Abstract
Excessive alcohol consumption is one of the leading causes of preventable death in the United States. Approximately 14% of those who use alcohol meet criteria during their lifetime for alcohol dependence, which is characterized by tolerance, withdrawal, inability to stop drinking, and continued drinking despite serious psychological or physiological problems. We explored genetic influences on alcohol dependence among 1,897 European-American and African-American subjects with alcohol dependence compared with 1,932 unrelated, alcohol-exposed, nondependent controls. Constitutional DNA of each subject was genotyped using the Illumina 1M beadchip. Fifteen SNPs yielded P < 10−5, but in two independent replication series, no SNP passed a replication threshold of P < 0.05. Candidate gene GABRA2, which encodes the GABA receptor α2 subunit, was evaluated independently. Five SNPs at GABRA2 yielded nominal (uncorrected) P < 0.05, with odds ratios between 1.11 and 1.16. Further dissection of the alcoholism phenotype, to disentangle the influence of comorbid substance-use disorders, will be a next step in identifying genetic variants associated with alcohol dependence.
Keywords: genetics, candidate genes
Excessive alcohol use is the third leading cause of preventable death in the United States (1). Although normative alcohol use is ubiquitous, alcohol dependence is a serious medical illness (2) that is experienced by ≈14% of alcohol users (3). Alcohol dependence constitutes a substantial health and economic burden, costing an estimated $184 billion in expenditures stemming from alcohol-related morbidity, accidents, lost productivity, and incarceration (4). These challenges underscore the importance of clarifying the etiology of alcohol dependence as a key public health priority.
Liability to alcohol dependence has both genetic and environmental influences, which act independently and in concert. First-degree relatives of affected individuals are at a 2- to 8-fold increased risk for alcohol dependence (5, 6). Adoption studies and twin studies have clarified that this familial clustering of alcohol dependence is attributable largely to genetic factors (7–11). In most recent studies, these heritable influences explain ≈50–80% of the individual differences in liability to alcohol dependence (12, 13).
In an effort to unmask specific genomic influences on alcohol dependence, scientists have brought a vast genomic toolkit to bear on this problem. Large-scale genome-wide association studies (GWAS) offer considerable promise. By genotyping a dense set of SNPs throughout the genome, investigators have the potential to identify with considerable precision genes that may lead to unknown biological pathways involved in alcohol dependence.
Candidate gene strategies frequently have identified significant associations between SNPs in the gene encoding the alpha2 subunit of the γ-aminobutyric acid A receptor (GABRA2), a major inhibitory neurotransmitter in the human nervous system that is involved in the behavioral effects of alcohol (14). Although some exceptions exist (15–17), there are multiple positive reports of association between SNPs in GABRA2 and alcohol- and other substance-related phenotypes (14, 18–22 and reviewed in 23).
We report on a large, well-characterized sample of 1,897 Diagnostic and Statistical Manual of Mental Disorders, edition 4 (DSM-IV) alcohol-dependent cases and 1,932 alcohol-exposed, nondependent controls from the Study of Addiction: Genetics and Environment (SAGE) analyzed at 948,658 SNPs that span the genome. First, using a hypothesis-free, genome-wide association strategy, we nominate SNPs associated with vulnerability to alcohol dependence. Second, we specifically examine the important role of SNPs in GABRA2 using a targeted, hypothesis-driven approach.
Results
The characteristics of the study participants are listed in Table 1, and further details are provided in Table S1. Based on self-report of race, the sample is 69% European descent and 31% African-American. A small number of subjects (3%) reported Hispanic ethnicity. Over 60% of alcohol-dependent cases are male. Comorbid drug dependence is common, with almost half of the alcohol-dependent sample diagnosed with comorbid marijuana or cocaine dependence.
Table 1.
