Abstract
The search to understand how genomes innovate in response to selection dominates the field of evolutionary biology. Powerful molecular evolution approaches have been developed to test individual loci for signatures of selection. In many cases, however, an organism's response to changes in selective pressure may be mediated by multiple genes, whose products function together in a cellular process or pathway. Here we assess the prevalence of polygenic evolution in pathways in the yeasts Saccharomyces cerevisiae and S. bayanus. We first established short-read sequencing methods to detect cis-regulatory variation in a diploid hybrid between the species. We then tested for the scenario in which selective pressure in one species to increase or decrease the activity of a pathway has driven the accumulation of cis-regulatory variants that act in the same direction on gene expression. Application of this test revealed a variety of yeast pathways with evidence for directional regulatory evolution. In parallel, we also used population genomic sequencing data to compare protein and cis-regulatory variation within and between species. We identified pathways with evidence for divergence within S. cerevisiae, and we detected signatures of positive selection between S. cerevisiae and S. bayanus. Our results point to polygenic, pathway-level change as a common evolutionary mechanism among yeasts. We suggest that pathway analyses, including our test for directional regulatory evolution, will prove to be a relevant and powerful strategy in many evolutionary genomic applications.
Keywords: gene regulation, adaptation, yeast
A main challenge of evolutionary biology is to understand the influence of selection on genetic variation within and between species. Classically, molecular evolution methods have targeted individual genes or loci for tests of selection. Their successes have uncovered both protein-coding and regulatory variants with signatures of non-neutral evolution (1–3). However, decades of quantitative genetic mapping studies indicate that in most cases, variation in phenotype between individuals is the result of multiple sequence changes at unlinked loci (4). The genetic response to changes in selective pressure is likely to follow the same pattern, but, to date, methods for identifying cases of polygenic evolution have been at a premium.
Some of the strongest evidence for polygenic adaptation has emerged from the study of protein-coding variants between phylogenetic lineages. Signatures of positive selection can be detected in the protein-coding sequences of groups of genes of related function (5–8), suggestive of a coherent series of genetic changes accumulated by a population in response to selection. Additionally, genetic variation in gene expression represents a rich data source for signatures of selection, both positive (9, 10) and purifying (11, 12), although the mechanisms that govern regulatory evolution are not fully understood. Regulatory variants can act in cis, to impact the expression of a neighboring gene, or in trans, targeting the expression of genes elsewhere in the genome. Hallmarks of positive selection have been observed at individually mapped cis-regulatory variants (1, 3); one appealing model predicts that such a locus is part of a suite of adaptive regulatory changes in downstream effectors of a pathway (13–16). More generally, in the face of either positive or relaxed selection, a population may accumulate multiple independent changes at effector loci, in cis-regulatory sequences or in protein-coding regions, avoiding the potentially pleotropic effects of variation in master regulators (1, 17).
In this work, we set out to test the polygenic evolutionary model in a simple eukaryotic system. We developed genome-scale sequencing methods to identify cis-regulatory variants between two Saccharomyces species, measuring allele-specific expression levels of a gene in a hybrid diploid (18, 19), and we used the results to uncover evidence of directional evolution of the expression of genes in pathways. To extend these findings, we independently tested for evidence of polygenic evolution using population resequencing data.
Results
Differential Allele-Specific Expression in an Interspecific Hybrid.
We sought to study directional evolution of cis-regulatory control of expression on a genomic scale in Saccharomyces cerevisiae and S. bayanus. As an experimental paradigm (20–22), we used genome-wide analysis of expression in a hybrid diploid formed from the mating of the two species. Because the orthologs of a given gene from both species are present in the same environment of trans-acting factors, differential expression of the orthologs reflects the presence of cis-acting differences between the species. Detecting expression variation within the hybrid requires techniques that recognize the alleles encoded by each ortholog in each species. For this purpose, we measured allele-specific expression with Illumina short-read sequencing (RNA-seq) (23–25). We mated the W303 strain of S. cerevisiae and the CBS 4001 isolate of S. bayanus to form a hybrid diploid and isolated RNA from two independent cultures of this strain as biological replicates. For each, we sequenced cDNA libraries, for a total of eight lanes. We used the set of sequencing reads mapping uniquely to either the S. cerevisiae or the S. bayanus genome to quantitate the expression of the allele of each gene from each species, yielding 4,238 ortholog pairs with observable allele-specific expression.
