Abstract
Aberrant methylation of promoter CpG islands in cancer is associated with silencing of tumor-suppressor genes, and age-dependent hypermethylation in normal appearing mucosa may be a risk factor for human colon cancer. It is not known whether this age-related DNA methylation phenomenon is specific to human tissues. We performed comprehensive DNA methylation profiling of promoter regions in aging mouse intestine using methylated CpG island amplification in combination with microarray analysis. By comparing C57BL/6 mice at 3-mo-old versus 35-mo-old for 3627 detectable autosomal genes, we found 774 (21%) that showed increased methylation and 466 (13%) that showed decreased methylation. We used pyrosequencing to quantitatively validate the microarray data and confirmed linear age-related methylation changes for all 12 genomic regions examined. We then examined 11 changed genomic loci for age-related methylation in other tissues. Of these, three of 11 showed similar changes in lung, seven of 11 changed in liver, and six of 11 changed in spleen, though to a lower degree than the changes seen in colon. There was partial conservation between age-related hypermethylation in human and mouse intestines, and Polycomb targets in embryonic stem cells were enriched among the hypermethylated genes. Our findings demonstrate a surprisingly high rate of hyper- and hypomethylation as a function of age in normal mouse small intestine tissues and a strong tissue-specificity to the process. We conclude that epigenetic deregulation is a common feature of aging in mammals.
In mammals, DNA methylation occurs almost exclusively within the context of CpG dinucleotides and an estimated 80% of all CpG sites are methylated. CpG islands are clusters of CpG dinucleotides that are often located near the 5′ end of genes (Bird 1986; Cross and Bird 1995). Methylation of promoter CpG islands is rare in normal tissues, frequent in cancer, and is usually associated with a closed chromatin structure and transcriptional silencing of the gene (Toyota and Issa 2005; Jones and Baylin 2007). DNA methylation profiles represent a more chemically and biologically stable source of molecular diagnostic information than mRNA or proteins. Therefore, CpG island hypermethylation has been suggested to be a good biomarker for tumorigenesis. A number of tumor suppressor genes are silenced by promoter CpG island methylation in colorectal cancer (Herman and Baylin 2003). In parallel, genome-wide DNA hypomethylation is also thought to play an important role in genomic instability and carcinogenesis (Ehrlich 2002; Estecio et al. 2007). Focal DNA hypomethylation is associated with activation of genes, for example, S100A4 gene in colorectal carcinoma and cyclin D2 in gastric carcinoma (Rosty et al. 2002; Oshimo et al. 2003).
Although most CpG islands are unmethylated in normal human tissues, methylation changes of a small subset of genes can be seen in normal healthy individuals in aging colon mucosa. This methylation involves tumor suppressor genes, which often become more substantially methylated in neoplastic cells (Issa et al. 1994, 2001; Ahuja et al. 1998; Ahuja and Issa 2000). The causes of initiation and spreading of age-related methylation remain to be elucidated. Because cancer is, for the most part, a disease of aging, it has been proposed that age-related epigenetic changes initiate tumorigenesis, and mouse studies of over/under expression of DNA methyltransferases are supportive of this concept (Laird et al. 1995; Linhart et al. 2007). It is not known whether age-related methylation is a phenomenon restricted to humans where life span is relatively long, or whether it is a common feature of physiologic aging.
Here, we studied age as a parameter that could potentially influence DNA methylation, and compared differential methylation by genome-wide CpG island methylation profiling in mouse small intestine. We find a surprisingly high rate of epigenetic deregulation with age (hypermethylation and hypomethylation), along with tissue specificity to the process.
Results
Methylation analysis of candidate genes
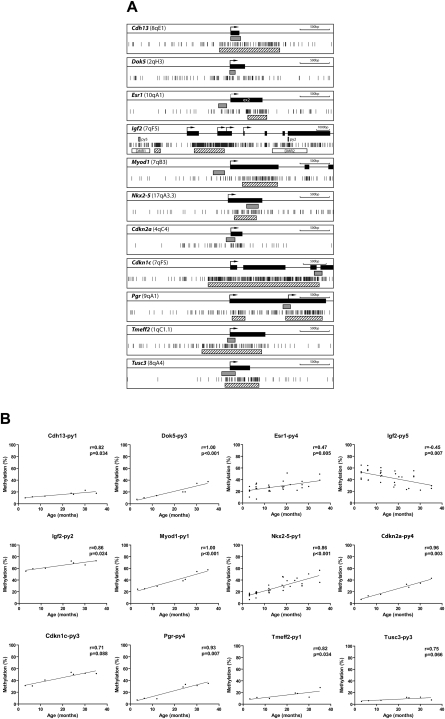
We first examined a total of 21 CpG genomic regions including Apc, Cdh13, Dok5, Egfr, Esr1, Hic1, Igf2-differentially methylated region (DMR) 1 and DMR2, Mgmt, Mlh1, Myod1, Nkx2-5, Cdkn2a (also known as p16), Cdkn1c (also known as p57Kip2), Pgr, Rarb, Rassf1, Timp3, Tmeff2, Tusc3, and Vcan, which had all previously been reported to be hypermethylated in human aging normal colon tissues (Kondo and Issa 2004) or hypermethylated in colon cancer (Kikuchi et al. 2002; Xu et al. 2004; Woodson et al. 2005; Kakar et al. 2008). To investigate DNA methylation as a function of age in normal mouse small intestine, we used PCR-pyrosequencing of bisulfite modified genomic DNA. Figure 1A and Supplemental Figure 1a show CpG maps of the genes analyzed, along with the location of the regions amplified. As a positive control, we used SssI methyltransferase treated DNA in each case. Nine out of 21 genomic regions (Cdh13, Dok5, Esr1, Igf2-DMR2, Myod1, Nkx2-5, Cdkn2a, Pgr, and Tmeff2) showed linear age-related hypermethylation in normal small intestine, in the age range from 3-mo-old to 35-mo-old mice (Fig. 1B). Three additional loci (Cdkn1c [CpG island edge], Tusc3, and Hic1) showed similar trends that did not reach statistical significance. In addition to hypermethylation in DMR2, we also detected hypomethylation in DMR1 of Igf2, an imprinted gene. The methylation levels of eight of 21 genes were not significantly different between young and aged mouse small intestine (Supplemental Fig. 1b; Supplemental Table 1).