Characteristics of alcohol-dependent cases and nondependent controls
| Characteristic | Cases n = 1,897 | Controls n = 1,932 | Total n = 3,829 |
| Sex, N (%) | |||
| Males | 1,155 (60.9) | 606 (31.4)* | 1,761 (46.0) |
| Females | 742 (39.1) | 1,326 (68.6) | 2,068 (54.0) |
| Age, years | |||
| Mean ± SD | 39.0 ± 9.3 | 39.3 ± 9.1 | 39.2 ± 9.2 |
| Range | 18.0–77.0 | 18.0–65.0 | 18.0–77.0 |
| Self-reported race, n (%) | |||
| European-American | 1,235 (65.1) | 1433 (74.2)* | 2,668 (69.5) |
| African-American | 662 (34.9) | 499 (25.8) | 1,161 (30.3) |
| Self-reported ethnicity, n (%) | |||
| Hispanic | 76 (4.0) | 56 (2.8) | 132 (3.4) |
| Alcohol dependence | |||
| Diagnosis, n (%) | 1,897 (100.0) | 0 (0.0)* | 1,897 (49.5) |
Number of symptoms, 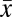 ± s ± s
|
5.2 ± 1.5 | 0.6 ± 0.9* | 2.9 ± 2.6 |
| Comorbid diagnoses, n (%) | |||
| Marijuana dependence | 663 (34.9) | 0 (0.0)* | 663 (17.3) |
| Cocaine dependence | 916 (48.2) | 0 (0.0)* | 916 (23.9) |
| Opioid dependence | 263 (13.8) | 0 (0.0)* | 263 (6.8) |
| Other dependence | 469 (24.7) | 0 (0.0)* | 469 (12.2) |
| Smoked 100+ cigarettes | 1,707 (89.9) | 1,139 (53.4)* | 2,846 (74.3) |
| Nicotine dependence† | 1,159 (61.0) | 95 (4.9)* | 1,254 (32.7) |
*Difference between cases and controls, P < 0.0001.
†Nicotine dependence defined by a score of 4 or greater on the Fagerström Test for Nicotine Dependence.
Primary genome-wide association analyses identified 15 SNPs with P < 10−5 (Table 2 and Figs. S1 and S2). Secondary analyses stratified by race demonstrated similar odds ratios (OR) in populations of European and African descent, although the allele frequencies are markedly different across the two groups (Table S2).
Table 2.
Genetic association testing for alcohol dependence: Adjusted odds ratios and confidence intervals for SNPs with P ≤ 1 × 10−6
| Frequency of test allele |
Overall SAGE Sample (n = 3,829) |
||||||||||
| SNP | Chr | Position (hg18) | Locus and context | Test allele | EA cases | EA controls | AA cases | AA controls | Adjusted OR (95% CI) | P | FDR |
| rs10893366 | 11 | 124,683,613 | PKNOX2, intronic | T | 0.19 | 0.15 | 0.31 | 0.26 | 1.39 (1.23–1.58) | 1.93E-07 | 0.178 |
| rs2039617 | 10 | 97,749,193 | CC2D2B, intronic | T | 0.06 | 0.10 | 0.33 | 0.40 | 0.69 (0.60–0.80) | 5.95E-07 | 0.274 |
| rs9302534 | 16 | 17,956,211 | Intergenic | C | 0.35 | 0.39 | 0.73 | 0.76 | 0.78 (0.70–0.86) | 2.73E-06 | 0.582 |
| rs1318937 | 3 | 15,270,368 | SH3BP5, intronic | G | 0.15 | 0.11 | 0.51 | 0.45 | 1.35 (1.19–1.52) | 3.54E-06 | 0.582 |
| rs2700648 | 3 | 100,590,373 | Intergenic | A | 0.29 | 0.24 | 0.45 | 0.40 | 1.29 (1.16–1.44) | 3.99E-06 | 0.582 |
| rs10803574 | 2 | 139,427,333 | Intergenic | A | 0.45 | 0.40 | 0.17 | 0.14 | 1.28 (1.15–1.43) | 4.41E-06 | 0.582 |
| rs6483362 | 11 | 88,052,099 | GRM5, intronic | A | 0.09 | 0.07 | 0.30 | 0.24 | 1.42 (1.22–1.66) | 4.53E-06 | 0.582 |
| rs2722650 | 19 | 49,593,366 | ZNF285A, intronic | A | 0.14 | 0.17 | 0.57 | 0.63 | 0.76 (0.67–0.86) | 7.14E-06 | 0.582 |
| rs10893365 | 11 | 124,681,647 | PKNOX2, intronic | C | 0.19 | 0.15 | 0.44 | 0.41 | 1.31 (1.17–1.48) | 7.20E-06 | 0.582 |