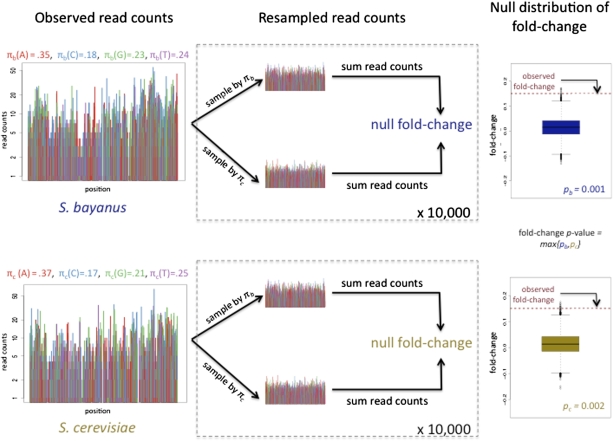
For a given ortholog pair, we used the average log-ratio of S. bayanus to S. cerevisiae per-base read counts as a statistic for differential allele-specific expression. However, such a measure reflects not only the biologically relevant difference in transcript abundance but also the inherent “sequenceability” of one allele relative to the other. In particular, GC-rich regions tend to be preferentially sequenced by the Illumina platform (26, 27), raising the possibility that comparison of raw allele-specific read counts from an ortholog pair might overestimate the abundance of the more GC-rich ortholog. We investigated the impact of allelic base composition on our RNA-seq read counts (Fig. S1) and found that, roughly, a 5% difference in GC content between alleles was associated with a 10% fold-change in their apparent expression. To account for such confounding, we developed a resampling-based method for analyzing differential allele-specific expression. In our approach, the significance of differences in read counts is evaluated by reference to a null distribution that incorporates differences in sequence composition. As illustrated in Fig. 1, we resampled the base-level read counts of the two orthologs to form “null” ortholog pairs with no differential expression and the same marginal nucleotide distributions as the original orthologs. In a given null pair, any apparent difference in the number of RNA-seq reads mapping to one ortholog rather than the other can be attributed to sequencing artifacts alone. When analyzing our actual data, to assert that the observed difference in RNA-seq reads between orthologs in a pair arose from variation in expression between the species and not from technical effects, we required that the observed RNA-seq fold-change lie outside the range of differences we expected under the null. This comparison between observed and null data resulted in a P value for each ortholog pair. The resampling strategy was stringent, identifying fewer cases of significant differential expression than did a standard statistical approach that does not account for GC bias (Table S1).
Fig. 1.
Schematic of RNA-seq method for inferring differential allele-specific expression in a hybrid diploid; resampling procedure for an example gene. (Left) Observed base-level read counts are displayed for the S. bayanus and S. cerevisiae orthologs, using color to represent the nucleotide at each base. The x axis gives the position of each base, and the y axis gives the number of allele-specific reads whose first nucleotide maps to a given position. Above each plot are the allele-specific marginal nucleotide frequencies πb = [πb(A),πb(C),πb(G),πb(T)] and πc = [πc(A),πc(C),πc(G),πc(T)], for S. bayanus and S. cerevisiae, respectively. (Center) For each ortholog, “null” counts are created by resampling base-level read counts according to the S. bayanus and S. cerevisiae nucleotide frequencies πb and πc, respectively. Null expression log-fold-changes are computed by averaging (across lanes for each of the two biological replicates) log-ratios of S. bayanus to S. cerevisiae null per-base read counts. The resampling procedure is repeated 10,000 times for each ortholog. (Right) Boxplots of the 10,000 null expression log-fold-changes for each ortholog. The observed log-fold-change from the original read counts is represented by dark red dashed lines and is compared with each null distribution to obtain two-sided P values, pb and pc. The significance of the observed allele-specific expression difference is summarized by the maximum P value, max(pb, pc).