Figure 1.
Methylation profiles by pyrosequencing analysis. (A) Gene structure and CpG sites analyzed. Maps represent 3.5 kb of sequence around CpG islands (hatched boxes) and exons (black boxes) of genes analyzed in this study. Short vertical bars represent CpG sites. Arrows point to transcriptional start sites. Gray boxes represent amplified regions for pyrosequencing (py). (B) Association of the percentages of methylated cytosines in the samples as obtained from pyrosequencing (y-axis) with age (x-axis) for 12 genes. The Spearman test was used to determine correlations, with significance set at P < 0.05. R represents a measure of the linear relationship between two variables, and varies from −1 to +1.
Genome-wide methylation analysis
To study age-related methylation on a genome-wide scale, we performed DNA methylation profiling of promoter regions by methylated CpG island amplification microarrays (MCAM), a restriction enzyme, ligation, and a PCR-based method that has high sensitivity and specificity for DNA methylation analysis (Shen et al. 2007). We cohybridized MCA DNA from 3-mo-old and 35-mo-old small intestines on a promoter oligonucleotide array (Agilent). In this analysis, a high Cy5 (old tissue) to Cy3 (young tissue) normalized signal intensity ratio indicates hypermethylation, while a low ratio indicates hypomethylation. We observed that most probes that have informative signal intensity were located within SmaI-digested DNA fragments that were <2 kb in length (Supplemental Fig. 2), which is consistent with a high rate of true positives.
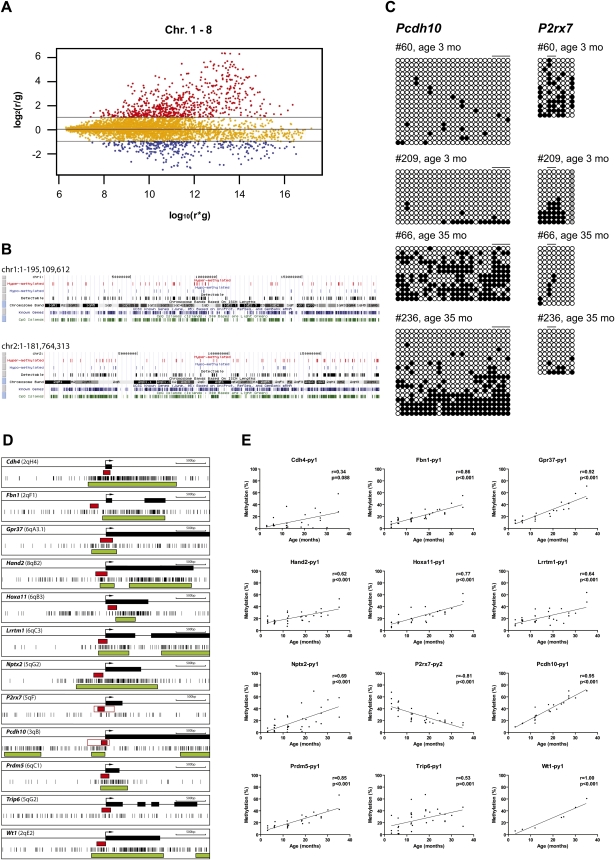
To analyze the microarray data, we first used Lowess normalization, then a beta-uniform mixture (BUM) model to fit the P-values and estimate the false discovery rate (FDR). We selected significant genes by controlling the FDR at 5%, and the fold change greater than two (based on our prior validation studies in other models). Quality control included ratio–intensity (R–I) plots that suggested technically adequate arrays (Fig. 2A; Supplemental Fig. 3a). To demonstrate, accurately measure, and compensate for methylation-dependent dye bias, we performed dye-swap experiments and found that the correlation between the two arrays was high (r = −0.95, P < 0.0001), indicating that the technique is reliable (Supplemental Fig. 3b).
Figure 2.
