Abstract
The SH3–SH2–kinase domain arrangement in nonreceptor tyrosine kinases has been conserved throughout evolution. For Src family kinases, the relative positions of the domains are important for enzyme regulation; they permit the assembly of Src kinases into autoinhibited conformations. The SH3 and SH2 domains of Src family kinases have an additional role in determining the substrate specificity of the kinase. We addressed the question of whether the domain arrangement of Src family kinases has a role in substrate specificity by producing mutants with alternative arrangements. Our results suggest that changes in the positions of domains can lead to specific changes in the phosphorylation of Sam68 and Cas by Src. Phosphorylation of Cas by several mutants triggers downstream signaling leading to cell migration. The placement of the SH2 domain with respect to the catalytic domain of Src appears to be especially important for proper substrate recognition, while the placement of the SH3 domain is more flexible. The results suggest that the involvement of the SH3 and SH2 domains in substrate recognition is one reason for the strict conservation of the SH3–SH2–kinase architecture.
Nonreceptor tyrosine kinases consist of kinase domains flanked by various modulatory domains. Src family kinases (SFKs)1 have a conserved domain architecture consisting of (from the N- to C-terminus) unique, SH3, SH2, and catalytic domains (1, 2). The SH3–SH2–kinase domain arrangement is conserved in other families of nonreceptor tyrosine kinases as well (3–5). In fact, across all species, whenever the three domains are present together they have the same arrangement (6). Recent genome analyses show that the SH3–SH2–kinase arrangement is also conserved in unicellular choanoflagellates, the most primitive organisms possessing SFKs; thus, this arrangement evolved before the origin of animals (7, 8). New signaling proteins arise by domain recombination, but clearly, selective pressure to maintain this arrangement of domains in nonreceptor TKs exists. The importance of this conserved domain arrangement has not been explicitly addressed by previous studies.
The SH3 and SH2 domains of SFKs play major roles in kinase regulation. The SH3 domain interacts intramolecularly with a polyproline type II helix in the SH2–kinase linker region, while the SH2 domain interacts with the C-terminal pTyr527 (9, 10). These two interactions occur on the face opposite the kinase active site, and they keep the kinase in an inhibited state until an activating signal is received (11–13). Thus, one purpose of the conserved SH3–SH2–kinase arrangement is to permit the assembly of SFKs into an autoinhibited conformation.
SH3 and SH2 domains also play important roles in targeting SFKs to cellular substrates (2, 14–18). Some SFK substrates, such as the EGF and PDGF receptors and RACK1, possess phosphorylation sites that bind the SH2 domain (19–22). Many Src substrates contain ligands for both the SH3 and SH2 domains (23–27). One such substrate is the adaptor protein p130Cas, which contains 15 potential tyrosine phosphorylation sites and C-terminal ligands for the SH3 and SH2 domains of Src (24, 28). Src binding to the C-terminus triggers processive phosphorylation of Cas (29, 30), followed by binding of the SH2 domains of the Crk adaptor protein (28, 31). Formation of the Cas–Crk complex leads to downstream Rac and Cdc42 activation and an increased level of cell migration (32–34).
Another SFK substrate that depends on SH3–SH2 interactions is the mitotic Src substrate Sam68 (26, 35). The SH3 binding site of Sam68 resides in the N-terminus, while the SH2 binding ligand and phosphorylation sites are in the C-terminus (35, 36). For both Cas and Sam68, SFK phosphorylation is driven primarily by SH3–SH2 interactions (30, 36). Because the intrinsic specificity of tyrosine kinase catalytic domains is not sufficiently high to explain selective signaling, it has been proposed that substrate specificity is governed by the flanking domains (2, 14, 37). Evidence for this hypothesis comes from studies on the Abl nonreceptor tyrosine kinase, where it was shown that replacing the SH2 domain with exogenous SH2 domains leads to alterations in substrate specificity (38).
In this study, we address the question of whether the evolutionarily conserved domain arrangement of SFKs plays a role in determining substrate specificity. We produced a series of Src mutants in which the positions of the domains were rearranged. We reasoned that these changes might create new substrate interactions or alter existing interactions. As expected, most of the mutants were more active than wild-type Src due to a loss of autoinhibition. In addition, we observed that rearranging the domains led to specific changes in substrate specificity. Furthermore, the relative positions of the domains and the positions of ligand sequences in potential substrates can modulate phosphorylation efficiency. Our results suggest that the ability of SFKs to recognize key cellular substrates is one explanation for the strict conservation of the SH3–SH2–kinase architecture.
MATERIALS AND METHODS
Reagents and Antibodies
DMEM, trypsin-EDTA, SF900II-SFM, penicillin, streptomycin, and amphotericin B were from Invitrogen. FBS, Polybrene, nocodazole, NADH, PEP, ATP, PK/LDH, anti-FlagHRP, and anti-tubulin antibodies were from Sigma. Cas C-20 and Sam68 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Crk antibody was purchased from BD Biosciences (San Jose, CA). Anti-phosphotyrosine 4G10 mouse monoclonal antibody was from Millipore, and anti-pY416 antibody was from Biosource. HRP-conjugated secondary anti-mouse and anti-rabbit antibodies and ECL and ECL+ kits were from Amersham.
Plasmid Construction
The domain-rearranged mutants were designed on the basis of a previously described strategy (38). Plasmid pUSEamp-cSrc (Upstate) was used as the parental template to generate PCR fragments of all the domains of Src. Each domain was flanked by unique restriction sites. The domains were ligated together in various combinations to generate the mutants. A spacer of four amino acids (similar to wild-type Src) was used whenever the SH3 and SH2 domains were adjacent in the proteins. The linker sequence (present in wild-type Src between the SH2 and kinase domains) was N-terminal to the catalytic domain in all mutants. In mutant UC32, the C-terminal tail (containing Y527) was maintained at the C-terminus of the protein. The mutants were cloned into the NotI and BamHI sites of p3XFLAG-CMV-7.1 (Sigma). Site-directed mutagenesis was performed on wild-type Src cloned into p3XFLAG-CMV-7.1 to generate the R175L/W118A mutant. Mutants 2UC3 and 3UC2 were also subcloned into the NotI and BamHI sites of pFastBacHTbM to generate baculovirus constructs. pFastBacHtbM was derived from pFastBacHtb (Gibco) by site-directed mutagenesis to create a BamHI site after NotI in the multiple cloning sequence. For retroviral expression, the mutants were subcloned into the EcoRI and XhoI sites of pMSCV-IRES-GFP (a kind gift from N. Carpino, Stony Brook University). All the constructs were confirmed by restriction digestion and sequencing.
Cell Culture and Transient Transfection
SYF cells were purchased from American Type Culture Collection (Manassas, VA) and gradually adapted for maintenance in DMEM containing 10% FBS and a 1× antibiotic/antimycotic solution at 37 °C in a humidified 5% CO2 incubator. Cells were transiently transfected using TransIT LT1 transfection reagent (Mirus) following the manufacturer′s recommendations. Typically, 0.5–1 μg of DNA was transfected using a DNA: TransIT ratio of 1:2.
Western Blotting and Immunoprecipitation
Cells were transiently transfected and harvested 40 h post-transfection in EB++ buffer (39). For Crk immunoprecipitations, cells were harvested in NP40 buffer (40). Protein concentrations were estimated using the Bradford assay (Bio-Rad). For analysis of whole cell lysates, lysates (10–50 μg) were separated via 10% SDS–PAGE and transferred onto a PVDF membrane. The membranes were probed with anti-Flag, anti-pY416, and 4G10 antibodies. For immunoprecipitation, lysates (500 μg) were precleared for 1 h at 4 °C, followed by incubation with antibody (1–5 μg) for 2 h or overnight at 4 °C on a rotator. Immunoprecipitates were washed three times with buffer, and proteins were eluted in 2.5× Laemmli buffer, separated via SDS–PAGE, transferred to PVDF, and analyzed by Western blotting. For Sam68 immunoprecipitations, 24 h post-transfection cells were serum-starved for 12–16 h, and then the medium was changed to serum containing DMEM and nocodazole (5 μg/100 cm plate) to arrest cells in mitosis (41). Cells were harvested ~16 h later with EB++ buffer.