We applied the resampling method to each of the two biological replicates and identified ortholog pairs for which both P values fell below a given cutoff. The results, given in Table S2, yielded 2,124 ortholog pairs with maximum P value < 0.05 (at which 212 genes would be expected under the null from sequenceability effects alone), 1,570 ortholog pairs with P < 0.005 (21 would be expected under the null), and 1,176 ortholog pairs with P < 0.0005 (2 would be expected under the null). To evaluate the resulting dataset, we designed quantitative RT-PCR assays to measure allele-specific expression for a subset of genes that spanned the range of P values and fold-changes from RNA-seq from among those significant at P < 0.05 (Table S3). As shown in Fig. S2, we observed good agreement between RT-PCR and RNA-seq measures of allelic expression differences in the hybrid. Of immediate relevance for our downstream analysis (see below), the two methods agreed on the sign of the fold-change in 20/22 genes tested, confirming the ability of our RNA-seq procedure to determine for a given ortholog pair whether the S. bayanus or S. cerevisiae allele was associated with higher expression.
Testing for Directional cis-Regulatory Evolution.
We hypothesized that some cis-regulatory changes between S. cerevisiae and S. bayanus could reflect a history of selection on gene expression when analyzed jointly over groups of genes of common function. For a given such pathway (Fig. 2), we expected to observe a preponderance of ortholog pairs in which the S. bayanus allele was up-regulated with respect to the S. cerevisiae allele or a preponderance in which the S. cerevisiae allele was up-regulated with respect to the S. bayanus allele. Such an imbalance reflects an accumulation of independent genetic changes acting in the same direction, which is unlikely under neutrality (28). Rather than assume a specific model for neutral evolution, we sought to base our null expectation on the genome-wide prevalence of differential allele-specific expression inferred in the hybrid.
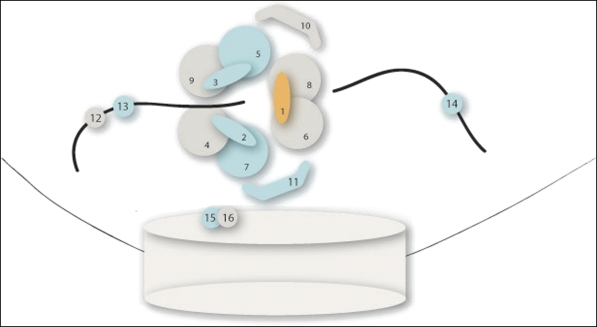
Fig. 2.
Evidence for directional cis-regulatory evolution, for the example of the nuclear exosome. Shown is a schematic of the proteins in the Gene Ontology terms “nuclear exosome” and “colocalizes_with: nuclear exosome,” colored to reflect expression differences between S. cerevisiae and S. bayanus alleles in the interspecific hybrid. Blue, up-regulation of the S. bayanus allele; orange, up-regulation of the S. cerevisiae allele; gray, no measured expression effect. The complex in the center of the cartoon represents the exosome. The thick black line indicates RNA in the act of processing, the beige cylinder represents a nuclear pore, and the thin line represents the nuclear envelope. 1: CSL4; 2: RRP4; 3: RRP40; 4: RRP41; 5: RRP46; 6: MTR3; 7: RRP42; 8: RRP43; 9: RRP45; 10: RRP6; 11: RRP44; 12: MMP6; 13: LRP1; 14: NRD1; 15: KAP95; 16: SRP1.
To test for directional cis-regulatory evolution between the species, we first used a previously curated set of gene groups defined by coregulation in S. cerevisiae (14). We assigned a direction score to each gene in our dataset: 1 if expression of the S. bayanus allele was significantly higher than expression of the S. cerevisiae allele at P < 0.05, −1 if expression of the S. cerevisiae allele was significantly higher than expression of the S. bayanus allele, and 0 otherwise. We took the sum of the direction scores of the genes in each regulon as a measure of the imbalance in the direction of cis-regulatory changes across the group. We then assessed the significance of each such statistic by reference to a resampling-based null distribution, yielding a two-sided P value, and we set a lenient threshold corresponding to ∼1 expected false-positive group out of all tested. Table 1A reports the set of regulons reaching this criterion, which included two amino acid biosynthesis pathways, a set of ribosomal and translation genes, a regulon of respiratory enzymes, and a group of transporters. The gene groups emerging from this analysis reflect a history of rare but detectable directional evolution across pathways during the divergence of S. cerevisiae and S. bayanus.
Table 1.
