Abstract
Mitochondria consist of four compartments, outer membrane, intermembrane space, inner membrane and matrix; each harboring specific functions and structures. In this study, we used mass spectrometry (LC-MS/MS) to characterize the protein composition of Trypanosoma brucei mitochondrial membranes, which were enriched by different biochemical fractionation techniques. The analyses identified 202 proteins that contain one or more transmembrane domain(s) and/or positive GRAVY scores. Of these, various criteria were used to assign 72 proteins to mitochondrial membranes with high confidence, and 106 with moderate to low confidence. The sub-cellular localization of a selected subset of 13 membrane assigned proteins was confirmed by tagging and immunofluorescence analysis. While most proteins assigned to mitochondrial membrane have putative roles in metabolic, energy generating, and transport processes, ~50% have no known function. These studies result in a comprehensive profile of the composition and sub-organellar location of proteins in the T. brucei mitochondrion thus, providing useful information on mitochondrial functions.
Keywords: Mass spectrometry, Membranes, Mitochondrion, Organelle fractionation, T. brucei
1. INTRODUCTION
Trypanosomatids (Order Kinetoplastida) are unicellular flagellates and the causative agents of several devastating diseases in humans including Human African Trypanosomiasis (HAT) or sleeping sickness (Trypanosoma brucei), Chagas disease (Trypanosoma cruzi), and leishmaniasis (Leishmania spp.). They possess a single prominent mitochondrion, which is composed of an outer membrane (OM), inner membrane (IM), intermembrane space (IMS), and matrix. The matrix harbors the uniquely structured mitochondrial (mt) DNA, termed kinetoplast DNA (kDNA) [1], which, as in other organisms, encodes a small number of proteins [2]. Recent studies show that the T. brucei mitochondrion contains over 1000 proteins [3], the vast majority of which are encoded by nuclear genes, synthesized in cytosol, and imported to their proper sub-mt destination [4]. Each of the four mt compartments harbors specific proteins and processes. The protein translocase machinery (TOM complex) is embedded in the mt OM [5], the respiratory chain complexes and the multi subunit protein translocases complex (TIM) are present in the mt IM [6], cytochrome c (cyt c) is in the IMS [7], and the multi-protein complexes that catalyze RNA editing are located within the matrix [8].
The availability of the genome sequences of T. brucei [9], T. cruzi [10] and L. major [11] has paved the way for transcriptome and proteome analyses of Trypanosomatids [12–19]. A previous mt proteome analysis of T. brucei procyclic form (PF) cells extrapolated to a total of 1000 mt proteins, of these specific assignments were made with varying levels of confidence for 880 proteins [3]. More complete information on mt and sub-mt protein composition and location is required for a more comprehensive understanding of the various sub-mt compartments, and the T. brucei mitochondrion as a whole. Such comprehensive proteomic analyses of sub-mt compartments have been performed in other systems such as the mt IM from mouse liver [20], and the mt OM from yeast [21] and N. crassa [22]. Membrane proteins in general are a key set of proteins as they are at a boundary between functional compartments and perform many important functions such as transport, reception, and trafficking. In addition, more than half of the known drug targets are membrane proteins [23]; thus their characterization would aid in drug target discovery. However, membrane proteins are some of the most challenging proteins to study due to their hydrophobic nature and relatively low abundance.
Here, we report a comprehensive analysis of T. brucei PF cells mt membrane proteome. We performed sub-cellular fractionation to enrich for mt membranes, and identified the proteins in these fractions by LC-MS/MS analysis. The assignment to mt membrane was based on selective enrichment in mitochondria versus whole cell lysate [3], at least one predicted transmembrane domain (TMD) and/or positive GRAVY (grand average hydropathy) score, association with known mt complexes, demonstrated or putative role in relevant biological processes, and /or homology to yeast mt membrane proteins. The localization of a subset of these proteins was validated by immunofluorescence analysis by expression of c-Myc epitope tagged proteins in the parasite.
2. MATERIALS AND METHODS
2.1 Trypanosome Growth
T. brucei PF cells IsTaR 1.7a were grown to density of 1–2 × 107 cells/ml in vitro at 27 °C in SDM-79 media containing hemin (7.5 mg/ml) (Sigma) and 10 % (v/v) FBS. PF T. brucei strain 29.13 [24], which contains integrated genes for T7 polymerase and the tetracycline repressor, was grown in the presence of G418 (15 µg/ml) and hygromycin (25 µg/ml) (Sigma). The cells were harvested by centrifugation at 6,000 × g for 10 min at 4°C. The transgenic PF cell lines expressing a TAP-tagged protein were supplemented with 2.5 µg/ml phleomycin (Sigma). Exogenous protein expression was induced by adding 0.1 µg/ml tetracycline (Sigma) and allowing the cultures to grow for 3 days prior to harvesting.
2.2 Sub-mt fractionation
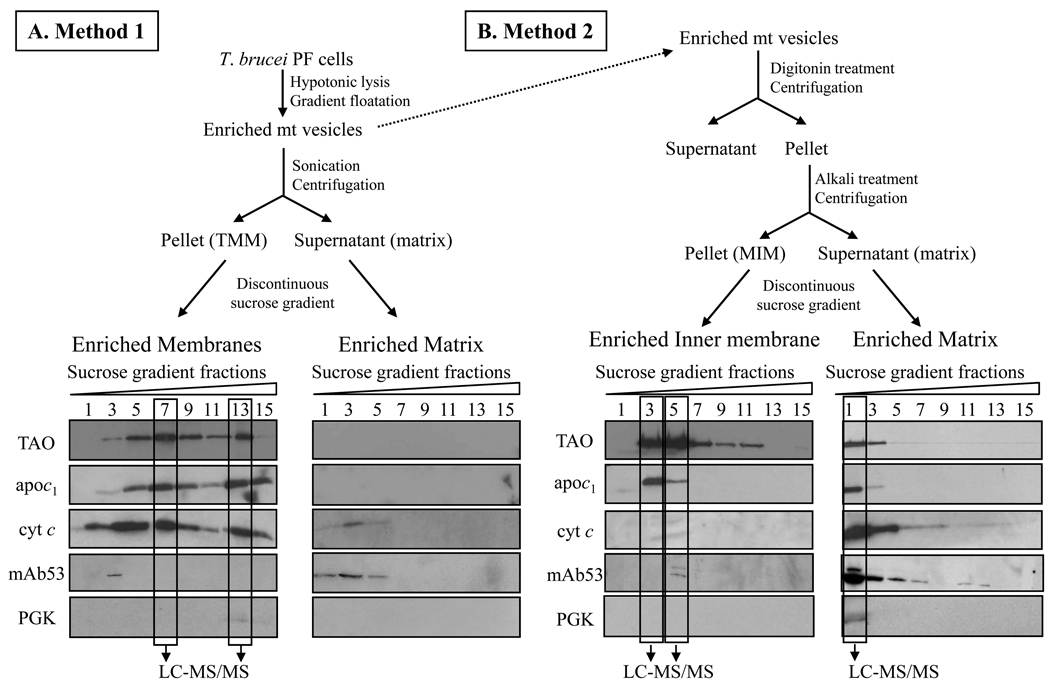
Mt vesicles were isolated by hypotonic lysis and enriched using Percoll gradients as described elsewhere [25]. The membrane and matrix fractions were generated by 2 different methods (Figure 1A/B). In Method 1, sub-mt membranes were isolated following sonication and step gradient purification according to [26,27]. Briefly, mt vesicles were resuspended at 10 mg/ml in breaking buffer (0.6 M Sorbitol, 20 mM Hepes/KOH, pH 7.4, 10 mM EDTA) and incubated for 30 min on ice in 9 vol of 20 mM Hepes/KOH, pH 7.4, 0.5 mM EDTA and 1 mM PMSF. After adjustment to a final sucrose concentration of 0.45 M and incubation for 10 min on ice, the sample was sonicated for 2 × 90 s (duty cycle 40 %). After a clarifying spin at 20,000 × g for 20 min at 4°C, the supernatant was centrifuged at 200,000 × g for 45 min at 4°C. Aliquots of the Total mt Membranes fraction (TMM) and supernatant (matrix) enriched fractions were subjected to a sucrose step gradient consisting of 1.5 ml of 1.6 M, 5.5 ml of 1.35 M, 2.5 ml of 1.1 M and 1.5 ml of 0.85 M sucrose prepared in 5 mM Hepes/KOH, pH 7.4, 10 mM KCL and 1 mM PMSF. Samples were centrifuged at 134,000 × g at 4°C for 16 h in a L7-55 Ultracentrifuge (SW55 Ti swinging bucket rotor) from Beckman. Fractions of 0.5 ml were collected from the top, and subjected to SDS-PAGE and Western blot analyses.
