Abstract
Population genetics has evolved from a theory-driven field with little empirical data into a data-driven discipline in which genome-scale data sets test the limits of available models and computational analysis methods. In humans and a few model organisms, analyses of whole-genome sequence polymorphism data are currently under way. And in light of the falling costs of next-generation sequencing technologies, such studies will soon become common in many other organisms as well. Here, we assess the challenges to analyzing whole-genome sequence polymorphism data, and we discuss the potential of these data to yield new insights concerning population history and the genomic prevalence of natural selection.
Population genetics originated in the first half of the 20th century as a field driven by theoretical insights but with very limited empirical data, and for several decades theory remained well ahead of the data available to test its predictions. This situation began to change with the emergence of protein electrophoretic variation (e.g., Harris 1966; Hubby and Lewontin 1966; Lewontin and Hubby 1966; Lewontin 1972). Since the introduction of polymerase chain reaction (PCR) technology, the scale of data has grown exponentially, as restriction fragment length polymorphisms, microsatellites, and small-scale DNA sequencing (e.g., Kreitman 1983) broadened the range of questions open to empirical investigation. With the recent flood of genome-wide single nucleotide polymorphism (SNP) data, and now the advent of fully sequenced population samples of genomes, population genetics has become a fundamentally data-driven discipline.
As the data-generating capacity of population genetics has grown, so has its importance in related disciplines. Population genetics is now at the core of analyses in molecular ecology and conservation biology, where it provides a framework for understanding the distribution of genetic variability among populations and for inferring the demographic histories of natural populations from molecular data. It is also central in studies of molecular evolution, providing a foundation for understanding the contributions of mutation, genetic drift, and natural selection in the evolution of genes and genomes. Finally, with the focus in human genetics on association mapping (Lander and Schork 1994; Risch and Merikangas 1996; Pritchard et al. 2000a), admixture mapping (Chakraborty and Weiss 1988; Stephens et al. 1994), relatedness mapping (Cheung and Nelson 1998; Albrechtsen et al. 2009), and related techniques, population genetics has found its way into medical genetics as a core analytical discipline.
Currently, large-scale next-generation sequencing projects are moving forward in a number of organisms including humans, Drosophila, and Arabidopsis. Before the availability of such data, several genome-wide studies have been completed using Sanger sequencing (e.g., Bustamante et al. 2005; Begun et al. 2007) or SNP genotyping (Hinds et al. 2005; The International HapMap Consortium 2005, 2007; Jakobsson et al. 2008; JZ Li et al. 2008). The low-coverage sequencing of six Drosophila simulans genomes by Begun et al. (2007) was an important step forward for population genomics, and yet today one Illumina Genome Analyzer run can produce substantially more data than were present in that study. This expanded data-generating capacity has led to the recent public release of more than 40 Drosophila melanogaster genomes (http://www.dpgp.org), along with the recent published analysis of 40 silkworm genomes (Xia et al. 2009).
The challenges associated with SNP data obtained by genotyping (particularly ascertainment bias) have been discussed extensively elsewhere (e.g., Kuhner et al. 2000; Nielsen 2000, 2004; Marth et al. 2004) and will not be a focus of this review. Instead, we focus on the analysis of next-generation sequencing data, which is likely to be the foundation of many future population genomic studies. Analysis of these data is currently in its infancy. And yet, if the cost of next-generation sequencing continues to decline, genome-wide population genetic data will likely be available not only for humans and the main model organisms, but for most organisms on which active research is being carried out in genetics, ecology, or evolution. Our ability to obtain samples and to propose good biological questions will be the limiting factor—instead of the sequencing costs. In the anticipation of this future, we review some of the fundamental issues relating to the analysis of genome-wide population genetic data.
Next-generation sequencing
Large-scale sequencing (for review, see Shendure and Ji 2008) is now possible using platforms such as Illumina sequencing (Bentley et al. 2008), 454 Life Sciences (Roche) pyrosequencing (Margulies et al. 2005), Applied Biosystems SOLiD sequencing (Fu et al. 2008), and cPAL sequencing (Drmanac et al. 2009). The declining cost of generating such data is transforming the field of population genetics, making large genomic data sets available to most researchers. While the technology has hitherto mostly been used by researchers working on humans and the main model organisms, next-generation sequencing is also emerging as an economical alternative to other methods for generating population genetic data from natural populations of other organisms. Various reduced-representation shotgun sequencing (RRSS) techniques can be used to select a subset of the genome for sequencing (Altshuler et al. 2000; Baird et al. 2008). When combined with techniques for labeling reads (e.g., Meyer et al. 2008), so that DNA from many individuals can be analyzed in the same pooled sequencing reaction, RRSS using next-generation sequencing provides an increasingly affordable means for generating population genetic data. Next-generation sequencing is therefore likely to become the standard choice for generating population genetic data in fields such as conservation genetics and molecular ecology, but it will carry new demands for computational infrastructure and statistical and bioinformatics training. While next-generation sequencing may not erase every advantage of genetic model organisms, it can allow for the construction of a genetic map (giving regional estimates of recombination rate) by collecting sequence data from laboratory crosses (Baird et al. 2008) or related wild-caught individuals. This strategy implies an added investment of resources, but knowledge about recombination rates is critical for many population genetic inferences (e.g., Thornton and Andolfatto 2006; Becquet and Przeworski 2007; O'Reilly et al. 2008; Pool and Nielsen 2009). Recombination is not a principal focus of this article, as it has been reviewed elsewhere (Coop and Przeworski 2007).
The special nature of the data produced by next-generation sequencing platforms may entail a new set of challenges for unbiased estimation of population genetic parameters. In contrast to traditional approaches, where a defined fragment is amplified by PCR and then sequenced, sequence reads from next-generation technologies stem from individual DNA molecules and are distributed across the genome in a largely random fashion (although regions with very high or very low GC content may be under-represented) (Ossowski et al. 2008). Data produced by these technologies are most comparable to single-pass whole-genome shotgun sequences, which suffer from three basic problems: sequence errors, assembly errors, and missing data. The severity of these problems will depend in part on the depth of sequencing, with higher coverage potentially minimizing many errors (e.g., Bentley et al. 2008). But for organisms with large genomes, the trade-off of coverage versus cost and sample size may justify dealing with the statistical complexities of low-coverage data sets (at least until further sequencing improvements and/or cost reductions are achieved). This trade-off may also depend on specific research goals (e.g., the optimal coverage for a study focused on linkage disequilibrium might be higher than for a study based on allele frequencies), but further work is needed to inform this aspect of experimental design.