MCAM analysis of age-related methylation. (A) R–I plot of the significant probes with FDR at 5% and fold change greater than two for MCAM. An R–I plot displays the log2(R/G) ratio for each element on the array as a function of the log10(R × G) product intensities and can reveal systematic intensity-dependent effects in the measured log2 (ratio) values. The red and blue spots indicate probes hypermethylated and hypomethylated in aged small intestine, respectively. (B) Chromosomal regions of age-related methylation. (Black vertical bars) Detectable regions by MCAM (DNA fragments < 2 kb); (blue vertical bars) regions showing hypomethylation with age (ratio < 2.0); (red vertical bars) regions showing hypermethylation with age (ratio > 2.0). (C) Bisulfite sequencing analysis in small intestine. Methylation profiles of Pcdh10 and P2rx7 in young and old small intestines. Each circle represents an individual CpG dinucleotide. (Filled circles) Methylated CpG; (open circles) unmethylated CpG; (gray circles) incomplete sequence. Orders of CpGs follow the direction of genomic DNA sequence shown in Fig. 1A. The drawing is not to scale. Each block of lines represents methylation data from sequencing of cloned PCR products. Each single line indicates the methylation profile detected by direct sequencing analysis from one clone. Horizontal bars represent the CpG sites used for pyrosequencing analysis. (D) Gene structure and CpG sites analyzed. Maps represent 3.5 kb of sequence around CpG islands (green boxes) and exons (black boxes) of genes analyzed in this study. Short vertical bars represent CpG sites. Arrows point to transcriptional start sites. Red boxes represent amplified regions for pyrosequencing. (E) Association of the percentages of methylated cytosines in the samples as obtained from pyrosequencing (y-axis) with age (x-axis) for 12 genes. The Spearman test was used to determine correlations, with significance set at P < 0.05. R represents a measure of the linear relationship between two variables, and varies from −1 to +1.
The normalized hybridization ratio is listed in the Supplemental Table 2 and Supplemental File 1. The P-value is listed in the Supplemental File 2 as score, which is defined as −10log10(P-value). Using both a FDR of 5% and a fold change of larger than twofold as the criteria for significant feature identification, there are 1958 probes (out of 8389) that are significantly changed. These changed probes mapped to 1206 unique genes. Overall, out of 3627 detectable autosomal genes, 774 (21%) showed age-related hypermethylation and 466 (13%) showed age-related hypomethylation. Autosomal genes without a CpG island in their respective 5′ untranslated regions (UTRs) were also detected as candidates for age-related hypermethylation (7%) and hypomethylation (11%). Among the list of differentially methylated genes, we found Myod1, Cdkn2a, and Tmeff2, all of which were found to be hypermethylated with age using bisulfite pyrosequencing (Fig. 1), thus confirming that the technique is sensitive enough to detect age-related methylation. Thirty-four genes (0.9% of the total) had mixed results in that one probe showed hypermethylation, while another showed hypomethylation category with age. This low number of probable false positives is consistent with our stringent criteria and FDR.
Distribution of age-related methylation on autosomal chromosomes
We mapped the distribution of hypermethylated and hypomethylated genes on autosomal chromosomes and show the results in Figure 2B and Supplemental Figure 4. When controlling for CpG island density, none of the chromosomes seemed to be preferentially affected by the process. To examine regional methylation in more detail, we divided each autosomal chromosome equally into three parts using the total number of nucleotides, and calculated the percentages of hyper- or hypomethylated genes in the different regions. Interestingly, there was a greater number of detectable genes (and hence CpG islands) in telomeric regions (43.4%), compared to centromeric (25.7%) and middle regions (30.9%). However, propensity to methylation was the same in each region. Thus, the average percent of hypermethylated genes was 20.7%, 21.5%, and 22.5%, while hypomethylated genes constituted 13.4%, 11.5%, and 11.8% in centromeric, middle, or telomeric regions, respectively.
Functional pathways affected
We examined the potential function of genes that showed hypermethylation, hypomethylation, and unchanged methylation with age by MCAM. Transcriptional network and pathway analysis was performed on pairwise comparisons between hypermethylated genes and unchanged genes, hypomethylated genes and unchanged genes, hypermethylated genes and hypomethylated genes, detectable genes and the rest of genome, respectively (Supplemental Table 3).
Functional class scoring analysis was performed on differentially methylated genes with age by using the Gene Ontology Tree Machine (GOTM) (Zhang et al. 2004). GOTM uses the hypergeometric test for enrichment analysis, and the method proposed by Benjamini and Hochberg is used for multiple test adjustment (Yoav and Yosef 1995). We analyzed biological processes, molecular functions, and cellular components that were relatively enriched by the gene lists of interest using the entire mouse genome as reference set. Supplemental Table 4 shows the top 10 enriched categories for hyper- and hypomethylated genes. Hypermethylated genes showed significant enrichment for various development and differentiation processes, while hypomethylated genes were not significantly enriched in any biological process.
Validation of age-related methylation
The surprisingly high rate of epigenetic variation detected by microarrays raised the possibility of artifacts, despite the demonstrated reproducibility. We therefore validated selected array data using pyrosequencing analysis. We examined 12 loci showing more than a twofold methylation ratio between old and young colon including Cdh4, Fbn1, Gpr37, Hand2, Hoxa11, Lrrtm1, Nptx2, P2rx7, Pcdh10, Prdm5, Trip6, and Wt1. We detected highly significant differences in percentage of methylation by age for all genes (Fig. 2D,E; Supplemental Table 1).
To validate the Pcdh10 and P2rx7 data further, we used bisulfite genomic sequencing of cloned PCR products, which provided detailed information on the pattern of CpG methylation of each CpG site along a single individual DNA strand. This approach confirmed extensive differences in DNA methylation by age (Fig. 2C).