Figure 1.
Schematics for the isolation of mt membranes and matrix fractions. (A) Method 1: mt membrane fraction was isolated by disruption of vesicles followed by differential centrifugation and enrichment on sucrose step gradients. (B) Method 2, MIM fraction was isolated by digitonin treatment, followed by alkali extraction, differential centrifugation and enrichment by gradient sedimentation. The gradient fractions were analyzed by SDS-PAGE and Western blot using antibodies against markers of the mt IM (TAO, apoc1), the IMS (cyt c), the matrix (mAb53) and the cytosol (PGK). The indicated fractions were analyzed by MS.
In Method 2, mt vesicles (10 mg/ml) were treated with digitonin (Sigma) at a final concentration of 80 µg/ml for 15 min at 4°C. Following centrifugation at 12,000 × g for 20 min at 4°C, the organellar pellet was incubated with 0.1 M sodium carbonate, pH 11.5 for 30 min on ice and centrifuged at 100,000 × g for 1h at 4°C. The pellet from this step was used as the mt Inner Membrane (MIM) fraction and the supernatant as the matrix fraction. Aliquots of the MIM and the matrix enriched fractions were layered on top of discontinuous sucrose gradients as described above.
2.3 Western blot analysis
The proteins in each of the mt fractions from above were resolved by 10 % SDS-PAGE (approximately 2–5µg was loaded in each lane) and transferred onto PVDF membranes. The blots were blocked with 5% non-fat milk, probed with mAbs against the trypanosome alternative oxidase (TAO) (1:100) [28], mAb53 (1:25) [29], and with rabbit polyclonal antibodies against T. brucei apocytochrome c1 (apoc1) (1:500) [30], cytochrome c (1:500) [31] and phosphoglycerate kinase (PGK) (1:5000) [32] followed by either goat anti-mouse or goat anti-rabbit IgG HRP-conjugated secondary antibodies (1:2000) (BioRad). The ECL enhanced chemiluminescence (Pierce) was used to visualize reactive bands. For quantitative Western analysis, fluorescent detection was performed using IRDye680 goat anti-rabbit antibody (LI-COR Biosciences, Lincoln, NE) at 1:2000 dilution. Bands were visualized and quantified by Odyssey™ Infrared Imaging System (LI-COR Biosciences, Lincoln, NE) using one-color fluorescence detection at 700. The corresponding SDS-PAGE gel was stained with Sypro Ruby (Invitrogen, Molecular Probes™), scanned using Storm™ imaging system (Molecular Dynamics, Inc.), and the fluorescence level of protein bands was analyzed using ImageQuant™ software.
2.4 Protein identification by LC-MS/MS
Proteins were analyzed by mass spectrometry as described [3]. Briefly, the proteins in enriched mt membrane and matrix fractions were separated on 10% SDS-PAGE gels and protein bands were visualized by Sypro Ruby Staining (Molecular Probes). Each gel lane was divided into 10 pieces, the proteins were digested in-gel with sequencing grade modified trypsin (Promega); the resulting peptides were extracted with 50% ACN (VWR), 5% formic acid (Sigma Aldrich) and dried in Speed Vac [3]. The peptides were fractionated by nano-flow liquid chromatography using a 7.5 cm long × 75 µm ID C18 capillary column at a flow rate of 200 nl/min. The peptides were analyzed using a LTQ Linear Ion Trap Mass Spectrometer. The CID spectra were compared with the T. brucei protein database downloaded from GeneDB using TurboSEQUEST software, and protein matches determined using PeptideProphet and ProteinProphet software [33,34].
2.5 Sequence analysis of identified proteins
The probable functions of the proteins were assigned based on GeneDB annotation and for proteins with unknown function possible motifs and/or domains were searched in InterPro (http://www.ebi.ac.uk/Tools/InterProScan/), Pfam (http://pfam.sanger.ac.uk/) and NCBI CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) databases. The average GRAVY [35] scores for all proteins were obtained using ProtParam (http://www.expasy.org.tools/protparam.html). The protein transmembrane topology prediction was conducted using the TMHMM 2.0 program (http://www.cbs.dtu.dk/services/TMHMM). MitoP2 yeast database (http://www.mitop.de:8080/mitop2/) was used for homology searches.
2.6 Plasmid constructs and transfections
To create constructs for the inducible expression of c-Myc epitope tagged proteins in T. brucei, the ORFs of interest were PCR amplified from genomic DNA of T. brucei strain Lister 427. The detailed list of primers used to amplify selected ORFs is in the supplemental material. The PCR products were cloned into pGEM-T easy vector (Promega), digested with Bam HI or Bgl II and Hind III enzymes (New England Biolabs), and ligated into the pLEW79-MHT vector [36,37]. The plasmids were linearized with Not I enzyme, transfected into PF T. brucei 29.13 cell line, phleomycin-resistant clones were selected, and checked for tetracyline-regulated expression.
2.7 Immunofluorescence microscopy
The expressed tagged proteins were localized by immunofluorescence analysis (IFA) using anti-c-Myc antibody (Sigma) as described [29]. Briefly, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 (Fisher) and blocked with 5% FBS. Incubation with primary and secondary antibodies in 3% BSA-PBS was done for 1 h followed by 10 min incubation with 4,6-Diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma) stain to visualize both nuclear and kinetoplast DNA. Phase contrast images of the cells and their fluorescence were captured with a Nikon fluorescence microscope equipped with camera and the appropriate filters. The primary antibodies used were: rabbit anti-c-Myc (Sigma) diluted 1:200; mouse supernatant anti-MRP1 diluted 1:2 [29]. The secondary antibodies were: Texas® Red-X goat anti-mouse IgG (Invitrogen) diluted 1:400 and goat anti-rabbit IgG-FITC (Sigma) diluted 1:500.
3. RESULTS AND DISCUSSION
3.1 Protein identification
LC-MS/MS analyses of the TMM, MIM, and matrix fractions in aggregate identified a total of 3,559 unique peptides corresponding to 1,134 unique gene products, i.e. proteins (Supplementary Table 1A). In these analyses, 643 proteins were matched by two or more peptides while 491 were identified by a single peptide match, of which 371 (76%) had a protein identification probability of ≥ 0.99, and the rest had probabilities between 0.9 and 0.98. All of the proteins identified with two or more peptide matches had protein identification probability of ≥ 0.99. Of the total 1,134 proteins identified in this study, 146 had not been previously detected in an extensive proteomic analysis of the T. brucei mitochondrion [3] (Supplementary Table 1B). Some of these newly and previously identified proteins were localized to mt membranes, as described below.
Comparison of the total acquired MS/MS data with polypeptides predicted from six-frame translation of nucleic acid (NA) database led to the identification of 15 new ORFs, with coding sequence that had not been recognized in the GeneDB annotation (Supplementary Table 2 and Supplementary Figure 1). This frequency of finding new potential protein coding sequences is similar to that previously observed [3]. Of these 15 polypeptides, two show homology to the retrotransposon hot spot (RHS) family and three to T. brucei proteins annotated as hypothetical. No homology to T. brucei was found for the other ten, but these show homology to peptides predicted from the T. cruzi and/or L. major genome sequences. The first five probably represent T. brucei protein coding genes that had not been recognized. These other ten ORFs may also represent such genes, or genes that diverged from the sequenced strains, or it is also possible that the strain used may have slight differences in genomic sequences.
3.2 Enriched mitochondrial membranes
Mt membranes, which were prepared by method 1, namely sonication of mt vesicles, collection by centrifugation, and fractionation in sucrose gradients were enriched for the TAO and apoc1 IM proteins, and the IMS protein cyt c (Figure 1A). The protein recognized by mAb53, which is specific to a protein of the MRB1 complex [29], was primarily detected in the matrix fraction. The membrane proteins that peaked in fractions 7 and 13 were designated as Total mt Membranes (TMM) fraction and selected for analysis by MS. LC- MS/MS analyses identified a total of 455 proteins in these enriched membrane fractions, of which 138 proteins possess at least one TMD, suggesting membrane localization (Tables 1A/B; Supplementary Table 3A/B and Supplementary Table 4).