IP Kinase Assays
The kinases were expressed in SYF cells and immunoprecipitated with Flag-agarose beads (Sigma) by incubation with lysates (0.5–1 mg of protein) overnight at 4 °C on a rotator. The IP samples were washed twice with RIPA buffer and once with PBS containing 0.1 mM sodium orthovanadate. The beads were then washed with kinase assay buffer [100 mM Tris (pH 7.5), 10 mM MgCl2, and 0.5 mM Na3VO4] and divided into three equal parts. Two samples were used for duplicate kinase reactions (4 min at 30 °C). The reaction mixtures consisted of enzyme bound to the beads, kinase assay buffer, 500 μM ATP, 0.1 μM [γ-32P]ATP, and 600 μM peptide substrate (AEEEIYGEFEAKKKKG). The reactions were stopped with 0.1% trifluoroacetic acid (TFA) and the mixtures spotted onto P-81 Whatman filter circles. The filters were washed with 0.1% H3PO4 three times for 10 min each. The filters were rinsed with acetone and air-dried. Radioactivity was determined in a scintillation counter. The remaining sample was used to analyze the levels of kinases by anti-Flag Western blotting.
Retrovirus Generation and Infection
Retroviruses were derived as described previously (42, 43). Phoenix cell producer cell lines were obtained from N. Carpino and were maintained in DMEM containing 10% FBS and a 1× antibiotic/antimycotic solution. Cells were transiently transfected with 10 μg of the retroviral constructs along with 10 μg of helper plasmid. Retroviruses were collected at 32 °C for 2 days at 12 h intervals. Aliquots were frozen at –80 °C. For infection, 2× 105 SYF cells were plated per six-well plate and spin-infected with the retroviruses along with Polybrene (4 μg/mL). Two days postinfection, >90% of the cells were GFP positive. The cells were harvested and used for wound healing assays.
Wound Healing Assay
Wound healing assays were performed essentially as described previously (44). Retrovirally infected SYF cells were plated on 60 mm dishes and grown to confluency. The plates were washed with 1× PBS, and multiple wounds were scratched using a P200 pipet tip across the plate. Phase contrast images of eight separate wounds were imaged at 10 min intervals for 10–12 h using a Carl Zeiss inverted microscope with a 20× objective. The width of the wound was measured at 30 min to 1 h intervals. Rates for all eight positions were determined.
Protein Purification and Kinase Assay
Hck was expressed in Sf9 cells and purified as described previously (45). Mutants 2UC3 and 3UC2 were expressed using the Bac-to-Bac baculovirus expression system using the manufacturer′s guidelines (Invitrogen). Sf9 cells (1 L) were infected with the viruses at a multiplicity of infection of 10. Cells were harvested 48 h postinfection. For purification, all steps were carried out at 4 °C. Cell pellets were lysed in buffer A [20 mM Tris (pH 8.0), 10% (v/v) glycerol, 5 mM β-mercaptoethanol (β-ME), 0.5 M NaCl, and 20 mM imidazole] with protease inhibitors using a French pressure cell at 750 psi. The lysate was centrifuged at 18000 rpm for 30 min at 4 °C. The lysate was then applied to a 2–3 mL column of nickel-nitrilotriacetic acid resin (Qiagen) pre-equilibrated in 3 column volumes of buffer A, and the column was washed with 10 column volumes of buffer A. The column was further washed with 2 column volumes of buffer B [20 mM Tris (pH 8.0), 10% (v/v) glycerol, 5 mM β-ME, 1 M NaCl, and 20 mM imidazole]. Before elution, the column was given a final wash with buffer C [20 mM Tris (pH 8.0), 10% (v/v) glycerol, and 5 mM β-ME]. The protein was eluted in 3 column volumes of buffer D [20 mM Tris (pH 8.0), 10% (v/v) glycerol, 5 mM β-ME, and 100 mM imidazole] and analyzed by 10% SDS–PAGE. The peak fractions were pooled and concentrated using an Amicon concentrator (15 mL), and buffer was exchanged with buffer C. The pure proteins were stored at –20 °C in 40% (v/v) glycerol. Protein concentrations were estimated using the Bradford assay. The proteins were >90% pure.
Kinase activity was measured using a continuous spec-trophotometric assay (46). In this assay, the production of ADP is coupled to the oxidation of NADH and measured as a reduction in absorbance at 340 nm. Reactions were carried out at 30 °C in a final volume of 50 μL. The SH3-binding and SH2-binding peptide substrates have been described previously (47, 48). The peptides were HPLC-purified and their identities confirmed by mass spectrometry. The kinase reaction mixtures contained 100 mM Tris (pH 7.5), 10 mM MgCl2, 500 μM ATP, 1 mM phosphoenolpyruvate (PEP), 2.5 mg/mL NADH, 89 units/mL pyruvate kinase, and 124 units/mL lactate dehydrogenase. The enzymes were used at a concentration ranging from 10 to 30 nM, and peptides were used at a concentration of 50 μM.
RESULTS
Generation of Mutants
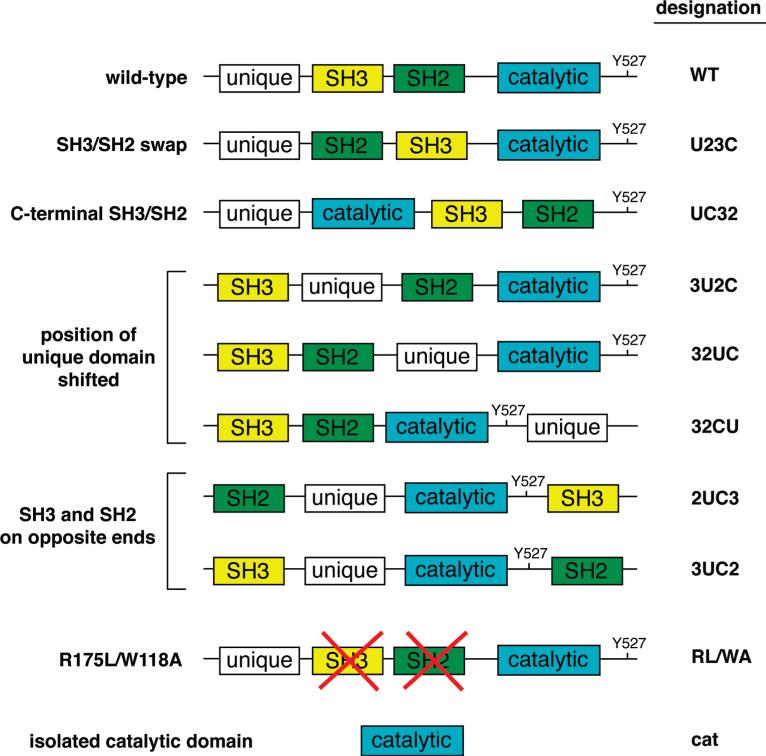
To study the role of Src domain arrangement in determining substrate specificity, we generated a panel of mutants in which the positions of the domains were altered. We introduced unique restriction sites into the sequences flanking the domains, to preserve the folded structures of the individual domains. We also maintained the natural four-amino acid distance found between the SH3 and SH2 domains of Src. The constructs were Flag-tagged at the amino terminus; thus, the N-terminal myristoylation signal was removed in all proteins analyzed. We made three groups of mutants. In the first group, the position of the unique domain was maintained at the N-terminus but other domains were rearranged. Thus, in mutant U23C, the position of the SH3 and SH2 domains is reversed with respect to wild-type Src, and in mutant UC32, the SH3 and SH2 domains are C-terminal to the kinase domain (Figure 1). [The mutants were named according to the positions of their domains from the N- to C-terminus, with unique (U), SH2 (2), SH3 (3), and catalytic (C). Thus, wild-type Src is U32C.] UC32 therefore resembles Ack family kinases, the only naturally occurring group of kinases in which the SH3 domain is immediately C-terminal to the kinase domain (4, 5). In the second group of mutants, we changed the position of the unique domain. The unique domain (residues 1–82) was positioned between the SH3 and SH2 domains (mutant 3U2C), N-terminal to the kinase domain (mutant 32UC) or C-terminal to the kinase domain (mutant 32CU) (Figure 1). In the third group of mutants, we placed the SH3 and SH2 domains on opposite ends of the protein (mutants 2UC3 and 3UC2). The SH3 and SH2 domains often cooperate to recognize substrates, so these mutants were designed to test the importance of proximity between the SH3 and SH2 domains. We also tested two control constructs. First, the mutant R175L/W118A has point mutations in the SH2 domain (R175L) and SH3 domain (W118A) of wild-type Src; these mutations have been previously reported to abrogate SH2 and SH3 domain-mediated interactions (48–50). We also used the catalytic domain alone with no flanking domains as a control. The amino acid sequences of all proteins used in these studies are given in Figure 1 of the Supporting Information.