Tissue specificity
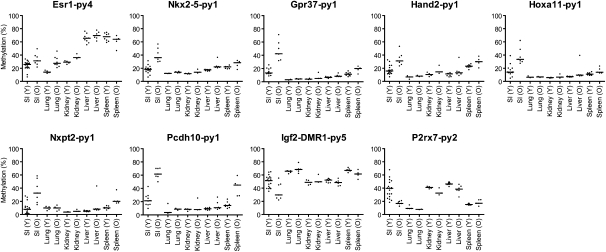
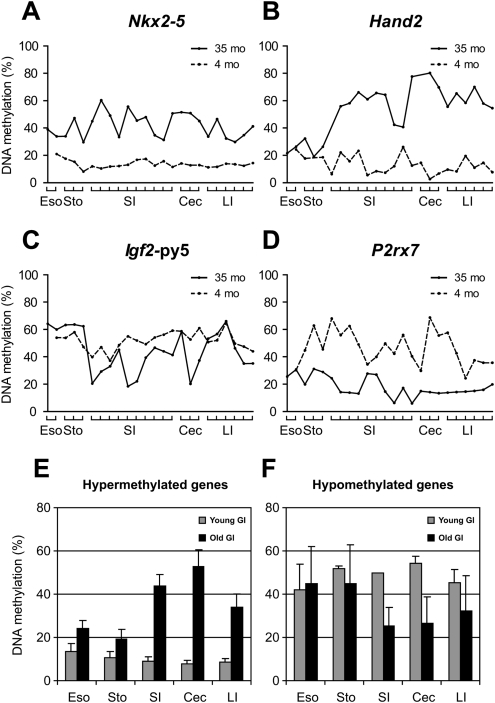
The process of age-related methylation shows tissue specificity in humans (Issa et al. 1994; Tsuchiya et al. 2000; Eads et al. 2001; Shen et al. 2002; Kang et al. 2003; Waki et al. 2003). Here, we identified considerable methylation changes with aging by analyzing young and old small intestine samples. To investigate whether this pattern is also tissue specific in mice, we examined samples obtained from aging lung, liver, and spleen (Fig. 3; Supplemental Fig. 5). The results are summarized in Supplemental Tables 5 and 6 with statistical analysis. We studied 11 genomic regions that show clear age-related methylation in small intestine. Of these, three of 11 showed similar changes in lung, seven of 11 changed in liver, and six of 11 changed in spleen, though to a lower degree than the changes seen in colon (Fig. 3; Supplemental Fig. 5). We also examined variation in methylation across the gastrointestinal tract. As shown in Figure 4, age-related increase and decrease in methylation was evident from the esophagus to the rectum, with the largest differences (for the genes analyzed) being seen in small intestine and cecum.
Figure 3.
Tissue specificity of age-related methylation. Comparison of methylation changes in small intestine, lung, kidney, liver, and spleen tissues with age. Each dot corresponds to one animal, grouped into young (Y, <12 mo) and old (O, >12 mo).
Figure 4.
Methylation profiles of young and aged gastrointestinal tract (esophagus, stomach, small intestine, cecum, and large intestine). (A–D) Methylation profiles of Nkx2-5, Hand2, Igf2-py5, and P2rx7, respectively. (X-axis) The percentage of methylation; (y-axis) regions analyzed in the GI tract from esophagus to large intestine; (dotted lines) young tissues (3 mo); (solid lines) aged tissues (35 mo). (E) Methylation profiles of hypermethylated genes with age. Average percentages of methylation of Hand2, Pgr, Nptx2, Prdm5, and Dok5. (F) Methylation profiles of hypomethylated genes with age. Average percentages of methylation of P2rx7 and Igf2-py5.
Repetitive element methylation
DNA hypomethylation in cancer typically occurs at repetitive sequences residing in satellite or pericentromeric regions, and can result in the reactivation of retrotransposons, leading to disruption of normal gene structure and function (Jones and Baylin 2002; Issa 2004). We studied global DNA methylation using bisulfite treatment of DNA and PCR of multiple DNA repetitive elements, such as long interspersed nuclear elements (LINE-1), major satellite repeats, and short interspersed nuclear elements (SINE B1). We did not find any statistically significant differential methylation of these repeats associated with age in small intestine (Supplemental Fig. 6; Supplemental Table 1).
Human–mouse comparison
The candidate gene analysis suggests some level of conservation between aging in human colon and mouse intestine. To study this more formally, we performed MCAM analysis on normal human colonic tissues, comparing mixed samples between four young tissues (29-, 30-, 41-, and 41-yr-old) and four old tissues (61-, 68-, 69-, and 72-yr-old) (Supplemental Table 7). In human colon, the proportion of autosomal genes showing hypermethylation was 884 out of 8821 (10%), while 1% of genes showed age-related hypomethylation. Because of a generally poor sequence conservation between mice and humans at promoter CpG islands (Antequera and Bird 1993; Matsuo et al. 1993), only 2108 genes could be compared directly (detectable on both human and mouse arrays). Of 276 genes hypermethylated in human colon and detectable on the mouse arrays, 116 (42%) also showed hypermethylation in mice. Of the 99 genes hypomethylated in human colon, three (3%) showed a similar pattern in mice. Thus, there is only partial conservation in the age-related methylation gene subset between mice and humans.
