Abstract
Isolated cleft lip with or without cleft palate (CL/P) is among the most common human birth defects, with a prevalence around 1 in 700 live births. The Runt-related transcription factor 2 (RUNX2) gene has been suggested as a candidate gene for CL/P based largely on mouse models; however, no human studies have focused on RUNX2 as a risk factor for CL/P. This study examines the association between markers in RUNX2 and isolated, nonsyndromic CL/P using a case-parent trio design, while considering parent-of-origin effects. Case-parent trios from four populations (77 from Maryland, 146 from Taiwan, 35 from Singapore, and 40 from Korea) were genotyped for 24 single nucleotide polymorphisms (SNPs) in the RUNX2 gene. We performed the transmission disequilibrium test on individual SNPs. Parent-of-origin effects were assessed using the transmission asymmetry test and the parent-of-origin likelihood ratio test (PO-LRT). When all trios were combined, the transmission asymmetry test revealed a block of 11 SNPs showing excess maternal transmission significant at the P < 0.01 level, plus one SNP (rs1934328) showing excess paternal transmission (P = 0.002). For the 11 SNPs showing excess maternal transmission, odds ratios of being transmitted to the case from the mother ranged between 3.00 and 4.00. The parent-of-origin likelihood ratio tests for equality of maternal and paternal transmission were significant for three individual SNPs (rs910586, rs2819861, and rs1934328). Thus, RUNX2 appears to influence risk of CL/P through a parent-of-origin effect with excess maternal transmission.
Keywords: RUNX2, oral cleft, maternal transmission effects, parent-of-origin
INTRODUCTION
Oral clefts are one of the most common birth defects in humans and represent a significant public health problem both in terms of the medical and economic burden for affected individuals and their families. Nonsyndromic cleft lip with or without palate (CL/P) is “complex” or “multifactorial” in its etiology, in that both genes and environmental risk factors control risk [Wyszynski et al., 1997; Cobourne, 2004]. Although several candidate genes have been extensively studied in different populations (transforming growth factor α (TGFA), IRF6, BCL3, RARA, etc.), only a few genes have been shown to contain mutations that appear causal (MSX1, PVRL1, etc.), and these are rare and often show incomplete penetrance [van den Boogaard et al., 2000; Zucchero et al., 2004; Carinci et al., 2007].
The Runt-related transcription factor 2 (RUNX2 [MIM 600211]) gene is located on chromosome 6p, where studies of multiplex families have yielded evidence for linkage [Eiberg et al., 1987; Marazita et al., 2004]. RUNX2 (CBFA1) is a critical transcription regulator involved in both bone and tooth formation [Aberg et al., 2004]. Mutations in this gene have been associated with cleidocranial dysplasia [CCD; Mundlos et al., 1997; Otto et al., 1997; Cooper et al., 2001]. Several studies have reported that RUNX2 may lead to cleft palate in mice [Ducy et al., 1999; Aberg et al., 2004]. However, to date no study has focused on whether the RUNX2 gene is a risk factor for CL/P in humans.
It is important to consider parent-of-origin effects when studying birth defects because maternal genotype controls the in utero environment of the developing fetus, and separating maternal genotypic effects from imprinting effects remains an important question [Weinberg and Umbach, 2005; Wilkins and Haig, 2003]. Maternal parent-of-origin effects have been suggested for several genes associated with non-syndromic CL/P [Mossey et al., 1998; Martinelli et al., 2001; van Rooij et al., 2003; Jugessur et al., 2003; Rubini et al., 2005]. In this paper, we tested for association between markers in RUNX2 and risk of CL/P in 298 case-parent trios specifically considering parent-of-origin effects.
METHODS
SAMPLE DESCRIPTION
As part of an international study of oral clefts, we collected data on case-parent trios recruited through treatment centers in Maryland (MD, Johns Hopkins and University of Maryland), the Chang Gung Memorial Hospital in Taiwan (TW), KK Women’s and Children’s Hospital in Singapore (SP), and Yonsei Medical Center in South Korea (KR). Research protocols were reviewed and approved by institutional review boards at each institution. Table I lists the gender of all CL/P probands. All parents of probands were unaffected in the SP, TW, and KR trios, but four parents among the 77 MD trios also had an oral cleft. All probands underwent clinical genetics evaluations (including assessing other congenital anomalies or major developmental delays) and were classified as having an isolated, nonsyndromic CL/P.
TABLE I.
Gender among 298 nonsyndromic CL/P cases from four populations
| CL/P cases |
|||
|---|---|---|---|
| Population | Total (n) | Male (n) | Female (n) |
| Taiwan | 146 | 95 | 51 |
| Singapore | 35 | 24 | 11 |
| Korea | 40 | 22 | 18 |
| Maryland | 77 | 44 | 33 |
| Total | 298 | 185 | 113 |
CL/P, cleft lip with or without cleft palate.
SNP SELECTION, DNA, AND GENOTYPING
Single nucleotide polymorphisms (SNPs) were selected in a region surrounding RUNX2 on chromosome 6p21, with a goal of identifying one SNP per 5 kb of physical distance. Variants with “SNP scores” (an assessment of design quality of the Illumina assay (San Diego, CA) based on a proprietary algorithm) above 0.6, high validation levels in dbSNP (this included validation levels where the submitter had validated the SNP on multiple platforms), and high heterozygosity levels (particularly in multiple populations) were given higher priority during the selection process. From 26 selected SNPs, 24 were polymorphic in all four populations (Table II).
TABLE II.
