gene Expression BeadChip (Illumina, Inc.), which contains 48,000 probes. Total RNA was purified using the MirVana RNA extraction kit (Ambion), quantified using the Ribogreen reagent (Lifetech, Inc.), and quality-controlled on a Bioanalyzer (Agilent). One hundred nanograms of input total RNA was amplified and labeled using the TotalPrep kit (Ambion). The labeled product was then hybridized to the array and scanned on a BeadArray Reader (Illumina, Inc.) according to the manufacturer's instructions. The data were quintiles-normalized, and unexpressed probes (detection P-value > 0.01) were removed.
Microarray analysis of DNA methylation
Microarray-based DNA methylation analysis was performed on two biological replicates for hESC, hESC-Fibro, and Fibro cells on the HumanMethylation27 BeadChip (Illumina, Inc.). This array interrogates 27,578 CpG sites representing about 14,000 genes. Five hundred nanograms of genomic DNA from biological duplicates of each cell line was bisulfite-converted using the EZ DNA Methylation Kit (Zymo Research). The bisulfite-converted DNA was processed and hybridized to the HumanMethylation27 BeadChip (Illumina, Inc.), according to the manufacturer's instructions, and scanned on a BeadArray Reader (Illumina, Inc.). The degree of methylation was expressed as the β-value, where β = methylated/(methylated + unmethylated). The β-values for each sample were normalized by range-scaling the data for each probe, using data from fully methylated (generated by treating genomic DNA with SssI DNA methyltransferase), fully unmethylated (generated by whole-genome amplification of genomic DNA), and partially methylated (generated by mixing equal amounts of fully methylated and fully methylated genomic DNA) control DNA samples run in triplicate.
Data release
The hESC, hESC-Fibro, and Fibro cells data generated from this project can be accessed through http://genome.gis.a-star.edu.sg. One can select the hg18 genome assembly and scroll down the GIS track section to select the methylation tracks from different cell lines. The raw sequences and processed data can be downloaded through ftp site ftp.gis.a-star.edu.sg (user id dna-methyl and password methyl2009) and from NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE19418. The monocyte mapping data were used in this publication for comparison purposes only, and these data, along with the raw data, will be released after publication of the detailed analysis of this cell type (Jun Wang, Beijing Genome Institute, pers. comm.).
Acknowledgments
We thank Jun Wang and Yingrui Li from the Beijing Genome Institute for mapping assistance and for sharing their unpublished monocyte methylome map for use as an adult cell reference standard. We thank Candace Lynch of TSRI and the Genome Technology and Biology Group at the Genome Institute of Singapore for technical support, and A. Shahab and C.S. Chen for computing support. We also thank Courtney Harper from Illumina for sequencing support, and Yubo Zheng for performing the GO analysis. Funding for this work was provided by A*STAR of Singapore (C.L.W.), NIH ENCODE grants (R01 HG004456-01, R01HG003521-01, and 1U54HG004557-01) (C.L.W.), CIRM grants (CL1-00502-1, RT1-1108, and TR1-01250) (J.FL.), the Esther B. O'Keeffe Foundation (J.F.L.), and an NICHD Career Development Award (L.C.L.). David Barker and Franz-Josef Mueller provided critical comments on the manuscript.
Footnotes
[Supplemental material is available online at http://www.genome.org. Sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE19418.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.101907.109.
References
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Chudin E, Wu B, Zhou L, Garcia EW, Liu Y, Shin S, Plaia TW, Auerbach JM, Arking DE, et al. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- Custodio N, Vivo M, Antoniou M, Carmo-Fonseca M. Splicing- and cleavage-independent requirement of RNA polymerase II CTD for mRNA release from the transcription site. J Cell Biol. 2007;179:199–207. doi: 10.1083/jcb.200612109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dindot SV, Person R, Strivens M, Garcia R, Beaudet AL. Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions. Genome Res. 2009;19:1374–1383. doi: 10.1101/gr.089185.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E. De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene. 2002;289:41–48. doi: 10.1016/s0378-1119(02)00469-9. [DOI] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R, Loring JF, Snyder EY. Preparation of autogenic human feeder cells for growth of human embryonic stem cells. Curr Protoc Stem Cell Biol. 2008;Chapter 1:Unit 1C.5. doi: 10.1002/9780470151808.sc01c05s4. [DOI] [PubMed] [Google Scholar]
- Haines TR, Rodenhiser DI, Ainsworth PJ. Allele-specific non-CpG methylation of the Nf1 gene during early mouse development. Dev Biol. 2001;240:585–598. doi: 10.1006/dbio.2001.0504. [DOI] [PubMed] [Google Scholar]
- Heard E, Clerc P, Avner P. X-Chromosome inactivation in mammals. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33:450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F, Whittaker AJ, Orr-Weaver TL, Jaenisch R. The putative Drosophila methyltransferase gene dDnmt2 is contained in a transposon-like element and is expressed specifically in ovaries. Mech Dev. 2000;95:215–217. doi: 10.1016/s0925-4773(00)00325-7. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, Park IH, Rao MS, Shamir R, Schwartz PH, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon–intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- Shann YJ, Cheng C, Chiao CH, Chen DT, Li PH, Hsu MT. Genome-wide mapping and characterization of hypomethylated sites in human tissues and breast cancer cell lines. Genome Res. 2008;18:791–801. doi: 10.1101/gr.070961.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, III, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, Held WA. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci. 2005;102:3336–3341. doi: 10.1073/pnas.0408436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Ting AH, McGarvey KM, Baylin SB. The cancer epigenome—components and functional correlates. Genes & Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Guo Y, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Rohde C, Tierling S, Jurkowski TP, Bock C, Santacruz D, Ragozin S, Reinhardt R, Groth M, Walter J, et al. DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet. 2009;5:e1000438. doi: 10.1371/journal.pgen.1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]