Relation to Polycomb targets
DNA hypermethylation in cancer is more frequent among genes targeted by Polycomb group (PcG) proteins in embryonic stem (ES) cells (Boyer et al. 2006; Lee et al. 2006; Ohm et al. 2007; Rauch et al. 2007; Schlesinger et al. 2007; Widschwendter et al. 2007). Given that age-related methylation is a precursor to cancer-related methylation, we reasoned there could be a relationship between ES cell PcG occupancy and age-related DNA methylation. There are reliable databases of PcG occupancy in mouse and human ES cells (Boyer et al. 2006; Lee et al. 2006); therefore, we evaluated whether PcG targets in ES cells are enriched in the group of genes showing age-related methylation in mouse and human, respectively. Indeed, of 497 SUZ12 targets detectable on the arrays, 45% showed age-related methylation. Similarly, of 338 EED targets detectable on the arrays, 48% showed age-related methylation. Finally, of 316 genes targeted by both SUZ12 and EED, 51% showed age-related methylation in mouse. All these demonstrated substantial enrichment compared to non-PcG targets. Conversely, of the 466 genes showing age-related hypomethylation, only 41 were targets of SUZ12, EED, or both. There was no PcG enrichment among hypomethylated genes (Supplemental Table 8). Similarly, PcG targets in human are also prone to age-related hypermethylation compared with all detectable genes (Supplemental Table 9).
Age-related gene expression
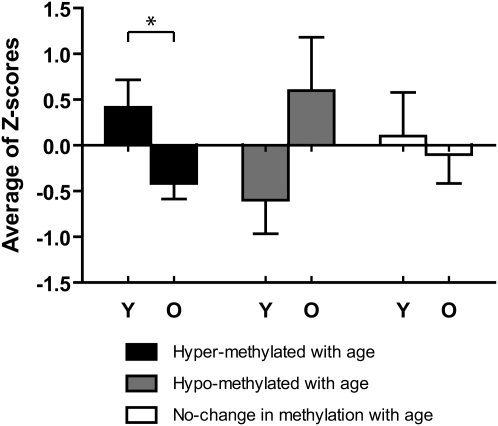
To test the impact of DNA methylation changes with age on gene expression, we performed real-time RT-PCR analysis on RNA isolated from young and old large intestines. We chose four age-related hypermethylated genes, Gpr37, Hoxa11, Pcdh10, and Prdm5, and expressed the data as average Z-score for all genes in young versus old mice. As can be seen in Figure 5, these genes showed on average significant down-regulation with age (P < 0.05). By contrast, P2rx7, a gene that showed age-related hypomethylation, was up-regulated in old tissues, while Tpm2 and Cpe, two genes chosen because they are expressed in normal colon and showed unchanged methylation with age, had no significant age-related change in gene expression (Fig. 5).
Figure 5.
Age-related changes in mRNA expression in large intestine. Expression was measured for four genes showing age-related hypermethylation (Gpr37, Hoxa11, Pcdh10, and Prdm5), one showing age-related hypomethylation (P2rx7) and two (Tmp2 and Cpe) showing unchanged methylation with age. The expression data for each gene was Z-score transformed, and we averaged values for hyper/hypo/unchanged methylation. Black and gray solid bars represent the expression level of hypermethylated and hypomethylated genes with age. (Y) Young tissues; (O) old tissues. Bars represent standard error. *P < 0.05
Discussion
In this study, we used MCAM for a comprehensive analysis of DNA methylation patterns in aging mouse small intestine and detected a remarkable rate of alterations, which were highly tissue-specific. There was partial conservation between aging events in human and mouse intestine, and PcG targets in ES cells were preferentially targeted by this process. The data shed new light on aging in mammals.
The validity of our microarray findings was confirmed by pyrosequencing analysis of 12 genes. There was no chromosome clustering of methylation with age. However, MCAM does not detect all genes because of its dependence on SmaI sites, and this clustering analysis is therefore limited. In those genes validated by pyrosequencing, methylation changes appear linear, and range from very tight associations (R = 0.95 for Pcdh10) to more variable associations (R = 0.47 for Esr1) suggesting that, for some genes, factors other than aging must be involved. Indeed, we have previously reported on an association between chronic inflammation and accelerated age-related methylation (Issa et al. 2001), and a recent mouse model came to a similar conclusion (Hahn et al. 2008). Presumably, stochastic inflammation events in individual mice may account for the variation observed in some genes. In fact, these may be useful markers of epigenetic variation in general (sometimes referred to as metastable alleles).
The causes of age-related methylation changes remain to be determined. Modest changes in DNA methyltransferase expression have been observed in aging tissues (Ray et al. 2006; Ding et al. 2008; Xiao et al. 2008), but this could relate to proliferative stimuli, and DNMT levels have not clearly been associated with hypermethylation in aging or cancer (Eads et al. 1999). The fact that PcG targets are enriched among hypermethylated genes suggests that baseline, tissue-specific patterns of gene expression may be a factor in predisposing to methylation. However, the significant mouse/human divergence in methylation suggests that other factors must also be important; indeed, this is consistent with the hypothesis that methylation changes are highly influenced by sequences around gene promoters, which are poorly conserved (Feltus et al. 2003; Shen et al. 2007). Because 3 yr of time is enough for similar age-related methylation changes in mice, as humans do over 80 yr of time, the age rather than time dependence of the process (along with published data on microdissected crypts) suggests that stem cell proliferation is an important factor as well (Yatabe et al. 2001; Kim and Shibata 2002; Siegmund et al. 2009), which could also account for the effects of inflammation. The tissue-specificity of age-dependent methylation remains mysterious. It may relate to tissue-specific exposures, stem cell proliferation patterns, or even the nature of the stem cells themselves. The tissue specificity may also explain why other investigators failed to find an effect of age on DNA methylation (Tra et al. 2002; Eckhardt et al. 2006).