SNP minor allele frequencies among parents of 298 CL/P cases from four populations
| Minor allele frequency |
|||||||
|---|---|---|---|---|---|---|---|
| No. | SNP Name | Physical locationa | Minor allele | Taiwan | Singapore | Korea | Maryland |
| 1 | rs7771980 | 45,497,267 | C | 0.041 | 0.068 | 0.086 | 0.069 |
| 2 | rs2677104 | 45,503,851 | C | 0.273 | 0.322 | 0.233 | 0.150 |
| 3 | rs2819855 | 45,508,372 | A | 0.446 | 0.457 | 0.480 | 0.395 |
| 4 | rs2819854 | 45,512,506 | A | 0.403 | 0.440 | 0.395 | 0.458 |
| 5 | rs910586 | 45,518,290 | T | 0.039 | 0.042 | 0.053 | 0.283 |
| 6 | rs2819853 | 45,520,635 | G | 0.036 | 0.042 | 0.039 | 0.283 |
| 7 | rs765724 | 45,525,096 | T | 0.039 | 0.042 | 0.053 | 0.289 |
| 8 | rs1343799 | 45,529,608 | G | 0.036 | 0.042 | 0.039 | 0.283 |
| 9 | rs2819861 | 45,532,918 | A | 0.036 | 0.042 | 0.039 | 0.293 |
| 10 | rs2790103 | 45,540,243 | T | 0.040 | 0.042 | 0.053 | 0.276 |
| 11 | rs2790093 | 45,545,462 | G | 0.036 | 0.042 | 0.039 | 0.279 |
| 12 | rs2790098 | 45,550,428 | C | 0.036 | 0.042 | 0.053 | 0.279 |
| 13 | rs4714854 | 45,555,912 | T | 0.036 | 0.042 | 0.053 | 0.279 |
| 14 | rs9472494 | 45,559,814 | T | 0.041 | 0.042 | 0.086 | 0.300 |
| 15 | rs2396442 | 45,564,959 | C | 0.049 | 0.042 | 0.086 | 0.274 |
| 16 | rs1934328 | 45,573,731 | T | 0.295 | 0.212 | 0.250 | 0.454 |
| 17 | rs7773875 | 45,580,276 | C | 0.253 | 0.186 | 0.178 | 0.189 |
| 18 | rs7771889 | 45,594,185 | G | 0.293 | 0.254 | 0.243 | 0.276 |
| 19 | rs10485422 | 45,597,170 | A | 0.031 | 0.017 | 0.072 | 0.226 |
| 20 | rs6904353 | 45,601,061 | T | 0.034 | 0.025 | 0.086 | 0.416 |
| 21 | rs13207392 | 45,604,434 | T | 0.034 | 0.025 | 0.087 | 0.264 |
| 22 | rs7748231 | 45,609,530 | G | 0.034 | 0.025 | 0.086 | 0.417 |
| 23 | rs10948237 | 45,615,272 | A | 0.034 | 0.025 | 0.086 | 0.417 |
| 24 | rs1928533 | 45,617,802 | C | 0.029 | 0.025 | 0.072 | 0.400 |
SNP, single nucleotide polymorphism; CL/P, cleft lip with or without cleft palate.
Based on NCBI Human Genome build 35.1.
Genomic DNA samples were prepared from peripheral blood by the protein precipitation method described previously [Bellus et al., 1995]. DNA concentration was determined using the PicoGreen® dsDNA Quantitation Kit (Molecular Probes, Inc., Eugene, OR) and all DNA samples were stored at −20°C. A 4 μg aliquot of each genomic DNA sample was dispensed into a bar-coded 96-well microtiter plate at a concentration of 100 ng/μl and genotyped for SNP markers using the Illumina Golden-Gate™ chemistry with Sentrix® Array Matrices from the manufacturer [Oliphant et al., 2002] at the SNP Center of the Genetic Resources Core Facility, a part of the McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins School of Medicine. Two duplicates and four Centre d’Etude du Polymorphisme Humain (CEPH) controls were included on each plate to evaluate genotyping consistency within and between plates and to insure correct orientation. Genotypes were generated on a BeadLab 1000 system (San Diego, CA) [Fan et al., 2003].
STATISTICAL ANALYSIS
Within each population, the minor allele frequency (MAF) was computed among parents. Pairwise linkage disequilibrium (LD) was computed as r2 for all SNPs using the Haploview program [Barrett et al., 2005]. LD blocks were identified from the pairwise LD in each population separately (Fig. 1). Four blocks of LD were identified, consisting of 4, 11, 3, and 6 SNPs, respectively. As an initial screening, single-SNP and two to five-SNP haplotypes were analyzed using the family-based association test (FBAT) program, where empirical P-values were calculated for each test [Laird et al., 2000; Rabinowitz and Laird, 2000]. For this FBAT analysis, each population was analyzed separately for individual SNPs and haplotypes. Clayton’s extension of the transmission disequilibrium test (TDT) incorporated into STATA 8.2 (College Station, TX) [Spielman et al., 1993; Cordell et al., 2004] was used on individual SNPs to test for evidence of linkage and LD in the combined sample of 298 trios.
Fig. 1.