| rs1386449 | 11 | 19,444,659 | Intergenic | T | 0.00 | 0.00 | 0.14 | 0.07 | 1.98 (1.47–2.67) | 7.29E-06 | 0.582 |
| rs750338 | 11 | 124,677,803 | PKNOX2, intronic | C | 0.26 | 0.20 | 0.47 | 0.45 | 1.28 (1.15–1.43) | 7.61E-06 | 0.582 |
| rs1505846 | 2 | 139,494,752 | Intergenic | A | 0.45 | 0.40 | 0.17 | 0.14 | 1.28 (1.15–1.42) | 8.01E-06 | 0.582 |
| rs9636231 | 2 | 139,388,425 | Intergenic | A | 0.32 | 0.26 | 0.14 | 0.12 | 1.30 (1.16–1.46) | 9.13E-06 | 0.582 |
| rs1363605 | 5 | 172,949,693 | Intergenic | A | 0.15 | 0.17 | 0.36 | 0.44 | 0.76 (0.68–0.86) | 9.62E-06 | 0.582 |
| rs10224675 | 7 | 143,923,695 | TPK1, intronic | G | 0.00 | 0.00 | 0.06 | 0.10 | 0.46 (0.33–0.65) | 9.75E-06 | 0.582 |
AA, African-American; Chr, chromosome; EA, European-American; FDR, false discovery rate.
Replication Studies.
The top associated SNPs were tested for replication in two independent datasets (Table S3). The first replication sample is the family-based study from the Collaborative Study on the Genetics of Alcoholism (COGA). In our family-based association analyses, none of the SNPs demonstrated association with a P value < 0.05; however, rs1386449 and rs10224675, which in our primary analyses are associated with alcohol dependence only in African-Americans, have a P value < 0.10 in the family-based analysis with a small number of African-American families. The top results also were examined in alcohol-dependent case and community-based comparison subjects of German ancestry (24). Of the seven SNPs that were genotyped and tested in the sample, none reached a significance level of P < 0.05.
We also show association results (Table S4) from our study for the SNPs recently reported in the independent GWAS of alcohol-dependent men by Treutlein and colleagues (24). Only one SNP, rs13160562, shows modest evidence of replication [0.88; 95% confidence interval (CI) 0.78–0.99, P = 0.03]. In a meta-analysis, this SNP did not reach genome-wide significance (meta-analysis OR = 0.83, 95% CI 0.77–0.90, P = 2.74 × 10−6). None of the other SNPs reported by Treutlein and colleagues were associated with alcohol dependence (P > 0.05).
Candidate Gene Findings.
The findings for SNPs genotyped in GABRA2 that overlap with SNPs reported by Edenberg et al. (14) are displayed in Table 3. Results for all GABRA SNPs in the entire sample and stratified by self-reported race are shown in Table S5 and Table S6, respectively. This analysis confirms the modest association of alcohol dependence with variants in GABRA2.
Table 3.
SAGE association results for GABRA2 SNPs also genotyped in the family-based COGA sample
| SAGE |
|||||||
| Frequency of risk allele |
|||||||
| SNP | Position | COGA*P | Risk allele | Cases | Controls | Adjusted odds ratio (95% CI) | P |
| rs572227 | 45,946,150 | 3.80E-02 | A | 0.376 | 0.366 | 1.15 (1.04–1.27) | 8.89E-03 |
| rs548583 | 45,958,101 | 1.20E-02 | T | 0.404 | 0.385 | 1.14 (1.03–1.26) | 1.05E-02 |
| rs279858 | 46,009,350 | 8.70E-03 | G | 0.375 | 0.366 | 1.16 (1.05–1.28) | 5.04E-03 |
| rs279843 | 46,019,961 | 4.90E-02 | T | 0.444 | 0.421 | 1.11 (1.00–1.22) | 4.24E-02 |
| rs279841 | 46,035,520 | 3.80E-02 | A | 0.368 | 0.364 | 1.13 (1.02–1.25) | 2.29E-02 |
*COGA family based association from ref. 14.