Sequence errors
Because next-generation sequence reads originate from a single DNA molecule, errors in the sequences can be due to DNA damage, errors introduced during amplification, and sequencing errors. The stage at which errors occur will determine the frequency of that error in the sequenced DNA pool. While it might be assumed that erroneous bases will occur on single reads only, evidence of nonrandom errors has been reported (Keightley et al. 2009), and so a statistical analysis of error probabilities will be important even for high-coverage data sets. If unaccounted for, errors will inflate nucleotide diversity and skew the allele frequency spectrum (AFS) toward rare alleles, which will mainly be visible as an excess of singletons (e.g., Johnson and Slatkin 2008).
Thus far, the processing of sequence data and especially the calling of SNPs has been focused on minimizing the false-positive rate, by introducing stringent quality criteria to call SNPs (e.g., Altshuler et al. 2000). Johnson and Slatkin (2006, 2008) noted that stringent SNP-calling criteria will bias diversity estimates by excluding many true SNPs (especially rare alleles) from the data. Therefore, they suggested incorporating quality values directly into the estimation of diversity instead of only using them as a pre-filter. However, this is only possible if the probability of a sequencing error is a known function of the sequence quality value. This relationship has been thoroughly investigated for ABI-Sanger sequencing (Ewing and Green 1998), but is currently much less clear for next-generation sequencing methods. Empirically validated error models for new sequencing platforms that incorporate sequence context and position within reads could improve the correlation between quality scores and error probabilities.
Once error probabilities can be estimated accurately, it is relatively easy to correct for the presence of sequencing errors statistically (e.g., Hellmann et al. 2008; Jiang et al. 2009). Lynch (2008) described a method to estimate error rates and nucleotide diversity in a mixed procedure, where the error rate and nucleotide diversity are first estimated from sites with high coverage using a maximum likelihood approach and then used in a method of moments estimation of nucleotide diversity across the genome. Lynch (2009) then extended this approach to also correct the AFS for missing data and errors, assuming either Hardy-Weinberg equilibrium or a known inbreeding coefficient. While the above methods focus on basic population genetic inferences at the genome-wide level, in the future they might be generalized to more complex demographic models or adapted to search for localized changes in diversity or allele frequencies.
Assembly errors
Next-generation sequence reads are hitherto shorter than in traditional Sanger sequencing (presently up to ∼75 bp for Illumina, and up to ∼450 bp for 454 Life Sciences sequencing), and this poses serious challenges for assembling reads (e.g., Sundquist et al. 2007; Chaisson and Pevzner 2008; Zerbino and Birney 2008; Bryant et al. 2009), as well as mapping reads to a reference genome (e.g., H Li et al. 2008; R Li et al. 2008; Langmead et al. 2009). These problems can be partially ameliorated via “paired-end” sequencing, which involves short sequence reads on each side of DNA fragments of a particular size class. However, assembly remains challenging in repetitive or highly polymorphic genomic regions, and it is worthwhile to consider the potential biases that imperfect assembly may introduce.
For some mapping algorithms, sequence reads with more than one or two differences from a reference genome will not be placed (e.g., H Li et al. 2008). This makes the mapping of alleles that are different from the reference genome less probable than for a reference-matching allele, causing a bias in allele frequency toward the allele found in the reference sequence. It may additionally reduce the number of SNPs discovered and bias estimates of nucleotide diversity toward smaller values. Moreover, if the reference genome itself is a consensus genome from multiple individuals, this approach will skew the AFS toward high-frequency alleles. The issue of reference sequence bias could be addressed via alignment tools that are more robust to polymorphism, and by incorporating known polymorphisms and their frequencies into the reference sequence. Assembly should ideally take into account the locations of transposable elements in the reference genome (many of which may not exist in other individuals), and allow for indel variation in general.
In the case of ambiguous placements it is common practice to discard those reads. Hence, repetitive and duplicated regions may have lower coverage. Finally, erroneous alignments of paralogous sequences will inflate nucleotide diversity and could push the AFS toward intermediate frequency alleles. Improved assembly and mapping remain very important and active areas of research, but the most significant improvement to assembly may come from sequencing technology: longer read lengths, and also “paired-end” reads that collect data from each end of fragments of a particular size class. Importantly for population genomic studies, these same advances will increase the haplotype information that can be empirically determined from diploid samples (Bansal et al. 2008; Kidd et al. 2008; Long et al. 2009), along with facilitating the identification of genome rearrangements (Korbel et al. 2007), including copy number variants.
Missing data
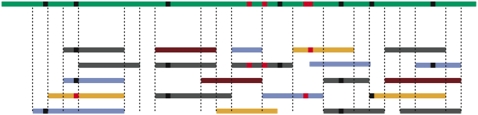
Another challenge for the analysis of whole-genome sequence polymorphism is missing data. Due to the stochastic placement of sequence reads across the genome, the sampled chromosomes at any particular site may not include all individuals (Figure 1). And unless all samples are sequenced at very high genomic coverage (i.e., >30×) (Bentley et al. 2008), it may not be clear whether both of a diploid individual's alleles have been sequenced. Sample sizes will therefore vary along the chromosome and will not be known with certainty. This uncertainty increases if the identity of the individual from which a read was sampled is unknown (i.e., for pooled samples) and decreases with coverage per individual. Ignoring missing data will introduce biases in the estimation of population genetic parameters. However, this problem can be circumvented by summing over all possible (unknown) chromosome sample sizes (Hellmann et al. 2008; Lynch 2008; Jiang et al. 2009).
Figure 1.
Reads from different individuals are aligned to a reference genome, and SNPs have been called. In this toy example there are seven true segregating sites (black dots) and four false ones (red dots). Also note that segments differ in sample size. In segment 4, for example, five reads from three individuals were sampled; thus, there were at least three chromosomes and at most five sampled in this segment.
In association studies it is common practice to impute missing data from the surrounding haplotype patterns (Marchini et al. 2007; Servin and Stephens 2007). This technique could be useful if the goal is to identify putative disease causing SNPs. However, imputation is likely to introduce bias in most population genetic analyses. For example, since singleton polymorphisms cannot be imputed, the use of imputation would lead to downwardly biased nucleotide diversity estimates and a bias against singletons in the AFS. Additional bias may result if the sampled alleles represent only a subset of the population's haplotype diversity (as found for human “tag-SNPs” by Bhangale et al. [2008]).
Next-generation sequencing technologies are evolving with great speed, but the development of appropriate analysis tools is lagging behind. It takes time to characterize the occurrences of sequencing errors and biases with respect to nucleotide content (for example) and then develop appropriate estimators that take such problems into account. Because population genetic inferences are particularly susceptible to sequencing errors and missing data, researchers who use next-generation sequencing data for inferences about demography and selection should always keep these problems in mind. Fortunately, most of the bias introduced by sequencing errors and missing data can be mitigated using appropriate statistical corrections.