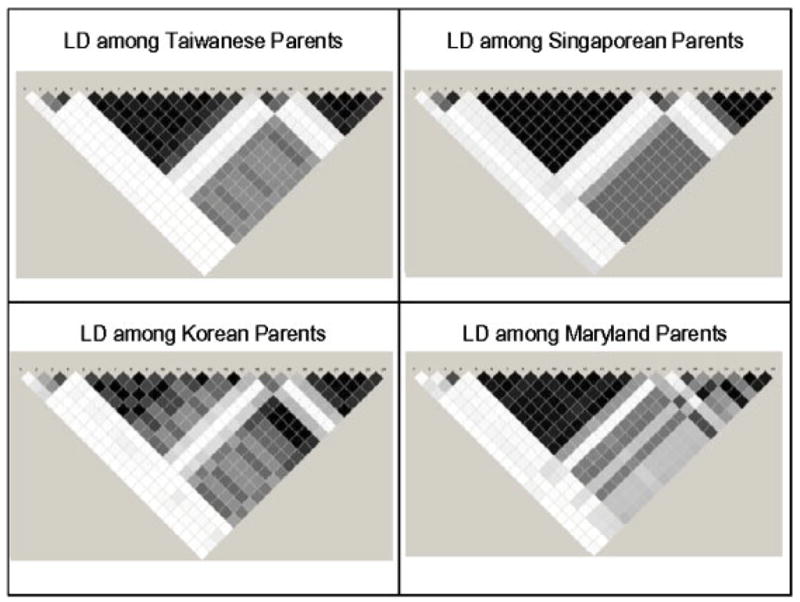
Linkage disequilibrium as measured by r2 in RUNX2 among parents of CL/P children from four populations. White: r2 = 0. Shades of gray: 0<r2<1. Black: r2 = 1. RUNX2, Runt-related transcription factor 2; CL/P, cleft lip with or without cleft palate.
Parent-of-origin analyses were conducted in the combined sample in several ways. Firstly, parent-of-origin effects were examined using the transmission asymmetry test (TAT) suggested by Weinberg et al. [1998], which is similar to the TDT but excludes matings between two heterozygotes (where transmission can be ambiguous). The TAT was stratified into separate allelic tests for fathers and mothers. Next, we used the likelihood-based approach proposed by Weinberg [1999] to test for parent-of-origin effects. This log-linear model considers the three mating types in which the mother and the father carry different numbers of variant alleles, with further stratification by the number of alleles inherited by the child. Like Weinberg, we will call this latter method the “parent-of-origin likelihood ratio test” (PO-LRT). This model considers maternally mediated in utero effects (maternal genotypic effects on the phenotype of the fetus), which could otherwise confound assessment of parent-of-origin effects, along with a separate term for imprinting [Weinberg, 1999]. Here, imprinting reflects a differential transmission of alleles to the affected child from mothers vs. fathers. This PO-LRT was executed using the LEM software (Tilburg, The Netherlands) [van Den Oord and Vermunt, 2000].
To assess the overall significance of these TAT findings for maternal and paternal transmission given the observed pattern of LD among SNPs in RUNX2 while considering multiple tests (see Fig. 1), we conducted a permutation test for each of the inferred LD blocks in this gene (consisting of k = 4, 11, 3, and 6 SNPs, respectively). Therefore, the block became the unit of analysis. A Bonferroni correction for each individual SNP P-value would be unacceptably conservative, in light of the high dependence between SNPs within a block. Generating P-values for each LD block permitted correcting for multiple comparisons with fewer and largely independent P-values (i.e. in RUNX2 there were four LD blocks to be tested, compared to 24 SNPs). We generated 10,000 data sets by randomly selecting the transmitted/nontransmitted status of the minor allele across all SNPs within a block for each trio. In this manner, LD structure was maintained, but any potential information regarding the transmission status was destroyed, as would occur under a true null hypothesis. Thus, this permutation test gives a clear assessment of the likelihood of the observed data under the null hypothesis of no preferential transmission of alleles. Many different test statistics can be devised to generate a block-specific P-value. In our analysis, the final inference was based on the arithmetic average of all of the −log10(P-values) within a block (i.e. a function of the geometric mean of these 10,000 P-values on simulated replicates). Histograms of these mean −log10(P-values) are plotted separately for the maternal and paternal transmissions from the 10,000 permutation tests along with the observed data.
RESULTS
Among these 24 SNPs, there was considerable variation in allele frequency among parents from MD and the three Asian populations (Table II). From the allele frequencies given in Table II, it is clear that some markers showed sharp distinctions between MD and Asian samples, while others did not. Measures of genetic distance based on either individual SNPs or haplotypes also varied considerably (data not shown). The three Asian populations (TW, SP, and KR) had very low MAFs for SNPs 5–15 and 19–24 compared to the MD trios, while SNPs 2–4 and 16–18 had similar MAFs in all four populations. For genes in the former group, obviously there will be fewer informative matings (e.g. matings involving heterozygotes) among Asian populations, but trios from all populations can still provide some information. Patterns of LD across the entire gene were calculated within each population, and Figure 1 shows that LD patterns among the 24 polymorphic SNPs were similar across all populations.
In conventional FBAT analysis of individual SNPs and haplotypes (ignoring parent-of-origin), only SNP 16 showed empirical significance at the 1% level among KR trios, and haplotypes including this marker were also significant. However, none of the other populations yielded evidence of linkage and association for SNPs in RUNX2 when parent-of-origin was not considered (data not shown). When all markers were screened in the combined data set of all four populations using the TDT without considering parent-of-origin, the odds ratio of transmission for the minor allele, OR(transmission), was significant for SNP 10 (OR = 1.62, P = 0.039), SNP 16 (OR = 1.40, P = 0.014), and SNP 18 (OR = 1.35, P = 0.032) (Table III).
TABLE III.