Discussion
Our study of a relatively large sample of alcohol-dependent cases and nondependent controls used a two-pronged approach to investigate the genetics of alcohol dependence: a GWAS with testing of previously identified genome-wide significant results supplemented by a targeted gene study of GABRA2. Advantages of the genome-wide design include its hypothesis-free strategy and its suitability for the discovery of novel genetic contributors to disease. However, the genome-wide examination requires correction for multiple testing, and the threshold for significance of GWAS findings is high. In contrast, targeted gene studies test specific hypotheses to provide validation of previously reported findings and therefore require a much lower threshold for significance.
In the GWAS arm of the study, we identified 15 SNPs associated with alcohol dependence using a significance threshold of P < 10−5. In two independent samples, one a large family-based study of 258 families with more than 2,000 genotyped individuals and the second a study of alcohol-dependent men and community-based comparison subjects of German descent, none of the association findings replicated using a significance threshold of P < 0.05. Two of the top SNPs identified in SAGE are common in African-American populations and are rare (minor allele frequency < 1%) in subjects of European origin. These SNPs trended toward significance (P < 0.10) in our family-based association tests. Overall, no newly identified variants were associated with alcohol dependence at the genome-wide significance threshold; however, this interpretation of our results is conservative.
Our top 15 association signals were not replicated in the independent datasets, nor did the present study replicate the two genome-wide significant results reported by Treutlein and colleagues (24), who examined alcohol-dependent men in treatment and compared them with individuals from the community. Their top two findings, rs7590720 and rs1344694, were not associated with alcohol dependence in our study in our primary analysis or in secondary analyses stratified by ancestry and sex. In comparing our findings for the top 15 signals reported by Treutlein et al. (24), our results modestly support one finding, rs13160562 in the endoplasmic reticulum aminopeptidase 1 (ERAP1) gene (meta-analysis OR = 0.83, 95% CI 0.77–0.90, P = 2.74 × 10−6).
Different analytic strategies can highlight various findings as the top results. We decided a priori that our primary analysis would examine men and women of European and African origin together to maximize our sample size and power to detect robust association. Although a substantial number of variants across the genome differ in frequency between these populations, this strategy posits that the underlying biologic contributions to disease risk act in a similar manner across populations. Likewise, although the prevalence of a disorder such as alcohol dependence differs between men and women, this strategy assumes that at least part of the underlying biologic risk is the same. This analytic strategy by design highlights SNPs that have similar effects across both populations and genders. However, we recognize that this strategy can decrease power if there is genetic heterogeneity and variants act in one group but not another. Results of secondary analyses performed separately in subjects of European and African descent are presented in Tables S7 and S8. None of the top SNPs overlap in these different analyses.
In our hypothesis-based approach to test association, we examined the GABRA2 gene, the genetic finding most consistently associated with alcohol dependence in the literature. We did see modest evidence of association (with P values comparable to the original reports) in GABRA2, albeit with an OR ∼1.1 (P ∼0.01). This modest genetic risk is consistent with a model whereby multiple genes of small effect contribute to the vulnerability to alcohol dependence. Similar levels of genetic risk have been identified by meta-analyses of other complex diseases such as diabetes (25) and traits such as height (26).
Discrepancies in findings between our results and other samples may be attributed to these inherent differences in study design, sampling strategies, gender, and ethnicity. The design of our study introduces certain caveats into the interpretation of findings. Subjects were drawn from three studies that were individually ascertained for alcohol (COGA), nicotine (Collaborative Genetic Study of Nicotine Dependence, COGEND), and cocaine (Family Study of Cocaine Dependence, FSCD) dependence. Although alcohol-dependent cases and nondependent controls were uniformly screened and defined in this study, the potential introduction of genetic heterogeneity exists because of the three distinct ascertainment designs. In addition to the possibility that some of our top signals are false positives, the high levels of comorbid substance-use disorders may have increased the likelihood that we would identify association to genes contributing broadly to addiction, potentially limiting our ability to replicate these associations in samples ascertained solely for alcohol dependence. Given a substantial genetic epidemiological literature (27–29) supporting considerable overlap of genetic influences of alcohol dependence and substance-use disorders, it is unlikely that the excess comorbidity has biased the findings.