Table 1.
| Table 1A. Proteins with known or putative function identified and assigned to mt membranes. They are grouped based on their association with demonstrated / predicted biological processes. The proteins were identified by indicated number of peptides (number of unique peptides in parenthesis) in TMM and MIM fractions. Blank columns indicate the proteins were not identified by MS in the analyzed samples. | |||||
|---|---|---|---|---|---|
| Protein_ID | GeneDB Description | TMD | GRAVY | TMM | MIM |
| Mitochondrial Respiration | |||||
| Complex III | |||||
| Tb09.211.4700 | RISP reiske iron-sulfur protein , mitochondrial precursor | 1 | −0.275 | 5(4) | 7(6) |
| Complex IV | |||||
| Tb11.01.3780 | cytochrome oxidase assembly factor, putative | 8 | 0.319 | 2(2) | |
| Tb09.160.1140 | electron transport protein SCO1/SCO2, putative | 1 | −0.489 | 1(1) | |
| Complex V | |||||
| Tb927.7.7420 | ATP synthase alpha chain, mitochondrial precursor | 0 | 0.082 | 11(11) | 11(11) |
| Tb927.3.1380 | ATP synthase beta chain, mitochondrial precursor | 0 | 0.083 | 9(9) | 10(10) |
| Other in aerobic respiration | |||||
| Tb927.7.5820 | monooxygenase, putative | 0 | 0.055 | 1(1) | |
| Electron transport | |||||
| Tb10.6k15.3640 | TAO alternative oxidase | 1 | −0.225 | 1(1) | 2(2) |
| Tb11.02.4485 | CYB5 cytochrome b5, putative | 1 | 0.071 | 1(1) | |
| Cation/Ion transport | |||||
| Tb09.160.2910 | tricarboxylate carrier, putative | 5 | 0.228 | 2(2) | 2(2) |
| Tb927.5.3400 | calcium-translocating P-type ATPase | 10 | 0.149 | 6(6) | 10(10) |
| Tb11.01.0720 | cation transporter, putative | 8 | 0.616 | 2(2) | |
| Amino acid transport | |||||
| Tb927.8.7600 | amino acid transporter, putative | 12 | 0.548 | 1(1) | 1(1) |
| Protein folding/modification/transport | |||||
| Tb11.01.4870 | inner membrane preprotein translocase Tim 17, putative | 2 | 1(1) | 1(1) | |
| Tb927.7.1340 | 10 kDa heat shock protein, putative | 0 | 0.181 | 2(2) | |
| Tb09.160.3090 | heat shock protein, putative | 1 | −0.388 | 2(2) | |
| Tb09.211.1550 | chaperone protein DNAJ, putative | 3 | −0.125 | 2(2) | 2(2) |
| Tb927.7.6200 | chaperone protein DNAJ, putative | 1 | −0.228 | 1(1) | |
| Tb11.03.0110 | chaperone protein DNAJ, putative | 1 | −0.559 | 1(1) | |
| Tb10.61.3100 | chaperone protein DNAJ, putative | 1 | −0.617 | 1(1) | 1(1) |
| Protein catabolic process | |||||
| Tb09.211.0680 | CAAX prenyl protease 1, putative | 5 | 0.051 | 2(2) | 1(1) |
| Tb11.01.6360 | metalloprotease, putative | 1 | −0.416 | 3(3) | 1(1) |
| Tb927.4.3300 | mitochondrial ATP-dependent zinc metallopeptidase, putative | 1 |
−0.194 |
1(1) |
3(3) |
| Tb927.4.4210 | ATP-dependent zinc metallopeptidase, putative | 2 | −0.319 | 3(3) | 2(2) |
| Tb09.211.0220 | rhomboid-like protein | 4 | 0.076 | 1(1) | 1(1) |
| Regulation | |||||
| Tb10.v4.0045 | prohibitin | 1 | −0.022 | 5(5) | 5(5) |
| Various Metabolic processes | |||||
| Tb927.3.1840 | 3-oxo-5-alpha-steroid 4-dehydrogenase, putative | 4 | 0.1 | 3(3) | 4(4) |
| Tb10.406.0290 | protein tyrosine phosphatase, putative | 7 | 0.613 | 1(1) | |
| Tb927.8.6410 | short-chain dehydrogenase, putative | 1 | −0.278 | 1(1) | |
| Tb927.5.1210 | short-chain dehydrogenase, putative | 2 | 0.156 | 4(4) | 5(5) |
| Tb10.6k15.3250 | succinyl-CoA ligase [GDP-forming] beta-chain, putative | 1 | −0.056 | 1(1) | |
| Tb927.6.4540 | 3-hydroxy-3-methylglutaryl-CoA reductase, putative | 1 | 0.086 | 1(1) | |
| Tb927.8.1420 | acyl-CoA dehydrogenase, mitochondrial precursor, putative | 0 | 0.167 | 1(1) | |
| Tb927.6.4210 | aldehyde dehydrogenase, putative | 0 | 0.144 | 3(3) | |
| Tb11.01.8470 | dihydrolipoyl dehydrogenase | 0 | 0.023 | 8(8) | 5(5) |
| Tb927.3.1790 | pyruvate dehydrogenase E1 beta subunit, putative | 0 | 0 | 7(7) | 6(6) |
| Tb10.70.4420 | methionine biosynthetic protein, putative | 0 | 0.123 | 1(1) | |
| Tb09.211.1960 | 3-demethylubiquinone-9 3-methyltransferase, putative | 0 | 0.089 | 1(1) | |
| Tb927.7.5680 | deoxyribose-phosphate aldolase, putative | 0 | 0.038 | 2(2) | |
| Tb10.70.5110 | mitochondrial malate dehydrogenase | 0 | 0.209 | 5(5) | 3(3) |
| Transport | |||||
| Tb09.211.1750 | mitochondrial carrier protein, putative | 1 | 0.079 | 6(6) | 5(5) |
| Tb10.389.0690 | mitochondrial carrier protein, putative | 4 | 0.144 | 1(1) | 2(2) |
| Tb10.61.1820 | mitochondrial carrier protein, putative | 3 | −0.021 | 4(4) | 5(5) |
| Tb11.02.2960 | mitochondrial carrier protein, putative | 1 | 0.297 | 2(2) | 2(2) |
| Tb09.211.2370 | mitochondrial carrier protein, putative | 0 | 0.27 | 1(1) | 1(1) |
| Tb927.2.2970 | mitochondrial carrier protein, putative | 0 | 0.173 | 3(3) | 1(1) |
| Tb927.3.2980 | mitochondrial carrier protein, putative | 0 | 0.161 | 1(1) | |
| Tb927.4.1660 | mitochondrial carrier protein, putative | 0 | 0.019 | 1(1) | |
| Tb11.03.0540 | ABC transporter, putative | 6 | 0.117 | 1(1) | |
| Table 1B. Proteins currently annotated as hypothetical assigned to mt membranes. | |||||
|---|---|---|---|---|---|
| Protein_ID | Domains/motifs | TMD | GRAVY | TMM | MIM |
| Complex IV | |||||
| Tb09.211.4740 | ubiE, ubiquinone/menaquinone biosynthesis methyltransferase | 1 | −0.511 | 3(3) | |
| Tb09.v1.0420 | 1 | −0.321 | 1(1) | ||
| Tb11.01.1900 | Macro domain, Appr-1-p processing | 1 | −0.127 | 1(1) | |
| Tb11.0400 | Tyrosine protein kinase | 1 | −0.227 | 2(2) | 2(2) |
|
Tb927.3.750 |
SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domain; Putative methyl transferase |
1 |
−0.102 |
1(1) |
3(3) |
| Tb927.7.6990 | 1 | −0.347 | 1(1) | ||
| Tb11.01.5390 | DnaJ domain | 1 | −0.565 | 1(1) | |
| Tb11.02.5660 | 1 | −0.033 | 1(1) | 2(2) | |
| Complex V | |||||
| Tb10.70.7760 | LETM1, inner mitochondrial membrane protein | 1 | −0.628 | 2(2) | 2(2) |
| Tb11.47.0022 | 2 | −0.527 | 1(1) | 3(3) | |
| Tb927.7.840 | 1 | −0.231 | 1(1) | ||
| Tb11.02.4120 | 0 | 0.188 | 3(3) | 3(3) | |
| Novel Complex | |||||
| Tb927.8.580 | 1 | −0.25 | 2(2) | ||
| Other mt proteins | |||||
|
Tb10.6k15.1800 |
ABC1 family; ubiB, putative ubiquinone biosynthesis |
2 |
−0.017 |
1(1) |
1(1) |
| Tb10.26.0100 | 2 | 0.61 | 1(1) | ||
| Tb10.70.3010 | 1 | −0.49 | 3(3) | ||
| Tb10.70.4500 | 1 | −0.163 | 1(1) | ||
| Tb11.01.4740 | 1 | −0.251 | 1(1) | 1(1) | |
| Tb11.12.0014 | 2 | 0.191 | 2(2) | 1(1) | |
| Tb927.2.4090 | GTP-binding protein, HSR1-related | 2 | 0.055 | 1(1) | 1(1) |
| Tb927.2.4700 | 2 | −0.327 | 1(1) | 1(1) | |
| Tb927.5.510 | 1 | 0.098 | 1(1) | ||
| Tb927.7.640 | 3 | −0.06 | 2(2) | ||
| Tb927.8.5660 | 3 | 0.303 | 1(1) | ||
The second method, namely sodium carbonate treatment of mt vesicles, which had been treated with digitonin to remove the OM prior to fractionation by sedimentation in sucrose gradients, resulted in much of the TAO and apoc1 mt IM proteins being enriched and primarily sediment in fractions 3 through 5 with slight cross-contamination from matrix fraction (mAb53) (Figure 1B). Some of these proteins were in the matrix fraction but sedimented at the top of the gradient. Based on the Western signal, fractions 3 and 5 were designated mt Inner Membrane (MIM) fraction and selected for LC-MS/MS analysis. Cytochrome c was almost undetectable in the MIM fraction when compared to TMM fraction (Figure 1A), suggesting TMM fraction contains some IMS proteins. Markers for other soluble IMS proteins were not used since they were not available to us. Three T. brucei IMS protein candidates, Tb927.3.1600, Tb11.02.3065 and Tb09.160.4440 were identified by sequence analysis, the first two are homologs of yeast and human small Tim chaperones [38] and the last one is a homolog of Erv1 protein [39]. All three proteins were identified by MS analyses of whole organelles, but were not detected in the TMM and MIM fractions, indicating that many IMS proteins are not present in these fractions and hence are probably lost during preparation.
