Abstract
The HpxO enzyme from Klebsiella pneumoniae was recently proposed, on the basis of genetic studies, to catalyze the hydroxylation of uric acid to 5-hydroxyisourate as part of the purine catabolic pathway. Its primary sequence suggests that the HpxO catalytic activity depends on a flavin cofactor (FAD), contrasting with all previously-studied urate oxidase enzymes, which have no cofactor requirement. Here we demonstrate biochemically that HpxO is an FAD-dependent urate oxidase. Our data are consistent with the proposal that HpxO-bound flavin hydroperoxide is the hydroxylating species. These results confirm the existence of a novel mechanistic paradigm in purine catabolism.
Catabolism of purine nucleobases is an important mechanism for nitrogen assimilation by many organisms when their growth is limited by the availability of ammonia or alternative nitrogen sources (1). The bases are converted to hypoxanthine 1 or xanthine 2, then oxidized to uric acid 3 (Figure 1A). The steps to this point are ubiquitous in Nature but humans, amongst other organisms, lack the ability to metabolize urate to allantoin 6 (2). In contrast, the pathway from urate, in a variety of microbes and plants, has been intensively studied and proceeds under aerobic conditions in three steps from urate to allantoin 6. The first step is the hydroxylation of urate, catalyzed by urate oxidase (1,3). In all previously-studied cases, the catalytic mechanism of urate oxidase is remarkable in that it does not require a cofactor (4). Detailed studies have implicated the electron-rich urate dianion as an enzyme-bound intermediate which can react spontaneously with molecular oxygen in its triplet ground state, forming a peroxide intermediate. Analogy has thus been made between urate and the isoalloxazine moiety of flavins which in its reduced form undergoes a similar oxidation. In extension of the analogy, EPR spin trapping experiments have suggested the presence of transient radical intermediates in the steps leading to the urate hydroperoxide intermediate (5).
Figure 1.
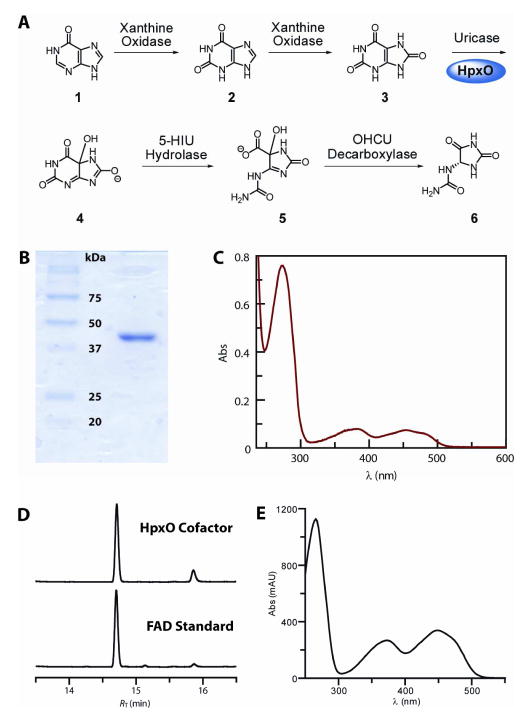
Isolation of Klebsiella pneumoniae HpxO. The recombinant enzyme with an N-terminal His6-tag was overexpressed in Escherichia coli BL21(DE3) using standard protocols. The enzyme was purified by Ni2+-nitrilotriacetic acid chromatography. (A) Purine catabolic pathway from hypoxanthine to allantoin. HpxO catalyzes hydroxylation of urate 3 to the unstable 5-hydroxyisourate 4, which is converted both enzymatically and non-enzymatically via 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU) 5 to allantoin 6; (B) SDS-PAGE analysis (12% acrylamide) of the purified enzyme. The molecular weight is ~45 kDa including the affinity tag; (C) UV-visible spectrum of HpxO, with absorbance maxima at 454, 381, 273 and <220 nm; (D) Reverse-phase (C18) HPLC analysis of the small molecule released from HpxO on heat-denaturation. Absorbance detection was at 250 nm. The mobile phase consisted of a gradient from 0–65% methanol in potassium phosphate buffer. A minor impurity (RT = 15.9 min) present in the commercial sample of FAD (RT = 14.7 min) was also found in the HpxO-derived sample; the increased proportion of this impurity in the HpxO sample was attributed to the effects of the heat denaturation; (E) UV-Vis spectrum of the HPLC purified FAD isolated from heat-denatured HpxO.