Directional evolution of gene expression in pathways
| Name | Statistic* | # Genes† | P‡ | Annotation | |
| A. Gene groups defined by coregulation in S. cerevisiae (14); 1.08 false-positive groups expected | |||||
| Cluster_Histidine | −7 | 8 | 9.00E-05 | Histidine biosynthesis | |
| Node 73 | 71 | 349 | 0.001 | Ribosome and translation | |
| Cluster_NRG1 | −7 | 12 | 0.002 | Stress-induced transport | |
| Cluster_Lysine | −5 | 8 | 0.006 | Lysine biosynthesis | |
| Node 45 | −9 | 45 | 0.007 | Respiration | |
| B. Gene groups defined as the Biological Process categories from Gene Ontology; 0.53 false-positive groups expected | |||||
| GO:0422540 | 79 | 269 | 1.0E-05 | Ribosome biogenesis | |
| GO:0016070 | 124 | 827 | 0.009 | RNA metabolic process | |
| GO:0006275 | −8 | 56 | 0.01 | Cellular aromatic compound metabolic process | |
Listed are the top-scoring gene groups with sign imbalance in cis-regulatory variation. Results for all groups are given in Table S8.
*Directional differential expression statistic measuring the imbalance in the signs of cis-regulatory variants within gene groups, defined as the sum of the directional scores of the group members (1 for a gene significantly up-regulated in S. bayanus for both biological replicates, −1 for a gene significantly up-regulated in S. cerevisiae, and 0 otherwise).
†Total number of genes in each group for which a directional score was available.
‡Resampling-based P value assessing the significance of directional cis-regulatory expression variation in each group.
We next repeated our procedure on Biological Process gene categories manually assembled by the Gene Ontology consortium (29). Table 1B lists the resulting categories, including ribosomal biogenesis, genes mediating aromatic compound metabolism, and a broad group of RNA-processing genes; a subset of the latter, the genes involved in the nuclear exosome, are illustrated in Fig. 2. We conclude that signatures of directional evolution can be detected in tightly defined biochemical pathways as well as in broad and heterogeneous gene classes.
Sequence-Based Tests for Selection Within and Between Species.
Given our evidence for directional selection during the divergence of S. cerevisiae and S. bayanus at the level of gene expression across pathways, we next sought to test more generally for polygenic evolution. The whole-genome sequences of 34 nonlaboratory S. cerevisiae isolates and one isolate of S. bayanus (30) comprise a dataset suitable for sequence-based tests of selection. We reasoned that, although currently existing sequence-based evolutionary genetic approaches are not designed to uncover directional regulatory evolution, they would allow the study of polygenic evolution more broadly. For this purpose, for each gene we applied the McDonald–Kreitman test (31) to compare the frequency of amino acid changes and synonymous changes within and between species. We also used a McDonald–Kreitman-like test to compare the prevalence of variants in regulatory regions with the prevalence of synonymous changes, again within and between species (9, 32). Nonsynonymous and upstream variants were more common within S. cerevisiae than between the species (30), and test results on individual genes primarily revealed cases of enrichment for polymorphism at candidate functional sites (Table S4). The ubiquitin gene UBI4 harbored an excess of species differences in the upstream region (nominal P = 2.7 × 10−9, at which <<1 gene was expected if all were evolving neutrally), but variation in upstream regions did not afford sufficient statistical power for a dataset of high-scoring genes in McDonald–Kreitman-like tests. However, in McDonald–Kreitman analysis of coding regions, at nominal P < 0.005 we detected 94 genes (12.6 expected if all genes were evolving neutrally), including three genes with an excess of nonsynonymous changes between species: the fatty acid oxidative enzyme TES1, the transcription factor YRM1, and the cell wall component CCW14.
Excluding the latter candidate cases of positive selection from further analysis, we next sought to identify pathways enriched for genes with excess nonsynonymous polymorphism. Because analysis of Gene Ontology categories gave modest but appreciable power (Table S5), we focused on this class of gene groups, which revealed that the accumulation of coding changes among S. cerevisiae isolates can be pathway-dependent. Among the top-scoring results, shown in Table 2, were categories annotated in stress response, cell division, and DNA metabolism, each enriched for genes with an excess of protein-coding variants within S. cerevisiae. In a given pathway, such enrichment is consistent with a history of relaxed constraint or balancing selection across the species, or with pathway-level divergence between S. cerevisiae populations. Given the known preponderance of alleles segregating at low frequency within S. cerevisiae (30), we repeated McDonald–Kreitman tests and pathway analyses on a dataset in which singleton sites were filtered out (Tables S6 and S7). Top-scoring pathways from these filtered data mirrored pathways emerging from analysis of all sites, albeit with weaker P values for enrichment of polymorphism (Table S7); thus, although singleton alleles drive much of the statistical power for this analysis, more common variants likely exhibit similar trends of enrichment in pathways. We conclude that an excess of fixed sequence differences between S. cerevisiae and S. bayanus, indicative of positive selection between the species, can be detected at a handful of individual loci and that genes of common function share patterns of sequence polymorphism within S. cerevisiae.
