Abstract
By virtue of its unique electrochemical properties, iron makes an ideal redox active cofactor for many biologic processes. In addition to its important role in respiration, central metabolism, nitrogen fixation, and photosynthesis, iron also is used as a sensor of cellular redox status. Iron-based sensors incorporate Fe-S clusters, heme, and mononuclear iron sites to act as switches to control protein activity in response to changes in cellular redox balance. Here we provide an overview of iron-based redox sensor proteins, in both prokaryotes and eukaryotes, that have been characterized at the biochemical level. Although this review emphasizes redox sensors containing Fe-S clusters, proteins that use heme or novel iron sites also are discussed. Antioxid. Redox Signal. 11, 1029–1046.
Introduction
The need for sensors of cellular redox status is critical in all organisms. Aerobic organisms produce an appreciable amount of reactive oxygen species (ROS) as part of respiration, necessitating careful monitoring of oxidative stress and concurrent regulation of antioxidant systems (76). Obligate anaerobes must activate stress-response pathways to survive exposure to toxic oxygen or ROS such as H2O2. Although the early Earth is thought to have been anaerobic, the development of photosynthesis gradually increased oxygen production to present-day levels. In response, Archaea, the contemporary "proxies" for progenitor, anaerobic life forms, evolved with peroxide-detoxifying miniferritins (or Dps proteins) that use Fe chemistry to detoxify H2O2 by making ferric oxy minerals (15, 20, 56, 97, 106, 110, 164). Facultative anaerobes must carefully balance aerobic, microaerobic, and anaerobic metabolism in response to changing oxygen concentrations. Furthermore, maintenance of a specific narrow window of redox potential within the cell or within a cellular organelle (in the case of eukaryotes) is essential for proper function of many cellular pathways.
To provide coordinated regulation of antioxidant systems and redox-dependent pathways in response to changes in the cellular redox environment requires a moiety that is exquisitely sensitive to such changes. In many redox-sensing proteins, reactive protein thiol residues are used for this purpose (84). Selective oxidation, reduction, or chemical modification of these sensor thiols results in a change in protein activity and signal transduction in response to redox fluctuations. Protein thiols are particularly sensitive to H2O2 and nitric oxide (NO) and are often used as sensors of H2O2 in vivo (84).
However, some transition metals also can serve this purpose if their oxidation state is sensitive to physiologically relevant redox fluctuations. Oxidation or reduction of some transition metals will alter their coordination number and preferred ligands, providing a potential mechanism for transducing the redox signal to a protein frame. In particular, iron is used for a variety of redox-related functions. Iron is the fourth most abundant element of the earth's crust. As a first-row transition element, iron has incompletely filled d orbitals and can form a range of oxidation states. The most common oxidation states of iron are II (d6) and III (d5) (referred to as ferrous and ferric iron ions, respectively), although higher oxidation states are seen as reaction intermediates during the catalytic cycle of some iron enzymes. By virtue of their abundance and redox properties, iron ions were readily incorporated for use in biologic processes early in evolution and today play a key role in biologic pathways such as respiration, photosynthesis, and nitrogen fixation.
Iron also reacts with oxygen and ROS such as superoxide and H2O2. O2 is in the triplet spin state at its lowest energy level and is therefore spin restricted from accepting electrons as pairs. Iron and other transition metals can donate or accept single electrons and overcome this spin restriction to oxidize O2. Ferrous iron can also directly reduce H2O2 to generate hydroxyl anion and the highly reactive hydroxyl radical, a reaction first described by Fenton >100 years ago. Reactions between iron and oxygen metabolites can be deleterious to organisms, especially when they result in hydroxyl radical production and subsequent damage to nucleic acids, proteins, and lipids. However, because of the sensitivity of iron to oxygen and ROS, iron has been co-opted for use as a sensor of such species. When used as a sensor, iron or an iron cofactor is carefully coordinated to a polypeptide chain to tune the reactivity of the metal center. The protein importantly provides a structural frame for translating the controlled iron-mediated redox reaction into a biochemical effect: a change in protein activity.
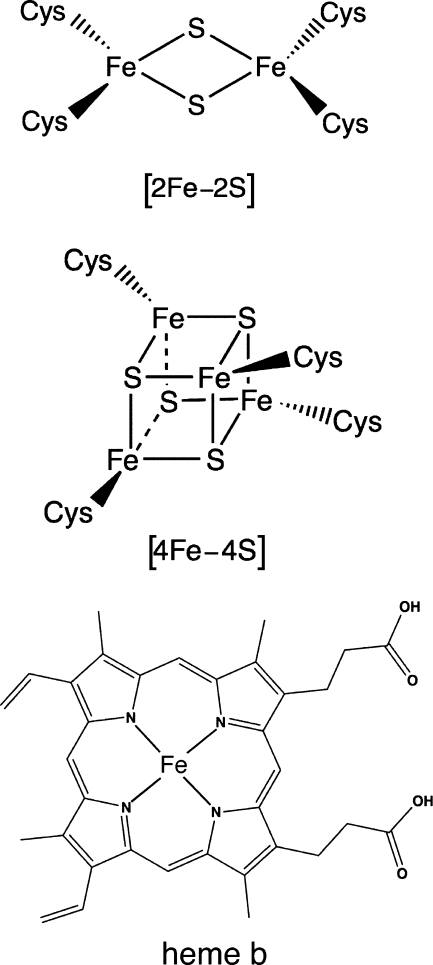
Before delving into the myriad examples of iron-based redox sensors, we must briefly define the various iron cofactors found in protein active sites (Fig. 1). Iron can be directly coordinated by amino acids in the polypeptide chain, forming mono- or dinuclear iron centers. The ligands for iron vary, based on iron oxidation state. Ferrous iron (Fe2+) is a borderline soft Lewis acid that prefers borderline soft-base ligands such as sulfur and nitrogen (from His, Cys, and Met amino acid side chains, for example). In contrast, ferric iron (Fe3+) is a hard Lewis acid and prefers hard-base ligands such as oxygen (found in Glu and Asp side chains, for example). In addition to the relatively simple iron-only centers, cells also can synthesize Fe-S cofactors that contain iron and sulfide organized into planar [2Fe-2S] or cuboidal [4Fe-4S] clusters. Iron is also incorporated into porphyrin structures (especially protoporphyrin IX) to form heme. Further discussion of how these various iron cofactors are bound to protein active sites is given in each subsequent section. It is also worth mentioning that iron can be incorporated into complex mixed metal clusters in a number of enzymes, although little is known about the potential role of these mixed metal clusters in redox sensing.
FIG. 1.
Iron cofactors commonly used in biology. From top to bottom, heme b, [2Fe-2S] cluster, [4Fe-4S] cluster, and heme b.
We focus this review primarily on Fe-S clusters as redox sensors, because they have been extensively studied, and, to a lesser degree, on recently characterized heme-based sensors of oxygen and nitric oxide (NO). We also briefly highlight some interesting discoveries on the use of mononuclear iron centers for redox sensing. This review is not meant to be a comprehensive list of all iron-containing redox sensors. Rather, we attempt to highlight interesting examples from each class of sensor with a focus on the redox-sensing mechanism of the iron cofactor and the subsequent transduction of signal to the accompanying polypeptide frame surrounding the metal center.
Fe-S Clusters as Redox Sensors
Unique properties of Fe-S clusters
Fe-S clusters are complex protein cofactors consisting of iron and inorganic sulfide. Fe-S clusters are typically (but not exclusively) bound to cysteinyl sulfur in protein active sites (104). The most common Fe-S clusters found in nature are tetranuclear [4Fe-4S] clusters with low potentials (approximately −300 mV) that undergo the [4Fe-4S]2+,+ redox transition. However, [2Fe-2S] clusters also are abundant, and a variety of more complex Fe-S clusters, such as the [8Fe-7S] P cluster of nitrogenase, can be found in some enzymes (11). Under physiologic conditions, Fe-S clusters have a hypothetical redox potential window of −600 to +500 mV, and a survey of Fe-S cluster proteins reveals that most of this redox potential range is covered by one or more Fe-S proteins. In the case of redox-sensor proteins, the Fe-S cluster redox potentials must be carefully tuned to elicit a response to specific oxidants at a physiologically relevant threshold of oxidant concentration. In view of this critical property of Fe-S clusters, we briefly discuss the various factors that influence protein-bound Fe-S cluster redox potentials [thoroughly reviewed in (142)].
Fe-S cluster ligands have a profound effect on cluster redox potential. For example, in the binuclear [2Fe-2S] Reiske proteins, two cysteinyl sulfur ligands are replaced by imidazolyl groups from two His residues (61). This change to more-neutral cluster ligands increases the redox potential of Reiske Fe-S proteins by ∼100 mV relative to [2Fe-2S] ferredoxins with all Cys cluster ligation. The extent of NH-S hydrogen bonding between sulfur ligands of the cluster and protein side chains or the protein backbone also can influence cluster redox potential. In general, an increased level of hydrogen bonding increases the reduction potential for a given cluster type, making it more difficult to oxidize that cluster (i.e., stabilizes the reduced form of the cluster). This effect stems from hydrogen bond competition with sulfur-to-iron charge transfer and an increase in the negative charge on the sulfur ligands (5, 50). Although a direct correlation between the number of hydrogen bonds and absolute change in cluster redox potential is not always apparent, in at least one case, a large change in redox potential between two highly similar [2Fe-2S] clusters has been attributed to the relative flipping of one main-chain NH-S hydrogen bond between the two protein structures (98).