Figure 1.
Domain arrangements in c-Src and mutants. The mutants are named according to the position of the domains from the N- to C-terminus. The unique domain is colored white, the SH3 domain yellow, the SH2 domain green, and the catalytic domain blue. The position of the Tyr527 sequence (normally present in the C-terminal tail of wild-type Src) is indicated. The R175L/W118A construct contains point mutations that block ligand binding to the SH2 and SH3 domains. The amino acid sequences of the constructs are given in Figure 1 of the Supporting Information.
Mutants Are Activated Relative to Wild-Type Src
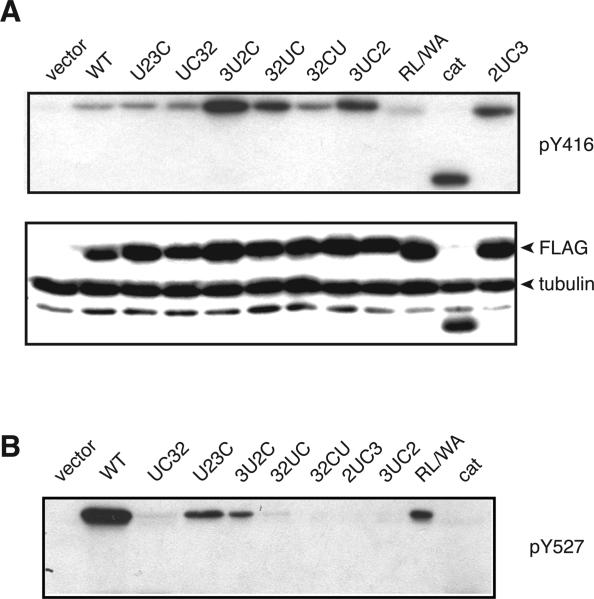
We expressed the mutants and wild-type c-Src in Src/Yes/Fyn triple knockout (SYF) cells, which lack all Src family kinases (39). We carried out anti-pY416 immunoblotting of cell lysates as a measure of kinase autophosphorylation. We observed an increased level of kinase autophosphorylation in several mutants, particularly those in which the position of the unique domain was altered (Figure 2A). Kinase autophosphorylation is presumably due to the disruption of the autoinhibitory contacts in Src. Consistent with this, the isolated catalytic domain exhibited high levels of Y416 phosphorylation (Figure 2A). The loss of autoinhibition was most pronounced when the distance between the SH3 and SH2 domains was altered or when they were not directly adjacent to the catalytic domain (Figure 2A). We examined phosphorylation of the mutants at the Y527 tail sequence that is normally present at the C-terminus of wild-type Src (Figure 2B). With one exception (mutant 32UC), mutants containing the catalytic domain and Y527 sequence at the C-terminus were phosphorylated, while mutants with the Y527 sequence between domains were not, presumably because Csk was unable to access Y527 in these contexts.
Figure 2.
Phosphorylation of c-Src and the mutants. (A) SYF cells were transiently transfected with wild-type c-Src or the mutants. The names of the constructs are given in Figure 1: the unique domain is denoted by U, the SH3 domain by 3, the SH2 domain by 2, and the kinase catalytic domain by C. The cells were harvested 40 h post-transfection, and whole cell lysates (10 μg) were separated via 10% SDS–PAGE and transferred onto PVDF membranes. The membranes were probed with anti-pY416 antibody. To assess expression, membranes were probed with anti-Flag antibody. Anti-tubulin antibody was included as a loading control. Vector represents SYF cells transfected with empty vector alone. The figure is representative of three separate experiments; a comparison of results from three different transfected cell lysates is shown in Figure 2 of the Supporting Information. (B) Transfected SYF cells were lysed and subjected to an immunoprecipitation reaction with anti-Flag antibody. The immunoprecipitates were separated by 10% SDS–PAGE, transferred to a PVDF membrane, and analyzed by anti-pY527 Western blotting.
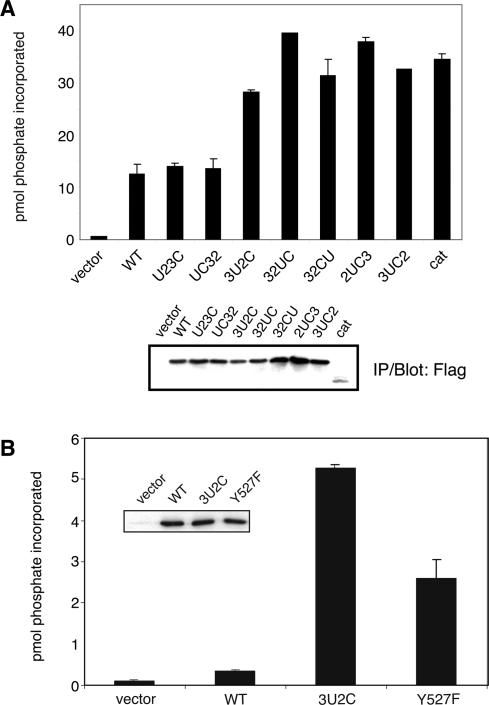
We also evaluated the kinase activity of the mutants by immunoprecipitating them from SYF cells and incubating the immunoprecipitates with a substrate peptide and [γ-32P]ATP. All of the mutants were active in this assay (Figure 3A). The 3UC2, 32UC, 32CU, 2UC3, and 3UC2 mutants all displayed higher activity than wild-type Src in this assay, as did the isolated catalytic domain. We used a similar assay to compare the activity of wild-type Src and the 3U2C mutant with that of the previously studied activated Y527F Src mutant (Figure 3B). In this experiment, carried out with transiently transfected SYF cells, the activity of 3U2C was much higher than that of WT Src, and higher than that of Y527F. The autophosphorylation and IP kinase results are summarized in Table 1, together with all of the data on the mutants. Mutants U23C and UC32 exhibited levels of autophosphorylation and kinase activity similar to those of wild-type c-Src (Figures 2 and 3). The common feature of these mutants is that the unique domain is present in its proper position, raising the possibility that the unique domain directly or indirectly (via SH3 or SH2 domains) modulates kinase activity. In general, our results suggest that domain rearrangement leads to loss of autoregulation of the kinase. Our data also support previous findings that the SH3 and SH2 domains of Src act as a unit to repress the kinase (51, 52).