Number of transmitted (T) or nontransmitted (NT) minor alleles in 298 CL/P cases (all populations combined) from TDT and estimated odds ratios of transmission OR(transmission) ignoring parent-of-origin
| TDT |
|||||
|---|---|---|---|---|---|
| No. | SNP name | T | NT | P-value | ORa |
| 1 | rs7771980 | 28 | 29 | 0.895 | 0.97 |
| 2 | rs2677104 | 91 | 92 | 0.941 | 0.99 |
| 3 | rs2819855 | 135 | 115 | 0.206 | 1.17 |
| 4 | rs2819854 | 137 | 122 | 0.351 | 1.12 |
| 5 | rs910586 | 47 | 32 | 0.091 | 1.47 |
| 6 | rs2819853 | 44 | 31 | 0.133 | 1.42 |
| 7 | rs765724 | 47 | 33 | 0.112 | 1.42 |
| 8 | rs1343799 | 44 | 31 | 0.133 | 1.42 |
| 9 | rs2819861 | 44 | 29 | 0.079 | 1.52 |
| 10 | rs2790103 | 47 | 29 | 0.039 | 1.62 |
| 11 | rs2790093 | 44 | 30 | 0.104 | 1.47 |
| 12 | rs2790098 | 45 | 31 | 0.108 | 1.45 |
| 13 | rs4714854 | 45 | 31 | 0.108 | 1.45 |
| 14 | rs9472494 | 50 | 36 | 0.131 | 1.39 |
| 15 | rs2396442 | 53 | 36 | 0.072 | 1.47 |
| 16 | rs1934328 | 125 | 89 | 0.014 | 1.40 |
| 17 | rs7773875 | 97 | 75 | 0.093 | 1.29 |
| 18 | rs7771889 | 120 | 89 | 0.032 | 1.35 |
| 19 | rs10485422 | 37 | 28 | 0.264 | 1.32 |
| 20 | rs6904353 | 43 | 37 | 0.502 | 1.16 |
| 21 | rs13207392 | 41 | 32 | 0.292 | 1.28 |
| 22 | rs7748231 | 44 | 37 | 0.437 | 1.19 |
| 23 | rs10948237 | 46 | 36 | 0.269 | 1.28 |
| 24 | rs1928533 | 41 | 36 | 0.569 | 1.14 |
Shading in the table indicates inferred LD blocks.
T, transmitted; NT, not transmitted; OR, odds ratio; CL/P, cleft lip with or without cleft palate; TDT, transmission disequilibrium test; LD, linkage disequilibrium; SNP, single nucleotide polymorphism.
OR(transmission): odds ratio of transmission for the minor allele.
Parent-of-origin effects were first investigated by stratifying informative transmissions and nontransmissions by parental source (paternal and maternal) for all SNPs in the combined data set (Table IV). TAT revealed 11 contiguous SNPs (SNPs 5–15) with excess maternal transmission significant at the P < 0.01 level, and SNP 16 that showed excess paternal transmission (P = 0.002). For SNPs 5–15, the estimated OR(transmission) from the mother was statistically significant (ranging from 3.00 to 4.00) in TAT analysis. However, the PO-LRTs were only significant for SNP 5 (P = 0.036, OR(transmission) = 3.59), SNP 9 (P = 0.036, OR(transmission) = 3.73), and SNP 16 (P = 0.029, OR (transmission) = 0.44; Table IV, last column). PO-LRTs gave estimated risk ratios for an imprinting effect ranging between 2.51 and 3.73 for the 11 contiguous SNPs (SNPs 5–15), suggesting excess maternal transmission of this 46.7 kb region.
TABLE IV.
Number of transmitted (T) or non-transmitted (NT) minor alleles to 298 CL/P cases (all populations combined)a from TAT and estimated odds ratios, plus parent-of-origin likelihood ratio test results about inequality of maternal vs. paternal transmission
| Paternal |
Maternal |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TAT |
TAT |
PO-LRTb |
|||||||||
| No. | SNP name | T | NT | P-value | ORc | T | NT | P-value | ORc | ORd | P-value |
| 1 | rs7771980 | 9 | 8 | 0.808 | 1.13 | 16 | 18 | 0.732 | 0.89 | 0.79 | 0.692 |
| 2 | rs2677104 | 25 | 30 | 0.500 | 0.83 | 22 | 24 | 0.768 | 0.92 | 1.10 | 0.811 |
| 3 | rs2819855 | 36 | 34 | 0.811 | 1.06 | 37 | 25 | 0.128 | 1.48 | 1.40 | 0.342 |
| 4 | rs2819854 | 35 | 36 | 0.906 | 0.97 | 37 | 29 | 0.325 | 1.28 | 1.32 | 0.417 |
| 5 | rs910586 | 15 | 13 | 0.705 | 1.15 | 20 | 5 | 0.003 | 4.00 | 3.59 | 0.036 |
| 6 | rs2819853 | 14 | 12 | 0.695 | 1.17 | 18 | 5 | 0.007 | 3.60 | 3.19 | 0.063 |
| 7 | rs765724 | 15 | 13 | 0.705 | 1.15 | 20 | 6 | 0.006 | 3.33 | 2.97 | 0.065 |
| 8 | rs1343799 | 14 | 12 | 0.695 | 1.17 | 18 | 5 | 0.007 | 3.60 | 3.19 | 0.063 |
| 9 | rs2819861 | 13 | 12 | 0.841 | 1.08 | 19 | 5 | 0.004 | 3.80 | 3.73 | 0.036 |