Electrostatic interactions also play a role in tuning redox potential. In particular, the overall protein fold, especially the orientation of amide dipoles relative to the cluster, can have a profound effect. The proximity of charged amino acid side chains to the cluster also will exert electrostatic effects. Finally, the level of cluster solvation or solvent exposure is a critical variable tuned by different proteins to control Fe-S cluster redox potential. One of the best-characterized examples of solvent effects emerges from comparison studies between high-potential iron-sulfur proteins (HiPIPs) and low-potential bacterial ferredoxins (Fds). In both enzymes, the initial status of the cluster is [4Fe-4S]2+, but HiPIP clusters oxidize to the [4Fe-4S]3+ state with reduction potentials of +100 to +400 mV, whereas Fd clusters reduce to the [4Fe-4S]+ state with reduction potentials of −300 mV or less. Structural analysis indicates that the HiPiP cluster is buried within the protein core and is largely solvent inaccessible, whereas the Fd cluster is much closer to the protein surface and is partially solvent exposed (138).
Careful monitoring of the covalency of the Fe-S bonds in each type of protein indicates decreasing covalency in the cluster of Fd relative to HiPIP. This effect was recently shown to be due to hydrogen bonding between H2O and the cluster in Fd (cluster solvation) (32). The same study demonstrated that the HiPIP cluster undergoes a similar decrease in Fe-S bond covalency if HiPIP is partially unfolded, exposing the cluster to solvent (32). Thus, controlling the solvation of a particular cluster within the protein fold can have dramatic effects on cluster redox potential, an idea that is certainly relevant in the context of redox sensing.
Transcriptional regulators
In this section, we examine several transcription factors that regulate gene expression in response to oxidation or reduction of an Fe-S cluster. By poising the Fe-S cluster redox potential to respond to a specific oxidant or reductant, these transcription factors are able to mediate the appropriate transcriptional response in the cell.
[2Fe-2S] SoxR and superoxide
SoxR is a member of the MerR family of transcriptional regulators. The MerR family has been shown to alter transcription at a particular subgroup of bacterial promoters that contain an unusually long spacer region between the −35 and −10 RNA polymerase (RNAP) binding sites. Such promoters are transcriptionally inactive because of the suboptimal spacing of the RNAP binding sites, such that they are offset on different faces of the DNA double helix. Once a MerR protein is activated, it initiates a conformational change in the DNA, resulting in helix unwinding, to place the RNAP binding sites in the correct spatial orientation for transcription initiation (recently reviewed in ref. 16). MerR proteins have been shown to interact directly with RNAP and may help stabilize RNAP interaction with the suboptimal promoter (19, 89). MerR family members are often used to regulate genes involved in the homeostasis or detoxification of metals such as Hg2+, Cu+, and Zn2+ and are typically activated by binding of metal to the variable C-terminal region of the protein. MerR family members typically occupy their binding sites between the −10 and −35 elements, regardless of the activating signal. Signal binding serves as an allosteric switch to activate the DNA-unwinding activity of the proteins.
In the case of SoxR, a stable [2Fe-2S]+/2+ cluster is bound to each 17-kDa. monomer of the SoxR dimer (66, 166). Regardless of cluster occupancy or oxidation state, the SoxR dimer displays a high affinity for its main target, the soxS promoter (67). SoxS is a concentration-dependent transcriptional activator that regulates a host of genes involved in oxidative stress defense once it is induced by SoxR (57, 154). Although both apoSoxR and the reduced [2Fe-2S]+ SoxR bind to DNA, neither is able to enhance transcription of soxS. In contrast, the oxidized [2Fe-2S]2+ SoxR stimulates transcription of soxS up to 100-fold. This effect is readily reversed by reduction of the [2Fe-2S]2+ cluster back to the [2Fe-2S]+ state (36, 46). It was established in vivo that SoxR is oxidized by superoxide-generating agents such as paraquat and menadione, resulting in activation of the soxS promoter. The oxidation of the SoxR [2Fe-2S]+ cluster during exposure to oxidants was confirmed in vivo in Escherichia coli by using whole-cell electron paramagnetic resonance (EPR) spectroscopy (35). EPR can be used to quantify levels of the paramagnetic [2Fe-2S]+ SoxR cluster and to monitor cluster oxidation to the EPR-silent diamagnetic [2Fe-2S]2+ form. The EPR-detectable [2Fe-2S]+ SoxR cluster disappears within 2 min of oxidant addition and quickly reappears after the removal of the oxidative stress. Complete disassembly of the SoxR cluster was ruled out by adding the strong reductant dithionite to permeabilized cells after paraquat treatment. Under these conditions, the [2Fe-2S]+ form of the SoxR cluster was immediately detected with EPR, indicating simple reduction of existing [2Fe-2S]2+ SoxR rather than de novo construction of new cluster (35).
A specific in vivo oxidant for [2Fe-2S]+ SoxR has not been clearly established. In vitro, the SoxR Fe-S cluster from various organisms has a redox potential of around −290 mV. It has been suggested that the SoxR cluster may be directly oxidized by the superoxide anion or that SoxR may become oxidized as the cellular NADPH/NADP+ ratio shifts during oxidative stress. In the second scenario, the NADPH/NADP+ pool (with a redox potential estimated to be −340 mV in vivo) provides the reducing power to keep the SoxR cluster in the reduced state under normal conditions, but depletion of NADPH under oxidative stress promotes SoxR oxidation. This model is further supported by the recent discovery of specific proteins, RseC and the RsxABCDGE system, that are required to maintain reduced SoxR in vivo (87, 88). The Rsx proteins are homologous to the NADPH-dependent enzymes involved in nitrogen fixation in other bacteria. However, the low redox potential of the SoxR cluster would theoretically allow other cellular constituents, such as glutathione, actually to oxidize the SoxR cluster, even under nonstress conditions (although Fe-S clusters are unlikely to be in equilibrium with GSH:GSSG in vivo).
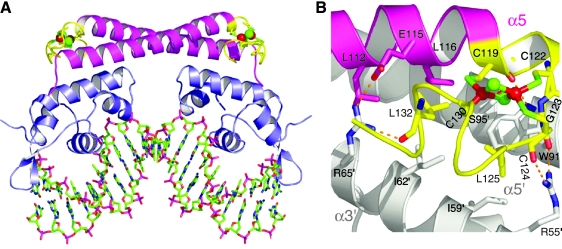
Recently this conundrum was addressed by measuring the redox potential of the SoxR cluster while SoxR is bound to DNA at its target promoter (55). This elegant study revealed that binding of SoxR to DNA increases the cluster redox potential to approximately +200 mV, an increase of 490 mV relative to the cluster redox potential of free SoxR. The increased redox potential ensures that the DNA-bound form of SoxR is kept in the inactive, reduced form under most cellular conditions. The recently published crystal structure of oxidized SoxR protein bound to DNA shows that the SoxR [2Fe-2S]2+ cluster is solvent exposed, even when SoxR is bound to DNA (Fig. 2) (162). The solvent exposure of the SoxR cluster would allow a quick response to oxidants in vivo and rapid reduction on alleviation of oxidative stress.
FIG. 2.
(A) Crystal structure of oxidized [2Fe-2S] SoxR dimer bound to DNA (stick representation). For SoxR, blue is the DNA-binding domain, magenta is the dimerization helix, and yellow is the Fe-S cluster-binding domain with red spheres as iron and green spheres as sulfur. (B) A close-up showing the interactions between the Fe-S cluster-binding domain of one SoxR monomer and the DNA-binding domain of the other monomer (shown in white for clarity). (Reproduced with permission from ref. 160). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
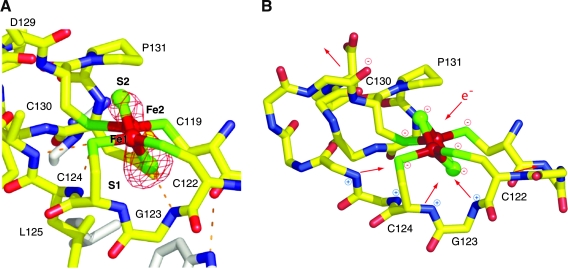
The SoxR-DNA structure also revealed that the [2Fe-2S]2+ cluster of SoxR is in an “asymmetric environment” (162). The lower (S1) bridging sulfur atom is surrounded by three partially positively charged main-chain amides and is hydrogen bonded to one of them. In contrast, the upper (S2) bridging sulfur atom is close to a partially negatively charged carbonyl oxygen atom (Fig. 3A). Both iron atoms and the S2 bridging sulfur atom are exposed to the solvent, allowing them to react with cellular oxidants. Because of the asymmetric protein charge distribution around the SoxR cluster, cluster reduction may transduce a protein conformational change (relative to active, oxidized SoxR cluster) by repulsing the carbonyl oxygen near the S2 sulfur while attracting the positively charged amides from the main chain near the S1 sulfur (Fig. 3B). These movements may push the C-terminal region of SoxR outward (away from the DNA) and alter the interactions between the SoxR monomers. Thus, electrostatic changes around the cluster could convey the redox switching to the larger protein frame.
FIG. 3.