Table 2.
Pathway enrichment of within-species sequence variation
| Name | MK enrichment* | P† | Annotation |
| GO:0007124 | 6.0 | 9 × 10−5 | Pseudohyphal growth |
| GO:0006950 | 2.2 | 0.0005 | Response to stress |
| GO:0006259 | 2.1 | 0.006 | DNA metabolic process |
| GO:0007126 | 2.5 | 0.03 | Meiosis |
Listed are the top-scoring pathways in a test for enrichment among genes with evidence for protein coding variation within S. cerevisiae according to the McDonald–Kreitman test. Results for all pathways are given in Table S5. Gene groups were defined as the Biological Process categories from Gene Ontology, with 1.2 false-positive groups expected.
*The ratio between the number of genes in the pathway scored as significant in the McDonald–Kreitman test and the number of genes in all pathways scored as significant, divided by the analogous ratio for all tested genes regardless of significance.
†Hypergeometric P value for pathway enrichment of genes significant in the McDonald–Kreitman test.
Discussion
Decades of work based on the study of individual genes with signatures of selection have shed light on the mechanisms of evolutionary change (1–3). However, any such locus may represent only one component of a suite of changes among genes of related function that have arisen in response to the same selective pressure over evolutionary time. The prevalence and mechanisms of polygenic evolution are unknown, and addressing the question requires genome-scale data sources and analytic methods that in many cases remain to be developed. For genomic analysis of the evolution of gene expression, cis-acting differences in a heterozygote diploid can be mapped at high resolution (20–22), but to date, methods for this purpose have required custom microarray tools. The RNA-seq platform we pioneer here enables study of cis-acting variation in expression in any organism and any strain (23–25). Likewise, we have demonstrated the power of extending the classical sequence-based McDonald–Kreitman test for selection to pathways, an approach that has increasing utility with the advent of population genomic sequencing data (5–8, 32).
We have developed an analytic strategy to detect nonneutral evolution in pathways, finding cis-regulatory changes with effects in the same direction that have accumulated in one phylogenetic lineage relative to another. Any case of directional evolution of gene expression in a pathway can be explained with either of two evolutionary models. A lineage can accumulate novel alleles that predominantly up-regulate or predominantly down-regulate the genes of a pathway because these changes result in a fitness advantage in a particular niche. Alternatively, coherent purifying selection can keep levels of pathway genes predominantly low or predominantly high in a particular lineage; if this selective force is relaxed in a second lineage, novel alleles accumulated by drift will change expression levels in a direction opposing the original selective constraint. Tests for directional selection have their origin in analyses of quantitative trait loci mapped to macroscopic phenotypes (28), but studies of a given trait rarely have sufficient power for the test. By contrast, genome-scale datasets of cis-regulatory variants of the kind reported here represent the result of hundreds to thousands of independent genetic events in the history of the two parent genomes. Harnessing these data, our test for coherent regulatory change complements previous strategies to observe the evolution of regulatory motifs in pathways (13–16), providing a significance estimate and a direction of inferred effects on pathway output for each candidate case of nonneutral evolution. Interestingly, among the hits were a number of metabolic pathways, including respiration and amino acid biosynthesis genes. Although the latter could be the result of adaptation to laboratory conditions, it is tempting to speculate that changes in nutrient availability may underlie cases of pathway evolution in Saccharomycetes, as would be expected from findings in other organisms (33–35).