| 10 | rs2790103 | 16 | 11 | 0.336 | 1.45 | 20 | 5 | 0.003 | 4.00 | 2.86 | 0.092 |
| 11 | rs2790093 | 15 | 12 | 0.564 | 1.25 | 18 | 5 | 0.007 | 3.60 | 2.99 | 0.079 |
| 12 | rs2790098 | 15 | 12 | 0.564 | 1.25 | 19 | 6 | 0.009 | 3.17 | 2.60 | 0.110 |
| 13 | rs4714854 | 15 | 12 | 0.564 | 1.25 | 19 | 6 | 0.009 | 3.17 | 2.60 | 0.110 |
| 14 | rs9472494 | 15 | 14 | 0.853 | 1.07 | 22 | 7 | 0.005 | 3.14 | 2.99 | 0.051 |
| 15 | rs2396442 | 17 | 14 | 0.590 | 1.21 | 24 | 8 | 0.005 | 3.00 | 2.51 | 0.086 |
| 16 | rs1934328 | 41 | 17 | 0.002 | 2.41 | 35 | 33 | 0.808 | 1.06 | 0.44 | 0.029 |
| 17 | rs7773875 | 33 | 21 | 0.102 | 1.57 | 32 | 32 | 1.000 | 1.00 | 0.65 | 0.245 |
| 18 | rs7771889 | 36 | 18 | 0.014 | 2.00 | 40 | 31 | 0.285 | 1.29 | 0.64 | 0.238 |
| 19 | rs10485422 | 15 | 13 | 0.705 | 1.15 | 17 | 6 | 0.022 | 2.83 | 2.42 | 0.135 |
| 20 | rs6904353 | 13 | 14 | 0.847 | 0.93 | 18 | 11 | 0.194 | 1.64 | 1.78 | 0.294 |
| 21 | rs13207392 | 16 | 15 | 0.857 | 1.07 | 19 | 7 | 0.019 | 2.71 | 2.50 | 0.102 |
| 22 | rs7748231 | 13 | 13 | 1.000 | 1.00 | 18 | 11 | 0.194 | 1.64 | 1.64 | 0.373 |
| 23 | rs10948237 | 13 | 14 | 0.847 | 0.93 | 18 | 11 | 0.194 | 1.64 | 1.78 | 0.294 |
| 24 | rs1928533 | 12 | 13 | 0.841 | 0.92 | 15 | 13 | 0.705 | 1.15 | 1.27 | 0.671 |
Shading in the table indicates inferred LD blocks.
T, transmitted; NT, not transmitted; OR, odds ratio; CL/P, cleft lip with or without cleft palate; TAT, transmission asymmetry test; LD, linkage disequilibrium; SNP, single nucleotide polymorphism.
TAT analysis was used on trios with matings between two heterozygous parents deleted.
Parent-of-origin likelihood ratio test (PO-LRT) includes separate terms for imprinting.
OR(transmission): odds ratio of transmission of the minor allele.
OR: odds ratio for imprinting effect (i.e. differential transmission from mothers and from fathers).
The overall significance of the findings for maternal and paternal transmissions in the TAT was assessed by permutation tests to minimize the problem of multiple comparisons while preserving all information about LD. Excess transmission from the mother to the affected child was present for almost all of the SNPs in the 11-SNP block containing SNPs 5–15, while no obvious pattern of excess paternal transmission existed relative to the permuted values generated under the H0. Using the arithmetic mean of the −log10(P-values) over all 11 SNPs in this block, only 37 out of the 10,000 permuted replicates exceeded the observed test statistic for maternal transmission (Fig. 2, left side), and thus, this result is statistically significant at a 5% level after correcting for the number of tests (four tests, one for each block). On the other hand, there was no evidence of deviation from independence among paternal transmissions (P-value = 0.645, without correction).
Fig. 2.
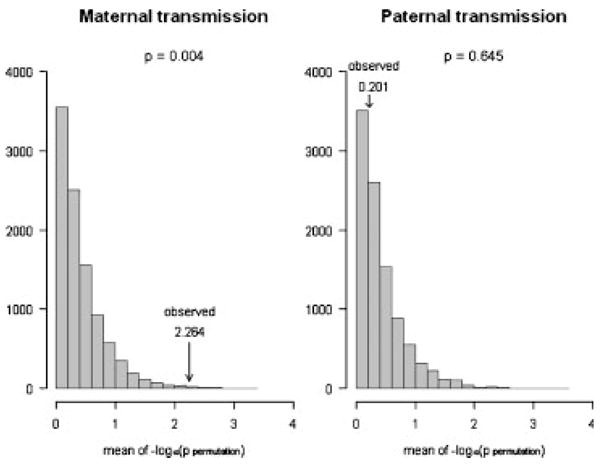
Permutation tests under H0 for LD block 2 for maternal and paternal transmission under TAT on 298 CL/P trios. Results of permutation tests for maternal and paternal transmissions in the largest inferred LD block (11 SNPs) in RUNX2. A total of 10,000 data sets were generated by randomly selecting transmitted/nontransmitted status of the minor allele for all SNPs within the LD block for each case. Using the arithmetic mean of the 11 −log10 P-values (calculated for each of the 10,000 data sets) as test statistic, we find that only 37 out of the 10,000 permuted data sets exceeded the test statistic for maternal transmission in the observed data (left), while 6,450 out of the 10,000 permutation-based data sets exceeded the test statistic for paternal transmission (right). LD, linkage disequilibrium; TAT, transmission asymmetry test; CL/P, cleft lip with or without cleft palate; RUNX2, Runt-related transcription factor 2; SNP, single nucleotide polymorphism.