There is an extensive body of literature demonstrating that the type of methylation observed (promoter CpG island) would be associated with an altered chromatin structure and silencing of gene expression. Indeed, we have demonstrated for a few selected genes that DNA methylation changes are associated with altered gene expression in aging colon. Across the genome, some of the genes affected are likely not expressed to start with, and methylation of those is function-neutral. Others, however, play roles in cellular physiology including growth, differentiation, apoptosis, etc. Mosaic methylation of these genes then creates gene expression diversity in the tissues which, in turn, may fuel the development of selection-dependent events, such as tumor growth. Indeed, it has been extensively documented that genes hypermethylated in aging tissues are very often also hypermethylated in cancer (Shen and Issa 2002). Recently, overexpression of DNMT3B in a mouse model has been shown to increase methylation in normal tissues and to accelerate tumorigenesis (Linhart et al. 2007), which is consistent with our model. Whether DNA methylation changes can account for the pathophysiology of other age-related phenotypes remains to be determined. It is attractive to consider the possibility that common aging events, such as insulin resistance and neuron degeneration, may have an epigenetic component, and this needs to be explored experimentally. An important feature of DNA methylation changes is potential reversibility by pharmacologic manipulation (Issa 2007), which might be worth exploring in diseases of aging if an important epigenetic component was demonstrated.
Our results will be useful to study the interactions between genetic variation, exposures, and epigenetic variation. For example, it might be of interest to compare different mouse strains with different disease susceptibilities to see if any variation there can be accounted for by DNA methylation. Similarly, it appears reasonable to test whether genetic, dietary, or pharmacologic interventions that modify life span in mice are associated with measurable differences in age-related methylation. A similar gene discovery approach could also be used to probe the possibility of an epigenetic component to various age-related diseases, as discussed above.
In summary, we have found a remarkable degree of epigenetic instability marked by DNA methylation changes in aging mouse intestine. This adds a new dimension to studying the molecular mechanisms of aging and associated diseases.
Methods
Mouse tissue samples
All normal tissues were obtained from C57BL/6 mice. We collected a total of 36 normal small intestine samples (age range, 3.1–35.2 mo old), 24 lung tissues (age range, 4–28-mo-old), 19 liver tissues (age range, 2.9–35.2-mo-old), and 16 spleen tissues (age range, 2.9–35.2-mo-old). In addition, we collected entire gastrointestinal tracts from 3-mo-old and 32-mo-old mice. We separated two pieces for esophagus, three pieces for stomach, 10 pieces for small intestine, three pieces for cecum, and six pieces for large intestine. All tissues were digested with proteinase K and SDS to extract genomic DNA.
MCAM
MCAM from young mouse small intestine (3.1-mo-old, female) and aged small intestine (35.2-mo-old, male) was performed as described previously (Shen et al. 2007). Mouse promoter arrays were purchased from Agilent Technologies. Microarray protocols, including labeling, hybridization, and washing procedures, can be found on the Agilent website (http://www.chem.agilent.com). After washing, arrays were scanned on an Agilent scanner and analyzed using Agilent Feature Extraction software at the M.D. Anderson Microarray Core Facility.
Computational analysis
We built a database to facilitate the performance of MCAM in detecting DNA methylation status using the SmaI/XmaI isoschizomers. Mouse genome sequences were downloaded from the UCSC Genome Database (http://genome.ucsc.edu/). The SmaI/XmaI site “CCCGGG” was searched along each chromosome in a case insensitive fashion. Fragments between two SmaI/XmaI sites were extracted. If the fragment length was between 20 b and 10 kb, the fragment was saved in FASTA format with the first line indicating chromosome number, the starting point of the fragment along chromosome (counting from CCCGGG), and the length of the fragment (including starting and ending CCCGGG).
Microarray hybridization analysis
We used normalized signal intensity based on Agilent software and verified the normalization using R–I plots. We then used probes located outside of SmaI/XmaI fragments (length up to 10 kb) for background calculation. The signal intensity for the probes within the SmaI/XmaI fragments was adjusted for background and analyzed for the ratio between Cy3 and Cy5 signals. The ratios of hybridization intensities were adjusted by using Lowess normalization (Cleveland 1979). In this method, the logarithm of hybridization ratio is plotted versus the sum of the logarithm of hybridization intensities (A) and the ratios of the majority of probes are assumed to be unchanged. The Lowess algorithm is used to fit a curve through the points in the R–I plot. This curve defines the zero for the logarithm of methylation ratio and is used to correct the methylation ratio for each probe.
To determine significantly altered probes, we compute P-values from Z-scores by assuming that the logarithm of the ratio of the two channels in the background follows a Gaussian distribution. Based on empirical observations, the width of the distribution, σ, depends on the signal intensity. We estimate the width σ using an iterative procedure.
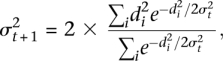 |
where di is the log ratio and is assumed to be properly centered by the normalization procedure. The sum is over all probes. The purpose of weighting is to downgrade the influence of the outliers (in this case the signal) outside of σ, so that the contribution to σ is dominated by the background. The formula becomes exact if di follows the Gaussian distribution with standard deviation σ. The noise in the background depends on A, the average of log intensity of red and green channels. Therefore, it is justified to compute standard deviation of the background by binning A. In each bin of A, a Z-score can be computed for each probe by the formula ζi = di/σ(Ai), where σ(Ai) is the standard deviation of the background for the bin the probe i is in. After computing the P-values from the Z-scores, we use the BUM model (Pounds and Morris 2003) to account for multiple testing and compute the FDR.