(A) Protein environment surrounding the [2Fe-2S]2+ cluster of SoxR when bound to DNA. Iron and sulfur are red and green spheres, and NH-S hydrogen bonds are dashed lines. (B) Proposed model of electrostatic changes that occur around the SoxR [2Fe-2S] cluster on reduction (addition of e-). (Reproduced with permission from ref. 160). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
The [2Fe-2S] cluster of SoxR is also sensitive to reactive nitrogen species such as nitric oxide. In vivo and in vitro, the SoxR cluster can react with NO to form dinitrosyl-iron-dithiol adducts (34). Nitrosylated SoxR is able to activate transcription of the soxS target promoter to a similar degree as oxidized SoxR, suggesting that the dinitrosyl-iron-dithiol modification triggers a conformational change similar to that of cluster oxidation (108). Strangely, despite SoxR modification and SoxS upregulation, most of the SoxS regulon is not strongly activated by NO in vivo (107). The reason for this discrepancy is unclear. Possibly SoxR has not evolved to sense NO and is only nonspecifically activated by reactive nitrogen species, or other regulatory factors may modulate the downstream SoxS-mediated response to NO.
[4Fe-4S] FNR and oxygen
Facultative anaerobic bacteria such as E. coli undergo complex physiological changes in response to oxygen levels. Many of these changes are controlled at the transcriptional level by the fumarate and nitrate reduction (FNR) regulator (80, 128). FNR is a member of the CRP subfamily of helix-turn-helix transcriptional regulators, in which the HTH motif is located in the C-terminal region of the protein. The N-terminus of FNR contains a tricysteine motif (Cx2Cx5C) between C20 and C29 that is reminiscent of ferredoxin-like Fe-S cluster binding sites. This N-terminal motif and a more distal cysteine (C120) are required for FNR activity in vivo (100, 136, 140). The active, DNA-binding form of FNR is a dimer containing one [4Fe-4S]2+ cluster per monomer (82, 92). The [4Fe-4S]2+ cluster is critical for FNR function and is required for FNR dimerization, DNA binding at specific sites, and transcriptional activation of target promoters [reviewed in (83)].
Unlike SoxR, the [4Fe-4S]2+ cluster of FNR is not stable to reduction or to oxygen exposure, and FNR is present largely in the apo-form under aerobic growth conditions (119). Stabilization of the [4Fe-4S]2+ cluster under anaerobic conditions activates FNR and leads to transcriptional changes in >100 FNR target genes (80). Assembly and disassembly of the [4Fe-4S]2+ cluster controls FNR activity, and this process is fundamentally more complex than the relatively simple redox conversion of the SoxR [2Fe-2S]+/2+ cluster.
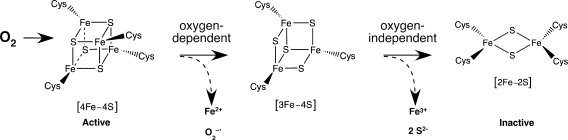
Mössbauer spectroscopy of FNR revealed that as soon as 2 min after in vitro exposure to oxygen, the [4Fe-4S]2+ cluster rapidly converts to a [2Fe-2S]2+ cluster (82). Further exposure to oxygen ultimately leads to complete disassembly of the [2Fe-2S]2+ cluster and formation of apoFNR. The switch from a cubane [4Fe-4S]2+ cluster to a planar [2Fe-2S]2+ cluster apparently leads to protein conformational changes that cause FNR to dissociate into monomers, thereby inactivating the protein. The external oxygen concentration required to inactivate half of FNR in vivo is in the range of 1–10 μM, a concentration range in which oxygen diffusion into the cell begins to outpace oxygen consumption by metabolism (10, 155).
The presence of a semistable [2Fe-2S]2+ intermediate during FNR cluster conversion may be relevant when E. coli is growing in oxygen concentrations that vary between microaerobic and fully anaerobic. Under such conditions, maintenance of the [2Fe-2S]2+ intermediate might allow more rapid activation of FNR when oxygen levels decrease to low enough levels to maintain the stable [4Fe-4S]2+ cluster. However, the in vivo conversion of the [2Fe-2S]2+ intermediate to the active [4Fe-4S]2+ cluster is not well defined. Conversion of the [2Fe-2S]2+ cluster to form the [4Fe-4S]2+ cluster or synthesis of the complete [4Fe-4S]2+ would both seem to require de novo Fe-S assembly (to generate either a [2Fe-2S] or a [4Fe-4S] cluster). It has not been established that one assembly process would necessarily be faster than another in vivo.
The mechanistic details of FNR cluster oxidation and disassembly have been the subject of intense study (24, 25, 82, 145). The most recent model to emerge is shown in Fig. 4. On exposure to oxygen, each [4Fe-4S]2+ cluster is oxidized to the [4Fe-4S]3+ state, simultaneously generating superoxide. However, the oxidized cluster is unstable and rapidly degrades to the [3Fe-4S]+ form, with the concurrent release of Fe2+. This initial oxidation reaction increases linearly with oxygen concentration and has an apparent rate constant of k1 = 250 ± 50 M/s at 25°C. The second step of FNR conversion is the oxygen-independent spontaneous decay of the [3Fe-4S]+ cluster to a [2Fe-2S]2+ cluster with the release of two S2- and one Fe3+ with an apparent rate constant of k2 = 0.008 per second. The exact oxidation state of each released iron atom has been the subject of some debate and seems to be strongly influenced by the iron chelator used to detect the released iron as well as the iron-binding potential of buffer components such as phosphate (23).
FIG. 4.
[4Fe-4S] cluster oxidation and disassembly on exposure of FNR to oxygen (see text for details).
As we have seen, organisms use both the reversible oxidation of an Fe-S cluster and the assembly/disassembly of an Fe-S cluster to translate cellular redox fluctuations into changes in gene expression. However, in some cases, transcriptional regulation may not be fast enough to protect the cell from redox changes. Next, we discuss a class of redox sensors in which the Fe-S cluster is used to modulate posttranscriptional regulation or to control enzyme activity, thereby providing a quick response to cellular redox conditions.
Posttranscriptional regulators
The [4Fe-4S]2+ cluster posttranscriptional regulator, cytosolic aconitase
Aconitase or citrate (isocitrate) hydro-lyase is an enzyme of the TCA cycle that catalyzes the isomerization of citrate to isocitrate via a cis-aconitate intermediate. Aconitase contains a [4Fe-4S]2+ cluster that is used for substrate binding and activation during the reaction cycle. Eukaryotes contain a mitochondrial aconitase (m-aconitase) dedicated to the TCA cycle. However, a distinct cytosolic aconitase (c-aconitase) was identified nearly 50 years after the initial discovery of m-aconitase. Since that time, it has become clear that the aconitase family includes TCA enzymes, mRNA regulatory proteins, and dual-function proteins. The c-aconitase uses a [4Fe-4S] cluster to sense iron availability and redox status, and our discussion focuses on this protein. Although the historical nomenclature of c-aconitase is complicated, where appropriate, we refer to the apo-protein (without its [4Fe-4S] cluster) as iron-regulatory protein 1 (IRP1) and refer to the [4Fe-4S] cluster form as c-aconitase. On loss of the [4Fe-4S] cluster, c-aconitase is converted to IRP1 and gains an additional function, mRNA regulation. IRP1 regulates mRNA by binding to a conserved iron-responsive element (IRE) encoded in the mRNA sequence. Depending on the location of the IRE structure, IRP1 binding can inhibit translation (5' UTR) or inhibit nuclease degradation (stabilize the target mRNA) (3' UTR). IRP2 is a second translational regulatory protein that is 50–60% homologous to IRP1. However, IRP2 is discussed later in the sections on heme and Fe regulation, because it lacks some of the residues necessary for binding a [4Fe-4S]2+ cluster and also lacks aconitase activity (64, 94, 95, 121, 161).
Evolutionary relations between aconitases and mRNA regulation are complex and may be dynamic, even now. Many animals, invertebrates and vertebrates, have aconitases with mRNA-binding activity. Animals have multiple IRE-containing mRNA targets for IRP1. Some of those mRNA targets are evolutionarily ancient, as in ferritin, and some are more recent, as in the heme biosynthetic enzyme aminolevulinic acid synthase and one of the transferrin receptors of higher animals (114). Invertebrates typically have only one IRP that also has aconitase activity, although Drosophila, in addition to IRP, has a second cytoplasmic aconitase (96, 167). Plants have no detectable IRE sequences or active IRP proteins, although they do contain c-aconitase (8, 117, 120). For example, an animal IRE fused to a plant ferritin mRNA was not functional in plants (86). Plants could have lost the IRP-IRE regulatory pair during the integration of plastid, nuclear, and mitochondrial genomes.
Bacterial aconitases contain Fe-S clusters and can be TCA-cycle enzymes, RNA-binding proteins, or both, depending on the specific organism. In E. coli, aconitase B (AcnB) is the most abundant aconitase and makes the largest contribution to the TCA cycle. Similar to IRP1 in higher eukaryotes, AcnB binds target mRNAs to regulate their stability (150, 158). The iron-dependent dimerization of AcnB switches it from a catalytically active dimer to a RNA-binding monomer when cellular iron levels decrease (151). E. coli also possesses a second stress-response aconitase, AcnA, which is oxidant resistant and regulated by soxS (158). In Bacillus subtilis, only one aconitase is found, regulated during sporulation and vegetative growth, which is both the TCA-cycle catalyst and an RNA-binding protein. The natural mRNA targets for B. subtilis aconitase encode a major cytochrome oxidase and several iron-uptake proteins. In vitro, the B. subtilis aconitase also recognizes the ferritin mRNA regulatory IRE structure (2).