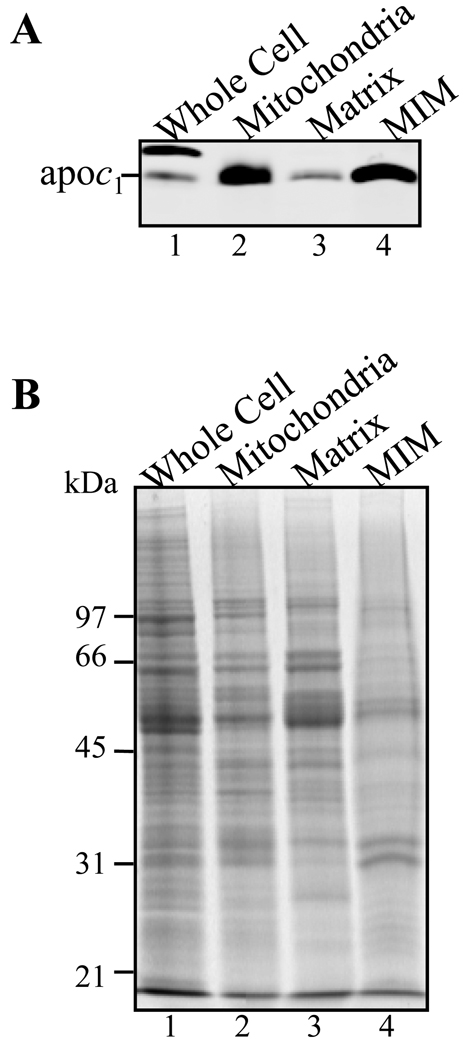
An approximate 17-fold enrichment of MIM using apoc1 antibody was found in mt fraction using a quantitative Western analysis. The signal for apoc1 was 8.5-fold higher than with whole cell lysate when half of the protein amount of Percoll purified mitochondria were examined (Figure 2B, lanes 1 and 2). Protein quantification analysis was done by staining parallel gels with Sypro Ruby and measuring the fluorescence levels in each lane. This method is very accurate for determining the relative amounts of specific proteins and is preferable to Bradford analysis for insoluble proteins. An additional 2.6-fold enrichment of apoc1 in MIM fraction was estimated upon further fractionation into sub-mt compartments. The result is based on 1.7-fold higher signal compared to the mt fraction using 0.6 times the relative amount of protein (Figure 2A, lanes 2 and 4). In addition, very little signal for apoc1 was detected in matrix fraction (Figure 2A, lane 3). Thus, the procedure chosen for purification is suitable for mt membrane enrichment although possibility of some loss of MIM fraction proteins during the purification cannot be excluded.
Figure 2.
SDS-PAGE and Western analysis showing enrichment of MIM using apoc1 as a marker for mt IM. (A) Proteins from whole cell (lane 1), Percoll gradient purified mitochondria (lane 2), mt matrix (lane 3) and MIM fraction 3 (lane 4) were analyzed by quantitative Western analysis using anti-apoc1 antibody. (B) Corresponding SDS-PAGE gel stained with SYPRO Ruby.
MS analyses of MIM fractions (3 and 5) led to the identification of 458 proteins, of which 158 have at least one TMD, suggesting membrane localization; 277 of the proteins had also been detected in the TMM fraction (Tables 1A/B; Supplementary Table 3A/B and Supplementary Table 4). Thus a total of 637 different proteins, of which 202 have at least one TMD, were found in the TMM and MIM fractions.
3.3 Sub-cellular assignment of proteins
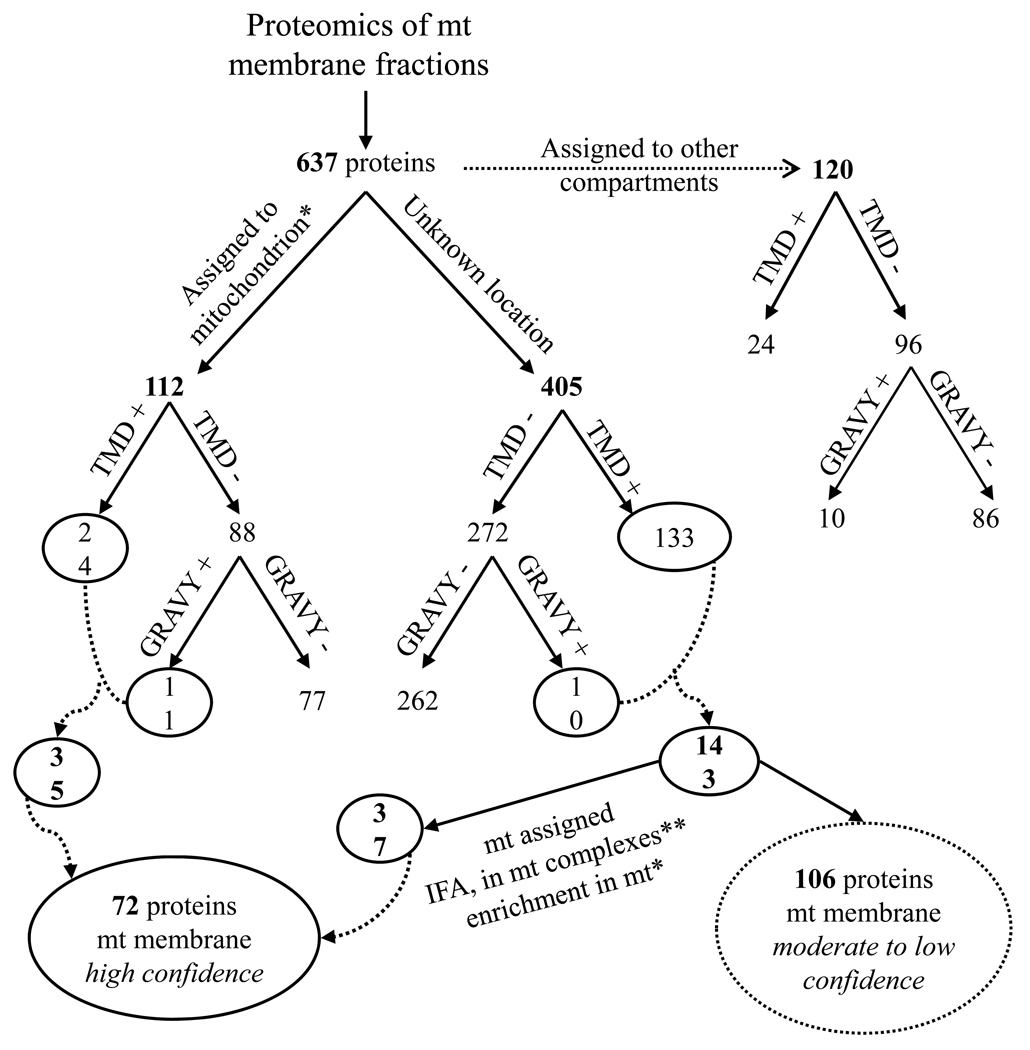
Of the total of 637 different proteins identified in the TMM and MIM fractions, some are likely localized in the organelle membranes, others in the mt matrix, and yet others are non-mt as indicated by the Western analyses above. Of these proteins, 120, including 24 that have at least one TMD, are likely contaminants of the enriched mt vesicles since they are assignable to non-mt locations, which include glycosomes, ER, the nucleus, Golgi and flagellum based on their GeneDB annotation, key-word and literature searches (Supplementary Table 4), although some genes may be misannotated. Glycosomal proteins are typically found in purified mt vesicles [3] just as mt proteins are in glycosome preparations [12,14]. However, the possibility of dual-localization cannot be eliminated in the absence of direct experimental data since about one quarter of the yeast mt proteome has a dual localization [40]. The remaining 178 proteins, which have predicted TMD and/or positive GRAVY scores are potential mt membrane proteins (see Fig. 3 for representative results). Of these, 35 were previously assigned to the mitochondrion [3], thus results from this study assign them to mt membrane location. The other 143 had not been assigned to the mitochondrion and 37 of those were assigned to the mitochondrion with high confidence based on several criteria including IFA, enrichment in the mt fraction [3], functional annotation, demonstrated or putative role in mt processes, and association with known mt complexes [29,41–43] (Table 1A/B). The presence of one or more TMD and/or hydrophobic domain (GRAVY) suggests that they are localized to mt membranes.