Finally, power is an important consideration in GWAS. This study included more than 3,800 subjects and had 80% power to detect a genetic variant with an OR of 1.3 and greater (for a risk locus with 30% minor allele frequency) at a genome-wide significance threshold of 5 × 10−8. A genetic effect of this magnitude can be considered strong, given that previous studies of GABRA2 report genetic risks in the 1.2 range. Significance levels are related to sample size and strength of the genetic effect. To increase the power to detect significant results, two strategies are possible: enlarge the sample or refine the phenotype to increase the detectable genetic effect. Increasing the sample size has been a common strategy to detect robust association results and has been used to identify association in diabetes (25) and schizophrenia (30). To this end, meta-analysis efforts are currently in progress. As noted by Zeggini and Ioannidis (31), single GWAS studies rarely have been successful at achieving genome-wide significance, and careful meta-analysis provides an avenue for systematically augmenting power to detect modest effects. However, the phenotypic precision often is reduced in these larger studies, and the potential for the introduction of genetic heterogeneity exists.
A second strategy is to narrow the phenotype and to analyze a more homogeneous sample. A GWAS analysis was performed on the COGA subset of European-American subjects (996 subjects overlap with this report) (32). This approach focuses the analysis on subjects recruited under a single ascertainment protocol for alcohol dependence, and the severity of illness in these subjects is high. No finding in this analysis reached a genome-wide statistically significant level of genetic risk, and different top SNPs were nominated in this approach.
Nonetheless, our results underscore the important contribution of GWAS by nominating genes that may play a role in the etiology of alcohol dependence. Continued efforts aimed at gene identification using complementary approaches and coupled with refinement of the phenotypes will be pivotal in illuminating the complex biological and environmental substrate in which alcohol dependence develops.
Methods
The Study of Addiction: Genetics and Environment (SAGE) is funded as part of the Gene Environment Association Studies (GENEVA) initiative supported by the National Human Genome Research Institute (dbGaP study accession phs000092.v1.p1). Alcohol-dependent cases and nondependent control subjects were selected from three large, complementary datasets, COGA, FSCD, and COGEND. Across all studies, case subjects were identified as having a lifetime history of alcohol dependence using DSM-IV criteria (2). Control subjects were required to report a history of drinking because alcohol use is required to develop alcohol dependence. Control subjects had no significant alcohol-dependence symptoms. Because of the likely genetic overlap between alcohol and drug dependence, a diagnosis of drug dependence was an exclusionary criterion for control subjects.
The Institutional Review Board at each contributing institution reviewed and approved the protocols for genetic studies under which all subjects were recruited. Subjects provided written informed consent for genetic studies and agreed to have their DNA and phenotypic information available to qualified investigators through National Institutes of Health repositories. Additional description of the studies is available at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1.
The datasets used for the analyses described in this paper can be obtained from the database of Genotypes and Phenotypes (dbGaP) at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1 through dbGaP accession number phs000092.v1.p1.
Collaborative Study on the Genetics of Alcoholism.
A case-control series of unrelated individuals was selected from more than 8,000 subjects who participated in the genetic arm of COGA. COGA systematically recruited families with multiple members affected with alcohol dependence and community-based comparison families from participating centers across the United States. COGA contributed 612 alcohol-dependent cases and 412 control subjects of European descent and 287 alcohol-dependent cases and 97 controls of African-American descent. Individuals in this case-control sample were independent from the COGA family linkage association sample that was genotyped previously (33, 34).
Family Study of Cocaine Dependence.
Cocaine-dependent subjects were recruited systematically from chemical dependency treatment units in the greater St. Louis, MO, metropolitan area (35). Community-based comparison subjects were identified through the Missouri Family Registry and matched by age, race, gender, and residential zip code. This study contributed 280 alcohol-dependent cases and 247 controls of European descent and 268 alcohol-dependent cases and 249 controls who self-identified as African-American. Because of the study design, alcohol-dependent case subjects also met criteria for cocaine dependence.
Collaborative Genetic Study of Nicotine Dependence.
COGEND was designed as a community-based study of nicotine dependence. Subjects were recruited from Detroit, MI, and St. Louis, MO. More than 53,000 subjects were screened by telephone, more than 2,800 were personally interviewed, and nearly 2,700 donated blood samples for genetic studies (36, 37). COGEND contributed 343 alcohol-dependent cases and 774 controls of European descent and 107 African-American alcohol-dependent cases and 153 African-American controls.