However, our test for directional regulatory evolution is not designed to detect cases of selection on amino acid changes, and it focuses on the divergence between a pair of isolates rather than divergence among many strains. Indeed, sequence-based tests detected cases of polygenic evolution in a set of pathways different from those identified by the expression-based approach, as expected given the distinctions between the two methods. In particular, McDonald–Kreitman tests revealed patterns of protein-coding polymorphism at the pathway level between S. cerevisiae isolates. Previous analyses of rare alleles in S. cerevisiae have suggested relaxed selection as a major evolutionary force in this system (30), and relaxation resulting from environmental change would be expected to drive an accumulation of derived alleles in particular response pathways. Thus, pathways enriched for nonsynonymous polymorphism across S. cerevisiae may shed light on the changes in the life history of this species since its divergence from S. bayanus. In addition, some cases of pathway-level polymorphism may result from divergence between S. cerevisiae populations, as the result of adaptation (36, 37) or of relaxed selection in particular niches. Improved sequence sampling of the well-defined populations in this species (30) ultimately will enable in-depth study of the changes in selective pressure at play during their divergence.
Emerging from our work and that of others (5–7, 13–16, 32) is a picture of evolutionary change within and between species mediated by polygenic suites of variants in pathways. With respect to adaptation, a more parsimonious mechanism could involve a single advantageous change in an upstream regulator that up- or down-regulates a pathway in one mutational step (2). However, in experimental organisms, changes in trans-acting transcriptional regulators are more prevalent in lines that have accumulated mutations than in wild isolates (12) and are more common within species than between species (17), suggesting that upstream effects are often deleterious and subject to purifying selection. As such, the time taken by a mutational search for an adaptive variant in an upstream regulator often may be comparable to that required for the acquisition of multiple changes in downstream effectors (1). By the same token, relaxed selection on the function of a pathway frequently would give rise to suites of downstream changes but less often to a major upstream variant. The prevalence of polygenic evolution may explain why detecting evidence for strong selection on single loci has proven challenging to date and will serve as continued motivation for the study of mechanisms by which genes in pathways coevolve.
Materials and Methods
Strains, RNA Preparation, and Sequencing.
Strain OZY27, a hybrid of S. cerevisiae (isogenic to W303; his3 leu2 lys2 trp1 ura3) and S. bayanus (derived from CBS 4001; ade2 his3 lys2) (38), was kindly provided by O. Zill, University of California, Berkeley. For each of two biological replicates, an OZY27 culture was grown to log phase at 25 °C in YPD medium (39). Total RNA was isolated by the hot acid phenol method (39) and treated with Turbo DNase (Ambion) according to the manufacturer's instructions. A Solexa/Illumina 1G Genome Analyzer library was constructed for each biological replicate as described (40). Each biological replicate was sequenced at both the Vincent J. Coates Genomic Sequencing Laboratory at the University of California, Berkeley, and the Core Instrumentation Facility at the University of California, Riverside. The former provided two lanes of 37-bp reads for each biological replicate; the latter provided three lanes of 40-bp reads for biological replicate 1 and one lane of 40-bp reads for replicate 2. All resulting reads were trimmed to 30 bp.
RNA-seq Preprocessing and Mapping.
We downloaded 4,853 ortholog pairs (http://www.broad.mit.edu/regev/orthogroups/orthologs.html) and in each pair converted the S. cerevisiae ortholog sequence to that of W303 using sequence from (30). For each lane, 30-bp reads were mapped to either strand of the ortholog pair set using Bowtie (41), allowing no mismatches between the read and the reference sequence. All further analyses were done in R (42). For each position in the combined orfeome, we determined whether the read starting at that location was uniquely-mappable (2), i.e., had an edit distance of 2 or greater to any other position in the combined (double-stranded) reference orfeome or genome. The combined orfeome consists of all 4,853 S. bayanus and S. cerevisiae ortholog pairs; the combined genome consists of the concatenation of the S. bayanus (http://downloads.yeastgenome.org/sequence/fungal_genomes/S_bayanus/WashU/contig/) and S. cerevisiae (ftp://ftp.yeastgenome.org/yeast/data_download/sequence/NCBI_genome_source/) reference genomes, converting the latter to that of the W303 strain as above. The ensuing analysis considered only uniquely-mappable (2) reads and the 4,238 ortholog pairs with a minimum of 200 uniquely mappable (2) bases in both species and a maximum difference in length of 100 bp. The correlation in read counts was greater between replicate lanes, sequencing centers, and biological replicates than between S. bayanus and S. cerevisiae orthologs (Fig. S3).
Identifying Ortholog Pairs with Differential Allele-Specific Expression.