Figure 3.
Grouping of proteins identified from the MS/MS analyses. The number of proteins designated to mitochondria, to another cellular compartment or to an unknown location is indicated. The putative membrane assignment was done by presence of predicted TMD and/or positive GRAVY score. The circled groups were assigned to mt membranes with high confidence, whereas the dotted-circled groups with moderate to low confidence.
* [3]
Of the 72 proteins assigned to mt membrane with high confidence, we were able to assign 24 proteins without a known function (“hypothetical” proteins in the T. brucei genome database) to mt membrane with high confidence. The assignment employed criteria including enrichment in mt fraction, presence of at least one TMD, homology to yeast mt proteins, and association with known mt complexes (Table 1B). The assignment to complexes took advantage of recent work that determined the composition of several mt complexes using affinity tag and mAb affinity purification combined with MS analyses [29,41–43]. Of these 24 proteins, LC-MS/MS analyses revealed the presence of 13 membrane-bound proteins, which are associated with complexes of the electron transport chain as well as the ATP synthase complex, all located in the mt IM. Eight are associated with complex IV [40], four with complex V [41], and one with a novel mt complex (A. Zíková, unpublished results). .While these proteins are currently annotated as hypothetical, conserved motifs/domains that are indicative of possible function(s) were identified in eight of these proteins (Table 1B). None of the mt encoded subunits in Complex III, IV or V were identified, possibly due to the characteristics of these proteins, which include non-migration into SDS-PAGE gels, and few potential trypsin cleavage sites, making their peptide identification by routine MS analysis difficult.
The remaining 48 proteins assigned to mt membrane with high confidence have known or predicted functions (Table 1A). Twenty seven of these (56%) were identified in both the TMM and MIM fractions, while 11 were only identified in TMM and ten were only in MIM, suggesting the latter ten may localize to the mt IM and former 11 to the mt OM. Identification of 3-hydroxy-3-methylglutaryl-CoA reductase and succinyl-CoA ligase in the MIM fraction and presence of a TMD in each of these proteins implies a membrane location. This differs from the matrix location as assigned in prior reports [44,45], although it does not preclude that these proteins could have substantial domains on the matrix side of the membrane.
In addition, 341 proteins, which have no TMD/GRAVY score, were detected in the TMM and/or MIM fractions, of which 30 are embedded in one of the mt membrane complexes (Supplementary Table 5). For the other proteins, some may not be localized in membrane but may be associated with membrane proteins, and others may have no association with mt membranes but be a mt protein contaminant of the membrane preparations.
The transport of a wide range of solutes as well as the import of critical proteins into the mitochondrion is performed by proteins residing in the mt IM and OM. A proportion (23%) of the membrane proteins identified in the TMM and MIM fractions that have one to 12 TMDs were found to have predicted functions as membrane transporters for amino acids, anions and cations, or other molecular mt carrier proteins (Table 1A). The GeneDB database has 26 products annotated as mt carrier family (MCF) proteins, of which our analyses identified ten, three of which map to the tandemly repeated genes Tb10.61.1810, Tb10.61.1820, and Tb10.61.1830. Members of the MCF transport ATP/ADP, various intermediates of the Krebs cycle as well as several other solutes through the mt IM thus linking cytosolic biochemical pathways with those in the mt matrix. We recently identified 12 other MCF proteins in a previous study [3]. The remaining four predicted MCF proteins may be located in another cellular compartment, since not all predicted MCF proteins are mt [46], or they are not expressed in the PF stage of T. brucei, or they were present but not detected.
Translocation of mt pre-proteins across the mt IM is facilitated by either of the two TIM complexes known as TIM23 and TIM22. The TIM23 core complex, which contain Tim23, Tim17 and Tim50, imports proteins that have N-terminal targeting sequences; the TIM22 complex inserts proteins that have multiple membrane spanning domains into IM [47]. The present analysis only identified the Tim17 protein member of the Tim23 complex proteins in T. brucei [5]. In yeast and humans, there are five small Tim chaperones, located in the IMS compartment, which assist the delivery of substrate proteins to either the sorting and assembly (SAM) complex or TIM22 complex [38]. In T. brucei, Tb927.7.2200 is a homolog of Tim9, Tb927.3.1600, is a homolog of Tim10, and Tb11.02.3065 is a homolog of Tim8 and Tim13. All three proteins were found in our MS analyses as well as Tb927.3.4380, which shows 26% identity with S. cerevisiae Sam50, the core component of SAM (Supplementary Table 1A).
The supernatant from the digitonin treatment fraction was considered as OM and therefore analyzed by MS. No suitable markers against T. brucei mt OM are available that can be used in Western blot analysis. Seventeen proteins with TMDs, which are present only in this fraction and not seen previously, were identified (Tb09.v1.0390; Tb09.211.4630; Tb10.389.0730; Tb10.389.1550; Tb10.6k15.2750; Tb10.61.0950; Tb10.70.5220; Tb11.01.0480; Tb11.01.4340; Tb11.01.7530; Tb11.02.0780; Tb11.02.5360; Tb927.3.1080; Tb927.3.2420; Tb927.5.3710; Tb927.7.220; Tb927.7.6920). Of these, 5 proteins (Tb10.389.0730, Tb11.02.0780, Tb927.3.1080, Tb927.5.3710 and Tb927.7.220) are annotated to be involved in the synthesis of phosphatidylcholine, cardiolipid and sterol, which compose the lipid bilayer of the mt OM, suggesting that at least some of these 17 proteins may be localized to this membrane. This is supported by the presence in this fraction of Tb927.2.2510, the ortholog of the voltage-dependent anion channel (VDAC) in T. brucei [48,49], which is the most abundant protein in the mt OM. We might have lost some mt OM proteins since breakage of the long tubular mitochondrion during cell disruption cannot be avoided and hence our beginning material is resealed vesicles. Thus, further investigations are required for definitive assignment of this group of proteins. Besides Tim17, the other components of the mt translocation machinery are yet to be discovered in T. brucei, indicating that the mt protein import machinery is divergent in this organism.