Thus, aconitases in single-cell organisms and in higher multicelled organisms can have both RNA and catalytic function or only catalytic function, and in the case of some higher organisms, the m-aconitase of the mitochondrial TCA cycle is distinct from the RNA regulatory protein IRP1/c-aconitase. Interestingly, the nuclear encoded m-aconitase gene in animals has acquired an IRE, although the m-aconitase IRE binds IRP1 relatively weakly, contains the initiator AUG, and evolved after the ferritin IRE (38, 81, 114). In contrast, the IRP1/c-aconitase gene has no IRE (122) and does not appear to regulate its cognate mRNA. Thus, mechanisms of regulation of mitochondrial aconitase vary from no known mRNA regulation in plants, to selective regulation of the mRNA by the cognate gene product in bacteria, to coordinated regulation of the mRNA with other IRE-mRNAs (e.g., ferritin and iron-trafficking protein mRNAs) mediated by IRP1 in animals.
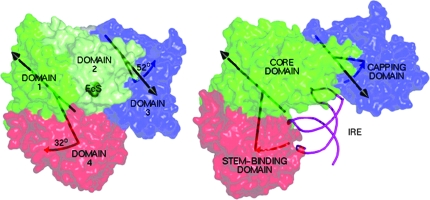
The structural basis for the dual functions of IRP1/c-aconitase has recently been advanced with crystal structures of both the [4Fe-4S] c-aconitase (37) and the IRP1 bound to the IRE of the ferritin mRNA (160). Earlier solution studies suggested that the redox switch was [4Fe-4S]2+cluster destruction by H2O2, which converted c-aconitase to IRP1 (14). The polypeptide structure of c-aconitase bound to a [4Fe-4S]2+ cluster looks very similar to mitochondrial aconitase structures (37). In contrast when the same polypeptide is bound to an IRE-RNA, the structure is very different from aconitase: three domains rather than four are found, and two of the domains are separated by large distances, rather than touching, as in c-aconitase [Fig. 5, taken from reference (160)]. The IRP1 without either Fe-S cluster or mRNA is rather disordered (14). However, alternate folding around the IRE-mRNA or [4Fe-4S]2+ cluster results in some of the same protein residues contacting mRNA in the IRP1-IRE structure or the [4Fe-4S]2+ cluster in c-aconitase (160). Given the differences between the charge and shape of the Fe-S cluster and the RNA, very large compensatory differences have to occur elsewhere in the folded polypeptide to allow this alternate folding, reflecting the plasticity of the polypeptide when either the RNA or [4Fe-4S]2+ cluster is absent (14).
FIG. 5.
A comparison of the structure of the polypeptide IRP1 bound to [4Fe-4S] (c-aconitase) on the left and bound to the IRE-RNA, noncoding regulatory structure on the right. Note the large difference in structure, indicating the possibility that the redox- sensitive “switch” occurs at or before [4Fe-4S]2+ transfer from the CIA cytosolic Fe-S assembly machinery to the apo-IRP1 polypeptide. (Reproduced with permission from ref. 158). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
The redox-sensing switch in IRP1/c-aconitase is different from that in the previously discussed FNR for two reasons. First, conversion of the [4Fe-4S]2+ cluster to a [2Fe-2S]2+ cluster triggers subunit dissociation and loss of DNA binding in FNR (92), whereas mammalian IRP1/c-aconitase is a single polypeptide. Second, for native IRP1/c-aconitase protein under some conditions, the [4Fe-4S] enzyme, [3Fe-4S] enzyme, and apo-protein all bind to target mRNA with similar affinities (170), whereas only the completely apo recombinant protein could bind mRNA in vitro (9). Possibly the actual [4Fe-4S]2+ switch occurs during IRP1/c-aconitase folding, in which a cytoplasmic chaperone that transfers the [4Fe-4S]2+ cluster to the nascent or partially folded IRP1/c-aconitase polypeptide is actually responsive to cellular redox and iron conditions. Cytoplasmic Fe-S cluster assembly and transfer machinery has been identified [for example, huNbp35 (141)] and could play a role in this process.
IRP1/c-aconitase is also phosphorylated in eukaryotes, and phosphorylation alters both Fe-S cluster binding and RNA binding (17, 129, 130, 153). How phosphorylation influences Fe-S sensing or may alter IRP1/c-aconitase folding choices is not clear. Interestingly, two IRE-containing target mRNAs regulated by IRP1 are involved in kinase/phosphatase activity or pathways (124, 125).
The selectivity of IRP2 binding to IRE RNA observed in vitro is matched by wide ranges of tissue-specific expression (39, 58, 153) and has been recently reviewed in (121) and (161). The important contributions of IRP2 to IRE-RNA regulation become apparent at the low oxygen concentrations found in most mammalian tissues that stabilize IRP1 Fe-S cluster formation, so that IRP1/c-aconitase exists as c-aconitase rather than in the mRNA-binding IRP1 form. Furthermore, IRP1 and IRP2 complexes with different IRE-RNAs show differential binding stability, indicating a possible divergence in the roles of IRP1 and IRP2 for mRNA regulation.
An additional role for c-aconitase in the cytoplasm may be balancing citrate and NADPH production for fatty acid biosynthesis in the cytoplasm (121). This hypothesis is supported by the observation that IRP1 expression levels are highest in brown fat, liver, and other tissues that carry out high levels of fatty acid synthesis. However, under conditions in which redox is perturbed (such as oxidative stress), conversion of c-aconitase to IRP1 may increase the importance of IRP1 mRNA regulation.
DNA repair enzymes and Fe-S clusters
Base excision repair (BER) glycosylases catalyze removal of chemically modified bases to repair DNA by breaking the N-glycosidic bond between the sugar-phosphate backbone of DNA and the damaged base. A plethora of BER enzymes, in organisms ranging from E. coli to humans, contain a [4Fe-4S] cluster (12). The role of the Fe-S cluster has been most well studied in the E. coli endonuclease III (EndoIII) and MutY glycosylases. Loss of the Fe-S cluster does not alter thermal stability of these enzymes and is not needed for their proper folding (52, 115). However, the Fe-S cluster is required for DNA binding and enzymatic activity of EndoIII and MutY.
In the absence of DNA, the EndoIII cluster is stable in the [4Fe-4S]2+ state, and oxidation or reduction does not occur at physiologically accessible redox potentials (27). However, on binding to DNA, the reduction potential of the [4Fe-4S]3+/2+ couple is shifted from approximately +250 mV to −30 mV, thereby stabilizing the oxidized form of the cluster (54). The crystal structures of EndoIII, both free and bound to DNA, show that on DNA binding, the Fe-S cluster binding site enters a more hydrophobic environment and is less solvent exposed (41, 90, 152). This change stems from the proximity of the cluster-binding site to the DNA-binding site of EndoIII, such that solvent would be excluded from the vicinity of the cluster on DNA binding. In a sense, DNA binding converts the Fe-S cluster in EndoIII from a low-potential ferredoxin-type to an HiPiP-type cluster (54).
Oxidation of the EndoIII Fe-S cluster also appears to increase DNA-binding activity approximately threefold over that of the reduced protein. Based on the interplay between cluster redox potential and DNA binding, a model has been proposed in which the EndoIII Fe-S cluster oxidation state is used as a sensor to control EndoIII activity (and perhaps localization) in response to oxidative stress (13, 102). Under nonstress conditions, [4Fe-4S]2+ EndoIII will bind transiently to DNA and become oxidized to [4Fe-4S]3+ EndoIII, thereby stabilizing its interaction with the DNA. The electron released during cluster oxidation will proceed through DNA by a charge-transport mechanism mediated by the DNA itself. The DNA base-pair π stack structure facilitates long-range charge transport. On encountering another [4Fe-4S]3+ EndoIII, this traveling electron will reduce the protein to [4Fe-4S]2+ EndoIII, causing it to dissociate from DNA because of the decreased DNA binding affinity on reduction. In this way, EndoIII is constantly sampling the charge-transport ability of the DNA as a measure of its integrity. On oxidative stress, DNA lesions caused by hydroxyl radicals and other ROS will disrupt the DNA-mediated charge transport. This results in accumulation of [4Fe-4S]3+ EndoIII, especially near sites where DNA lesions have occurred. Elegant studies have clearly demonstrated that such a process can occur in vitro on short stretches of DNA bound to an electrode surface and incubated with EndoIII (54).
Although compelling, this model does have some drawbacks. For instance, a number of transcription factors and other regulatory proteins bind and distort DNA in ways that might disrupt charge transport. Many of these DNA-binding proteins themselves have metal cofactors (SoxR, FNR) that may be sensitive to adventitious reduction by charge transport. It is not entirely clear how this complexity fits into the model of DNA-damage sensing by EndoIII. It also seems reasonable that EndoIII (and analogous cluster-containing glycosylases) could be activated through direct cluster oxidation by superoxide or hydrogen peroxide in the DNA-bound form. Direct oxidation by ROS would stabilize transient EndoIII interaction with DNA. Furthermore, such a high local concentration of ROS also would presumably damage DNA in the vicinity of EndoIII oxidation, resulting in colocalization of active EndoIII near damaged DNA. Because DNA binding stabilizes the oxidized form of EndoIII, [4Fe-4S]2+ EndoIII transiently interacting with DNA might be exquisitely sensitive to ROS oxidation in contrast to unbound [4Fe-4S]2+ EndoIII. Future studies to test these models further should clarify the stepwise route of in vivo activation of EndoIII.
Ferrochelatase and NO
Ferrochelatase is the terminal enzyme in the heme biosynthesis pathway in animals and catalyzes the insertion of ferrous iron into the protoporphyrin IX macrocycle to generate heme. Ferrochelatase in animals contains one [2Fe-2S]+/2+ cluster per protein monomer (29, 40). The [2Fe-2S] clusters are all Cys-ligated, with the ligands arranged in a Cx2Cx4C motif with a more distal Cys residue (26, 134). The clusters are located near the homodimer interface in the human ferrochelatase crystal structure and are approximately 15 Å from the ferrochelatase active site (165). The midpoint potential of the cluster was measured at −450 mV, and the spectral features of the cluster are quite similar to those of the 2Fe ferredoxin proteins (18, 26, 29). Importantly, loss of the [2Fe-2S] cluster abolishes ferrochelatase activity in vitro (29).