Prospects for demographic inference from whole-genome sequence polymorphism
Inference of population history is a central aim of population genetic studies, whether this knowledge is sought for its own sake or to strengthen the conclusions of genome-wide scans for positive selection or genotype–phenotype associations. Currently, demographic analysis of genome-wide SNP data sets often focuses on clustering methods that assign individuals' genomes to one or more populations, or methods that analyze genetic distances between individuals and/or populations (e.g., Jakobsson et al. 2008; JZ Li et al. 2008; Novembre et al. 2008). In some sense, such studies are less ambitious than some traditional methods based on a single or a few loci (e.g., Kuhner et al. 1998; Nielsen and Wakeley 2001; Beaumont et al. 2002) in that rather than estimating demographic parameters directly, they merely aim to quantify the relationship between individuals without a population genetic model or an explicit demographic context. Methods that do infer population parameters from large data sets often focus on the AFS or the genomic means of summary statistics and their variances across the genome. However, many uniquely informative aspects of genome-wide data—such as long-range haplotype patterns—have not been fully utilized. Analysis of whole-genome sequence polymorphism is clearly no less computationally intensive, but compared to SNP data, its advantages for demographic inference include better haplotype information, inclusion of rare population- and region-specific variants, and an unbiased AFS.
Historical inference from allele frequencies and summary statistics
One of the simplest ways to summarize population genetic data is via the AFS. Examples of the use of SNP allele frequency data for demographic inference are provided by Nielsen (2000), Wooding and Rogers (2002), Polanski and Kimmel (2003), Marth et al. (2004), and Williamson et al. (2005), who all modeled the expected AFS under different models of changing population sizes. These methods can also be applied to more than one population and more complex demographic models using the so-called multidimensional frequency spectrum (e.g., Caicedo et al. 2007; Gutenkunst et al. 2009; Nielsen et al. 2009). Although some of the early analyses were limited to a relatively small data set, inference based on the AFS is also computationally tractable for larger analyses. For example, Williamson et al. (2005) used a genome-wide data set of directly sequenced human protein-coding regions. However, while the AFS does contain significant information about past changes in population size, it fails to capture much of the relevant information from population genetic data (such as haplotype structure and variance across the genome), and it may not contain sufficient information for historical inference in more complicated models (Adams and Hudson 2004; Myers et al. 2008).
Several studies have used multiple statistics to compare empirical data against simulations with varying demographic histories. For instance, Schaffner et al. (2005) used several summary statistics (based on allele frequencies, linkage disequilibrium, and population differentiation) to jointly infer historical and recombination models for human populations. Voight et al. (2005) and Thornton and Andolfatto (2006) each used three different statistics to fit population bottleneck models for non-African populations of humans and D. melanogaster, respectively. Examining a different type of model—that of a population split with subsequent migration—Becquet and Przeworski (2007) used numbers of shared variants between populations, private alleles, and fixed differences to estimate demographic parameters for ape populations.
In addition to using different summary statistics, the studies cited above illustrate different methods for comparing summary statistics from empirical and simulated data, including a root mean squared error approach (Schaffner et al. 2005), combining summary statistic P-values (Voight et al. 2005), an approximate Bayesian rejection sampling approach (Thornton and Andolfatto 2006), and an approximate Bayesian Markov chain Monte Carlo likelihood approach (Becquet and Przeworski 2007). None of these methods were applied to genome-scale polymorphism data, and certainly one key to their potential scalability will be computational efficiency. Another issue is the transition from short, independent loci to full genomic coverage. At the simplest, this could be achieved by slicing chromosomes into mostly independent windows of some arbitrary length; but preferably, analyses should account for the nonindependent nature of sequence variation by statistically correcting for the effect of autocorrelation on P-values (Hahn 2006) and confidence intervals (e.g., Keinan et al. 2007).
When genome-scale polymorphism data are available, historical inference can be improved by accounting for both autosomal and X-linked patterns of diversity. The X chromosome will typically have a different effective population size than the autosomes, and will thus operate on a different population genetic time scale. Because the X chromosome will therefore be affected differently by events such as population size changes (Fay and Wu 1999; Hey and Harris 1999; Wall et al. 2002; Pool and Nielsen 2007), it represents a complementary source of information for demographic inference. For example, although a bottleneck model can be fitted to X-linked diversity data for non-African D. melanogaster (e.g., Thornton and Andolfatto 2006), Hutter et al. (2007) found that no simple bottleneck scenario could account for both X-linked and autosomal data, and Pool and Nielsen (2008) then suggested an alternate demographic model that was more compatible with X-linked and autosomal diversity levels. Relatively few genome-wide demographic analyses have incorporated both X-linked and autosomal variation, but in light of the above example, joint consideration of these data sources should produce more accurate inferences of population history.
Population structure and historical inference from haplotypes
One goal of population genetic analysis is to identify the genetic structure that exists within a set of genotyped individuals, which may give insight into population relationships and help to minimize false-positive results in association mapping studies. Principle components analysis (PCA) was introduced to population genetics more than 30 yr ago (Menozzi et al. 1978) but experienced renewed interest following its implementation by Patterson et al. (2006) in a form allowing statistical validation of inferred structure. The computational tractability of PCA makes it applicable to large data sets, as demonstrated by Novembre et al. (2008), who found that principle components inferred from genome-wide SNP data essentially reconstructed the geographic map of Europe. However, interpretation of principle components in terms of population history is far from clear (Novembre and Stephens 2008). PCA is therefore typically a first analysis aimed at defining the genetic relationships among groups.
Population structure can also be analyzed using clustering methods such as STRUCTURE (Pritchard et al. 2000b; Falush et al. 2003). STRUCTURE is relatively computationally intensive, and care must be taken to verify that results have converged, but it has been applied to fairly large data sets. Faster-converging MCMC methods for analyzing genetic structure are now available (Huelsenbeck and Andolfatto 2007; Corander et al. 2008; Alexander et al. 2009). Jakobsson et al. (2008) applied STRUCTURE to more than 500,000 SNPs in worldwide human populations. Supporting the demographic utility of linkage information, this study found that haplotypes were far more likely than individual SNPs to be geographically region-specific, and STRUCTURE analysis of haplotypes enabled detection of additional genetic structure within Africa.
The linkage model of STRUCTURE (Falush et al. 2003) uses “admixture linkage disequilibrium” to estimate ancestry along chromosomes, and a recent method (Price et al. 2009) accounts for local linkage disequilibrium as well. This type of information opens up new possibilities for demographic inference, as demonstrated by methods that infer both ancestry along chromosomes and parameters relevant to recent admixture history (e.g., Hoggart et al. 2004; Patterson et al. 2004), and by a method that uses the lengths of migrant DNA tracts to test for a recent change in migration rate (Pool and Nielsen 2009). By extension, methods that infer genomic tracts of relatedness between individuals (e.g., Purcell et al. 2007; Albrechtsen et al. 2009; Gusev et al. 2009) may also provide relevant information for inferring recent demographic events.