The proteolytic enzymes of mitochondrion belong to the superfamily of AAA+ proteins (ATPases associated with a variety of cellular activities), which comprises many proteins with chaperone activity [50]. The m-AAA proteolytic system is active at the matrix side of the mt IM and comprising, in S. cerevisiae, multiple copies of homologous subunits Yta10 and Yta12. Our MS analyses revealed three Yta10/Yta12 homologs annotated as ATP-dependent Zinc metallopeptidase in GeneDB. In addition, the m-AAA proteases activity in yeast is modulated by another membrane-protein complex composed of prohibitins (Phb1p and Phb2p). Our MS analyses identified one related prohibitin homolog in T. brucei, which shows 45% identity with Phb1, and is identical in sequence to Tb10.70.2920, another prohibitin homolog annotated in GeneDB, indicating possible gene duplication.
3.4 Sub-cellular localization of thirteen membrane assigned proteins
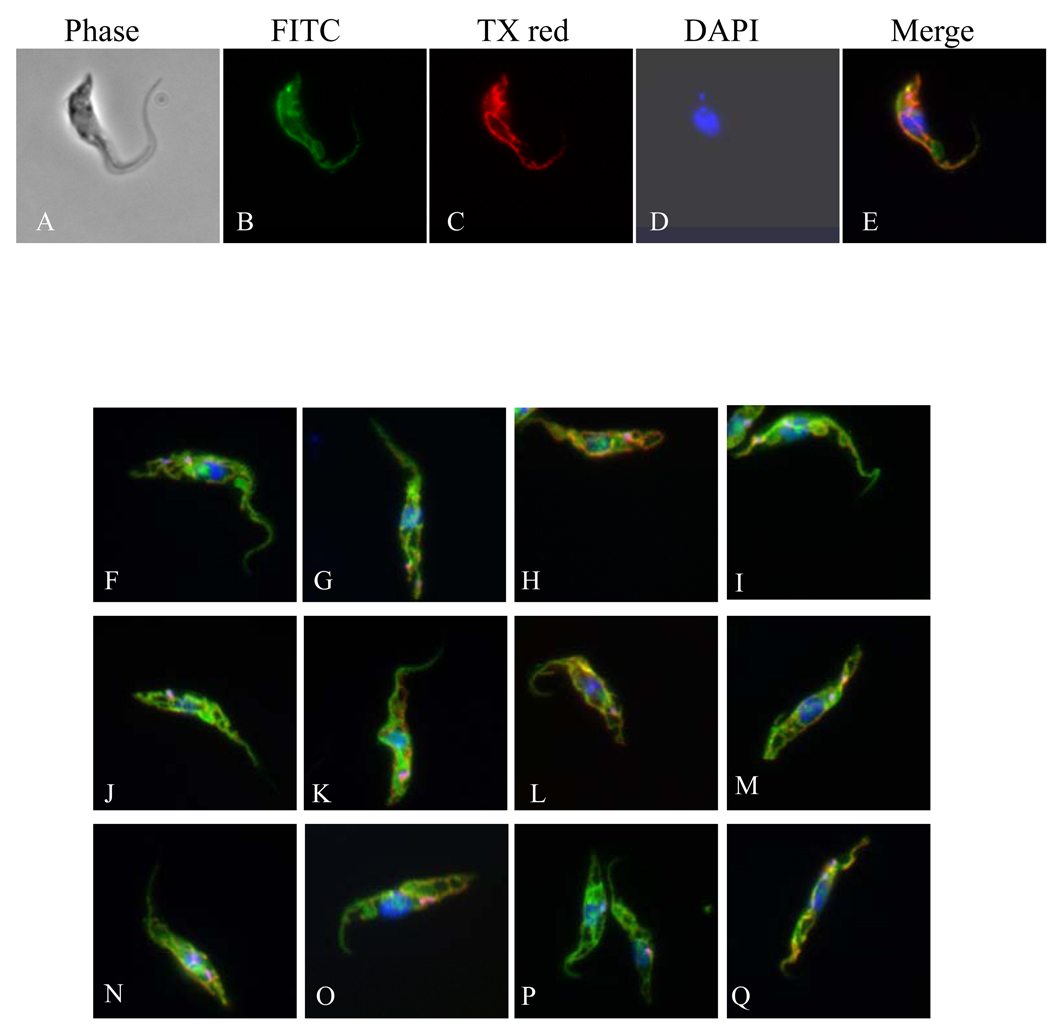
Thirteen membrane assigned proteins of various amino acids (aa) sequence lengths (from 363 to 631 aa), and displaying one to 12 predicted TMDs were selected in order to validate their sub-cellular localization by IFA (Supplementary Figure 2). These proteins were fused to c-Myc epitope tag and their sub-cellular localization was assessed after induced expression in PF T. brucei cells. Co-localization IFA analyses were performed with a mAb against the mt RNA-binding protein 1 (MRP1) [29]. As shown in Figure 4, all fusion proteins were evenly distributed throughout the reticulated mitochondrion, but do not intensely label the kinetoplast area, and do not co-localize completely with MRP 1, probably due to a different localization between MRP1 and the tagged protein inside the mitochondrion. Furthermore, these data indicated that the tag as well as overexpression did not alter the mt localization of the tagged proteins [51].
Figure 4.
Representative figures showing sub-cellular localization of mt membrane assigned proteins. Panel (A–E): Tb11.47.0022: (A) Phase contrast light microscopy showing PF T. brucei cells, (B) anti-c-Myc antibody coupled with FITC conjugated secondary antibody showing mt localization of target protein, (C) anti-MRP1 coupled with Texas® Red-X conjugated secondary antibody, (D) DAPI staining of nucleus and kDNA, and (E) merge. Panels F-U only the merge is shown; F: Tb10.70.3010, G: Tb927.8.7600, H: Tb10.6k15.1800, I: Tim17 (Tb11.01.4870), J: Tb11.01.4740, K: Tb11.02.5420, L: Tb927.5.1210, M: Tb10.70.1640, N: Tb11.01.2650, O: Tb927.8.5660, P: Tb11.12.0014, Q: Tb927.2.4090.
3.5 Other possible mt membrane proteins
In addition to the 72 mt membrane assigned proteins, 77 proteins, of these 40 with known function and 37 hypothetical proteins, were assigned to mt membranes with moderate confidence (Supplementary Table 3A/B), based on identification in membrane fractions, presence of TMD and/or positive GRAVY score and enrichment in mt fraction [3]. These have not been included in the above high confidence list, but it is likely that a substantial proportion of them localize to mt membranes. Among the 40 proteins with a functional annotation (Supplementary Table 3A), a P1 type nucleoside transporter was identified. Besides their role in providing essential nutrients to the parasite, some nucleoside transporters also mediate the uptake of widely employed anti-trypanosomal drugs such as pentamidine and melarsoprol [52]. It has been reported that pentamidine accumulated within the mitochondrion of susceptible compared to drug-resistant L. donovani parasites, implying that mt membranes must contain appropriate transporters [53]. A number of putative mt membrane proteins have also been identified in the plasma membrane proteome of T. brucei bloodstream form (BF) [18], and based on our results the plasma membranes fractions often are contaminated by membranes from the mitochondria. Therefore further investigations are required for definitive assignment of this group of proteins.
The protein sequences of the 37 proteins with unknown function (Supplementary Table 3B) and assigned to mt membrane with moderate confidence were subjected to motif searches against Pfam, NCBI, and InterPro databases. Putative domain(s) were identified for nine proteins, whereas the remainder had no recognizable motif (Supplementary Table 3B). Some motifs have a clear functional implication, for example Tb11.1390 has an iron sulfur binding domain, suggesting a possible function of this protein in iron–sulfur (Fe-S) clusters. Fe-S proteins are important cofactors for proteins that are involved in mt electron transport as well as in iron homeostasis. Tb09.211.1220 possesses a pentatricopeptide repeat (PPR). Mt PPR-proteins are essential for oxidative phosphorylation, some of them are required for the stabilization of mt rRNAs, and are associated with mt membranes [54]. Additionally, 29 hypothetical proteins can be assigned to mt membrane with low confidence (Supplementary Table 3B), based on identification in membrane fractions, presence of TMD and/or positive GRAVY score, and low confidence assignment to mitochondria in our earlier report [3].
A comprehensive analysis of the composition of T. brucei mt membranes was performed. All evident protein components of T. brucei mt complex II, III, IV and V [41,42,55–57] were compiled to assess the coverage of the protein detection, especially for the mt IM. The data suggest 46 proteins are present in these complexes. We identified 30 (~65%) of these proteins in our membrane fraction analyses (Supplementary Table 5), suggesting while coverage was high, some mt membrane proteins were lost during the purification procedures, or they were present but not detected by MS perhaps due to their small size.