Source of DNA.
All subjects deposited a blood sample in the Rutgers University Cell and DNA Repository (RUCDR), a central biologic repository for the National Institute on Alcohol Abuse and Alcoholism and National Institute on Drug Abuse (http://www.rucdr.org). DNA was extracted from the blood sample, and cell lines were developed as an additional DNA source.
Assessment.
A common assessment was performed across all three studies and was based on the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) (38). This shared methodology of interview administration, question format, and queried domains allowed harmonization of phenotypic data across all studies.
Genotyping and Data Cleaning.
Samples were genotyped at the Johns Hopkins Center for Inherited Disease Research (CIDR). Data were released for 4,189 study samples. Study samples, including 49 study duplicates, were plated and genotyped together with 135 HapMap controls (86 CEU; 49 YRI). Genotyping was performed using Illumina Human1Mv1_C BeadChips and the Illumina Infinium II assay protocol (39). Allele cluster definitions for each SNP were determined using Illumina BeadStudio Genotyping Module version 3.1.14 and the combined intensity data from the samples. Strict quality-control standards were implemented, and genotypes were released by CIDR for 1,040,106 SNPs (99.15% of attempted). The mean non-Y SNP call rate and mean sample call rate was 99.7% for the released CIDR dataset. Study duplicate reproducibility was 99.98%. Further extensive cleaning was undertaken to ensure high-quality genotyping by examining batch effects, potential chromosomal anomalies, and Mendelian errors. Further details are provided in the comprehensive data cleaning report posted at dbGaP http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/document.cgi?study_id=phs000092.v1.p1&phv=22928&phd=2274&pha=&pht=116&phvf=&phdf=20&phaf=&phtf=&dssp=1&consent=&temp=1.
SNPs with an allele frequency > 1% in either the European- or African-descent populations were analyzed (948,658 SNPs). A SNP call rate of 98% was required. Hardy–Weinberg equilibrium (HWE) was tested, and SNPs that deviated from HWE (P < 10−4) were excluded. The final number of subjects included in analyses was 3,829. Individuals were dropped if there was potential sample misidentification, sample relatedness, or other misspecification (n = 171).
Statistical Analyses.
We used the software package EIGENSTRAT (40) with all SNPs to calculate principal components reflecting continuous variation in allele frequencies representing ancestral differences in subjects. Two principal components were identified; the first distinguished African-American participants from European-American participants and the second distinguished Hispanic and non-Hispanic subjects. Each individual received scores on each principal component. These scores, representing continuous variation in race and ethnicity, can be used to control for effects of population stratification.
Genome-wide association analysis was conducted using logistic regressions in PLINK (41). Genotypes were coded log-additively (0, 1, 2 copies of the minor allele). Covariates represented sex, age [defined, using quartiles, as 34 years and younger (reference), 35–39 years, 40–44 years, and 45 years and older] and two principal components indexing continuous variation in race/ethnicity. We repeated analyses using self-reported race (European-American, African-American) as categorical variables. Similar results were seen with both analyses, and we present results using self-reported race. The false-discovery rate was calculated using the method of Storey and Tibshirani (42).
Replication Samples.
Family sample from COGA.
A set of 258 genetically informative, multiplex alcohol-dependence pedigrees in COGA have been studied previously in linkage- and association-based analyses (33, 34). Families report European and African ancestry (n = 219 European American, n = 35 African American, and n = 4 other ancestry). These pedigrees do not overlap with any of the case-control subjects used in the SAGE GWAS sample. Affected individuals were defined as those meeting criteria for DSM-IV alcohol dependence, and unaffected subjects reported no symptoms of alcohol dependence. The SNPs with the most significant evidence of association from SAGE were selected for genotyping in this family-based sample. SNPs had a genotyping call rate > 99%. All SNPs passed an HWE threshold of P > 0.05 as calculated independently in African-American and European-American samples. Pedigree errors were cleaned using PEDCHECK (43). Family-based association analyses were performed using a Family-Based Association Test (FBAT) (44), with alcohol-dependent and nondependent phenotypes adjusted for age, gender, and ethnicity. Because FBAT is robust to population stratification, analyses were not performed independently by ethnicity.
Case-control sample.