Figure 3.
Kinase activity of immunoprecipitated proteins. (A) SYF cells expressing wild-type Src or the mutants were lysed in RIPA buffer. Kinases were isolated by immunoprecipitation with Flag-agarose beads, and the samples were divided into three equal parts. Two samples were used in duplicate kinase assays with [γ-32P]ATP and the substrate peptide AEEEIYGEFEAKKKKG (top). One sample was analyzed by anti-Flag Western blotting to compare the amounts of kinases (bottom). (B) SYF cells were transiently transfected with the indicated constructs. Kinases were isolated by immunoprecipitation with Flag-agarose beads and assayed as described for panel A. The inset shows IP samples were analyzed by anti-Flag Western blotting.
Table 1.
Comparison of Properties of Domain-Rearranged Mutantsa
| activity | auto-P | Cas-P | migration | Sam68-P | |
|---|---|---|---|---|---|
| vector | – | – | – | – | – |
| WT | + | + | + | + | + |
| U23C | + | + | + | + | + |
| UC32 | + | + | + | + | + |
| 3U2C | +++ | +++ | +++ | +++ | +++ |
| 32UC | +++ | +++ | +++ | +++ | +++ |
| 32CU | +++ | ++ | +++ | +++ | +++ |
| 2UC3 | ++ | +++ | – | ± | +++ |
| 3UC2 | ++ | +++ | – | ± | – |
| R175L/W118A | ND | + | – | – | – |
| catalytic domain | +++ | +++ | – | – | – |
The following activities of wild-type and mutant forms of Src are summarized: kinase activity (Figure 3A), autophosphorylation (auto-P) (Figure 2A), Cas phosphorylation (Cas-P) (Figure 5A), migration (Figure 6), and Sam68 phosphorylation (Sam68-P) (Figure 7). A minus sign (–) indicates a signal that was either undetectable or similar to the vector control. A plus sign (+) indicates a signal that was similar to that of the WT, and increasing numbers of plus signs indicate signals stronger than that of the WT. The symbol ± indicates a signal that was weaker than that of the WT but higher than the vector control. ND, not determined. Mutants 2UC3 and 3UC2 exhibited changes in substrate specificity, as described in the text.
Mutants Display No Global Changes in Substrate Specificity
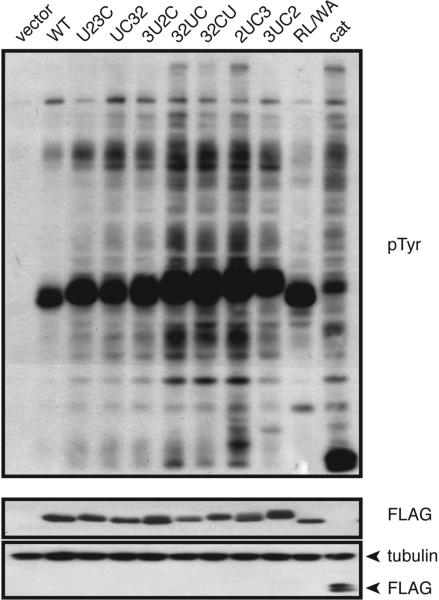
To examine phosphorylation of cellular proteins by the mutants, we expressed them in SYF cells and assessed overall tyrosine phosphorylation by anti-phosphotyrosine Western blotting of lysates. We observed that the overall tyrosine phosphorylation pattern of the mutants was very similar to that of wild-type c-Src (Figure 4). Some unique phosphoproteins were present, such as the low-molecular weight band observed for mutant 2UC3. Consistent with the results in Figures 2 and 3, most of the mutants showed enhanced phosphorylation as compared to c-Src (Figure 4). The R175L/W118A mutant was defective in recognition of several proteins, consistent with an earlier study in which the R175L mutant was unable to promote malignant transformation of SYF cells (53). The overexpressed catalytic domain was highly active, suggesting that many of the phosphoproteins visible in this experiment can be phosphorylated by SH3- and SH2-independent mechanisms. Because the patterns were not radically altered with rearrangement of the domains, the data suggested that for many proteins, the positions of the domains did not play a dominant role in determining substrate specificity.
Figure 4.
Global tyrosine phosphorylation analysis of Src and mutants. SYF cells were transiently transfected with c-Src or mutants. Cells were harvested 40 h post-transfection, and lysates (50 μg) were separated via 10% SDS–PAGE and transferred onto a PVDF membrane. The membrane was probed with anti-pTyr antibody and reprobed with anti-Flag antibody to check expression. The membrane was probed with anti-tubulin antibody as a loading control. Vector represents SYF cells transfected with empty vector alone. The figure is representative of three separate experiments.
Domain Rearrangement Leads to Changes in Substrate Specificity toward Cas
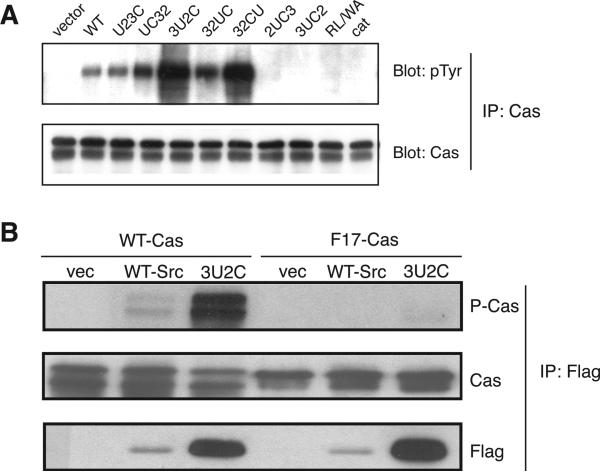
To test for any specific changes in phosphorylation that might not be visible in the overall analysis of whole cell lysates, we studied the Src substrate Cas. We expressed the mutants and c-Src in SYF cells and immunoprecipitated Cas and examined its phosphorylation using anti-pTyr Western blotting. SH3 and SH2 interactions are crucial for Src recognition of Cas (24, 30). Consistent with this, the R175L/W118A mutant and the isolated catalytic domains were unable to phosphorylate Cas (Figure 5A). We observed that altering the positions of the domains led to specific changes in Cas phosphorylation (Figure 5A). The U23C and UC32 mutants phosphorylated Cas to a level similar to that of wild-type Src (Figure 5A). We observed an increased level of phosphorylation of Cas by the three mutants in which the unique domain was repositioned (3U2C, 32UC, and 32CU) (Figure 5). Enhanced Cas phosphorylation could be due to an enhanced rate of phosphorylation of previously described sites, or alternatively to the phosphorylation of additional sites by the mutants. To distinguish between these possibilities, we carried out experiments with a Cas mutant (F17-Cas) in which all 17 known Src phosphorylation sites were changed to Phe (29). Src (WT or 3U2C) was immunoprecipitated from SYF cells and incubated with purified WT Cas or the F17-Cas mutant. Mutant 3U2C exhibited enhanced phosphorylation of WT Cas (Figure 5B) but not appreciable phosphorylation of the F17-Cas mutant, suggesting that 3U2C did not target additional sites.
Figure 5.
Phosphorylation of Cas by Src and the mutants. (A) SYF cells were transiently transfected with wild-type Src or the mutants. The cells were harvested 40 h post-transfection, and lysates were subjected to immunoprecipitation using anti-Cas antibody. The immunoprecipitates were separated via 7.5% SDS–PAGE, and Western blotting was carried out with anti-pTyr antibody. The membrane was stripped and reprobed with anti-Cas antibody to ensure equivalent Cas immunoprecipitation. The figure is representative of three separate experiments; a comparison of results from three different transfected cell lysates is shown in Figure 3 of the Supporting Information. (B) SYF cells expressing WT Src, mutant 3U2C, or vector control were lysed and subjected to immunoprecipitation reactions using anti-Flag antibody. Purified Cas (WT or F17-Cas) was added to the immunoprecipitates, and kinase reactions proceeded for 30 min at 30 °C. Reactions were stopped with SDS sample buffer, and mixtures were analyzed by anti-phosphotyrosine Western blotting. The membranes were stripped and reprobed with anti-Cas and anti-Flag antibodies.