Hellenthal et al. (2008) also used linkage patterns to infer population relationships, implementing an approach based on the copying model of Li and Stephens (2003) to estimate the ancestry sources of human populations. Rather than directly modeling the ancestry process that gives rise to haplotypes along recombining chromosomes, the copying model (also referred to as the “product of approximate conditionals” or the PAC likelihood model) builds samples sequentially by copying segments of existing chromosomes. A second demographic application of the PAC model is provided by Davison et al. (2009), who used it to estimate parameters of a population split model. Because it does not deal with the complexity of ancestral recombination graphs, the copying model is computationally much faster than coalescent-based approaches with recombination. However, the need to correct parameter estimates obtained by this approach (Davison et al. 2009) emphasizes that the PAC model is an approximation that may have significant differences from the true evolutionary process.
The Davison et al. (2009) study also illustrates that linkage patterns carry historic information beyond recent migration events. A second example is provided by Lohmueller et al. (2009), who used the joint distribution of haplotype number and major haplotype frequency in empirical and simulated data to estimate population size changes from human SNP data. In addition, Plagnol and Wall (2006) used linked clusters of mutations to detect signals of archaic structure in human populations. Thus, while long-range haplotype patterns carry unique information about the history of recent migration, short-range haplotype patterns can be strong signals of more ancient gene flow and other demographic events. In light of these studies, the haplotype information provided by next-generation sequencing data will offer a significant advantage over SNP data for detecting historical population events and fine population structure. The ability to detect rare population- or region-specific polymorphisms (which will often be missed in SNP studies) may also improve such inferences.
A final illustration of the potential demographic informativeness of haplotype patterns is shown in Box 1. The particular enrichment of long haplotypes shared between European and African humans could reflect a relatively high rate of recent migration between continents. However, we point out that haplotype patterns, like all population genetic summaries, are potentially influenced by other evolutionary processes such as selection and recombination. Some progress has been made in jointly analyzing natural selection and population history (e.g., Williamson et al. 2005; Wright et al. 2005; Li and Stephan 2006), but the development of realistic evolutionary models for population genomic analysis remains largely an unsolved problem.
Box 1. Haplotype sharing within and between populations.
An underutilized evolutionary signal in whole-genome diversity data is the frequency of long shared haplotypes. Localized excess of long shared haplotypes has been used to identify targets of positive selection (e.g., Sabeti et al. 2002), but the genome-wide abundance of long identical tracts shared across population boundaries may also shed light on recent rates of gene flow. This is similar to the logic underlying methods that infer admixture parameters (e.g., Falush et al. 2003) or changes in migration rate (Pool and Nielsen 2009) based on the sizes of introgressed chromosomal segments, except that here no inference of population ancestry along chromosomes is required.
To examine this pattern in the human genome, we compared the long shared haplotypes found within and between the African and European HapMap populations (SNP data with known phasing from HapMap release 23) against that predicted by simulations from the demographic and recombination model estimated by Schaffner et al. (2005), using the coalescent simulation program COSI. Data were simulated for 10,000 regions of length 1 Mb. Ascertainment correction was made using the two-dimensional frequency spectrum (for both populations together), by retaining variable sites from the simulated data with a probability equal to the ratio of the per-base-pair frequencies of the 2D frequency class between the HapMap and unfiltered simulated data. Because the Schaffner et al. (2005) model uses regional recombination rates drawn from the genetic map of Kong et al. (2002), only HapMap data within the bounds of this map were included, and centromeric regions were excluded. Singletons (polymorphisms observed in only one allele) were excluded from both data sets. Finally, to conservatively eliminate regions of low SNP coverage from the HapMap data, gaps of >10 kb between non-singleton SNPs were excluded from the analyzed regions.
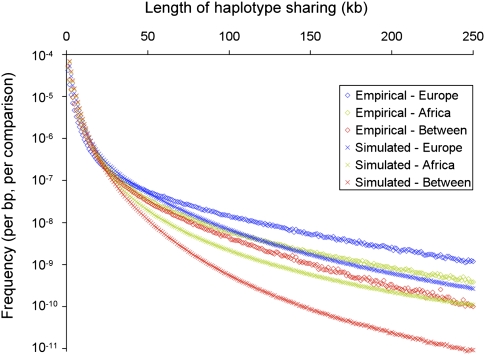
Results of this comparison (see Fig. 3) show at least two notable patterns. First, long shared haplotypes within populations are more abundant in the HapMap data than predicted by the Schaffner et al. (2005) model. For example, tracts within a 10-kb range centered on 200 kb are 3.7 times more abundant within the African HapMap data, and 3.9 times more abundant in Europe. Evolutionary processes that could account for this difference include (1) recombination rates more heterogeneous than modeled, (2) additional recent bottlenecks in both populations, and (3) selective sweeps. Second, it is apparent that long haplotypes shared between populations are more greatly enriched (by a factor of 12.6 in the same window) than within-population tracts. This pattern could be a signal of recently elevated migration between continents, but further analysis is needed to evaluate this and other hypotheses.
Figure 3.
Lengths of haplotypes shared by pairs of alleles within African (green) and European (blue) human populations, and between populations (red), in HapMap SNP data (open boxes) and simulations (×).
Identifying locus-specific and genome-wide effects of selection
One of the most exciting prospects of whole-genome polymorphism data is the increased power to characterize not only the recent adaptive history of natural populations, but also the genomic prevalence of positive and negative natural selection. Negative selection reduces variation in the genome by eliminating some mutations, holding others to low frequency, and also causing the loss of variants linked to deleterious alleles (background selection) (Charlesworth et al. 1993). Positive selection leads to local reductions in genetic diversity via the “genetic hitchhiking” effect of Smith and Haigh (1974). As a favorable mutation increases in frequency in a population, linked neutral variants will either become fixed along with it or be lost from the population. The size of the region of the genome affected by such a “selective sweep” is determined mainly by the strength of selection and the rate of recombination (Smith and Haigh 1974; Hudson and Kaplan 1988; Stephan et al. 1992).
A large literature has arisen characterizing the expected polymorphism patterns resulting from selective sweeps—ranging from a deficit of variation and an excess of rare alleles around the selected site (Hudson and Kaplan 1988; Tajima 1989; Braverman et al. 1995; Fu 1997), to an excess of high-frequency derived alleles in flanking regions (Fay and Wu 2000), to effects on linkage disequilibrium (e.g., Przeworski 2002; Kim and Nielsen 2004; McVean 2007). These signals have been incorporated into methods that scan population genomic data for loci affected by recent selective sweeps. For example, several studies (e.g., Carlson et al. 2005; Williamson et al. 2007; Nielsen et al. 2009) have used the distribution of human SNP frequencies along chromosomes to scan for completed sweeps. Whole-genome sequence polymorphism data should include many rare SNPs absent from previous data sets, and may thus increase the power of these methods to detect selection.