Although a great deal of evidence suggests that the [2Fe-2S] cluster is important for in vivo ferrochelatase function, its exact role is still unclear. The cluster does not appear to be necessary for iron reduction during insertion into protoporphyrin IX, nor does the cluster itself directly donate iron for heme formation. Because the clusters are present near the dimer interface of the human ferrochelatase, it is possible that the cluster helps stabilize the homodimer. The C-terminal region of mammalian ferrochelatase is the site of cluster formation and dimer interaction. In contrast, other ferrochelatase family members in most prokaryotic organisms that lack this C-terminal extension are monomeric, cluster-free enzymes. However, the C-terminal region of human ferrochelatase also contributes 18 hydrogen bonds to the dimer interface, so untangling the relative contributions of the hydrogen bonds and the Fe-S cluster to dimer formation is required to test this hypothesis. Recently, careful analysis of mutant forms of bacterial [2Fe-2S] ferrochelatases showed that loss of the [2Fe-2S] cluster increases the apparent Km of the enzyme for iron (137). Whether the decreased binding affinity for iron results from destabilization of the active homodimer or from a direct role for the cluster in iron acquisition is not clear.
More important (in relation to the topic of this review), it was shown in vivo that exposure of cells to nitric oxide or activation of NO-generating pathways leads to decreased ferrochelatase activity (42, 85). Careful in vitro studies subsequently established that physiologic levels of NO can specifically disrupt the [2Fe-2S] ferrochelatase cluster (133). The NO sensitivity of the ferrochelatase [2Fe-2S] cluster is in contrast to other eukaryotic [2Fe-2S] enzymes, such as plant ferredoxin, that do not show cluster loss on NO exposure. These results led to a model in which NO accumulation may deactivate ferrochelatase by disrupting the [2Fe-2S] cluster. NO regulation of the Fe-S cluster in IRP1/c-aconitase also has been observed, providing further evidence that NO signaling may be used to modulate global iron metabolism. Interestingly, [2Fe-2S] clusters of a limited number of bacterial ferrochelatases apparently are more stable and less sensitive than the mammalian ferrochelatase enzymes and thus may not be regulated by NO in vivo (137). A common thread for all cluster containing ferrochelatases is their obvious dependence on the available iron supply for both [2Fe-2S] cluster formation and heme formation. Thus, the cluster also may function as a sensor of cellular iron, such that heme biosynthesis is repressed under low-iron conditions or when iron trafficking is disrupted by oxidative stress, including NO exposure.
Heme-Based Redox Sensors
Heme-based sensors are found in every kingdom of life and are typically involved in sensing O2, NO, and CO (48). In most cases, heme in the inactive or resting state of the sensor protein is six-coordinate with two protein ligands binding in the proximal and distal positions relative to the prophyrin plane. One protein ligand to the heme iron often is sufficiently labile that binding of the signal ligand (e.g., O2) displaces the protein ligand and results in a structural change that regulates the activity of the protein. However, other examples (such as FixL, mentioned later) exist in which the resting heme is 5-coordinate, and ligand binding causes a rearrangement within the heme-binding pocket as it coordinates the sixth position. Binding of the signal to the heme may transduce the signal directly or may indirectly alter the redox state of the heme iron, which in turn alters protein conformation.
Heme-based sensors can be divided into four subfamilies based on the specific heme-binding domain used [for a recent review see (48)]. These heme-binding domains are themselves matched with a variety of functional domains that are responsible for transduction of the signal. For example, heme-binding domains are found fused to histidine kinase, phosphodiesterase, and DNA-binding domains. An exhaustive overview of heme-based sensors is beyond the scope of this review, and we do not discuss the globin-coupled sensor (GCS) or CooA heme sensor subfamilies (6). However, two subfamilies, the heme-PAS domain family and the H-NOX (for heme-nitric oxide/oxygen binding) family, are directly involved in redox sensing by virtue of their interactions with oxygen and nitric oxide. We briefly discuss two members of the heme-PAS domain family of sensors and their role in oxygen sensing: FixL and Ec DOS (E. coli direct oxygen sensor). We also examine the iron-dependent H-NOX enzyme, soluble guanylate cyclase (sGC), that uses heme for nitric oxide sensing.
Heme-PAS domain proteins
FixL is an oxygen sensor that regulates anaerobic (and microaerobic) respiration and nitrogen fixation in symbiotic, soil-dwelling bacteria (30, 31). In particular, nitrogen fixation must be carefully regulated in response to oxygen levels, as various nitrogenase cofactors and intermediates in nitrogen fixation are sensitive to oxidation. FixL contains a heme-binding PAS domain and a histidine kinase domain. Oxygen binding to the heme PAS domain represses the kinase activity of FixL, thereby blocking phosphorylation of the downstream regulatory partner protein, FixJ (1, 47, 105). In the absence of oxygen, the heme iron of FixL is five-coordinate, high-spin with a His residue providing the axial ligand (47, 105). On oxygen binding, the heme iron shifts to six-coordinate and low-spin (149). The dissociation rates of FixL-O2 are similar to those of the oxygen transporter myoglobin-O2, but the association rate is much slower (two orders of magnitude) for FixL (49). The decreased stability of the FixL-O2 compared with myoglobin-O2 is likely tied to its use as an oxygen sensor rather than an oxygen transporter.
Crystal structures of the oxygenated and deoxygenated PAS domain from Bradyrhizibium japonicum FixL (BjFixL) have revealed a possible mechanism for regulation of FixL activity by oxygen binding (53, 63). The heme-binding region of the FixL PAS domain is defined by a helix containing the proximal His ligand for heme attachment (helix Fα) and a distal antiparallel ß strand (Gß). These two structures are connected by a flexible loop (the so-called FG loop). Deoxygenated FixL shows extensive hydrogen bonding between the propionate side chain of the heme porphyrin and residues in both helix Fα and strand Gß. Oxygen binding to FixL causes alterations in the hydrogen-bond network in the vicinity of the heme pocket by generating a highly polarized bond to the heme iron. On oxygen binding, a nearby, highly conserved Arg220 residue (part of the Gß strand) shifts from a hydrogen bond with the heme propionate side chain to a new hydrogen-bond interaction with oxygen. The presence of the stable hydrogen-bond interaction likely allows FixL to discriminate between O2 and other diatomic gases (such as CO) that will form less polar bonds with the heme. These oxygen-induced rearrangements of the hydrogen-bond network around the heme pocket cause a 1.6 Å displacement of the FG loop and a flattening of the heme. Together, these structural changes could alter the activity of the FixL kinase domain. Unfortunately, this model has not been clearly confirmed, as a crystal structure of the full-length FixL protein (with both PAS and kinase domains) has not been forthcoming.
Ec DOS is somewhat homologous to FixL but contains two PAS domains in the N-terminus fused to a phosphodiesterase domain in the C-terminal region (127). Only one PAS domain in Ec DOS, PAS-A, actually binds heme. Ferrous heme in the PAS-A domain is in the six-coordinate low-spin state. The two axial ligands identified in the PAS-A domain are His77 and Met95 (91). Oxygen binding to ferrous Ec DOS activates the cAMP phosphodiesterase activity of the protein and consequently reduces cellular cAMP levels (126). Because cAMP is an important cellular second messenger, Ec DOS activation alters cAMP-regulated signaling pathways in an oxygen-responsive manner.
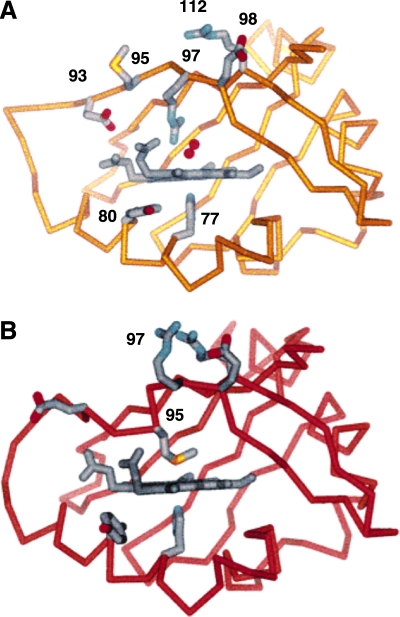
Recent crystal structures of ferrous Ec DOS PAS domain with either Met95 or oxygen as the distal ligand reveal extensive alteration around the heme-binding pocket on oxygen binding (Fig. 6) (91, 113). The Met95 ligand is displaced by oxygen, and Arg97 rotates nearly 180 degrees. Oxygen-dependent Arg97 rotation disrupts a salt bridge with Arg112 and Glu98 and brings the Arg97 into a hydrogen bond with the oxygen ligand. Displacement of the Met95 ligand causes it to swing away from the heme and toward the protein surface, resulting in distortion of the FG loop and nearby HI turn (Fig. 6). This significant structural change on oxygen binding to the PAS domain is postulated to transduce an intramolecular signal to the phosphodiesterase domain of Ec DOS, leading to its activation. Similar structural changes are also observed in oxidized ferric Ec DOS, in which the Met95 ligation to the heme iron is replaced by a water molecule. Possibly, a redox conversion of the heme iron may be involved in signal transduction. However, Ec DOS is thought to exist primarily in the reduced ferrous state in vivo, and clearly, oxygen sensing requires the heme to be in the ferrous state.