Similar examination of the other LD blocks in RUNX2 showed no evidence of excess transmission, either maternal or paternal for blocks 1 and 4 (data not shown). However, in the third LD block there was evidence of excess paternal transmission yielding a statistically significant empirical P-value (0.011 corrected for multiple tests). SNPs within this block had higher heterozygosity values and showed relatively little variation in allele frequencies among populations (see Table II). Furthermore, two of the three SNPs in this block (rs1934328 and rs7771889) showed evidence of linkage and LD when parent-of-origin was ignored (see Table III). SNP rs1934328 also yielded a nominally significant value from the PO-LRT with an estimated OR(imprinting) = 0.44 (P-value = 0.029), rejecting the null hypothesis of equal transmission from mothers and fathers (see Table IV).
DISCUSSION
Our study of case-parent trios from different populations (comprising a total of 298 case-parent trios) showed consistent evidence of linkage in the presence of association (LD) for multiple SNPs in RUNX2 only when parent-of-origin effects were considered. In a phenotypic study of RUNX2 mutants in mice, Aberg et al. [2004] reported cleft palate and early opening of the eyelids among mutant mice embryos. Yamachika et al. [2001] also reported a heterozygous C⃗ T transition in exon 3 of the RUNX2 gene in a patient with CCD and a cleft lip. In a cohort study of CCD patients, Cooper et al. [2001] found a significantly increased prevalence of submucosal cleft palate. Recently, Khan et al. [2006] suggested that CBFA2, a gene related to RUNX2, may be associated with enlarged cranial sutures with or without cleft palate.
The RUNX2 gene encodes a nuclear protein with a Runt DNA-binding domain and is a member of the RUNX2 family of transcription factors. RUNX2 contains nine alternatively spliced exons. Exons 1 and 2 contain alternatively utilized promoter regions and an ATG translational start codon. There are three alternative versions of exon 5 (exons 5, 5.1, and 5.2) and two alternative versions of exon 6 [exons 6 and 6.1; [Terry et al., 2004]. A sporadic case of CCD was found to be caused by heterozygosity at a G⃗ A transition at codon 283 in exon 5 of RUNX2 [Mundlos et al., 1997].
In our study, we analyzed 24 SNPs spanning 120.5 kb of the RUNX2 gene. We screened for parent-of-origin effects and 11 adjacent SNPs in RUNX2 showed evidence of excess maternal transmission. This region with excess maternal transmission falls within the same LD block in all four populations (Fig. 1), located between exons 4 and 5. The PO-LRT for inequality between maternal and paternal transmissions seldom exceeded marginal levels of significance for these 11 SNPs, likely due to the limited number of informative matings. The total number of informative fathers and mothers is modest, but after deleting heterozygous × heterozygous matings there were equal numbers of informative mothers and fathers (29 of each) for SNP rs910586, which showed the most significant result. Complete confirmation, of course, still requires larger samples of trios, larger numbers of informative heterozygotes, and more complete coverage with additional polymorphic SNPs.
Excess maternal transmission could reflect genomic imprinting or maternal genotype effects. Maternal genotypic effects for nonsyndromic CL/P have also been reported for several other candidate genes. van Rooij et al. [2003] reported that mothers carrying the TT genotype at the C677T variant in MTHFR showed an increased risk of having a child with CL/P. Martinelli et al. [2001] also observed an increase in TT homozygotes among mothers of CL/P subjects, with a risk ratio of 2.51 (1.00–6.14). However, Jugessur et al. [2003] found that children of mothers carrying the T variant allele had a slightly lower risk of CL/P. Interplay between maternal and fetal genotypes has been suggested for the gene coding for the TGFA [Mossey et al., 1998]. However, in a study of 134 Italian CL/P trios, Rubini et al. [2005] reported an association between the c844ins68 variant in the cystathionine β-synthase gene and nonsyndromic CL/P, and showed an 18.7-fold increase in risk for CL/P when the child received the c844ins68 allele from the mother rather than the father.
In the present study, log-linear models suggested by Weinberg [1999] to discriminate between maternal genotype and child genotype revealed a possible maternal imprinting effect for multiple SNPs in RUNX2. Maternal genotype effects were not individually significant for all 11 of these contiguous SNPs. Recently, Young et al. [2007] reported that RUNX2 transcription factors reinforce cell fate through an epigenetic mechanism that retains phenotypic gene expression patterns after cell division. RUNX2 also regulates expression of GNAS, a maternally imprinted gene [Bertaux et al., 2006; Weinstein et al., 2004]. In an analysis by real-time polymerase chain reaction of zebrafish RUNX2 genes, Flores et al. [2004] reported that RUNX2b was expressed only as a maternal transcript. They also detected maternal expression of RUNX2b during early embryogenesis in zebrafish.
Even though this candidate gene study involved a modest number of SNPs, addressing the issue of multiple comparisons is imperative before an overall statement about the significance of findings can be made. Since SNPs in strong LD typically have highly correlated P-values, adjusting significance levels via the overly conservative Bonferroni correction is particularly undesirable. Therefore, we conducted permutation tests for each of the four LD blocks and adjusted empirical P-values for the number of blocks, resulting in largely independent P-values. In the second LD block with 11 SNPs, we found strong evidence against the null hypothesis only for maternal transmission (the empirical P-value of 0.0037 would still be significant after correcting for multiple comparisons); but no such evidence for excess paternal transmission was seen. Thus, this 47 kb region of the gene seems to show excess maternal transmission to offspring with CL/P.
Interestingly, the next LD block of three SNPs with higher allele frequencies (and more informative) showed evidence of excess paternal transmission in these data. One SNP (rs1934328) in this block yielded significant evidence of imprinting when analyzed alone with the PO-LRT, giving an estimated OR(-imprinting) = 0.44, which was significantly less than what would be expected under the null hypothesis (H0:OR = 1). Since this model included separate terms for maternal genotype effects, this raises the possibility of imprinting in RUNX2.