The urate oxidase product, 5-HIU 4, is unstable in aqueous buffer, hydrolyzing spontaneously to 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU) 5 (6). Notwithstanding the operation of this spontaneous reaction under physiological conditions, all known organisms which express the cofactorless urate oxidase also possess a hydrolase enzyme which accelerates the conversion of 4 to 5. The decarboxylation of OHCU 5 to allantoin 6 is also spontaneous under physiological conditions, but nonenzymatically produces racemic allantoin (3a). A stereospecific decarboxylating enzyme, which produces only (S)-allantoin, is found in organisms with urate oxidase and 5-HIU hydrolase (7); some organisms also possess a racemase enzyme (1, 8). Allantoin can be futher catabolized to ammonia and carbon dioxide, thus utilizing the remaining nitrogen atoms in the imidazoline ring (1,9).
While the enzymology of the pathway outlined above has been thoroughly investigated, two very recent reports (10) have suggested that the pathway from hypoxanthine to allantoin, involving the same intermediates but different catalytic strategies for their interconversion, exists in the facultative anaerobe Klebsiella pneumoniae, a ubiquitous human pathogen (11). In particular, the enzyme proposed to catalyze the conversion of urate to 5-hydroxyisourate 4 shows sequence similarity to FAD-dependent monooxygenases catalyzing the hydroxylation of aromatic compounds. It was noted that the enzyme sequence includes residues which are conserved across this group of enzymes that contain an FAD-binding domain and a small NADH-binding domain (Figure S1 of the Supporting Information) (12). Both groups proposed that HpxO catalyzes the hydroxylation of urate by a mechanism similar to the well-studied p-hydroxybenzoate hydroxylase (13), and that HpxO is the first example of an FAD-dependent urate oxidase. However, de la Riva et al. also reported that in their study, cells with hpxO, expressed from a high-copy plasmid, lacked discernible urate oxidase activity (10b).
To test the prediction that HpxO is an FAD-dependent urate oxidase, we cloned, heterologously overexpressed and purified the recombinant, N-terminally His6-tagged protein (see Supporting Information for details), and then reconstituted its activity in vitro. The isolated protein was more than 95% pure, as determined by SDS-PAGE analysis (Figure 1B), with a molecular weight (including the N-terminal tag) of ~45 kDa. The HpxO protein solution, after gel filtration, was yellow, suggesting the presence of a tightly-bound cofactor. The UV-visible spectrum of the protein solution (Figure 1C) had maxima at 454, 381, 273 and <220 nm. Reverse-phase HPLC analysis of the small molecule released from the protein upon heat denaturation confirmed that the cofactor was FAD (Figure 1D, E). We determined the HpxO:FAD stoichiometry by measurement of the FAD-derived absorbance in a solution of HpxO denatured by the addition of urea to a concentration of 6 M. The 450 nm absorbance (ε450 = 9.7 mM−1 cm−1, Figure S2) was used to compute the FAD- derived 280 nm absorbance (A280/A450 = 1.84), which in turn was subtracted from the total absorbance at 280 nm to give the protein absorbance at that wavelength. The protein absorbance was then used to determine its concentration based on the theoretical HpxO molar extinction coefficient (57.4 mM−1 cm−1) at 280 nm (14). The FAD concentration and the protein concentration agreed to within 10%, and also agreed with the protein concentration determined by the Bradford method (15). Taken together, these results suggest that the FAD:HpxO stoichiometry is 1:1.