Replication analyses were performed in the GWAS sample of alcohol-dependent subjects reported by Treutlein et al. (24). Alcohol-dependent men (n = 487) were recruited from consecutive admissions to treatment facilities as part of the German Addiction Research Network (GARN; http://www.bw-suchtweb.de). Controls subjects (n = 1,358) were recruited through population-based epidemiologic studies. SNPs reported in this replication phase passed the standard quality-control measures, and analyses were performed in PLINK. See Treutlein et al. (24) for further details on the sample description, analyses, and methods.
Supplementary Material
Acknowledgments
Funding support for SAGE was provided through the National Institutes of Health (NIH) Genes, Environment and Health Initiative (GEI) Grant U01 HG004422. SAGE is one of the GWAS funded as part of the GENEVA under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (Grant U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by COGA (Grant U10 AA008401), COGEND (Grant P01 CA089392), and FSCD (Grant R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by NIH GEI Grant U01HG004438, the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). Funding support for the work by Treutlein et al., 2009 (24) was provided by the National Genome Research Network (NGFNplus).
Footnotes
Conflict of Interest Statement: L.J.B., J.P.R., A.M.G., S.F.S., and J.C.W. are inventors on the patent “Markers for Addiction” (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. N.L.S. is the spouse of S. F.S., who is listed as an inventor on the patent. L.J.B. served as a consultant for Pfizer Inc. in 2008.
This article is a PNAS Direct Submission.
Data deposition: Data can be obtained from dbGaP at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1 through dbGaP accession number phs000092.v1.p1.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911109107/DCSupplemental.
References
- 1.Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–1245. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- 2.American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. 4th Ed. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 3.Grant BF, Stinson FS, Harford TC. Age at onset of alcohol use and DSM-IV alcohol abuse and dependence: A 12-year follow-up. J Subst Abuse. 2001;13:493–504. doi: 10.1016/s0899-3289(01)00096-7. [DOI] [PubMed] [Google Scholar]
- 4.Harwood H. Updating estimates of the economic costs of alcohol abuse in the United States: Estimates, update methods, and data. Report prepared by The Lewin Group for the National Institute on Alcohol Abuse and Alcoholism, 2000. Based on estimates, analyses, and data reported in Harwood, H.; Fountain, D.; and Livermore, G. The Economic Costs of Alcohol and Drug Abuse in the United States 1992. Report prepared for the National Institute on Drug Abuse and the National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health, Department of Health and Human Services. Rockville, MD: National Institutes of Health; 2000. [Google Scholar]
- 5.Bierut LJ, et al. Familial transmission of substance dependence: Alcohol, marijuana, cocaine, and habitual smoking: A report from the Collaborative Study on the Genetics of Alcoholism. Arch Gen Psychiatry. 1998;55:982–988. doi: 10.1001/archpsyc.55.11.982. [DOI] [PubMed] [Google Scholar]
- 6.Merikangas KR, et al. Familial transmission of substance use disorders. Arch Gen Psychiatry. 1998;55:973–979. doi: 10.1001/archpsyc.55.11.973. [DOI] [PubMed] [Google Scholar]
- 7.Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;156:34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- 8.Pickens RW, et al. Heterogeneity in the inheritance of alcoholism. A study of male and female twins. Arch Gen Psychiatry. 1991;48:19–28. doi: 10.1001/archpsyc.1991.01810250021002. [DOI] [PubMed] [Google Scholar]
- 9.Goodwin DW, Schulsinger F, Hermansen L, Guze SB, Winokur G. Alcohol problems in adoptees raised apart from alcoholic biological parents. Arch Gen Psychiatry. 1973;28:238–243. doi: 10.1001/archpsyc.1973.01750320068011. [DOI] [PubMed] [Google Scholar]
- 10.Goodwin DW, et al. Drinking problems in adopted and nonadopted sons of alcoholics. Arch Gen Psychiatry. 1974;31:164–169. doi: 10.1001/archpsyc.1974.01760140022003. [DOI] [PubMed] [Google Scholar]
- 11.Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ. A twin-family study of alcoholism in women. Am J Psychiatry. 1994;151:707–715. doi: 10.1176/ajp.151.5.707. [DOI] [PubMed] [Google Scholar]
- 12.Heath AC, et al. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: Consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- 13.Knopik VS, et al. Genetic effects on alcohol dependence risk: Re-evaluating the importance of psychiatric and other heritable risk factors. Psychol Med. 2004;34:1519–1530. doi: 10.1017/s0033291704002922. [DOI] [PubMed] [Google Scholar]