The enhanced phosphorylation of Cas by mutants 3U2C, 32UC, and 32CU could be due to the increased overall activity of the mutants (Figures 2 and 3) or to a specific enhancement of Cas phosphorylation. On the other hand, while the 2UC3 and 3UC2 mutants had higher overall activity than wild-type Src (Figures 2 and 3), they exhibited less activity toward Cas (Figure 5A). This suggests that separation of the SH3 and SH2 domains resulted in a decreased level of Cas recognition.
Mutants Restore the Migration Defect of SYF Cells
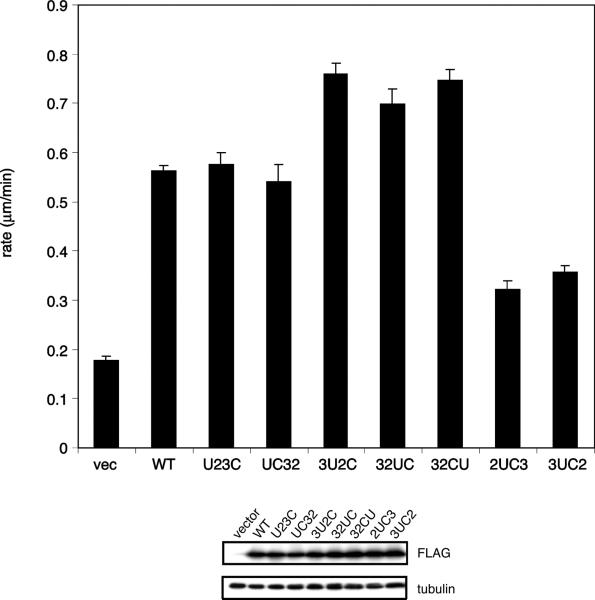
Since Cas was phosphorylated by the mutants, we tested for activation of downstream signaling. SYF cells show low levels of Cas phosphorylation and are deficient in cell migration (39). We carried out retroviral expression of the mutants in SYF cells and tested whether they could restore functional migration signaling by wound healing assays. Mutants that were able to phosphorylate Cas (Figure 5A) were active in these assays (Figure 6). Two of the mutants in which the unique domain was repositioned (3U2C and 32CU) were more active than wild-type c-Src, consistent with their high levels of Cas phosphorylation (Figure 5A). Mutants 2UC3 and 3UC2, which phosphorylated Cas poorly compared to c-Src (Figure 5A), were also not as efficient as wild-type Src in wound healing (Figure 6). Thus, the mutants exhibited changes in Cas phosphorylation, and there were corresponding changes in their function in this cell signaling pathway.
Figure 6.
Wound healing assays. In the top panel, SYF cells were infected with retroviruses expressing c-Src or the domain-rearranged mutants. The cells were more than 90% infected as measured by GFP fluorescence (data not shown). Multiple wounds were scratched on the 60 mm dishes, and eight positions were photographed for 10–12 h. The rate of wound closure was calculated by plotting the width of the wound vs time. The rate of closure shown above is the average of eight separate positions. The standard errors are those for all eight positions. In the bottom panel, expression of the constructs in SYF cells was tested by anti-Flag Western blotting.
Specific Changes in Substrate Specificity toward Sam68
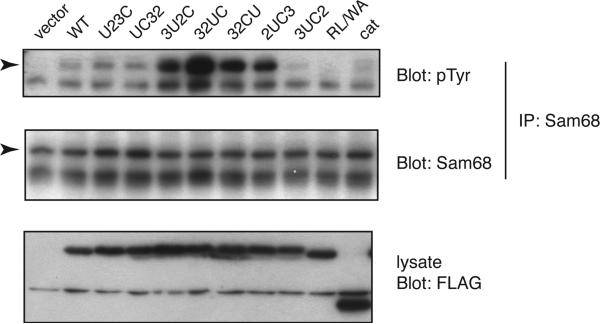
Next, we tested for specific changes in recognition of another protein, the mitotic Src substrate Sam68. We expressed mutant or wild-type forms of c-Src in SYF cells and arrested them in mitosis with nocodazole. We immunoprecipitated Sam68 from the mitotic cells and examined tyrosine phosphorylation using anti-pTyr Western blotting. The R175L/W118A mutant and isolated catalytic domain were deficient in Sam68 phosphorylation. Rearrangement of the domains led to changes in phosphorylation of Sam68 as compared to that of c-Src (Figure 7). The U23C and UC32 mutants were comparable to c-Src in their ability to phosphorylate Sam68. Mutants in which the unique domain was shifted (3U2C, 32UC, and 32CU) showed enhanced phosphorylation of Sam68 compared to c-Src (Figure 7). Since these three mutants also showed an increased level of phosphorylation of Cas (Figure 5A), the enhanced phosphorylation of Sam68 by this group of mutants is probably due to their increased activity rather than any change in specificity. On the other hand, although mutant 2UC3 phosphorylated Sam68 more strongly than c-Src, mutant 3UC2 showed very weak activity (Figure 7). This contrasts with results for Cas, in which both 2UC3 and 3UC2 had lower activity than wild-type c-Src (Figure 5A). The enhancement of Sam68 phosphorylation by 2UC3 points to a change in substrate specificity, suggesting that the arrangement of domains can influence Src substrate recognition.
Figure 7.
Phosphorylation of Sam68 by Src and the mutants. SYF cells were transiently transfected with wild-type Src or the mutants and arrested in mitosis using nocodazole 24 h after transfection. The mitotic cells were harvested 40 h post-transfection, and lysates were subjected to immunoprecipitation using anti-Sam68 antibody. The immunoprecipitates were separated by 10% SDS–PAGE, and Western blotting was carried out with anti-pTyr antibody. The membranes were stripped and reprobed with anti-Sam68 antibody to ensure equivalent Sam68 immunoprecipitation. The figure is a representative of three separate experiments; a comparison of results from three different transfected cell lysates is shown in Figure 4 of the Supporting Information.
In Vitro Kinase Assays with Mutants
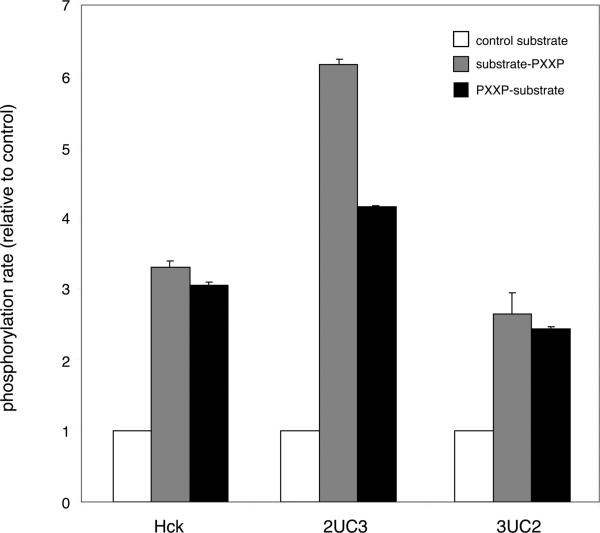
We tested whether the changes we observed in Sam68 phosphorylation by 2UC3 and 3UC2 could be observed in vitro. We expressed mutants 2UC3 and 3UC2 in Sf9 insect cells using baculovirus expression vectors. We purified the proteins and compared their properties to those of the Src family kinase Hck. Our initial studies showed that the kinetic parameters kcat and Km(ATP) were similar for the mutants and Hck (data not shown). Next, we compared phosphorylation of a synthetic peptide containing a Src phosphorylation site and the SH3 binding sequence from Sam68 (substrate-PXXP; the sequences of the peptides used are presented in Table 2). Consistent with our previous findings (54), Hck phosphorylated this peptide 3 times more efficiently than a control peptide with amino acid substitutions in the SH3-binding sequence (Figure 8). Mutant 2UC3 exhibited a more pronounced effect: it phosphorylated the SH3 substrate 6 times better than the control peptide. Mutant 3UC2 phosphorylated the SH3 substrate 2 times better than the control, a slightly smaller effect than Hck (Figure 8). This trend is consistent with Sam68 phosphorylation in SYF cells (Figure 7). We also tested a peptide containing a Src phosphorylation site and the SH3 binding sequence in the reverse orientation (PXXP-substrate). Wild-type Hck and mutants 2UC3 and 3UC3 all phosphorylated this peptide more efficiently than the control.