Having demonstrated that HpxO is a flavoprotein, we investigated its catalytic activity towards uric acid. Treatment of uric acid with HpxO in the presence of NAD(P)H resulted in a decrease in the 340 nm absorbance, consistent with the oxidation of the NAD(P)H to NAD(P)+. The rate of this decrease in A340 depended linearly on the enzyme concentration and could be saturated at high concentrations of urate or NADH. At saturating concentrations of NADH (1.5 mM, see below) the 385 nm absorbance was monitored (ε385 = 0.745 mM−1 cm−1). When urate was omitted there was a detectable, enzyme-dependent rate of NAD(P)H oxidation, but this was not significant relative to the rates observed in the presence of urate. A fit of the kinetic data where NADH was used as the nucleotide co-substrate to the Michaelis–Menten equation gave values for Km and kcat of 42 ± 8 μM and 42 ± 2 s−1 (kcat/Km = 1.0 ± 0.2 μM−1 s−1) for urate (Figure S3). This is similar to the cofactor-independent urate oxidase (kcat/Km = 0.21 μM−1 s−1). However, HpxO is considerably more efficient with respect to utilization of molecular oxygen. We estimated a Km value for molecular oxygen of at most ~50 μM (Figure S4) and hence a value of kcat/Km of at least ~0.7 mM−1 s−1, which appears to be at least an order of magnitude greater than the equivalent value for the cofactor-independent urate oxidase (0.034 mM−1 s−1).
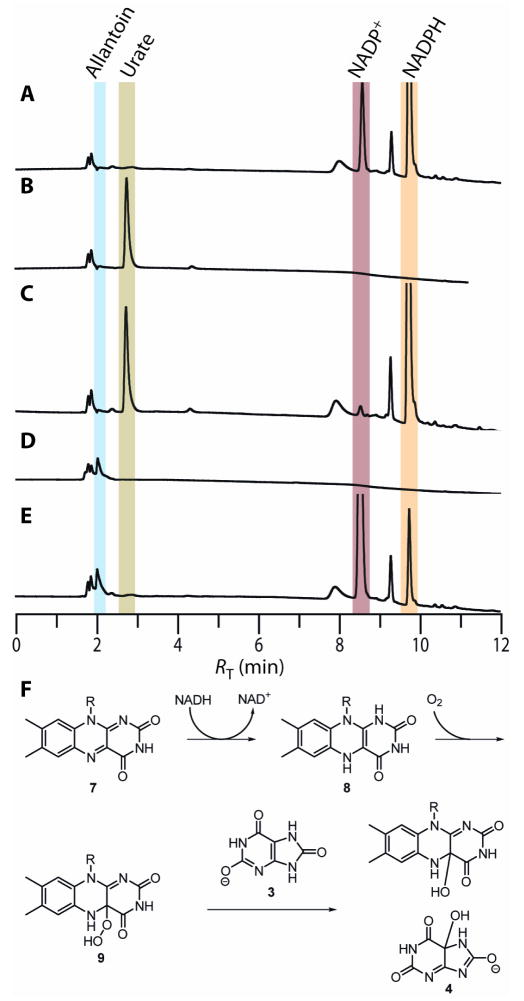
Previous studies on the stability of the proposed HpxO product, 5-HIU 4, have shown that it should react spontaneously under our assay conditions to form allantoin; full conversion should occur in ~1 h. HPLC analysis of samples of urate, incubated with HpxO for 1 h in the presence of NADPH followed by removal of the enzyme, confirmed the presence of allantoin and NADP+ (Figure 2E). Assignment of the product as allantoin was confirmed by comparison with an authentic standard and also with the reaction products from a commercially available, FAD-independent urate oxidase (Figure 2D). Control experiments where urate was omitted confirmed that HpxO can catalyze the slow conversion of NADPH to NADP+ in the absence of urate (Figure 2A–C). These results suggest 5-HIU 4 to be the immediate product of the HpxO-catalyzed reaction. This proposal is further supported by the presence of genes in the hpx cluster which are proposed to encode 5-HIU hydrolase and OHCU decarboxylase. Analysis of the time course of the HpxO-catalyzed reaction from uric acid to allantoin by UV-visible spectrophotometry also indicated that 5-HIU and OHCU accumulated and decayed on the expected time scales (Figure S5).