- 14.Edenberg HJ, et al. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matthews AG, Hoffman EK, Zezza N, Stiffler S, Hill SY. The role of the GABRA2 polymorphism in multiplex alcohol dependence families with minimal comorbidity: Within-family association and linkage analyses. J Stud Alcohol Drugs. 2007;68:625–633. doi: 10.15288/jsad.2007.68.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drgon T, D'Addario C, Uhl GR. Linkage disequilibrium, haplotype and association studies of a chromosome 4 GABA receptor gene cluster: Candidate gene variants for addictions. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:854–860. doi: 10.1002/ajmg.b.30349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Covault J, Gelernter J, Jensen K, Anton R, Kranzler HR. Markers in the 5′-region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology. 2008;33:837–848. doi: 10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2004;129:104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- 19.Fehr C, et al. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16:9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- 20.Lappalainen J, et al. Association between alcoholism and gamma-amino butyric acid alpha2 receptor subtype in a Russian population. Alcohol Clin Exp Res. 2005;29:493–498. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- 21.Lind PA, et al. The role of GABRA2 in alcohol dependence, smoking, and illicit drug use in an Australian population sample. Alcohol Clin Exp Res. 2008;32:1721–1731. doi: 10.1111/j.1530-0277.2008.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soyka M, et al. GABA-A2 receptor subunit gene (GABRA2) polymorphisms and risk for alcohol dependence. J Psychiatr Res. 2008;42:184–191. doi: 10.1016/j.jpsychires.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Enoch MA. The role of GABA(A) receptors in the development of alcoholism. Pharmacol Biochem Behav. 2008;90:95–104. doi: 10.1016/j.pbb.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Treutlein J, et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prokopenko I, McCarthy MI, Lindgren CM. Type 2 diabetes: New genes, new understanding. Trends Genet. 2008;24:613–621. doi: 10.1016/j.tig.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gudbjartsson DF, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 27.Tsuang MT, Bar JL, Harley RM, Lyons MJ. The Harvard Twin Study of Substance Abuse: What we have learned. Harv Rev Psychiatry. 2001;9:267–279. [PubMed] [Google Scholar]
- 28.Xian H, et al. Genetic and environmental contributions to nicotine, alcohol and cannabis dependence in male twins. Addiction. 2008;103:1391–1398. doi: 10.1111/j.1360-0443.2008.02243.x. [DOI] [PubMed] [Google Scholar]
- 29.Kendler KS, Myers J, Prescott CA. Specificity of genetic and environmental risk factors for symptoms of cannabis, cocaine, alcohol, caffeine, and nicotine dependence. Arch Gen Psychiatry. 2007;64:1313–1320. doi: 10.1001/archpsyc.64.11.1313. [DOI] [PubMed] [Google Scholar]
- 30.Purcell SM, et al. International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeggini E, Ioannidis JP. Meta-analysis in genome-wide association studies. Pharmacogenomics. 2009;10:191–201. doi: 10.2217/14622416.10.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edenberg HJ, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. doi: 10.1111/j.1530-0277.2010.01156.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reich T, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- 34.Foroud T, et al. Alcoholism susceptibility loci: Confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24:933–945. [PubMed] [Google Scholar]
- 35.Bierut LJ, Strickland JR, Thompson JR, Afful SE, Cottler LB. Drug use and dependence in cocaine dependent subjects, community-based individuals, and their siblings. Drug Alcohol Depend. 2008;95:14–22. doi: 10.1016/j.drugalcdep.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bierut LJ, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saccone SF, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bucholz KK, et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: A report on the reliability of the SSAGA. J Stud Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- 39.Gunderson KL, et al. Whole-genome genotyping. Methods Enzymol. 2006;410:359–376. doi: 10.1016/S0076-6879(06)10017-8. [DOI] [PubMed] [Google Scholar]
- 40.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 41.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Connell JR, Weeks DE. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19(Suppl 1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