Table 2.
Peptides Used for in Vitro Kinase Assaysa
| name | sequence |
|---|---|
| control substrate | AEEEIYGEFGGRGAAAAAAAVARGRG |
| substrate-PXXP | AEEEIYGEFGGRGAAPPPPPVPRGRG |
| PXXP-substrate | KKRPLPSPPKF(G)6EEEIYGEFG |
| substrate-FEEI | RRLEDAIYAAGGGGGEPPQFEEIG |
| substrate-pYEEI | RRLEDAIYAAGGGGGEPPQpYEEIG |
| FEEI-substrate | EPQFEEIGGGGGEDAIYARRG |
| pYEEI-substrate | EPQpYEEIGGGGGEDAIYARRG |
The top three peptides were used to study SH3-dependent targeting, and the bottom four peptides were used to study SH2-dependent targeting. The phosphorylation sites are underlined, and the SH3 and SH2 ligand sequences are in bold type.
Figure 8.
In vitro kinase assays using substrates with SH3 ligands. Kinase assays were performed using purified proteins and the indicated peptides at 50 μM (sequences given in Table 2). Initial rates were measured in triplicate, and they are plotted relative to the control substrate ± the standard deviation.
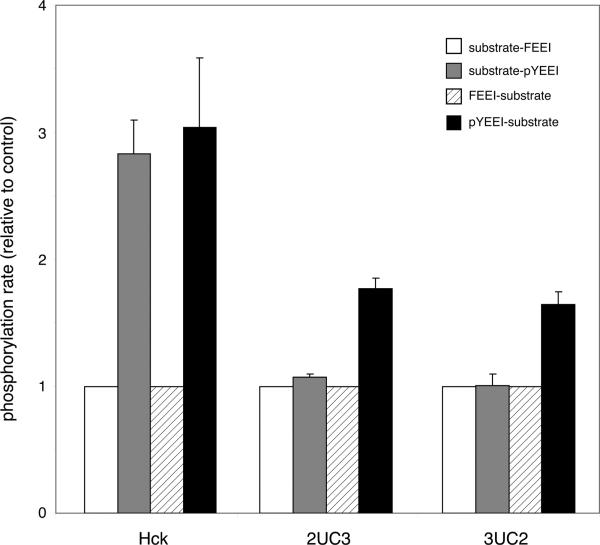
Next, we carried out similar studies on a synthetic Src substrate containing the pYEEI recognition sequence for the SH2 domain. As reported previously (47), the activity of Hck was higher toward the SH2-binding substrate (substrate-pYEEI) than a control substrate lacking p-Tyr (substrate-FEEI) (Figure 9). In contrast, 2UC3 and 3UC2 showed no difference between the SH2-binding and control substrates. We tested a peptide in which the SH2 ligand was N-terminal to the substrate sequence (pYEEI-substrate). Hck, 2UC3, and 3UC2 all phosphorylated this peptide more strongly than a control (FEEI-substrate); for Hck, the rate was 3-fold higher, while for the mutants, the rate was 1.5-2-fold higher (Figure 9). Thus, for Hck, SH2-mediated targeting occurs regardless of the orientation, while the mutants displayed targeting only for a peptide with an N-terminal pTyr. This suggests that the positioning of the SH2 domain relative to the kinase domain is especially important for proper substrate recognition.
Figure 9.
In vitro kinase assays using substrates with SH2 ligands. Kinase assays were performed using purified proteins and the indicated peptides at 50 μM (sequences given in Table 2). Initial rates were measured in triplicate. The rates of phosphorylation of substrate-pYEEI and pYEEI-substrate are presented relative to the appropriate control peptide.
DISCUSSION
Our results suggest that the SH3–SH2–kinase architecture of nonreceptor tyrosine kinases has been maintained through evolution in part because this position of domains is important in dictating specificity. Thus, we postulate that the adjacent placement of domains in the correct orientation can lead to simultaneous interactions with potential substrates and enhance phosphorylation. These findings are consistent with earlier studies on the Src kinase Fyn, in which it was demonstrated that the presence of the N-terminal region and SH3 domain enhanced the binding of cellular phosphoproteins to the SH2 domain (55). In another study, repositioning the domains of Abl led to changes in transformation potential without changes in activity, which might be explained by a change in the specificity of the kinase (38).
In our study, repositioning the domains of Src led to differential effects in recognition of Src substrates. For example, the mutants 2UC3 and 3UC2 exhibited very little phosphorylation of Cas (Figure 5A). These two mutants were also less effective than c-Src at promoting cell migration, a downstream effect of Cas (Figure 6). In contrast, 2UC3 phosphorylated Sam68 more effectively than c-Src, while 3UC2 had decreased activity toward Sam68 (Figure 7). This may be due to the different placement of the Src SH3 and SH2 binding sequences in Cas and Sam68. In Cas, the SH3 and SH2 ligands are clustered in the C-terminus while the substrate region is in the central portion of the molecule. Because the SH3 and SH2 domains are distant from each other in the 2UC3 and 3UC2 mutants, these proteins might not be able to benefit from the cooperative binding to Cas seen in wild-type c-Src (24, 56). In contrast, Sam68 possesses an N-terminal SH3 ligand, and the substrate and SH2-binding sites are in the C-terminus of the protein. It has previously been shown that SH3–polyproline interactions are the major determinant of Src phosphorylation for Sam68 (36). Phosphorylation of Sam68 by 2UC3 suggests that the SH3 domain can still target the kinase domain, even when present on the C-terminal side of the kinase domain. The lower activity of 3UC2 could be due to the fact that the SH3 and kinase domains are separated by the unique domain in that construct.
Our in vitro kinase assay data (Figure 8) support the results from cellular assays. Mutant 2UC3 exhibited strong phosphorylation of Sam68 (Figure 7), and 2UC3 had the highest activity toward peptide substrates containing a Sam68-derived polyproline sequence (Figure 8). Neither 2UC3 nor 3UC2 phosphorylated Cas significantly (Figure 5A). In Cas, the SH2-binding sequence is C-terminal to the phosphorylation sites. This is the same orientation as the substrate-pYEEI peptide, which was not phosphorylated better than controls by 2UC3 or 3UC2 (Figure 9). In contrast, a substrate with the reverse orientation (pYEEI-substrate) showed enhanced phosphorylation by both 2UC3 and 3UC2 relative to a control (Figure 9). Thus, the placement of the SH2-binding sequence relative to the phosphorylation sites seems to be particularly important for targeting. The reason that both 2UC3 and 3UC2 displayed this tendency is not clear at present. In natural NRTKs, SH2 domains are invariably found immediately N-terminal to the kinase domain, and the two domains appear to have coevolved (6). There is a correlation between the substrate specificity of NRTKs and the specificity of the associated SH2 domains (57); this correlation may be crucial for the SH2-mediated processive multisite phosphorylation of proteins. In the case of the 3UC2 and 2UC3 mutants, the presence of a C-terminal SH2-binding sequence may make bound proteins inaccessible to the kinase domain. Such proteins might also be more readily dephosphorylated by cellular phosphatases (37).