FIG. 6.
Crystal structure of EcDOS heme showing rearrangments in heme pocket on oxygen binding. (A) EcDOS-O2 (yellow) with His77 (below the heme plane) in the proximal position and O2 as two red spheres above the heme. (B) Deoxygenated EcDOS (red). Amino acids are numbered according to EcDOS sequence. (Reproduced with permission from ref. 111). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
The H-NOX family
Soluble guanylate cyclase is part of an NO-regulated signal-transduction cascade in mammals, although bacterial homologues of sGC have recently been identified and characterized. Nitric oxide synthase (NOS) is the upstream activator of sGC. The various endothelial and neuronal isoforms of NOS produce NO from oxidation of l-arginine. The released NO can then activate the downstream sGC to perpetuate the cascade. On activation by NO, sGC converts GTP to cyclic GMP (cGMP), which is a potent regulator of multiple pathways in the cell.
sGC forms an αβ heterodimer that can be present in several stoichiometries [see recent review on sGC in (116)]. The N-terminal region of the β monomer contains a regulatory heme domain, whereas both α and β monomers together form the catalytic GTP cyclase domain in the C-terminus of sGC. In the resting state, sGC contains a five-coordinate ferrous high-spin heme, with H105 from the β monomer providing the axial ligand (163, 169). In contrast to the oxygen sensors Ec DOS and FixL, when the signal ligand (NO) binds to ferrous heme in sGC, the bond between Fe2+ and the proximal His ligand appears to break (75, 143). This is a consequence of the unique properties of NO as compared with O2 or other diatomic gases. NO binding to heme tends to lengthen bonds trans to the NO (in this case, the axial His ligand) because of movement of the unpaired electron of NO into the dz2 orbital. Thus, NO binding weakens the His-Fe bond, allowing it to rupture and alter the protein conformation around the heme-binding site. Because of contribution of the His-Fe3+-OO− resonance structure to Fe2+-OO, the highly polarizable O2 ligand does not have this effect, explaining why the axial His ligand to the heme iron is not adversely perturbed by O2 binding to FixL and Ec DOS (116). Although it seems clear that formation of the NO-Fe complex with disruption of the His-Fe bond is required for sGC activation, the full mechanism is still unclear and may involve additional steps and/or other factors (such as the reaction products cGMP and pyrophosphate). For recent and thorough discussion of the various models of NO-dependent sGC activation, see (116, 123).
For both heme-PAS and H-NOX family members, reversible binding of the ligand can alter the hydrogen-bond network around the heme pocket, thereby translating the binding event into a change in protein activity. Moreover, by tuning the heme pocket, some selectivity for a particular ligand can be achieved, although many of these sensors can clearly bind a range of similar ligands in vitro. As is clear from our earlier discussion, heme can be a versatile sensor of diatomic gases, including O2 and NO. Because oxygen concentration is a critical determinant of cellular redox status and nitric oxide is a potential generator of oxidative stress, the heme-based sensor proteins play a critical role in redox sensing.
Heme as a signal ligand to control redox pathways
Because of its functional importance in respiration as part of the electron-transport chain, heme production is often closely correlated with oxygen availability. For that reason, levels of cellular heme are sometimes used as a proxy to control gene expression in response to oxygen. Several eukaryotic transcription factors directly bind heme, and their activities are controlled by cellular heme levels. Although these proteins do not use the heme to sense redox directly, cellular heme levels are responsive to redox changes.
Bach1
Bach1 is the first mammalian transcription factor found that binds heme (111). The protein is a BTB, basic leucine zipper, transcriptional repressor in which the BTB domain is N-terminal to the bZip, DNA-binding domain (71, 74, 101, 111). Bach 1 forms heterodimers with Maf proteins (MafK, MafF, MafG). The DNA target sequences for Bach1 are called MARE (Maf recognition elements) or ARE (antioxidant/phase II response elements) and are found upstream of genes regulated by iron, oxygen, or both. Some examples of MARE/ARE-regulated genes are those encoding NADPH quinone oxidoreductase, thioredoxin reductase, heme oxygenase (135), hemoglobin (74, 148), and ferritin (H and L) (33, 68, 69). Although Bach1 binds to all the MARE/ARE sequences, the binding stabilities vary over a significantly wide range, suggesting that the DNA/Bach1 interaction itself can influence regulation (68). In addition to binding the Bach1 repressors, Maf proteins also bind Nrf2 transcriptional activators (111).
Heme has both direct and indirect effects on Bach 1 that decrease Bach1 gene repression (leading to upregulation of Bach1 target genes). First, heme increases nuclear export of Bach1 proteins, so that the majority of Bach 1 is in the cytoplasm in heme-treated cells (144, 147), and less is available to bind DNA in the nucleus. Second, the Bach1-heme complex does not bind MARE/ARE DNA (68). The combination of direct molecular effects and changes in subcellular distribution of Bach1 provides increased access for Nrf2 to bind Maf proteins at MARE/ARE sequences, leading to increased transcription. Bach1 has multiple HRM (CP; cysteine-proline) motifs that contribute to heme binding (109, 111). The stoichiometry of heme binding is five, and based on mutagenesis studies, four hemes are bound in the C-terminal, B-zip domain (71), and one is in the N-terminal, BTB domain. At least three of the HRM-binding domains contribute to heme-induced nuclear export of Bach1 (146, 147). Sorting out the contributions of the individual Bach1-heme interactions to nuclear export of Bach 1, DNA binding, and/or Maf binding will help elucidate the functional role of heme in this process.
IRP1 and IRP2
Heme interacts directly with both IRP1 and IRP2 and increases protein turnover (77). This effect is resistant to iron chelators, indicating that it is heme itself, rather than iron released by heme degradation, that regulates IRP turnover (51, 99). Effects of heme binding to IRP regulators is complicated by IRP1-Fe-S cluster sensing and IRP1 and IRP2 sensing of excess environmental iron salts such as ferrous sulfate or ferric citrate (21, 59, 60, 121, 161). However, when iron chelators are used, the heme-specific effects on IRP turnover are readily observed (51). At least one heme-binding sequence in IRP2 is absent in IRP1, but the specific heme-binding sequence that regulates protein turnover is not clearly identified (79). The role of IRP1 and IRP2 phosphorylation (129, 130) on heme regulation of IRP stability is unknown.
A physiological consequence of heme regulation of IRP was recently demonstrated in the case of regulation of ferritin-L expression. Ferritin L contains an IRE element for regulating mRNA translation but is also controlled at the transcriptional level by Bach1-sensitive MARE/ARE elements in the promoter. When the ferritin promoter was studied with and without the IRE-RNA sequence in a reporter construct, it was shown that induction of ferritin L expression by heme was greater than the sum of the induction of the MARE/ARE-DNA or IRE-RNA regulators alone. Thus, in vivo, increased expression of ferritin L by heme binding to Bach1 is further enhanced by heme-mediated degradation of IRP2, leading to increased mRNA translation (70).
Other Iron-Based Redox Sensors
Although most iron-based redox-sensor proteins require an Fe-S cluster or heme, in principle, mono- or dinuclear iron centers can respond to redox status in a similar manner. Recently several mononuclear iron proteins were shown in bacteria to use this alternative iron site for sensing the strong oxidants H2O2 and NO. Careful biochemical and spectroscopic studies of these mononuclear iron proteins have uncovered several new and interesting mechanisms of iron-based redox sensing and signal transduction.
PerR and H2O2
PerR is a member of the Fur family of metalloregulatory proteins. In most cases, the Fur transcription factor regulates expression of iron homeostasis genes (transporters, siderophore biosynthesis enzymes, etc.) (4). In some organisms, Fur contains two metal-binding sites, a structural Zn2+ site and a regulatory Fe2+ site (Fur:Zn,Fe) (3). Iron binding to Fur:Zn activates its DNA-binding activity, allowing it to repress target promoters. As iron levels decrease, Fur:Zn can no longer repress transcription, and iron homeostasis genes are upregulated. However, some Fur family members, such as PerR, instead regulate genes involved in the response to H2O2 stress (65). The PerR repressor is quite similar to Fur and contains a structural Zn site (consisting of Cys4Zn) and a second more labile Fe2+ binding site with a mixture of potential O- and N-containing ligands (H37, D85, H91, H93, and D104) (93). Most characterized proteins that sense H2O2 stress rely on peroxide-mediated oxidation of protein thiols as the signaling event. In contrast, PerR uses a distinct H2O2-sensing mechanism that uses bound Fe2+ to oxidize adjacent His residues rather than the standard Cys oxidation-sensing mechanism (93). On exposure to H2O2 stress, PerR:Zn,Fe is rapidly oxidized with a second-order rate constant of approximately 105 M/s, which is quite comparable to other H2O2 sensors controlled by Cys oxidation. This oxidation event abolishes PerR DNA binding and relieves PerR repression of antioxidant genes such as catalase and alkylhydroperoxide reductase. Replacement of the Fe2+ atom in PerR with Mn2+ decreases PerR oxidation by 104-fold, highlighting the importance of the Fe for PerR oxidation.