The improved haplotype information of next-generation sequencing data will also augment efforts to detect selection. Selective sweeps produce a distinct spatial pattern of linkage disequilibrium (Stephan et al. 2006) that may represent a unique signal of hitchhiking as opposed to stochastic patterns from population bottlenecks (for example, see Jensen et al. 2007). Linkage patterns can also provide a clear signal of partial selective sweeps, based on the imbalance of haplotype homozygosity between a favored allele class and other variants in the same sample (Sabeti et al. 2002; Voight et al. 2006). By comparing haplotype homozygosity between samples, this approach can also identify population-specific selective sweeps (Sabeti et al. 2007).
The addition of interspecies divergence data to polymorphism within species can allow detection of recurrent selective fixations. For example, comparison of polymorphism and divergence at synonymous versus nonsynonymous sites (McDonald and Kreitman 1991) has been used to identify coding sequences subject to recurrent positive selection (e.g., Bustamante et al. 2005) and to establish the importance of regulatory sequences in adaptive evolution (e.g., Andolfatto 2005). The future availability of genome-wide polymorphism data from multiple closely related species will expand the range of possible analyses and improve our basic understanding of molecular evolution.
Characterizing genomic parameters of adaptation
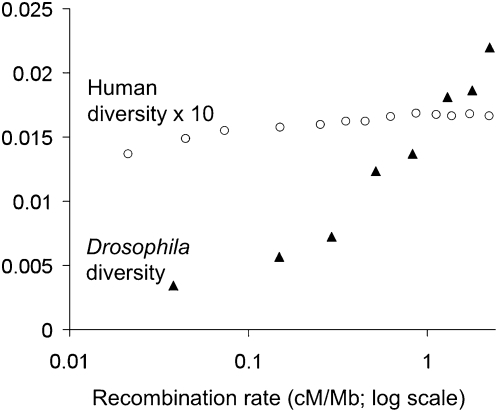
While many studies have identified specific loci with evidence for positive selection (reviewed extensively elsewhere; e.g., Nielsen et al. 2007; Kelley and Swanson 2008), it is increasingly possible to analyze genome-wide signals of hitchhiking. One example is the correlation between recombination and diversity, which was originally observed in D. melanogaster (Begun and Aquadro 1992), and suggested the influence of linked selection. While such a correlation could result from “background selection” against linked deleterious variation (Charlesworth et al. 1993), subsequent analyses have favored the genetic hitchhiking model as a primary explanation (Andolfatto and Przeworski 2001; Innan and Stephan 2003). Hellmann et al. (2008) recently verified that this correlation exists for human data (beyond the effect of mutation rate differences), and likewise found that a hitchhiking model fit the data best. However, the human and Drosophila correlations are of strikingly different magnitudes (Fig. 2), which may reflect the larger effective population size of Drosophila enabling a more pervasive influence of linked selection, and perhaps also a greater density of functional sites in the more compact Drosophila genome. And in general, it has become clear that in Drosophila, the assumption of selective neutrality in random portions of the genome is unlikely to hold (for review, see Sella et al. 2009).
Figure 2.
The relationship between recombination rate and nucleotide diversity in data from D. melanogaster and humans. The data points shown represent average nucleotide diversity for recombination rate bins. The D. melanogaster data (Shapiro et al. 2007) are from 349 loci with ≥50 synonymous sites sequenced in up to 15 African lines (as analyzed by Sella et al. 2009). The human data are from the whole-genome shotgun sequence data analyzed by Hellmann et al. (2008), analyzed in 100-kb windows and processed as described in that study. Human nucleotide diversity is corrected for differences in interspecific divergence (to account for differences in mutation rate) (Hellmann et al. 2003). Drosophila diversity is not corrected for divergence—the correlation between recombination and divergence was weakly negative for this data set (Sella et al. 2009). For both data sets, recombination rate bins were defined manually based on data availability and log-scale bin width.
With larger genome-wide data sets, it will become increasingly possible to move beyond qualitative conclusions about selection in the genome and obtain quantitative estimates of parameters such as the rate of selective sweeps and the strength of selection. Several recent polymorphism-based inference methods of this type have been developed and applied to data from Drosophila (Li and Stephan 2006; Andolfatto 2007; Macpherson et al. 2007; Jensen et al. 2008). These estimators differ statistically (likelihood vs. Bayesian), by the type of data analyzed (polymorphism and/or divergence) and in general framework, with some depending on the genomic variance created on differing spatial scales between models (e.g., Macpherson et al. 2007), and others taking a McDonald and Kreitman (1991)–based approach (e.g., Andolfatto 2007). Perhaps because of differences in methodology and the spatial scale of analysis, published estimates using these different approaches have been far from consistent. In particular, the mean estimates of average genomic selection coefficients for beneficial mutations in Drosophila range from very weak (s = 0.00001) to strong selection (s = 0.01). Whole-genome sequence polymorphism data will be instrumental in differentiating between these scenarios, since weak sweeps should leave narrow footprints (e.g., a high variance in diversity on a fine chromosomal scale) that may only be detectable from the densest data.
While the distribution of selection coefficients for adaptive mutations remains unclear (aside from a few microbial experimental evolution studies) (for review, see Eyre-Walker and Keightley 2007), the distribution of fitness effects for deleterious mutations can be inferred based on comparisons of allele frequencies at synonymous and nonsynonymous sites. Keightley and Eyre-Walker (2007) and Boyko et al. (2008) found that roughly half of human nonsynonymous mutations were neutral or weakly deleterious, while in Drosophila the vast majority were more strongly deleterious (Nes > 10) (Keightley and Eyre-Walker 2007). This difference may again reflect the larger Ne and increased efficiency of selection in Drosophila. Population sizes may also vary within species, as suggested by Lohmueller et al. (2008) to explain the higher proportion of deleterious variants inferred for European Americans relative to African-Americans (as expected if Europeans have had historically smaller population sizes). Because the study of deleterious variation often focuses on rare alleles, generation of whole-genome sequence polymorphism data from large population samples will be instrumental in refining our understanding of selective constraint in the genome and the genetic load of natural populations.