Our method for detecting significant differential expression between orthologs in a pair is described in detail in SI Text. Briefly, for a given ortholog pair we sampled the base-level read counts of the S. bayanus ortholog to create null S. bayanus data; to create null S. cerevisiae counts, we resampled, from the S. bayanus ortholog, base-level read counts according to the nucleotide frequencies in the S. cerevisiae ortholog. Computing differential expression statistics for each of 10,000 such null orthologs yielded a null distribution to which we compared the observed differential expression statistic to obtain a P value. Repeating the procedure with the base-level read counts of the S. cerevisiae ortholog yielded a second P value, and we conservatively retained the maximum of the two.
Testing for Directional Imbalance in cis-Regulatory Effects Across Pathways.
Our test for directional regulatory evolution is described in detail in SI Text. Briefly, for pathway analyses we used the regulons defined in (14) and GO_slim Biological Process categories (http://downloads.yeastgenome.org/literature_curation/go_slim_mapping.tab). For a given pathway, we added directional differential expression scores across all genes in the group (1 if the expression in the S. bayanus allele was significantly higher than in the S. cerevisiae allele, −1 if the expression in the S. cerevisiae allele was significantly higher than in the S. bayanus allele, and 0 otherwise) and obtained a P value by comparing the resulting statistic with a null distribution of 100,000 such statistics obtained by resampling genes at random, with replacement. At a given P value threshold P0, we estimated the expected number of false-positive pathways as the product of P0 and the number of tests; we set P0 to attain as close to one false-positive pathway as possible. For comparison, results from the Benjamini–Hochberg multiple testing procedure (43), which controls the false-discovery rate, are given in Table S8.
McDonald–Kreitman Analyses.
We downloaded S. cerevisiae strain sequences from (30), eliminating the laboratory strains W303, S288C, and SK1 from further analysis. For each gene, we aligned the amino acid sequences of S. cerevisiae strains and the reference sequence of S. bayanus (http://downloads.yeastgenome.org/sequence/fungal_genomes/S_bayanus/other) using MUSCLE (44) and regenerated DNA alignments with tranalign (http://emboss.sourceforge.net/). For a total of 2,511 genes, we applied the McDonald–Kreitman test using software from (32). For upstream regions, we used alignments from (30) for the region starting 200 bp upstream of each gene and ending at coding start, and tabulated polymorphic and divergent sites; for polymorphic and divergent silent sites, we used ORF alignments of nonlaboratory strains from (30). Given the 2 × 2 table from these data, for a total of 1,910 genes, we calculated single-gene McDonald–Kreitman-like P values using Fisher's exact test. For filtering of singletons in both coding and upstream analyses, we identified polymorphic sites at which exactly one S. cerevisiae strain harbored the minor allele and eliminated these sites from the analysis, using P < 0.005 as a cutoff for significance in the McDonald–Kreitman test.
For pathway analyses, we used 38 GO_slim categories as above, as well as gene regulons from (14), filtered as described in SI Text; to minimize the number of the latter with poor sampling, we considered a regulon only if ≥10 of its component genes had available McDonald–Kreitman data, for a total of 78 regulons tested. In each pathway, we calculated the overrepresentation of top-scoring McDonald–Kreitman genes using the R function phyper. We chose significance thresholds to attain as close to one false-positive pathway as possible, as above. For comparison, results from the Benjamini–Hochberg multiple testing procedure are given in Tables S5 and S7.
Supplementary Material
Acknowledgments
The authors thank O. Zill for strains; L. Tonkin, C. Preston, and O. K. Yoon for expertise and assistance with Illumina sequencing; H. Lee for assistance with population sequence data; C. Ellison, M. Eisen, R. Nielsen, J. W. Taylor, and M. Slatkin for helpful discussions; and C. Ellison, R. Lusk, D. Scannell, and T. Speed for comments on the manuscript. This work was supported by a Burroughs Wellcome Fund Career Award at the Scientific Interface to R.B.B.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: GEO data set GSE19837.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912959107/DCSupplemental.