Mass spectrometry was used to map the specific oxidative modifications on PerR after both in vitro and in vivo exposure to 18O-labeled H2O2 (93). These studies revealed that 18O was incorporated into H37 and H91 of PerR. Both these His residues are hypothesized to form part of the Fe2+-binding site. No Cys oxidation at the structural Zn2+ site was observed. Importantly, similar experiments showed no oxidation of the corresponding His residues in an iron-sensing Fur homologue of PerR, suggesting that PerR is distinct from other Fur proteins in its use of Fe2+ for H2O2 sensing. The current model for iron-catalyzed oxidation of H37 and H91 proceeds as PerR:Zn,Fe reduces H2O2 to produce hydroxyl radical through a classic Fenton reaction. Because •OH reacts in a diffusion-limited manner, it quickly oxidizes H37 or H91 or both in the vicinity of the Fe2+ site. Based on metal-catalyzed His oxidation in other proteins and model compounds, the •OH likely forms an adduct with the C-2 position of the imidazole ring (132). The C-2–•OH adduct (a 2-oxo-histidinyl radical) discharges an electron, possibly reducing the oxidized iron back to Fe2+. After deprotonation, a stable 2-oxo-histidine modification is formed, presumably altering the protein conformation in some way to inactivate DNA binding activity. PerR demonstrates how organisms have coopted metal-catalyzed oxidation of His as a way to sense H2O2 stress.
NorR and NO
NorR is a sigma 54–dependent transcriptional regulator in E. coli that activates flavorubredoxin and its associated flavoprotein (encoded by the norVW locus) in response to NO stress (44, 73, 107). The NorV and NorW proteins are responsible for NO detoxification by converting NO to nitrous oxide (45, 159). In contrast to the relatively small PerR, NorR is a large, multidomain protein. Sequence analysis of NorR shows a three-domain structure with an N-terminal GAF domain followed by a catalytic AAA+ ATPase domain required for sigma 54 interaction and promoter DNA melting during transcription (7, 72, 118, 156, 168). Binding of NorR to target promoters is dependent on the C-terminal DNA-binding HTH domain. The GAF domain appears to regulate NorR activation, as deletion of this domain results in constitutive NorR activity (44).
NorR purified anaerobically contains a mononuclear iron center bound to the GAF domain that appears to be in an EPR-silent ferrous state. On exposure of NorR to NO in vitro or in vivo, NorR forms a reversible mononitrosyl-iron complex. This paramagnetic species has an EPR signal with apparent g values at g = 4.19 and g = 3.82 (28). NO binding to the NorR mononuclear iron center stimulates the ATPase activity of the adjacent AAA+ domain, presumably enhancing NorR transcriptional activity (28). No amino acid residues on NorR were altered by NO, indicating that the iron center is poised to sense NO by formation of the mononitrosyl-iron complex rather than by stimulating other chemical modifications to the protein. The Kd of NorR for NO was measured at 50 ± 10 nM at pH 8.5 at 30°C. By using myoglobin as a competitor for NO binding, it was shown that the apparent rate constants for NO binding by NorR are koff > 1 s−1 and kon > 2 × 107 M/s (28). These observations support a role for NorR as a sensitive and rapid sensor of NO stress in vivo. Comparison of sGC kinetic parameters with those of NorR show that sGC has a kon > 1 × 108 M/s, possibly reflecting the use of mammalian sGC for NO signaling, as opposed to sensing of toxic NO accumulation.
The exact coordination site for the mononuclear ferrous iron site has been intensively probed by site-directed mutagenesis, EPR, MCD, and UV-Vis spectroscopy for the native iron and Co2+ substituted NorR, as well as structural modeling approaches (157). The most current model synthesized from this data suggests that the Fe2+ is six-coordinate in a distorted octahedral environment. The iron is likely bound by only one sulfur ligand (C113) but with an assortment of oxygen ligands from D96, D99, and D131 and a nitrogen ligand from R75. However, a five-coordinate iron site lacking the R75 ligand cannot be completely ruled out. Because release of NO would be facilitated by ligand replacement (say from R75), it seems reasonable that the resting state of NorR is likely to be six-coordinate, with one ligand primed for displacement on NO binding. Detailed structural analysis is required to address the NO binding and release mechanism as well as the NO-dependent structural changes that activate NorR ATPase activity.
IRP1 and IRP2
IRP1 and IRP2 are both sensitive to increasing cellular concentrations of iron, indicated by increased proteosomal degradation of the proteins when cells or animals are given excess iron (ferrous sulfate or ferric ammonium citrate) (21, 59, 121). In a recent set of observations, IRP1 without a [4Fe-4S]2+ cluster binding site (cysteine ligands replaced by serine) was regulated by iron normally in cultured cells, as was wild-type IRP1 in mice deficient in [4Fe-4S]2+ cluster assembly, indicating that iron-induced IRP1 degradation was independent of the [4Fe-4S]2+ cluster content (21). No obvious iron-binding sites are found in the IRP sequences, nor were single metals ions particularly notable in the crystal structure of IRP1 with RNA or with an Fe-S cluster (37, 160). It is unlikely that free Fe3+ ions are available as signals because of low solubility (10−18 M under physiologic conditions) or free Fe2+ ions because of the toxic (Fenton) chemistry with H2O2. More likely, the Fe signals for IRP1 and IRP2 are cytoplasmic Fe chaperones, such as the one recently observed for ferritin (139). Alternatively, iron-mediated IRP degradation could be initiated by an unidentified iron-dependent kinase or by hydroxylation of IRP by iron- or oxygen-dependent hydroxylases (62, 103, 112).
Roles of Iron-Based Redox Switches in Human Health
Many of the iron-based redox switches discussed here regulate important physiological pathways in response to changes in redox status; mutations that disrupt or alter the activity of the sensor proteins can, thus, lead to disease. Disruption of IRP regulation of iron-homeostasis genes has been linked to a number of human diseases (121). Mutations in the IRE of ferritin L lead to loss of IRP binding to ferritin L mRNA and increased translation of ferritin L. The result is a disease, hereditary hyperferritinemia cataract syndrome, characterized by elevated serum ferritin and early development of cataracts. Deletion of IRP2 in mice leads to altered iron distribution and anemia and, in some mouse strains, extensive neurodegeneration with overexpression of ferritin and decreased expression of transferrin receptor 1 (22, 43). Deletion of both IRP1 and IRP2 is embryonic lethal. Anemia also is associated with abnormalities in the [2Fe-2S] cluster protein, ferrochelatase; when mutation disrupts two of the cluster ligands (C406 and C411), this leads to diminished ferrochelatase activity in vivo and to protoporphyria (131).
sGC is part of a critical, NO-dependent pathway that promotes vasodilation while inhibiting platelet aggregation and leukocyte adhesion in the cardiovascular system. Disruption of the vascular endothelium can inhibit these protective pathways and aggravate cardiovascular disease. Therefore, stimulation of sGC by pharmacologic means has become an important therapy for patients with cardiovascular disease (78). Some of the pharmacologic agents require heme to be present in sGC, whereas others are NO and heme independent, but all are designed to activate sGC.
The molecular arms race between host and pathogen includes defense strategies that center around iron and oxidant stress. Host macrophages release damaging peroxide, nitric oxide, and active oxygen while sequestering available iron. In response, invading pathogens synthesize potent antioxidant enzymes and iron-scavenging pathways. Several of the iron-based redox sensors mentioned here (such as SoxR and PerR) control the expression of genes involved in the oxidative stress response of bacterial pathogens. Targeting these regulators could provide novel ways to sensitize microbes to killing by macrophages and neutrophils.
Future Directions
A number of important questions about the function of iron-based redox sensors remain to be answered. In some cases, the exact binding site for the iron cofactor is not firmly identified. Clearly establishing the biochemical and bioinorganic mechanisms used to modulate iron-cofactor sensitivity to specific redox signals also is critical for understanding in vivo function of the sensor proteins. Such studies must include more structural characterization of fully active sensor proteins, especially those with labile iron cofactors like Fe-S clusters.
It also has become apparent that study of many iron-based redox sensors must be extended to more physiological settings. The clear effects of DNA binding on the Fe-S redox potentials of SoxR and EndoIII are prime examples in which the simpler, isolated system can miss key, physiologically important interactions. Similarly, more whole-organism studies in higher eukaryotes are needed to complement cultured cell experiments to define the global and/or tissue-specific importance of individual iron-redox switches, as illustrated by the cases of IRP1 and IRP2. IRP2 is an especially pertinent example, as tissue-specific regulation by phosphorylation, heme binding, and iron binding is likely to occur. It is also likely that additional iron-dependent redox sensors with new functionalities will be identified in the coming years, exemplified by the nonheme dioxygenases (112). In conclusion, we hope the examples detailed here have clearly demonstrated the importance of iron, as both a potential mediator of redox stress and a critical sensor of redox perturbation used to regulate gene expression and protein activity.
Acknowledgments
The work of the authors described was supported by NIH GM81796 (F.W.O.) and NIH DK20251 (E.C.T.). We are grateful to all our students, postdoctoral fellows, and colleagues for their contributions to the work described.
Abbreviations
ARE, antioxidant/phase II response elements; BER, base excision repair; BTB, bric-a-brac, tramtrack, broad complex; cAMP, cyclic adenosine monophosphate; DOS, direct oxygen sensor; EPR, electron paramagnetic resonance; Fd, ferredoxin; Fe-S, iron-sulfur; FNR, fumarate and nitrate reduction regulator; GAF, cGMP-regulated cyclic nucleotide PDEs, adenylyl cyclases, and FhlA; GSH, glutathione; GSSG, oxidized glutathione; HiPIP, high-potential iron protein; HTH, helix-turn-helix; H-NOX, heme-nitric oxide/oxygen binding; HRM, heme regulatory motif; IRE, iron-response element; IRP, iron-regulatory protein; MARE, Maf recognition elements; MCD, magnetic circular dichroism; mRNA, messenger ribonucleic acid; NADP, nicotinamide adenine dinucleotide phosphate; NO, nitric oxide; NOS, nitric oxide synthase; PAS, Per-Arnt-Sim; RNAP, RNA polymerase; ROS, reactive oxygen species; sGC, soluble guanylate cyclase; TCA, tricarboxylic acid; UTR, untranslated region; UV-Vis, ultraviolet-visible.