The case-parent trio design offers the advantage of testing directly for maternal vs. paternal effects, and allows separating these from effects of the fetal genotype vs. parental origin in a robust manner [Cordell et al., 2004; Starr et al., 2005; Sinsheimer et al., 2003]. Another advantage of this design is that it minimizes issues of confounding that plague traditional case-control designs. This feature permitted pooling trios from four diverse populations into a combined test of allelic effects on the OR(transmission) to the affected child, while testing for parent-of-origin effects. The present study suggests maternal transmission effects for markers in RUNX2 and risk of nonsyndromic CL/P, although further work is still needed to confirm its ultimate impact on risk.
WEB RESOURCES
HAPLOVIEW: http://www.broad.mit.edu/mpg/haploview/index.php/
Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/Omim/
Acknowledgments
Contract grant sponsor: National Institute of Dental and Craniofacial Research; Contract grant numbers: R21-DE-013707, R01-DE-014581; Contract grant sponsor: NIH; Contract grant numbers: R01 HL 090577; Contract grant sponsor: Korean Research Foundation; Contract grant numbers: 2004-041-E00104.
This research was supported by R21-DE-013707 and R01-DE-014581 from the National Institute of Dental & Craniofacial Research, NIH R01 HL 090577, and Korean Research Foundation (2004-041-E00104 and 2005-214-E00042). We thank all participants who donated samples for this multicenter study of oral clefts, as well as the staff at each participating site and institution. We also thank Gerald Raymond for his assistance in screening patients at Hopkins, and Katherine Zarfas for her technical help in the analysis.
References
- Aberg T, Cavender A, Gaikwad JS, Bronckers AL, Wang X, Waltimo-Sirén J, Thesleff I, D’Souza RN. Phenotypic changes in dentition of Runx2 homozygote-null mutant mice. J Histochem Cytochem. 2004;52:131–139. doi: 10.1177/002215540405200113. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bellus GA, Hefferon TW, Ortiz de Luna RI, Hecht JT, Horton WA, Machado M, Kaitila I, McIntosh I, Francomano CA. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am J Hum Genet. 1995;56:368–373. [PMC free article] [PubMed] [Google Scholar]
- Bertaux K, Broux O, Chauveau C, Hardouin P, Jeanfils J, Devedjian JC. Runx2 regulates the expression of GNAS on SaOs-2 cells. Bone. 2006;38:943–950. doi: 10.1016/j.bone.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Carinci F, Scapoli L, Palmieri A, Zollino I, Pezzetti F. Human genetic factors in nonsyndromic cleft lip and palate: an update. Int J Pediatr Otorhinolaryngol. 2007;71:1509–1519. doi: 10.1016/j.ijporl.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Cobourne MT. The complex genetics of cleft lip and palate. Eur J Orthod. 2004;26:7–16. doi: 10.1093/ejo/26.1.7. [DOI] [PubMed] [Google Scholar]
- Cooper SC, Flaitz CM, Johnston DA, Lee B, Hecht JT. A natural history of cleidocranial dysplasia. Am J Med Genet. 2001;104:1–6. doi: 10.1002/ajmg.10024. [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Barratt BJ, Clayton DG. Case/pseudocontrol analysis in genetic association studies: a unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet Epidemiol. 2004;26:167–185. doi: 10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V, Amling M, Karsenty G. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999;13:1025–1036. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiberg H, Bixler D, Nielsen LS, Conneally PM, Mohr J. Suggestion of linkage of a major locus for nonsyndromic orofacial cleft with F13A and tentative assignment to chromosome 6. Clin Genet. 1987;32:129–132. doi: 10.1111/j.1399-0004.1987.tb03340.x. [DOI] [PubMed] [Google Scholar]
- Fan JB, Oliphant A, Shen R, Kermani BG, Garcia F, Gunderson KL, Hansen M, Steemers F, Butler SL, Deloukas P, Galver L, Hunt S, McBride C, Bibikova M, Rubano T, Chen J, Wickham E, Doucet D, Chang W, Campbell D, Zhang B, Kruglyak S, Bentley D, Haas J, Rigault P, Zhou L, Stuelpnagel J, Chee MS. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- Flores MV, Tsang VW, Hu W, Kalev-Zylinska M, Postlethwait J, Crosier P, Crosier K, Fisher S. Duplicate zebrafish runx2 orthologues are expressed in developing skeletal elements. Gene Expr Patterns. 2004;4:573–581. doi: 10.1016/j.modgep.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Jugessur A, Wilcox AJ, Lie RT, Murray JC, Taylor JA, Ulvik A, Drevon CA, Vindenes HA, Abyholm FE. Exploring the effects of methylenetetrahydrofolate reductase gene variants C677T and A1298C on the risk of orofacial clefts in 261 Norwegian case-parent triads. Am J Epidemiol. 2003;157:1083–1091. doi: 10.1093/aje/kwg097. [DOI] [PubMed] [Google Scholar]