In comparison to mt IM from mouse liver [20], we identified 25 of 35 (71%) proteins that have at least one TMD and/or positive GRAVY score and show homology to unique T. brucei proteins. Of the remaining 10 proteins, 2 were not assigned to mitochondrion in our previous study [3], and the other 8 were not detected by MS, suggesting that they may localize to other cellular compartments in T. brucei. Compared to yeast mt OM proteins [21], only 18 homologous proteins in T. brucei have at least one TMD and/or positive GRAVY score. Of these, 7 (39%) were identified in our membrane preparation and assigned to mt membranes, 5 were detected only in the whole cell fraction [3], and the other 6 were not identified in our MS analyses. In T. brucei mt OM proteins are probably too diverged to be recognized as homologs, which is supported by the fact that a TOM complex is yet to be identified in Trypanosomatids, and that VDAC is currently the only mt OM protein characterized in T. brucei [48,49]. Thus, OM assignment in T. brucei is most likely under-represented.
In a previous proteomic study, a total of 2897 proteins have been identified and 880 specific proteins were assigned to the mitochondrion with progressively diminishing stringent criteria for 401, 196, and 283 proteins, respectively [3]. Here, we identified 146 additional proteins, indicating that all together 3043 proteins represent the cellular proteome of T. brucei PF. Taking into account our results, we assigned 945 specific proteins to the mitochondrion; 434 proteins with high, 228 with moderate and 283 with low confidence. Of the 945, 178 proteins with at least one TMD and/or positive GRAVY score were identified in the TMM and MIM fractions and assigned to mt membranes; 69 with high confidence, 80 and 29 with moderate and low confidence, respectively that would require further follow up for definitive assignment. The results showed that a large proportion of proteins assigned to mt membranes at various level of confidence have unknown function, thus reflecting the complexity, and structural and functional divergence of biological processes in trypanosome mitochondria.
4. CONCLUDING REMARKS
Defining membrane proteomes is not only key to understanding the role of membrane proteins in fundamental biological processes but also is of significant interest for drug discovery. This first proteomic analysis of the mt membranes of T. brucei represents an important step towards determining the complete set of proteins residing within membranes. It is likely that the current set covers a substantial proportion of the mt membrane proteome, and it will serve as a framework for future studies to better understand the mt membrane physiology of trypanosomatids parasites.
Supplementary Material
Acknowledgments
We are grateful to Alena Zíková and Rachel A. Dalley for providing us with transgenic parasites; to Steve L. Hajduk (University of Georgia, Athens), Minu Chaudhuri (Meharry Medical College, Nashville), and Andre Schneider (University of Berne, Switzerland) for antibodies. This work was supported by NIH grant AI065935 to KS. Research conducted using equipment made possible by Economic Development Administration - US Department of Commerce and the M.J. Murdock Charitable Trust.
Abbreviations
- IM
inner membrane
- IMS
intermembrane space
- MIM fraction
mitochondrial Inner Membrane fraction
- mt
mitochondrial
- OM
outer membrane
- PF
procyclic form
- TMD
α-helical transmembrane domain
- TMM fraction
Total mitochondrial Membranes fraction
REFERENCES
- 1.Stuart K. Mitochondrial DNA of an African trypanosome. J. Cell. Biochem. 1983;23:13–26. doi: 10.1002/jcb.240230103. [DOI] [PubMed] [Google Scholar]
- 2.Stuart K, Feagin JE. Mitochondrial DNA of kinetoplastids. Int. Rev. Cytol. 1992;141:65–88. doi: 10.1016/s0074-7696(08)62063-x. [DOI] [PubMed] [Google Scholar]
- 3.Panigrahi AK, Ogata Y, Zikova A, Anupama A, et al. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics. 2009;9(2):434–450. doi: 10.1002/pmic.200800477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mokranjac D, Neupert W. Thirty years of protein translocation into mitochondria: Unexpectedly complex and still puzzling. Biochim. Biophys. Acta. 2008 doi: 10.1016/j.bbamcr.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 5.Schneider A, Bursac D, Lithgow T. The direct route: a simplified pathway for protein import into the mitochondrion of trypanosomes. Trends Cell Biol. 2008;18(1):12–18. doi: 10.1016/j.tcb.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Van Hellemond JJ, Opperdoes FR, Tielens AG. The extraordinary mitochondrion and unusual citric acid cycle in Trypanosoma brucei. Biochem Soc Trans. 2005;33(Pt 5):967–971. doi: 10.1042/BST20050967. [DOI] [PubMed] [Google Scholar]
- 7.Esseiva AC, Chanez AL, Bochud-Allemann N, Martinou JC, et al. Temporal dissection of Bax-induced events leading to fission of the single mitochondrion in Trypanosoma brucei. EMBO Rep. 2004;5(3):268–273. doi: 10.1038/sj.embor.7400095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stuart K, Panigrahi AK, Salavati R. In: RNA Editing: Frontiers in Molecular Biology. Bass BL, editor. Oxford: Oxford University Press; 2000. pp. 1–19. [Google Scholar]
- 9.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, et al. The genome of the African trypanosome, Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 10.El-Sayed NMA, Myler PJ, Bartholomeu D, Nilsson D, et al. The genome sequence of Trypanosoma cruzi, etiological agent of Chagas' disease. Science. 2005;309(5733):409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 11.Ivens AC, Peacock CS, Worthey EA, Murphy L, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309(5733):436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vertommen D, Van RJ, Szikora JP, Rider MH, et al. Differential expression of glycosomal and mitochondrial proteins in the two major life-cycle stages of Trypanosoma brucei. Mol Biochem Parasitol. 2008;158(2):189–201. doi: 10.1016/j.molbiopara.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Jones A, Faldas A, Foucher A, Hunt E, et al. Visualisation and analysis of proteomic data from the procyclic form of Trypanosoma brucei. Proteomics. 2006;6(1):259–267. doi: 10.1002/pmic.200500119. [DOI] [PubMed] [Google Scholar]
- 14.Colasante C, Ellis M, Ruppert T, Voncken F. Comparative proteomics of glycosomes from bloodstream form and procyclic culture form Trypanosoma brucei brucei. Proteomics. 2006;6(11):3275–3293. doi: 10.1002/pmic.200500668. [DOI] [PubMed] [Google Scholar]
- 15.Ferella M, Nilsson D, Darban H, Rodrigues C, et al. Proteomics in Trypanosoma cruzi--localization of novel proteins to various organelles. Proteomics. 2008;8(13):2735–2749. doi: 10.1002/pmic.200700940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNicoll F, Drummelsmith J, Muller M, Madore E, et al. A combined proteomic and transcriptomic approach to the study of stage differentiation in Leishmania infantum. Proteomics. 2006;6(12):3567–3581. doi: 10.1002/pmic.200500853. [DOI] [PubMed] [Google Scholar]
- 17.Foucher AL, McIntosh A, Douce G, Wastling J, et al. A proteomic analysis of arsenical drug resistance in Trypanosoma brucei. Proteomics. 2006;6(9):2726–2732. doi: 10.1002/pmic.200500419. [DOI] [PubMed] [Google Scholar]
- 18.Bridges DJ, Pitt AR, Hanrahan O, Brennan K, et al. Characterisation of the plasma membrane subproteome of bloodstream form Trypanosoma brucei. Proteomics. 2008;8(1):83–99. doi: 10.1002/pmic.200700607. [DOI] [PubMed] [Google Scholar]
- 19.Broadhead R, Dawe HR, Farr H, Griffiths S, et al. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 2006;440(7081):224–227. doi: 10.1038/nature04541. [DOI] [PubMed] [Google Scholar]
- 20.Da CS, Xenarios I, Langridge J, Vilbois F, et al. Proteomic analysis of the mouse liver mitochondrial inner membrane. J Biol Chem. 2003;278(42):41566–41571. doi: 10.1074/jbc.M304940200. [DOI] [PubMed] [Google Scholar]
- 21.Zahedi RP, Sickmann A, Boehm AM, Winkler C, et al. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol Biol Cell. 2006;17(3):1436–1450. doi: 10.1091/mbc.E05-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitt S, Prokisch H, Schlunck T, Camp DG, et al. Proteome analysis of mitochondrial outer membrane from Neurospora crassa. Proteomics. 2006;6(1):72–80. doi: 10.1002/pmic.200402084. [DOI] [PubMed] [Google Scholar]