References
- 1.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 2.Hoekstra HE, Coyne JA. The locus of evolution: Evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- 3.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 4.Weiss KM. Tilting at quixotic trait loci (QTL): An evolutionary perspective on genetic causation. Genetics. 2008;179:1741–1756. doi: 10.1534/genetics.108.094128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeffares DC, et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet. 2007;39:120–125. doi: 10.1038/ng1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berglund J, Pollard KS, Webster MT. Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 2009;7:e26. doi: 10.1371/journal.pbio.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kosiol C, et al. Patterns of positive selection in six mammalian genomes. PLoS Genet. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark AG, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- 9.Holloway AK, Begun DJ, Siepel A, Pollard KS. Accelerated sequence divergence of conserved genomic elements in Drosophila melanogaster. Genome Res. 2008;18:1592–1601. doi: 10.1101/gr.077131.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 2008;4:e1000271. doi: 10.1371/journal.pgen.1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rifkin SA, Houle D, Kim J, White KP. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature. 2005;438:220–223. doi: 10.1038/nature04114. [DOI] [PubMed] [Google Scholar]
- 12.Denver DR, et al. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet. 2005;37:544–548. doi: 10.1038/ng1554. [DOI] [PubMed] [Google Scholar]
- 13.Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: The evolution of ribosomal regulation in yeast. Proc Natl Acad Sci USA. 2005;102:7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasch AP, et al. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2004;2:e398. doi: 10.1371/journal.pbio.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ihmels J, et al. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- 16.Borneman AR, et al. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- 17.Wittkopp PJ, Haerum BK, Clark AG. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet. 2008;40:346–350. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- 18.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 19.Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- 20.Tirosh I, Reikhav S, Levy AA, Barkai N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science. 2009;324:659–662. doi: 10.1126/science.1169766. [DOI] [PubMed] [Google Scholar]
- 21.Serre D, et al. Differential allelic expression in the human genome: A robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4:e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge B, et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet. 2009;41:1216–1222. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 23.Bloom JS, Khan Z, Kruglyak L, Singh M, Caudy AA. Measuring differential gene expression by short read sequencing: Quantitative comparison to 2-channel gene expression microarrays. BMC Genomics. 2009;10:221. doi: 10.1186/1471-2164-10-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Main BJ, et al. Allele-specific expression assays using Solexa. BMC Genomics. 2009;10:422. doi: 10.1186/1471-2164-10-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, et al. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS One. 2008;3:e3839. doi: 10.1371/journal.pone.0003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullard JH, Purdom EA, Hansen KD, Durinck S, Dudoit S. Statistical inference in mRNA-seq: Exploratory data analysis and differential expression. Technical Report #247. Berkeley, CA: Division of Biostatistics, University of California-Berkeley; 2009. [Google Scholar]
- 27.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 28.Orr HA. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics. 1998;149:2099–2104. doi: 10.1093/genetics/149.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 32.Torgerson DG, et al. Evolutionary processes acting on candidate cis-regulatory regions in humans inferred from patterns of polymorphism and divergence. PLoS Genet. 2009;5:e1000592. doi: 10.1371/journal.pgen.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley JL, Swanson WJ. Positive selection in the human genome: From genome scans to biological significance. Annu Rev Genomics Hum Genet. 2008;9:143–160. doi: 10.1146/annurev.genom.9.081307.164411. [DOI] [PubMed] [Google Scholar]
- 34.Sezgin E, et al. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics. 2004;168:923–931. doi: 10.1534/genetics.104.027649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Zhang YP, Rosenberg HF. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat Genet. 2002;30:411–415. doi: 10.1038/ng852. [DOI] [PubMed] [Google Scholar]
- 36.Smith EN, Kruglyak L. Gene-environment interaction in yeast gene expression. PLoS Biol. 2008;6:e83. doi: 10.1371/journal.pbio.0060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerke J, Lorenz K, Cohen B. Genetic interactions between transcription factors cause natural variation in yeast. Science. 2009;323:498–501. doi: 10.1126/science.1166426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zill OA, Rine J. Interspecies variation reveals a conserved repressor of alpha-specific genes in Saccharomyces yeasts. Genes Dev. 2008;22:1704–1716. doi: 10.1101/gad.1640008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ausubel F, et al. Short Protocols in Molecular Biology. 3rd Ed. New York, NY: Wiley; 1995. [Google Scholar]
- 40.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Development Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2009. Vienna, Austria. ISBN 3-900051-07-0. Available at: http://www.R-project.org.
- 43.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methododl. 1995;57:289–300. [Google Scholar]
- 44.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.