Figure 2.
Reverse-phase (C18) HPLC analysis of the urate oxidase activity of HpxO. All reactions were performed in 0.1 M potassium phosphate, pH 8.0 at room temperature (~22°C). Reaction components, where present, were at the following concentrations: Uric acid, 0.1 mM; NADPH, 0.25 mM; HpxO, 0.13 μM; commercially-available urate oxidase (Sigma-Aldrich) from Candida sp., 2.2 units/mL (one unit is defined as the quantity of enzyme which oxidizes 1.0 μmol of uric acid to allantoin at pH 8.5 and at 25 °C); (A) HpxO control reaction omitting urate; (B) Control omitting NADPH; (C) Control omitting HpxO; (D) NADPH-independent reaction of urate catalyzed by urate oxidase from Candida sp.; (E) HpxO reaction including urate, NADPH and HpxO; (F) Proposed mechanism for HpxO-catalyzed formation of 5-HIU.
HpxO shows selectivity (V/K ratio of ~10) for NADH over NADPH. The HpxO steady-state rate showed no saturation at concentrations of NADPH up to 1 mM, suggesting this value as a lower limit for Km,NADPH. A linear fit to the rate-concentration data gave an estimate of 9.7 ± 0.3 mM−1 s−1 for the kcat/Km value (Figure S6A). In contrast the steady-state rate showed saturation at NADH concentrations above 0.5 mM, and a hyperbolic fit to this data gave 125 ± 30 mM−1 s−1 as an estimate of kcat,NADH/Km,NADH, (Figure S6B).
We also sought to demonstrate the formation of the dihydroflavin intermediate 8 by the reaction of NADPH with HpxO under anaerobic conditions. HpxO was incubated in the presence of glucose, glucose oxidase and catalase for 15 minutes in a sealed spectrophotometer cuvette prior to the introduction of an excess of NADPH. The absorbance spectrum in the 400–550 nm region was recorded after a further incubation of 10 minutes to allow for consumption of residual oxygen. The spectrum showed complete removal of the 454 nm-centered absorbance, consistent with the formation of dihydro-FAD 8 (Figure S7). Considered along with the evidence that molecular oxygen is a substrate and the precedent available from previously well-studied systems, this result suggests that the mechanism presented in Figure 2F is in operation during the HpxO catalytic cycle.
In summary, we have confirmed the recent proposals that HpxO is an FAD-dependent urate hydroxylase. The reaction produces 5-hydroxyisourate which is spontaneously converted to allantoin in two nonenzymatic steps. Our data suggests a mechanism for urate hydroxylation similar to that observed for the well-studied p-hydroxybenzoate hydroxylase. The results illustrate a previously-uncharacterized mechanistic motif for urate oxidation and thus for microbial purine metabolism.
Supplementary Material
Acknowledgments
We thank Dr. Cynthia Kinsland of the Protein Production Facility at the Department of Chemistry and Chemical Biology, Cornell University for the molecular cloning and construction of the vector encoding the recombinant HpxO protein.
Abbreviations
- EDTA
ethylenediamine tetraacetic acid
- EPR
electron paramagnetic resonance
- FAD
flavin adenine dinucleotide
- 5-HIU
5-hydroxyisouric acid
- UA
uric acid
- NAD(P)H
reduced b-nicotinamide adenine dinucleotide (phosphate)
- NAD(P)+
b-nicotinamide adenine dinucleotide (phosphate)
- Ni-NTA
nickel-nitrilotriacetic acid
- OHCU
2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline
Footnotes
SUPPORTING INFORMATION AVAILABLE
Details of the molecular cloning, experimental procedures and Figures S1–S7. This material is available free of charge over the Internet at http://pubs.acs.org.