The need for improved models of selection
Understanding the relative roles of natural selection and neutral forces in shaping genetic diversity is a central but unresolved issue in population genetics. However, our ability to accurately model the joint effects of demography and both positive selection and negative selection in a recombining genome is largely restricted to simulations. Most models of positive selection make strong simplifying assumptions, such as constant selection pressure over time, and/or all selection acting on new variants. There has been some progress in developing alternative models of selection, such as the case of a sweep from standing variation (Orr and Betancourt 2001; Innan and Kim 2004; Hermisson and Pennings 2005; Przeworski et al. 2005; Pennings and Hermisson 2006). Other potential departures from the basic recurrent hitchhiking model (Kaplan et al. 1989; Stephan et al. 1992) include variation in selection coefficients through space and/or time (e.g., Ohta 1972; Gillespie 1973; Takahata et al. 1975; Mustonen and Lässig 2007; Huerta-Sanchez et al. 2008). Even with growing population genomic data sets, testing among alternative models of selection in the presence of nonequilibrium demography will present a formidable challenge. Instead of generating very complex parametric models, it may be useful to concentrate on specific aspects of the data that can help distinguish between models, such as the effect of recombination rate on summary statistics, comparisons of markers with different modes of inheritance, and the distribution of shared haplotype lengths. While population genetic theory once far exceeded the data available to test it, today it is the models and methods that must catch up with the data.
Conclusions
Genome-wide data are becoming readily available in a number of organisms. It is clear that population genetics is increasingly moving toward genome-wide analyses, especially in organisms such as humans and Drosophila. But even ecological and evolutionary studies of natural populations may increasingly turn to genome-wide sequencing based on RRSS to cheaply and effectively generate large data sets. Analyses of genome-wide data will allow us to use new tools for understanding the ecology and evolution of natural populations. For example, we may use shared haplotypes to make inferences about very recent migration between populations. The study of genome-wide patterns of variability may also greatly improve our understanding of molecular evolution and the relative contributions of mutation, recombination, genetic drift, and natural selection. However, it will be important in such studies to take the special nature of the data into account: a high sequencing error rate, possible assembly errors, and missing data. While several of these problems can be addressed by using very high coverage, this is usually not cost-effective. Instead, we must increasingly rely on a statistical analysis of the data that takes all of these challenges into account.
Acknowledgments
This research was supported by a National Institutes of Health (NIH) Kirschstein-NRSA postdoctoral fellowship (F32 HG004182) to J.E.P., a Human Frontier Science Program postdoctoral fellowship (LT00794/2006-L) to I.H., and a NIH research grant (UO1HL084706) to R.N.
Footnotes
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.079509.108.
References
- Adams A, Hudson RR. Maximum-likelihood estimation of demographic parameters using the frequency spectrum of unlinked single-nucleotide polymorphisms. Genetics. 2004;168:1699–1712. doi: 10.1534/genetics.104.030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtsen A, Sand Korneliussen T, Moltke I, van Overseem Hansen T, Nielsen FC, Nielsen R. Relatedness mapping and tracts of relatedness for genome-wide data in the presence of linkage disequilibrium. Genet Epidemiol. 2009;33:266–274. doi: 10.1002/gepi.20378. [DOI] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, Linton L, Lander ES. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature. 2000;407:513–516. doi: 10.1038/35035083. [DOI] [PubMed] [Google Scholar]
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- Andolfatto P. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 2007;17:1755–1762. doi: 10.1101/gr.6691007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P, Przeworski M. Regions of lower crossing over harbor more rare variants in African populations of Drosophila melanogaster. Genetics. 2001;158:657–665. doi: 10.1093/genetics/158.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal V, Halpern AL, Axelrod N, Bafna V. An MCMC algorithm for haplotype assembly from whole-genome sequence data. Genome Res. 2008;18:1336–1346. doi: 10.1101/gr.077065.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becquet C, Przeworski M. A new approach to estimate parameters of speciation models with application to apes. Genome Res. 2007;17:1505–1519. doi: 10.1101/gr.6409707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh YP, Hahn MW, Nista PM, Jones CD, Kern AD, Dewey CN, et al. Population genomics: Whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhangale TR, Rieder MJ, Nickerson DA. Estimating coverage and power for genetic association studies using near-complete variation data. Nat Genet. 2008;40:841–843. doi: 10.1038/ng.180. [DOI] [PubMed] [Google Scholar]
- Boyko AR, Williamson SH, Indap AR, Degenhardt JD, Hernandez RD, Lohmueller KE, Adams MD, Schmidt S, Sninsky JJ, Sunyaev SR, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman JM, Hudson RR, Kaplan NL, Langley CH, Stephan W. The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics. 1995;140:783–796. doi: 10.1093/genetics/140.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Wong W, Mockler T. QSRA—a quality-value guided de novo short read assembler. BMC Bioinformatics. 2009;10:69. doi: 10.1186/1471-2105-10-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Todd Hubisz M, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Fledel-Alon A, York TL, Polato NR, Olsen KM, Nielsen R, McCouch SR, et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007;3:e163. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Thomas DJ, Eberle MA, Swanson JE, Livingston RJ, Rieder MJ, Nickerson DA. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 2005;15:1553–1565. doi: 10.1101/gr.4326505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, Pevzner PA. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18:324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty R, Weiss KM. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc Natl Acad Sci. 1988;85:9119–9123. doi: 10.1073/pnas.85.23.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Nelson SF. Genomic mismatch scanning identifies human genomic DNA shared identical by descent. Genomics. 1998;47:1–6. doi: 10.1006/geno.1997.5082. [DOI] [PubMed] [Google Scholar]
- Coop G, Przeworski M. An evolutionary view of human recombination. Nat Rev Genet. 2007;8:23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- Corander J, Marttinen P, Sirén J, Tang J. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics. 2008;9:539. doi: 10.1186/1471-2105-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison D, Pritchard J, Coop G. An approximate likelihood for genetic data under a model with recombination and population splitting. Theor Popul Biol. 2009;75:331–345. doi: 10.1016/j.tpb.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2009;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8:610–618. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu C-I. A human population bottleneck can account for the discordance between patterns of mitochondrial versus nuclear DNA variation. Mol Biol Evol. 1999;16:1003–1005. doi: 10.1093/oxfordjournals.molbev.a026175. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu C-I. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y-X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Peckham HE, McLaughlin SF, Rhodes MD, Malek JA, McKernan KJ, Blanchard AP. The Biology of Genomes Meeting, Cold Spring Harbor Laboratory. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2008. SOLID sequencing and Z-Base encoding. [Google Scholar]