Bisulfite pyrosequencing for promoter and global DNA methylation analysis
Bisulfite treatment using the EpiTect Bisulfite Kit was performed according to the manufacturer's instructions (Qiagen). We used a quantitative bisulfite pyrosequencing method for all DNA methylation analyses. Repetitive element DNA methylation was measured for the long interspersed nucleotide element-1 (LINE-1), the B1 element (one of the short SINEs), and major satellite DNA. Primer sequences and PCR conditions for bisulfite pyrosequencing assays are listed in Supplemental Table 10. The methylation levels at different C sites measured by pyrosequencing were averaged to represent the degree of methylation in each sample for each gene. For each assay, we used fully methylated DNA prepared by treating genomic DNA with SssI methylase (New England BioLabs) as a positive control. Pyrosequencing was done using the PSQ HS96 Gold SNP Reagents on a PSQ 96HS machine (Biotage). Each pyrosequencing reaction was done at least twice, and the values averaged.
Bisulfite sequencing
Bisulfite sequencing of cloned PCR products was used to confirm methylation of CpG sites. The oligonucleotide primers used for the amplification of the promoter region of Pcdh10 and P2rx7 were as follows; Pcdh10-bs1-F, ATTTGTTGATGTAAATAGGGGAATTTTT; Pcdh10-bs1-R, CTCTCAATCCATCCTACCTCCTACTTCA; and P2rx7-bs1-F, GTTTTGTTTAGGTTGGATTTGGGTTAGA; P2rx7-bs1-R, CCCTTTTCTCTCAAATAAAATAAATTCCA. Sequencing was performed at the M.D. Anderson Core Sequencing Facility. We cloned the PCR products into the TA vector pCR2.1 (Invitrogen) and extracted plasmid DNA from the resulting clones with the use of a QIAprep Spin Miniprep kit (Qiagen). Sequencing was performed at the M.D. Anderson Core Sequencing Facility.
Real-time RT-PCR assay for age-related gene expression
Total RNA from young and old gastrointestinal tracts was prepared by using the TRIzol reagent (Invitrogen) and reverse transcribed into cDNA using the high capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer's directions. The expression of Gpr37, Hoxa11, P2rx7, Pcdh10, Prdm5, Cpe, and Tpm2 was quantified using a TaqMan gene expression assay and an ABI Prism 7300 sequence detection system (Applied Biosystems). Gapdh was used as an internal control. Gene expression was normalized to Gapdh using the formula 2−ΔCt.
Statistics
Correlation between age and methylation was calculated using Spearman's correlation analysis for all tissues except lung, where only two points were available and a t-test was used instead. For gene expression analysis, we combined data on the genes in each category by deriving a Z-score for each sample/time-point. The Z-score was calculated as follows: Z-score = (expression level of each sample − mean value of expression levels)/standard deviation of expression levels. When analyzing multiple genes at a single time-point, we used the average of the Z-scores for that sample. In this analysis, a Z-score greater than zero means expression greater than the population mean. All calculations were done using GraphPad Prism 4.0 (GraphPad Software Inc.). All P-values are two-sided.
Acknowledgments
This work was supported by National Institutes of Health grants CA100632, CA098006, and CA105346. J.-P.J.I. is an American Cancer Society Clinical Research professor and was supported by a generous gift from the F. M. Kirby Foundation. The M.D. Anderson Cancer Center Core Sequencing Facility is supported by Core Grant CA16672 from the National Institutes of Health.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.096826.109.
References
- Ahuja N, Issa JP. Aging, methylation and cancer. Histol Histopathol. 2000;15:835–842. doi: 10.14670/HH-15.835. [DOI] [PubMed] [Google Scholar]
- Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Cleveland WS. Robust locally weighted regression and smoothing scatterplots. J Am Stat Assoc. 1979;74:829–836. [Google Scholar]
- Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- Ding WJ, Fang JY, Chen XY, Peng YS. The expression and clinical significance of DNA methyltransferase proteins in human gastric cancer. Dig Dis Sci. 2008;53:2083–2089. doi: 10.1007/s10620-007-0145-2. [DOI] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Danenberg PV, Laird PW. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 1999;59:2302–2306. [PubMed] [Google Scholar]
- Eads CA, Lord RV, Wickramasinghe K, Long TI, Kurumboor SK, Bernstein L, Peters JH, DeMeester SR, DeMeester TR, Skinner KA, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–3418. [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. DNA methylation in cancer: Too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- Estecio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. Predicting aberrant CpG island methylation. Proc Natl Acad Sci. 2003;100:12253–12258. doi: 10.1073/pnas.2037852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, Chu FF, Pfeifer GP. Methylation of Polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–10289. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- Issa JP. DNA methylation as a therapeutic target in cancer. Clin Cancer Res. 2007;13:1634–1637. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61:3573–3577. [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakar S, Deng G, Cun L, Sahai V, Kim YS. CpG island methylation is frequently present in tubulovillous and villous adenomas and correlates with size, site, and villous component. Hum Pathol. 2008;39:30–36. doi: 10.1016/j.humpath.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Kang GH, Lee HJ, Hwang KS, Lee S, Kim JH, Kim JS. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am J Pathol. 2003;163:1551–1556. doi: 10.1016/S0002-9440(10)63511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi T, Toyota M, Itoh F, Suzuki H, Obata T, Yamamoto H, Kakiuchi H, Kusano M, Issa JP, Tokino T, et al. Inactivation of p57KIP2 by regional promoter hypermethylation and histone deacetylation in human tumors. Oncogene. 2002;21:2741–2749. doi: 10.1038/sj.onc.1205376. [DOI] [PubMed] [Google Scholar]