The structures of nonreceptor tyrosine kinases make it clear that the positions of the domains are critical for enzyme regulation. Our studies point to the importance of domain arrangement in determining substrate specificity. This may also be relevant to the aberrant signaling observed in some cancers that express mutant forms of tyrosine kinases. Many insertions, mutations, or deletions lead to constitutively active kinases. These alterations could also produce changes in substrate specificity. For example, fusion of the BCR segment to Abl in chronic myelogenous leukemia leads to phosphorylation of Tyr177 in BCR. This phosphotyrosine residue binds to the Grb2 SH2 domain; mutation of Tyr177 decreases the transforming potential of BCR-Abl (58). It will be important to understand how the proper positioning of domains in tyrosine kinases confers the ability to recognize key cellular substrates.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Nick Carpino for providing the retroviral constructs and Phoenix cell line and Chris Gordon (Stony Brook University) for assistance with microscopy.
Footnotes
This work was supported by National Institutes of Health Grant CA58530 to W.T.M.
Abbreviations: SH3, Src homology 3; SH2, Src homology 2; SFKs, Src family kinases; EGF, epidermal growth factor; PDGF, platelet-derived growth factor; RACK1, receptor for activated C-kinase; DMEM, Dulbecco's modified Eagle's medium; EDTA, ethylenediaminetetraacetate; FBS, fetal bovine serum; PEP, phosphoenolpyruvate; PK/LDH, pyruvate kinase/lactate dehydrogenase; HRP, horseradish peroxidase; ECL, enhanced chemiluminescence; CMV, cytomegalovirus; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; PVDF, polyvinylene difluoride; PBS, phosphate-buffered saline; β-ME, β-mercaptoethanol; HPLC, high-performance liquid chromatography; Sf9, Spodoptera frugiperda; NRTKs, nonreceptor tyrosine kinases; BCR, B-cell receptor; Grb2, growth factor receptor-bound protein 2.
SUPPORTING INFORMATION AVAILABLE
Amino acid sequences of all constructs used and additional Western blotting experiments on activation loop, Cas, and Sam68 phosphorylation. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Robinson DR, Wu YM, Lin SF. The protein tyrosine kinase family of the human genome. Oncogene. 2000;19:5548–5557. doi: 10.1038/sj.onc.1203957. [DOI] [PubMed] [Google Scholar]
- 2.Brown MT, Cooper JA. Regulation, substrates and functions of src. Biochim. Biophys. Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 3.Shiu SH, Li WH. Origins, lineage-specific expansions, and multiple losses of tyrosine kinases in eukaryotes. Mol. Biol. Evol. 2004;21:828–840. doi: 10.1093/molbev/msh077. [DOI] [PubMed] [Google Scholar]
- 4.Hubbard SR, Till JH. Protein Tyrosine Kinase Structure and Function. Annu. Rev. Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- 5.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 6.Nars M, Vihinen M. Coevolution of the domains of cytoplasmic tyrosine kinases. Mol. Biol. Evol. 2001;18:312–321. doi: 10.1093/oxfordjournals.molbev.a003807. [DOI] [PubMed] [Google Scholar]
- 7.Segawa Y, Suga H, Iwabe N, Oneyama C, Akagi T, Miyata T, Okada M. Functional development of Src tyrosine kinases during evolution from a unicellular ancestor to multicellular animals. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12021–12026. doi: 10.1073/pnas.0600021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J, Miller WT. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 10.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 11.Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J, Miller WT. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 12.Sicheri F, Kuriyan J. Structures of Src-family tyrosine kinases. Curr. Opin. Struct. Biol. 1997;7:777–785. doi: 10.1016/s0959-440x(97)80146-7. [DOI] [PubMed] [Google Scholar]
- 13.Bjorge JD, Jakymiw A, Fujita DJ. Selected glimpses into the activation and function of Src kinase. Oncogene. 2000;19:5620–5635. doi: 10.1038/sj.onc.1203923. [DOI] [PubMed] [Google Scholar]
- 14.Miller WT. Determinants of substrate recognition in nonreceptor tyrosine kinases. Acc. Chem. Res. 2003;36:393–400. doi: 10.1021/ar020116v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pawson T, Nash P. Protein-protein interactions define specificity in signal transduction. Genes Dev. 2000;14:1027–1047. [PubMed] [Google Scholar]
- 16.Mayer BJ, Baltimore D. Signalling through SH2 and SH3 domains. Trends Cell Biol. 1993;3:8–13. doi: 10.1016/0962-8924(93)90194-6. [DOI] [PubMed] [Google Scholar]
- 17.Weng Z, Thomas SM, Rickles RJ, Taylor JA, Brauer AW, Seidel-Dugan C, Michael WM, Dreyfuss G, Brugge JS. Identification of Src, Fyn, and Lyn SH3-binding proteins: Implications for a function of SH3 domains. Mol. Cell. Biol. 1994;14:4509–4521. doi: 10.1128/mcb.14.7.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kypta RM, Goldberg Y, Ulug ET, Courtneidge SA. Association between the PDGF receptor and members of the src family of tyrosine kinases. Cell. 1990;62:481–492. doi: 10.1016/0092-8674(90)90013-5. [DOI] [PubMed] [Google Scholar]
- 19.Chang BY, Harte RA, Cartwright CA. RACK1: A novel substrate for the Src protein-tyrosine kinase. Oncogene. 2002;21:7619–7629. doi: 10.1038/sj.onc.1206002. [DOI] [PubMed] [Google Scholar]
- 20.Stover DR, Becker M, Liebetanz J, Lydon NB. Src phosphorylation of the epidermal growth factor receptor at novel sites mediates receptor interaction with Src and P85α. J. Biol. Chem. 1995;270:15591–15597. doi: 10.1074/jbc.270.26.15591. [DOI] [PubMed] [Google Scholar]
- 21.Mori S, Ronnstrand L, Yokote K, Engstrom A, Courtneidge SA, Claesson-Welsh L, Heldin CH. Identification of two juxtamembrane autophosphorylation sites in the PDGF β-receptor; involvement in the interaction with Src family tyrosine kinases. EMBO J. 1993;12:2257–2264. doi: 10.1002/j.1460-2075.1993.tb05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen K, Johnell M, Siegbahn A, Rorsman C, Engstrom U, Wernstedt C, Heldin CH, Ronnstrand L. Mutation of a Src phosphorylation site in the PDGF β-receptor leads to increased PDGF-stimulated chemotaxis but decreased mitogenesis. EMBO J. 1996;15:5299–5313. [PMC free article] [PubMed] [Google Scholar]
- 23.Guappone AC, Flynn DC. The integrity of the SH3 binding motif of AFAP-110 is required to facilitate tyrosine phosphorylation by, and stable complex formation with, Src. Mol. Cell. Biochem. 1997;175:243–252. doi: 10.1023/a:1006840104666. [DOI] [PubMed] [Google Scholar]
- 24.Nakamoto T, Sakai R, Ozawa K, Yazaki Y, Hirai H. Direct binding of C-terminal region of p130Cas to SH2 and SH3 domains of Src kinase. J. Biol. Chem. 1996;271:8959–8965. doi: 10.1074/jbc.271.15.8959. [DOI] [PubMed] [Google Scholar]
- 25.Richard S, Yu D, Blumer KJ, Hausladen D, Olszowy MW, Connelly PA, Shaw AS. Association of p62, a multifunctional SH2- and SH3-domain-binding protein, with src family tyrosine kinases, Grb2, and phospholipase C γ-1. Mol. Cell. Biol. 1995;15:186–197. doi: 10.1128/mcb.15.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor SJ, Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- 27.Thomas JW, Ellis B, Boerner RJ, Knight WB, White GC, II, Schaller MD. SH2- and SH3-mediated interactions between focal adhesion kinase and Src. J. Biol. Chem. 1998;273:577–583. doi: 10.1074/jbc.273.1.577. [DOI] [PubMed] [Google Scholar]