- Gillespie JH. Natural selection with varying selection coefficients—a haploid model. Genet Res. 1973;21:115–120. [Google Scholar]
- Gusev A, Lowe JK, Stoffel M, Daly MJ, Altshuler D, Breslow JL, Friedman JM, Pe'er I. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 2009;19:318–326. doi: 10.1101/gr.081398.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. Accurate inference and estimation in population genomics. Mol Biol Evol. 2006;23:911–918. doi: 10.1093/molbev/msj094. [DOI] [PubMed] [Google Scholar]
- Harris H. Enzyme polymorphisms in man. Proc R Soc Lond B Biol Sci. 1966;164:298–310. doi: 10.1098/rspb.1966.0032. [DOI] [PubMed] [Google Scholar]
- Hellenthal G, Auton A, Falush D. Inferring human colonization history using a copying model. PLoS Genet. 2008;4:e1000078. doi: 10.1371/journal.pgen.1000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I, Ebersberger I, Ptak SE, Pääbo S, Przeworski M. A neutral explanation for the correlation of diversity with recombination rates in humans. Am J Hum Genet. 2003;72:1527–1535. doi: 10.1086/375657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I, Mang Y, Gu Z, Li P, de la Vega FM, Clark AG, Nielsen R. Population genetic analysis of shotgun assemblies of genomic sequences from multiple individuals. Genome Res. 2008;18:1020–1029. doi: 10.1101/gr.074187.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J, Pennings PS. Soft sweeps: Molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Harris E. Population bottlenecks and patterns of human polymorphism. Mol Biol Evol. 1999;16:1423–1426. doi: 10.1093/oxfordjournals.molbev.a026054. [DOI] [PubMed] [Google Scholar]
- Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- Hoggart C, Shriver M, Kittles R, Clayton D, McKeigue P. Design and analysis of admixture mapping studies. Am J Hum Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubby JL, Lewontin RC. A molecular approach to the study of genic heterozygosity in natural populations. I. The number of alleles at different loci in Drosophila pseudoobscura. Genetics. 1966;54:577–594. doi: 10.1093/genetics/54.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. The coalescent process in models with selection and recombination. Genetics. 1988;120:831–840. doi: 10.1093/genetics/120.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Andolfatto P. Inference of population structure under a Dirichlet process model. Genetics. 2007;175:1787–1802. doi: 10.1534/genetics.106.061317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Sanchez E, Durrett R, Bustamante CD. Population genetics of polymorphism and divergence under fluctuating selection. Genetics. 2008;178:325–337. doi: 10.1534/genetics.107.073361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter S, Li H, Beisswanger S, De Lorenzo D, Stephan W. Distinctly different sex ratios in African and European populations of Drosophila melanogaster inferred from chromosomewide single nucleotide polymorphism data. Genetics. 2007;177:469–480. doi: 10.1534/genetics.107.074922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kim Y. Pattern of polymorphism after strong artificial selection in a domestication event. Proc Natl Acad Sci. 2004;101:10667–10672. doi: 10.1073/pnas.0401720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Stephan W. Distinguishing the hitchhiking and background selection models. Genetics. 2003;165:2307–2312. doi: 10.1093/genetics/165.4.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung H-C, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Jensen JD, Thornton KR, Bustamante CD, Aquadro CF. On the utility of linkage disequilibrium as a statistic for identifying targets of positive selection in nonequilibrium populations. Genetics. 2007;176:2371–2379. doi: 10.1534/genetics.106.069450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JD, Thornton KR, Andolfatto P. An approximate Bayesian estimator suggests strong, recurrent selective sweeps in Drosophila. PLoS Genet. 2008;4:e1000198. doi: 10.1371/journal.pgen.1000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Tavare S, Marjoram P. Population genetic inference from resequencing data. Genetics. 2009;181:187–197. doi: 10.1534/genetics.107.080630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PLF, Slatkin M. Inference of population genetic parameters in metagenomics: A clean look at messy data. Genome Res. 2006;16:1320–1327. doi: 10.1101/gr.5431206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PLF, Slatkin M. Accounting for bias from sequencing error in population genetic estimates. Mol Biol Evol. 2008;25:199–206. doi: 10.1093/molbev/msm239. [DOI] [PubMed] [Google Scholar]
- Kaplan NL, Hudson RR, Langley CH. The ‘hitchhiking effect’ revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics. 2007;177:2251–2261. doi: 10.1534/genetics.107.080663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Trivedi U, Thomson M, Oliver F, Kumar S, Blaxter ML. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 2009;19:1195–1201. doi: 10.1101/gr.091231.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A, Mullikin JC, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JL, Swanson WJ. Positive selection in the human genome: From genome scans to biological significance. Annu Rev Genomics Hum Genet. 2008;9:143–160. doi: 10.1146/annurev.genom.9.081307.164411. [DOI] [PubMed] [Google Scholar]
- Kidd JM, Cheng Z, Graves T, Fulton B, Wilson RK, Eichler EE. Haplotype sorting using human fosmid clone end-sequence pairs. Genome Res. 2008;18:2016–2023. doi: 10.1101/gr.081786.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Nielsen R. Linkage disequilibrium as a signature of selective sweeps. Genetics. 2004;167:1513–1524. doi: 10.1534/genetics.103.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature. 1983;304:412–417. doi: 10.1038/304412a0. [DOI] [PubMed] [Google Scholar]
- Kuhner MK, Yamato J, Felsenstein J. Maximum likelihood estimation of population growth rates based on the coalescent. Genetics. 1998;149:429–434. doi: 10.1093/genetics/149.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhner MK, Beerli P, Yamato J, Felsenstein J. Usefulness of single nucleotide polymorphism data for estimating population parameters. Genetics. 2000;156:439–447. doi: 10.1093/genetics/156.1.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC. The apportionment of human diversity. In: Dobzhansky TH, et al., editors. Evolutionary biology. Kluwer Academic Publishers; New York: 1972. pp. 381–398. [Google Scholar]
- Lewontin RC, Hubby JL. A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics. 1966;54:595–609. doi: 10.1093/genetics/54.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Stephan W. Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genet. 2006;2:e166. doi: 10.1371/journal.pgen.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Stephens M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 2003;165:2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J. SOAP: Short Oligonucleotide Alignment Program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, Boyko AR, Hernandez RD, Hubisz MJ, Sninsky JJ, White TJ, Sunyaev SR, Nielsen R, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Bustamante CD, Clark AG. Methods for human demographic inference using haplotype patterns from genomewide single-nucleotide polymorphism data. Genetics. 2009;182:217–231. doi: 10.1534/genetics.108.099275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, MacArthur D, Ning Z, Tyler-Smith C. HI: Haplotype Improver using paired-end short reads. Bioinformatics. 2009;25:2436–2437. doi: 10.1093/bioinformatics/btp412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Estimation of nucleotide diversity, disequilibrium coefficients, and mutation rates from high-coverage genome-sequencing projects. Mol Biol Evol. 2008;25:2409–2419. doi: 10.1093/molbev/msn185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Estimation of allele frequencies from high-coverage genome-sequencing projects. Genetics. 2009;182:295–301. doi: 10.1534/genetics.109.100479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson JM, Sella G, Davis JC, Petrov DA. Genomewide spatial correspondence between nonsynonymous divergence and neutral polymorphism reveals extensive adaptation in Drosophila. Genetics. 2007;177:2083–2099. doi: 10.1534/genetics.107.080226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y-J, Chen Z, et al. Genomes sequencing in open microfabricated high density picoliter reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth GT, Czabarka E, Murvai J, Sherry ST. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics. 2004;166:351–372. doi: 10.1534/genetics.166.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McVean G. The structure of linkage disequilibrium around a selective sweep. Genetics. 2007;175:1395–1406. doi: 10.1534/genetics.106.062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menozzi P, Piazza A, Cavalli-Sforza L. Synthetic maps of human gene frequencies in Europeans. Science. 1978;201:786–792. doi: 10.1126/science.356262. [DOI] [PubMed] [Google Scholar]