This work was supported by National Institutes of Health Grants AI066244-02 (to T.P.B.) and GM73220 (to S.E.E.).
References
- 1.(a) Vogels GD, van der Drift C. Bacteriol Rev. 1976;40:403–468. doi: 10.1128/br.40.2.403-468.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Brondino C, Romão MJ, Moura I, Moura JJG. Curr Opin Chem Biol. 2006;10:109–114. doi: 10.1016/j.cbpa.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 2.Oda M, Satta Y, Takenaka O, Takenaka N. Mol Biol Evol. 2002;19:640–653. doi: 10.1093/oxfordjournals.molbev.a004123. [DOI] [PubMed] [Google Scholar]
- 3.(a) Kahn K, Serfozo P, Tipton PA. J Am Chem Soc. 1997;119:5435–5442. [Google Scholar]; (b) Imhoff R, Power NP, Borrok MJ, Tipton PA. Biochemistry. 2003;42:4094–4100. doi: 10.1021/bi027377x. [DOI] [PubMed] [Google Scholar]
- 4.Kahn K, Tipton PA. Biochemistry. 1997;36:4731–4738. doi: 10.1021/bi963184w. [DOI] [PubMed] [Google Scholar]
- 5.Busi E, Terzuoli L, Basosi R, Porcelli B, Marinello E. Nucleosides, Nucleotides Nucleic Acids. 2004;23:1131–1134. doi: 10.1081/NCN-200027390. [DOI] [PubMed] [Google Scholar]
- 6.Ramazzina I, Folli C, Secchi A, Berni R, Percudani R. Nat Chem Biol. 2006;2:144–148. doi: 10.1038/nchembio768. [DOI] [PubMed] [Google Scholar]
- 7.(a) Kim K, Park J, Rhee S. J Biol Chem. 2007;282:23457–23464. doi: 10.1074/jbc.M703211200. [DOI] [PubMed] [Google Scholar]; (b) Cendron L, Berni R, Folli C, Ramazzina I, Percudani R, Zanotti G. J Biol Chem. 2007;282:18182–18189. doi: 10.1074/jbc.M701297200. [DOI] [PubMed] [Google Scholar]
- 8.van der Drift L, Vogels GD, van der Drift C. Biochim Biophys Acta. 1975;391:240–248. doi: 10.1016/0005-2744(75)90170-9. [DOI] [PubMed] [Google Scholar]
- 9.Mulrooney SB, Hausinger RP. J Bacteriol. 2003;185(1):126–134. doi: 10.1128/JB.185.1.126-134.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Pope SD, Chen L-L, Stewart V. J Bacteriol. 2008 doi: 10.1128/JB.01281-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) de la Riva L, Badia J, Aguilar J, Bender RA, Baldoma L. J Bacteriol. 2008;190(24):7892–7903. doi: 10.1128/JB.01022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Podschun R, Ullmann U. Clin Microbiol Rev. 1998;11(4):589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Keynan Y, Rubinstein E. Int J Antimicrob Agents. 2007;30(5):385–389. doi: 10.1016/j.ijantimicag.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 12.Harayama S, Kok M, Neidle EL. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 13.(a) Ortiz-Maldonado M, Entsch B, Ballou DP. Biochemistry. 2004;43:15246–15257. doi: 10.1021/bi048115t. [DOI] [PubMed] [Google Scholar]; (b) Entsch B, vanBerkel WJH. FASEB J. 1995;9:476–483. doi: 10.1096/fasebj.9.7.7737455. [DOI] [PubMed] [Google Scholar]
- 14.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradford MM. Anal Biochem. 1976;131:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.