- Kim KM, Shibata D. Methylation reveals a niche: Stem cell succession in human colon crypts. Oncogene. 2002;21:5441–5449. doi: 10.1038/sj.onc.1205604. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29–39. doi: 10.1023/a:1025806911782. [DOI] [PubMed] [Google Scholar]
- Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Li E, Weinberg RA, Jaenisch R. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart HG, Lin H, Yamada Y, Moran E, Steine EJ, Gokhale S, Lo G, Cantu E, Ehrich M, He T, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes & Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Clay O, Takahashi T, Silke J, Schaffner W. Evidence for erosion of mouse CpG islands during mammalian evolution. Somat Cell Mol Genet. 1993;19:543–555. doi: 10.1007/BF01233381. [DOI] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimo Y, Nakayama H, Ito R, Kitadai Y, Yoshida K, Chayama K, Yasui W. Promoter methylation of cyclin D2 gene in gastric carcinoma. Int J Oncol. 2003;23:1663–1670. [PubMed] [Google Scholar]
- Pounds S, Morris SW. Estimating the occurrence of false positive and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19:1236–1242. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, Kernstine KH, Riggs AD, Pfeifer GP. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Wu A, Wilkinson JE, Murphy HS, Lu Q, Kluve-Beckerman B, Liepnieks JJ, Benson M, Yung R, Richardson B. Aging in heterozygous Dnmt1-deficient mice: Effects on survival, the DNA methylation genes, and the development of amyloidosis. J Gerontol A Biol Sci Med Sci. 2006;61:115–124. doi: 10.1093/gerona/61.2.115. [DOI] [PubMed] [Google Scholar]
- Rosty C, Ueki T, Argani P, Jansen M, Yeo CJ, Cameron JL, Hruban RH, Goggins M. Overexpression of S100A4 in pancreatic ductal adenocarcinomas is associated with poor differentiation and DNA hypomethylation. Am J Pathol. 2002;160:45–50. doi: 10.1016/S0002-9440(10)64347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Shen L, Issa JP. Epigenetics in colorectal cancer. Curr Opin Gastroenterol. 2002;18:68–73. doi: 10.1097/00001574-200201000-00012. [DOI] [PubMed] [Google Scholar]
- Shen L, Ahuja N, Shen Y, Habib NA, Toyota M, Rashid A, Issa JP. DNA methylation and environmental exposures in human hepatocellular carcinoma. J Natl Cancer Inst. 2002;94:755–761. doi: 10.1093/jnci/94.10.755. [DOI] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Marjoram P, Woo YJ, Tavare S, Shibata D. Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proc Natl Acad Sci. 2009;106:4828–4833. doi: 10.1073/pnas.0810276106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Issa JP. Epigenetic changes in solid and hematopoietic tumors. Semin Oncol. 2005;32:521–530. doi: 10.1053/j.seminoncol.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Tra J, Kondo T, Lu Q, Kuick R, Hanash S, Richardson B. Infrequent occurrence of age-dependent changes in CpG island methylation as detected by restriction landmark genome scanning. Mech Ageing Dev. 2002;123:1487–1503. doi: 10.1016/s0047-6374(02)00080-5. [DOI] [PubMed] [Google Scholar]
- Tsuchiya T, Tamura G, Sato K, Endoh Y, Sakata K, Jin Z, Motoyama T, Usuba O, Kimura W, Nishizuka S, et al. Distinct methylation patterns of two APC gene promoters in normal and cancerous gastric epithelia. Oncogene. 2000;19:3642–3646. doi: 10.1038/sj.onc.1203704. [DOI] [PubMed] [Google Scholar]
- Waki T, Tamura G, Sato M, Motoyama T. Age-related methylation of tumor suppressor and tumor-related genes: An analysis of autopsy samples. Oncogene. 2003;22:4128–4133. doi: 10.1038/sj.onc.1206651. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Woodson K, Weisenberger DJ, Campan M, Laird PW, Tangrea J, Johnson LL, Schatzkin A, Lanza E. Gene-specific methylation and subsequent risk of colorectal adenomas among participants of the Polyp Prevention Trial. Cancer Epidemiol Biomarkers Prev. 2005;14:1219–1223. doi: 10.1158/1055-9965.EPI-04-0726. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Word B, Starlard-Davenport A, Haefele A, Lyn-Cook BD, Hammons G. Age and gender affect DNMT3a and DNMT3b expression in human liver. Cell Biol Toxicol. 2008;24:265–272. doi: 10.1007/s10565-007-9035-9. [DOI] [PubMed] [Google Scholar]
- Xu XL, Yu J, Zhang HY, Sun MH, Gu J, Du X, Shi DR, Wang P, Yang ZH, Zhu JD. Methylation profile of the promoter CpG islands of 31 genes that may contribute to colorectal carcinogenesis. World J Gastroenterol. 2004;10:3441–3454. doi: 10.3748/wjg.v10.i23.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatabe Y, Tavare S, Shibata D. Investigating stem cells in human colon by using methylation patterns. Proc Natl Acad Sci. 2001;98:10839–10844. doi: 10.1073/pnas.191225998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoav B, Yosef H. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): A web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;18:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]