- 28.Bouton AH, Riggins RB, Bruce-Staskal PJ. Functions of the adapter protein Cas: Signal convergence and the determination of cellular responses. Oncogene. 2001;20:6448–6458. doi: 10.1038/sj.onc.1204785. [DOI] [PubMed] [Google Scholar]
- 29.Patwardhan P, Shen Y, Goldberg GS, Miller WT. Individual Cas phosphorylation sites are dispensable for processive phosphorylation by Src and anchorage-independent cell growth. J. Biol. Chem. 2006;281:20689–20697. doi: 10.1074/jbc.M602311200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pellicena P, Miller WT. Processive phosphorylation of p130Cas by Src depends on SH3-polyproline interactions. J. Biol. Chem. 2001;276:28190–28196. doi: 10.1074/jbc.M100055200. [DOI] [PubMed] [Google Scholar]
- 31.Vuori K, Hirai H, Aizawa S, Ruoslahti E. Introduction of p130cas signaling complex formation upon integrin-mediated cell adhesion: A role for Src family kinases. Mol. Cell. Biol. 1996;16:2606–2613. doi: 10.1128/mcb.16.6.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Playford MP, Schaller MD. The interplay between Src and integrins in normal and tumor biology. Oncogene. 2004;23:7928–7946. doi: 10.1038/sj.onc.1208080. [DOI] [PubMed] [Google Scholar]
- 33.Chodniewicz D, Klemke RL. Regulation of integrin-mediated cellular responses through assembly of a CAS/Crk scaffold. Biochim. Biophys. Acta. 2004;1692:63–76. doi: 10.1016/j.bbamcr.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Klemke RL, Leng J, Molander R, Brooks PC, Vuori K, Cheresh DA. CAS/Crk coupling serves as a “molecular switch” for induction of cell migration. J. Cell Biol. 1998;140:961–972. doi: 10.1083/jcb.140.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukong KE, Richard S. Sam68, the KH domain-containing superSTAR. Biochim. Biophys. Acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Shen Z, Batzer A, Koehler JA, Polakis P, Schlessinger J, Lydon NB, Moran MF. Evidence for SH3 domain directed binding and phosphorylation of Sam68 by Src. Oncogene. 1999;18:4647–4653. doi: 10.1038/sj.onc.1203079. [DOI] [PubMed] [Google Scholar]
- 37.Mayer BJ, Hirai H, Sakai R. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr. Biol. 1995;5:296–305. doi: 10.1016/s0960-9822(95)00060-1. [DOI] [PubMed] [Google Scholar]
- 38.Mayer BJ, Baltimore D. Mutagenic analysis of the roles of SH2 and SH3 domains in regulation of the Abl tyrosine kinase. Mol. Cell. Biol. 1994;14:2883–2894. doi: 10.1128/mcb.14.5.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klinghoffer RA, Sachsenmaier C, Cooper JA, Soriano P. Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J. 1999;18:2459–2471. doi: 10.1093/emboj/18.9.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldberg GS, Alexander DB, Pellicena P, Zhang ZY, Tsuda H, Miller WT. Src phosphorylates Cas on tyrosine 253 to promote migration of transformed cells. J. Biol. Chem. 2003;278:46533–46540. doi: 10.1074/jbc.M307526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bagrodia S, Taylor SJ, Shalloway D. Myristylation is required for Tyr-527 dephosphorylation and activation of pp60c-src in mitosis. Mol. Cell. Biol. 1993;13:1464–1470. doi: 10.1128/mcb.13.3.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mikhailik A, Ford B, Keller J, Chen Y, Nassar N, Carpino N. A phosphatase activity of Sts-1 contributes to the suppression of TCR signaling. Mol. Cell. 2007;27:486–497. doi: 10.1016/j.molcel.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valster A, Tran NL, Nakada M, Berens ME, Chan AY, Symons M. Cell migration and invasion assays. Methods. 2005;37:208–215. doi: 10.1016/j.ymeth.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 45.LaFevre-Bernt M, Sicheri F, Pico A, Porter M, Kuriyan J, Miller WT. Intramolecular regulatory interactions in the Src family kinase Hck probed by mutagenesis of a conserved tryptophan residue. J. Biol. Chem. 1998;273:32129–32134. doi: 10.1074/jbc.273.48.32129. [DOI] [PubMed] [Google Scholar]
- 46.Barker SC, Kassel DB, Weigl D, Huang X, Luther MA, Knight WB. Characterization of pp60c-src tyrosine kinase activities using a continuous assay: Autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry. 1995;34:14843–14851. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
- 47.Pellicena P, Stowell KR, Miller WT. Enhanced phosphorylation of Src family kinase substrates containing SH2 domain binding sites. J. Biol. Chem. 1998;273:15325–15328. doi: 10.1074/jbc.273.25.15325. [DOI] [PubMed] [Google Scholar]
- 48.Qiu H, Miller WT. Role of the Brk SH3 domain in substrate recognition. Oncogene. 2004;23:2216–2223. doi: 10.1038/sj.onc.1207339. [DOI] [PubMed] [Google Scholar]
- 49.Bibbins KB, Boeuf H, Varmus HE. Binding of the Src SH2 domain to phosphopeptides is determined by residues in both the SH2 domain and the phosphopeptides. Mol. Cell. Biol. 1993;13:7278–7287. doi: 10.1128/mcb.13.12.7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayer BJ, Jackson PK, Van Etten RA, Baltimore D. Point mutations in the abl SH2 domain coordinately impair phosphotyrosine binding in vitro and transforming activity in vivo. Mol. Cell. Biol. 1992;12:609–618. doi: 10.1128/mcb.12.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol. Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 52.Young MA, Gonfloni S, Superti-Furga G, Roux B, Kuriyan J. Dynamic coupling between the SH2 and SH3 domains of c-Src and Hck underlies their inactivation by C-terminal tyrosine phosphorylation. Cell. 2001;105:115–126. doi: 10.1016/s0092-8674(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 53.Yeo MG, Partridge MA, Ezratty EJ, Shen Q, Gundersen GG, Marcantonio EE. Src SH2 arginine 175 is required for cell motility: Specific focal adhesion kinase targeting and focal adhesion assembly function. Mol. Cell. Biol. 2006;26:4399–4409. doi: 10.1128/MCB.01147-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott MP, Miller WT. A peptide model system for processive phosphorylation by Src family kinases. Biochemistry. 2000;39:14531–14537. doi: 10.1021/bi001850u. [DOI] [PubMed] [Google Scholar]
- 55.Panchamoorthy G, Fukazawa T, Stolz L, Payne G, Reedquist K, Shoelson S, Songyang Z, Cantley L, Walsh C, Band H. Physical and functional interactions between SH2 and SH3 domains of the Src family protein tyrosine kinase p59fyn. Mol. Cell. Biol. 1994;14:6372–6385. doi: 10.1128/mcb.14.9.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yadav SS, Miller WT. Cooperative activation of Src family kinases by SH3 and SH2 ligands. Cancer Lett. 2007;257:116–123. doi: 10.1016/j.canlet.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Songyang Z, Carraway KL, III, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 58.Million RP, Van Etten RA. The Grb2 binding site is required for the induction of chronic myeloid leukemia-like disease in mice by the Bcr/Abl tyrosine kinase. Blood. 2000;96:664–670. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.