- Khan A, Hyde RK, Dutra A, Mohide P, Liu P. Core binding factor beta (CBFB) haploinsufficiency due to an interstitial deletion at 16q21q22 resulting in delayed cranial ossification, cleft palate, congenital heart anomalies, and feeding difficulties but favorable outcome. Am J Med Genet A. 2006;140:2349–2354. doi: 10.1002/ajmg.a.31479. [DOI] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19:S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Marazita ML, Murray JC, Lidral AC, Arcos-Burgos M, Cooper ME, Goldstein T, Maher BS, Daack-Hirsch S, Schultz R, Mansilla MA, Field LL, Liu YE, Prescott N, Malcolm S, Winter R, Ray A, Moreno L, Valencia C, Neiswanger K, Wyszynski DF, Bailey-Wilson JE, Albacha-Hejazi H, Beaty TH, McIntosh I, Hetmanski JB, Tunçbilek G, Edwards M, Harkin L, Scott R, Roddick LG. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32–35. Am J Hum Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli M, Scapoli L, Pezzetti F, Carinci F, Carinci P, Stabellini G, Bisceglia L, Gombos F, Tognon M. C677T variant form at the MTHFR gene and CL/P: a risk factor for mothers? Am J Med Genet. 2001;98:357–360. doi: 10.1002/1096-8628(20010201)98:4<357::aid-ajmg1108>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Mossey PA, Arngrimsson R, McColl J, Vintiner GM, Connor JM. Prediction of liability to orofacial clefting using genetic and craniofacial data from parents. J Med Genet. 1998;35:371–378. doi: 10.1136/jmg.35.5.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, Owen MJ, Mertelsmann R, Zabel BU, Olsen BR. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Oliphant A, Barker DL, Stuenlpnagel JR, Chee MS. BeadArray™ Technology: enabling an accurate, cost-efficient approach to high-throughput genotyping. Biotechniques. 2002;32:S56–S61. [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50:211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- Rubini M, Brusati R, Garattini G, Magnani C, Liviero F, Bianchi F, Tarantino E, Massei A, Pollastri S, Carturan S, Amadori A, Bertagnin E, Cavallaro A, Fabiano A, Franchella A, Calzolari E. Cystathionine beta-synthase c.844ins68 gene variant and non-syndromic cleft lip and palate. Am J Med Genet A. 2005;136:368–372. doi: 10.1002/ajmg.a.30812. [DOI] [PubMed] [Google Scholar]
- Sinsheimer JS, Palmer CG, Woodward JA. Detecting genotype combinations that increase risk for disease: maternal-fetal genotype incompatibility test. Genet Epidemiol. 2003;24:1–13. doi: 10.1002/gepi.10211. [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Starr JR, Hsu L, Schwartz SM. Assessing maternal genetic associations: a comparison of the log-linear approach to case-parent triad data and a case-control approach. Epidemiology. 2005;16:294–303. doi: 10.1097/01.ede.0000158223.98649.eb. [DOI] [PubMed] [Google Scholar]
- Terry A, Kilbey A, Vaillant F, Stewart M, Jenkins A, Cameron E, Neil JC. Conservation and expression of an alternative 3′ exon of Runx2 encoding a novel proline-rich C-terminal domain. Gene. 2004;336:115–125. doi: 10.1016/j.gene.2004.04.015. [DOI] [PubMed] [Google Scholar]
- van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet. 2000;24:342–343. doi: 10.1038/74155. [DOI] [PubMed] [Google Scholar]
- van Den Oord EJ, Vermunt JK. Testing for linkage disequilibrium, maternal effects, and imprinting with (in)-complete case-parent triads, by use of the computer program LEM. Am J Hum Genet. 2000;66:335–338. doi: 10.1086/302708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij IA, Vermeij-Keers C, Kluijtmans LA, Ocké MC, Zielhuis GA, Goorhuis-Brouwer SM, van der Biezen JJ, Kuijpers-Jagtman AM, Steegers-Theunissen RP. Does the interaction between maternal folate intake and the methylenetetrahydrofolate reductase polymorphisms affect the risk of cleft lip with or without cleft palate? Am J Epidemiol. 2003;157:583–591. doi: 10.1093/aje/kwg005. [DOI] [PubMed] [Google Scholar]
- Weinberg CR. Methods for detection of parent-of-origin effects in genetic studies of case-parents triads. Am J Hum Genet. 1999;65:229–235. doi: 10.1086/302466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg CR, Umbach DM. A hybrid design for studying genetic influences on risk of diseases with onset early in life. Am J Hum Genet. 2005;77:627–636. doi: 10.1086/496900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg CR, Wilcox AJ, Lie RT. A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet. 1998;62:969–978. doi: 10.1086/301802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein LS, Liu J, Sakamoto A, Xie T, Chen M. Minireview: GNAS: normal and abnormal functions. Endocrinology. 2004;145:5459–5464. doi: 10.1210/en.2004-0865. [DOI] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. What good is genomic imprinting: the function of parent-specific gene expression. Nat Rev Genet. 2003;4:359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- Wyszynski DF, Duffy DL, Beaty TH. Maternal cigarette smoking and oral clefts: a meta-analysis. Cleft Palate Craniofac J. 1997;34:206–210. doi: 10.1597/1545-1569_1997_034_0206_mcsaoc_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- Yamachika E, Tsujigiwa H, Ishiwari Y, Mizukawa N, Nagai N, Sugahara T. Identification of a stop codon mutation in the CBFA1 runt domain from a patient with cleidocranial dysplasia and cleft lip. J Oral Pathol Med. 2001;30:381–383. doi: 10.1034/j.1600-0714.2001.300610.x. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci U S A. 2007;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, Christensen K, Suzuki Y, Machida J, Natsume N, Yoshiura K, Vieira AR, Orioli IM, Castilla EE, Moreno L, Arcos-Burgos M, Lidral AC, Field LL, Liu YE, Ray A, Goldstein TH, Schultz RE, Shi M, Johnson MK, Kondo S, Schutte BC, Marazita ML, Murray JC. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]