- Meyer M, Stenzel U, Hofreiter M. Parallel tagged sequencing on the 454 platform. Nat Protoc. 2008;3:267–278. doi: 10.1038/nprot.2007.520. [DOI] [PubMed] [Google Scholar]
- Mustonen V, Lässig M. Adaptations to fluctuating selection in Drosophila. Proc Natl Acad Sci. 2007;104:2277–2282. doi: 10.1073/pnas.0607105104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Fefferman C, Patterson N. Can one learn history from the allelic spectrum? Theor Popul Biol. 2008;73:342–348. doi: 10.1016/j.tpb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Estimation of population parameters and recombination rates from single nucleotide polymorphisms. Genetics. 2000;154:931–942. doi: 10.1093/genetics/154.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Population genetic analysis of ascertained SNP data. Hum Genomics. 2004;3:218–224. doi: 10.1186/1479-7364-1-3-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Wakeley J. Distinguishing migration from isolation: A Markov chain Monte Carlo approach. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. Recent and ongoing selection in the human genome. Nat Rev Genet. 2007;8:857–868. doi: 10.1038/nrg2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Hubisz MJ, Hellmann I, Torgerson D, Andrés AM, Albrechtsen A, Gutenkunst R, Adams MD, Cargill M, Boyko A, et al. Darwinian and demographic forces affecting human protein coding genes. Genome Res. 2009;19:838–849. doi: 10.1101/gr.088336.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, Indap A, King KS, Bergmann S, Nelsen MR, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. Population size and rate of evolution. J Mol Evol. 1972;1:305–314. [PubMed] [Google Scholar]
- O'Reilly PF, Birney E, Balding DJ. Confounding between recombination and selection, and the Ped/Pop method for detecting selection. Genome Res. 2008;18:1304–1313. doi: 10.1101/gr.067181.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Betancourt AJ. Haldane's sieve and adaptation from standing genetic variation. Genetics. 2001;157:875–884. doi: 10.1093/genetics/157.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O'Brien SJ, Altshuler D, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennings PS, Hermisson J. Soft sweeps II—molecular population genetics of adaptation from recurrent mutation or migration. Mol Biol Evol. 2006;23:1076–1084. doi: 10.1093/molbev/msj117. [DOI] [PubMed] [Google Scholar]
- Plagnol V, Wall JD. Possible ancestral structure in human populations. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanski A, Kimmel M. New explicit expressions for relative frequencies of single-nucleotide polymorphisms with application to statistical inference on population growth. Genetics. 2003;165:427–436. doi: 10.1093/genetics/165.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Nielsen R. Population size changes reshape genomic patterns of diversity. Evolution. 2007;61:3001–3006. doi: 10.1111/j.1558-5646.2007.00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Nielsen R. The impact of founder events on chromosomal variability in multiply mating species. Mol Biol Evol. 2008;25:1728–1736. doi: 10.1093/molbev/msn124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Nielsen R. Inference of historical changes in migration rate from the lengths of migrant tracts. Genetics. 2009;181:711–719. doi: 10.1534/genetics.108.098095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, Barnes KC, Rafaels N, Ruczinski I, Beaty TH, Mathias R, Reich D, Myers S. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000a;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000b;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M. The signature of positive selection at randomly chosen loci. Genetics. 2002;160:1179–1189. doi: 10.1093/genetics/160.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M, Coop G, Wall JD. The signature of positive selection on standing genetic variation. Evolution. 2005;59:2312–2323. [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZP, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G, Petrov DA, Przeworski M, Andolfatto P. Pervasive natural selection in the Drosophila genome? PLoS Genet. 2009;5:e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin B, Stephens M. Imputation-based analysis of association studies: Candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA, Huang W, Zhang C, Hubisz MJ, Lu J, Turissini DA, Fang S, Wang H-Y, Hudson RR, Nielsen R, et al. Adaptive genic evolution in the Drosophila genomes. Proc Natl Acad Sci. 2007;104:2271–2276. doi: 10.1073/pnas.0610385104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- Stephan W, Wiehe THE, Lenz MW. The effect of strongly selected substitutions on neutral polymorphism: Analytical results based on diffusion theory. Theor Popul Biol. 1992;47:237–254. [Google Scholar]
- Stephan W, Song YS, Langley CH. The hitchhiking effect on linkage disequilibrium between linked neutral loci. Genetics. 2006;172:2647–2663. doi: 10.1534/genetics.105.050179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens JC, Briscoe D, O'Brien SJ. Mapping by admixture linkage disequilibrium in human populations: Limits and guidelines. Am J Hum Genet. 1994;55:809–824. [PMC free article] [PubMed] [Google Scholar]
- Sundquist A, Ronaghi M, Tang H, Pevzner P, Batzoglou S. Whole-genome sequencing and assembly with high-throughput, short-read technologies. PLoS One. 2007;2:e484. doi: 10.1371/journal.pone.0000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N, Ishii K, Matsuda H. Effect of temporal fluctuation of selection coefficient on gene frequency in a population. Proc Natl Acad Sci. 1975;72:4541–4545. doi: 10.1073/pnas.72.11.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K, Andolfatto P. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics. 2006;172:1607–1619. doi: 10.1534/genetics.105.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Adams AM, Frisse LA, Qian Y, Hudson RR, Di Rienzo A. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Andolfatto P, Przeworski M. Testing models of selection and demography in Drosophila simulans. Genetics. 2002;162:203–216. doi: 10.1093/genetics/162.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SH, Hernandez R, Fledel-Alon A, Zhu L, Nielsen R, Bustamante CD. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc Natl Acad Sci. 2005;102:7882–7887. doi: 10.1073/pnas.0502300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding SA, Rogers A. The matrix coalescent and application to human single-nucleotide polymorphisms. Genetics. 2002;161:1641–1650. doi: 10.1093/genetics/161.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- Xia Q, Guo Y, Zhang Z, Li D, Xuan Z, Li Z, Dai F, Li Y, Cheng D, Li R, et al. Complete resequencing of 40 genomes reveals domestication events and genes in Silkworm (Bombyx) Science. 2009;326:433–436. doi: 10.1126/science.1176620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]