- 23.Russell RB, Eggleston DS. New roles for structure in biology and drug discovery. Nat Struct Biol. 2000;7 Suppl:928-30:928–930. doi: 10.1038/80691. [DOI] [PubMed] [Google Scholar]
- 24.Wirtz E, Leal S, Ochatt C, Cross GAM. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 25.Panigrahi AK, Schnaufer A, Stuart KD. In: Methods in Enzymology. Gott JM, editor. Elsevier Inc.; 2007. pp. 3–24. [DOI] [PubMed] [Google Scholar]
- 26.Zahedi RP, Sickmann A, Boehm AM, Winkler C, et al. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol Biol Cell. 2006;17(3):1436–1450. doi: 10.1091/mbc.E05-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pon L, Moll T, Vestweber D, Marshallsay B, et al. Protein import into mitochondria: ATP-dependent protein translocation activity in a submitochondrial fraction enriched in membrane contact sites and specific proteins. J Cell Biol. 1989;109(6 Pt 1):2603–2616. doi: 10.1083/jcb.109.6.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaudhuri M, Ajayi W, Temple S, Hill GC. Identification and partial purification of a stage-specific 33 kDa mitochondrial protein as the alternative oxidase of the Trypanosoma brucei brucei bloodstream trypomastigotes. J Eukaryot Microbiol. 1995;42(5):467–472. doi: 10.1111/j.1550-7408.1995.tb05892.x. [DOI] [PubMed] [Google Scholar]
- 29.Panigrahi AK, Zikova A, Dalley RA, Acestor N, et al. Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol. Cell. Proteomics. 2008;7(3):534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Priest JW, Hajduk SL. Trypanosoma brucei cytochrome c1 is imported into mitochondria along an unusual pathway. J Biol Chem. 2003;278(17):15084–15094. doi: 10.1074/jbc.M212956200. [DOI] [PubMed] [Google Scholar]
- 31.Rinehart J, Horn EK, Wei D, Soll D, et al. Non-canonical eukaryotic glutaminyl- and glutamyl-tRNA synthetases form mitochondrial aminoacyl-tRNA in Trypanosoma brucei. J Biol Chem. 2004;279(2):1161–1166. doi: 10.1074/jbc.M310100200. [DOI] [PubMed] [Google Scholar]
- 32.Parker HL, Hill T, Alexander K, Murphy NB, et al. Three genes and two isozymes: Gene conversion and the compartmentalization and expression of the phosphoglycerate kinases of Trypanosoma (Nannomonas) congolense. Mol. Biochem. Parasitol. 1995;69(2):269–279. doi: 10.1016/0166-6851(94)00208-5. [DOI] [PubMed] [Google Scholar]
- 33.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 34.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 35.Moller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17(7):646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 36.Jensen BC, Kifer CT, Brekken DL, Randall AC, et al. Characterization of protein kinase CK2 from Trypanosoma brucei. Mol. Biochem. Parasitol. 2006;151(1):28–40. doi: 10.1016/j.molbiopara.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panigrahi AK, Schnaufer A, Ernst NL, Wang B, et al. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9(4):484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gentle IE, Perry AJ, Alcock FH, Likic VA, et al. Conserved motifs reveal details of ancestry and structure in the small TIM chaperones of the mitochondrial intermembrane space. Mol Biol Evol. 2007;24(5):1149–1160. doi: 10.1093/molbev/msm031. [DOI] [PubMed] [Google Scholar]
- 39.Allen JW, Ferguson SJ, Ginger ML. Distinctive biochemistry in the trypanosome mitochondrial intermembrane space suggests a model for stepwise evolution of the MIA pathway for import of cysteine-rich proteins. FEBS Lett. 2008;582(19):2817–2825. doi: 10.1016/j.febslet.2008.07.015. %20. [DOI] [PubMed] [Google Scholar]
- 40.nur-Mills M, Tal M, Pines O. Dual targeted mitochondrial proteins are characterized by lower MTS parameters and total net charge. PLoS ONE. 2008;3(5):e2161. doi: 10.1371/journal.pone.0002161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zikova A, Schnaufer A, Dalley RA, Panigrahi AK, et al. The F(0)F(1)-ATP synthase complex contains novel subunits and is essential for procyclic Trypanosoma brucei. PLoS Pathog. 2009;5(5):e1000436. doi: 10.1371/journal.ppat.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zikova A, Panigrahi AK, Uboldi AD, Dalley RA, et al. Structural and functional association of Trypanosoma brucei MIX protein with cytochrome c oxidase complex. Eukaryot Cell. 2008;7(11):1994–2003. doi: 10.1128/EC.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zikova A, Panigrahi AK, Dalley RA, Acestor N, et al. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol. Cell. Proteomics. 2008;7(7):1286–1296. doi: 10.1074/mcp.M700490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riviere L, van Weelden SW, Glass P, Vegh P, et al. Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei. Gene identification and role in carbohydrate metabolism. J Biol Chem. 2004;279(44):45337–45346. doi: 10.1074/jbc.M407513200. [DOI] [PubMed] [Google Scholar]
- 45.Heise N, Opperdoes FR. Localisation of a 3-hydroxy-3-methylglutaryl-coenzyme A reductase in the mitochondrial matrix of Trypanosoma brucei procyclics. Z. Naturforsch. [C] 2000;55(5–6):473–477. doi: 10.1515/znc-2000-5-626. [DOI] [PubMed] [Google Scholar]
- 46.Haferkamp I. The diverse members of the mitochondrial carrier family in plants. FEBS Lett. 2007;581(12):2375–2379. doi: 10.1016/j.febslet.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 47.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. 723–749. [DOI] [PubMed] [Google Scholar]
- 48.Singha UK, Sharma S, Chaudhuri M. Down regulation of mitochondrial Porin inhibits cell growth and alters respiratory phenotype in Trypanosoma brucei. Eukaryot. Cell. 2009 Jul 17; doi: 10.1128/EC.00132-09. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pusnik M, Charriere F, Maser P, Waller RF, et al. The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol Biol Evol. 2009;26(3):671–680. doi: 10.1093/molbev/msn288. [DOI] [PubMed] [Google Scholar]
- 50.Major T, von JB, Ruppert T, Mogk A, et al. Proteomic analysis of mitochondrial protein turnover: identification of novel substrate proteins of the matrix protease pim1. Mol Cell Biol. 2006;26(3):762–776. doi: 10.1128/MCB.26.3.762-776.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sickmann A, Reinders J, Wagner Y, Joppich C, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A. 2003;100(23):13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carter NS, Berger BJ, Fairlamb AH. Uptake of diamidine drugs by the P2 nucleoside transporter in melarsen-sensitive and -resistant Trypanosoma brucei brucei. J Biol Chem. 1995;270(47):28153–28157. doi: 10.1074/jbc.270.47.28153. [DOI] [PubMed] [Google Scholar]
- 53.Mukherjee A, Padmanabhan PK, Sahani MH, Barrett MP, et al. Roles for mitochondria in pentamidine susceptibility and resistance in Leishmania donovani. Mol. Biochem. Parasitol. 2006;145(1):1–10. doi: 10.1016/j.molbiopara.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 54.Pusnik M, Small I, Read LK, Fabbro T, et al. Pentatricopeptide Repeat Proteins in Trypanosoma brucei Function in Mitochondrial Ribosomes. Mol Cell Biol. 2007;27(19):6876–6888. doi: 10.1128/MCB.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allen JW, Ginger ML, Ferguson SJ. Maturation of the unusual single-cysteine (XXXCH) mitochondrial c-type cytochromes found in trypanosomatids must occur through a novel biogenesis pathway. Biochem J. 2004;383(Pt. 3):537–542. doi: 10.1042/BJ20040832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horvath A, Kingan TG, Maslov DA. Detection of the mitochondrially encoded cytochrome c oxidase subunit I in the trypanosomatid protozoan Leishmania tarentolae. Evidence for translation of unedited mRNA in the kinetoplast. J Biol Chem. 2000;275(22):17160–17165. doi: 10.1074/jbc.M907246199. [DOI] [PubMed] [Google Scholar]
- 57.Speijer D, Muijsers AO, Dekker H, de HA, et al. Purification and characterization of cytochrome c oxidase from the insect trypanosomatid Crithidia fasciculata. Mol Biochem Parasitol. 1996;79(1):47–59. doi: 10.1016/0166-6851(96)02648-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.