References
- 1.Agron PG. Ditta GS. Helinski DR. Oxygen regulation of nifA transcription in vitro. Proc Natl Acad Sci U S A. 1993;90:3506–3510. doi: 10.1073/pnas.90.8.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alen C. Sonenshein AL. Bacillus subtilis aconitase is an RNA-binding protein. Proc Natl Acad Sci U S A. 1999;96:10412–10417. doi: 10.1073/pnas.96.18.10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Althaus EW. Outten CE. Olson KE. Cao H. O'Halloran TV. The ferric uptake regulation (Fur) repressor is a zinc metalloprotein. Biochemistry. 1999;38:6559–6569. doi: 10.1021/bi982788s. [DOI] [PubMed] [Google Scholar]
- 4.Andrews SC. Robinson AK. Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 5.Anxolabehere-Mallart E. Glaser T. Frank P. Aliverti A. Zanetti G. Hedman B. Hodgson KO. Solomon EI. Sulfur K-edge X-ray absorption spectroscopy of 2Fe-2S ferredoxin: covalency of the oxidized and reduced 2Fe forms and comparison to model complexes. J Am Chem Soc. 2001;123:5444–5452. doi: 10.1021/ja010472t. [DOI] [PubMed] [Google Scholar]
- 6.Aono S. Metal-containing sensor proteins sensing diatomic gas molecules. Dalton Trans. 2008;24:3137–3146. doi: 10.1039/b802070c. [DOI] [PubMed] [Google Scholar]
- 7.Aravind L. Ponting CP. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 8.Arnaud N. Ravet K. Borlotti A. Touraine B. Boucherez J. Fizames C. Briat JF. Cellier F. Gaymard F. The iron-responsive element (IRE)/iron-regulatory protein 1 (IRP1)-cytosolic aconitase iron-regulatory switch does not operate in plants. Biochem J. 2007;405:523–531. doi: 10.1042/BJ20061874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basilion JP. Kennedy MC. Beinert H. Massinople CM. Klausner RD. Rouault TA. Overexpression of iron-responsive element-binding protein and its analytical characterization as the RNA-binding form, devoid of an iron-sulfur cluster. Arch Biochem Biophys. 1994;311:517–522. doi: 10.1006/abbi.1994.1270. [DOI] [PubMed] [Google Scholar]
- 10.Becker S. Holighaus G. Gabrielczyk T. Unden G. O2 as the regulatory signal for FNR-dependent gene regulation in Escherichia coli. J Bacteriol. 1996;178:4515–4521. doi: 10.1128/jb.178.15.4515-4521.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beinert H. Holm RH. Munck E. Iron-sulfur clusters: nature's modular, multipurpose structures. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 12.Boal AK. Yavin E. Barton JK. DNA repair glycosylases with a [4Fe-4S] cluster: a redox cofactor for DNA-mediated charge transport? J Inorg Biochem. 2007;101:1913–1921. doi: 10.1016/j.jinorgbio.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boon EM. Livingston AL. Chmiel NH. David SS. Barton JK. DNA-mediated charge transport for DNA repair. Proc Natl Acad Sci U S A. 2003;100:12543–12547. doi: 10.1073/pnas.2035257100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brazzolotto X. Gaillard J. Pantopoulos K. Hentze MW. Moulis JM. Human cytoplasmic aconitase (iron regulatory protein 1) is converted into its [3Fe-4S] form by hydrogen peroxide in vitro but is not activated for iron-responsive element binding. J Biol Chem. 1999;274:21625–21630. doi: 10.1074/jbc.274.31.21625. [DOI] [PubMed] [Google Scholar]
- 15.Brenot A. King KY. Caparon MG. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol Microbiol. 2005;55:221–234. doi: 10.1111/j.1365-2958.2004.04370.x. [DOI] [PubMed] [Google Scholar]
- 16.Brown NL. Stoyanov JV. Kidd SP. Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 17.Brown NM. Anderson SA. Steffen DW. Carpenter TB. Kennedy MC. Walden WE. Eisenstein RS. Novel role of phosphorylation in Fe-S cluster stability revealed by phosphomimetic mutations at Ser-138 of iron regulatory protein 1. Proc Natl Acad Sci U S A. 1998;95:15235–15240. doi: 10.1073/pnas.95.26.15235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burden AE. Wu C. Dailey TA. Busch JL. Dhawan IK. Rose JP. Wang B. Dailey HA. Human ferrochelatase: crystallization, characterization of the [2Fe-2S] cluster and determination that the enzyme is a homodimer. Biochim Biophys Acta. 1999;1435:191–197. doi: 10.1016/s0167-4838(99)00196-x. [DOI] [PubMed] [Google Scholar]
- 19.Caslake LF. Ashraf SI. Summers AO. Mutations in the alpha and sigma-70 subunits of RNA polymerase affect expression of the mer operon. J Bacteriol. 1997;179:1787–1795. doi: 10.1128/jb.179.5.1787-1795.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiancone E. Ceci P. Ilari A. Ribacchi F. Stefanini S. Iron and proteins for iron storage and detoxification. Biometals. 2004;17:197–202. doi: 10.1023/b:biom.0000027692.24395.76. [DOI] [PubMed] [Google Scholar]
- 21.Clarke SL. Vasanthakumar A. Anderson SA. Pondarre C. Koh CM. Deck KM. Pitula JS. Epstein CJ. Fleming MD. Eisenstein RS. Iron-responsive degradation of iron-regulatory protein 1 does not require the Fe-S cluster. EMBO J. 2006;25:544–553. doi: 10.1038/sj.emboj.7600954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooperman SS. Meyron-Holtz EG. Olivierre-Wilson H. Ghosh MC. McConnell JP. Rouault TA. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood. 2005;106:1084–1091. doi: 10.1182/blood-2004-12-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crack JC. Gaskell AA. Green J. Cheesman MR. Le Brun NE. Thomson AJ. Influence of the environment on the [4Fe-4S]2+ to [2Fe-2S]2+ cluster switch in the transcriptional regulator FNR. J Am Chem Soc. 2008;130:1749–1758. doi: 10.1021/ja077455+. [DOI] [PubMed] [Google Scholar]
- 24.Crack JC. Green J. Cheesman MR. Le Brun NE. Thomson AJ. Superoxide-mediated amplification of the oxygen-induced switch from [4Fe-4S] to [2Fe-2S] clusters in the transcriptional regulator FNR. Proc Natl Acad Sci U S A. 2007;104:2092–2097. doi: 10.1073/pnas.0609514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crack JC. Green J. Le Brun NE. Thomson AJ. Detection of sulfide release from the oxygen-sensing [4Fe-4S] cluster of FNR. J Biol Chem. 2006;281:18909–18913. doi: 10.1074/jbc.C600042200. [DOI] [PubMed] [Google Scholar]
- 26.Crouse BR. Sellers VM. Finnegan MG. Dailey HA. Johnson MK. Site-directed mutagenesis and spectroscopic characterization of human ferrochelatase: identification of residues coordinating the [2Fe-2S] cluster. Biochemistry. 1996;35:16222–16229. doi: 10.1021/bi9620114. [DOI] [PubMed] [Google Scholar]
- 27.Cunningham RP. Asahara H. Bank JF. Scholes CP. Salerno JC. Surerus K. Munck E. McCracken J. Peisach J. Emptage MH. Endonuclease III is an iron-sulfur protein. Biochemistry. 1989;28:4450–4455. doi: 10.1021/bi00436a049. [DOI] [PubMed] [Google Scholar]
- 28.D'Autreaux B. Tucker NP. Dixon R. Spiro S. A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature. 2005;437:769–772. doi: 10.1038/nature03953. [DOI] [PubMed] [Google Scholar]
- 29.Dailey HA. Finnegan MG. Johnson MK. Human ferrochelatase is an iron-sulfur protein. Biochemistry. 1994;33:403–407. doi: 10.1021/bi00168a003. [DOI] [PubMed] [Google Scholar]
- 30.David M. Daveran ML. Batut J. Dedieu A. Domergue O. Ghai J. Hertig C. Boistard P. Kahn D. Cascade regulation of nif gene expression in Rhizobium meliloti. Cell. 1988;54:671–683. doi: 10.1016/s0092-8674(88)80012-6. [DOI] [PubMed] [Google Scholar]
- 31.de Philip P. Batut J. Boistard P. Rhizobium meliloti Fix L is an oxygen sensor and regulates R. meliloti nifA and fixK genes differently in Escherichia coli. J Bacteriol. 1990;172:4255–4262. doi: 10.1128/jb.172.8.4255-4262.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dey A. Jenney FE., Jr Adams MW. Babini E. Takahashi Y. Fukuyama K. Hodgson KO. Hedman B. Solomon EI. Solvent tuning of electrochemical potentials in the active sites of HiPIP versus ferredoxin. Science. 2007;318:1464–1468. doi: 10.1126/science.1147753. [DOI] [PubMed] [Google Scholar]
- 33.Dhakshinamoorthy S. Jain AK. Bloom DA. Jaiswal AK. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J Biol Chem. 2005;280:16891–16900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 34.Ding H. Demple B. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc Natl Acad Sci U S A. 2000;97:5146–5150. doi: 10.1073/pnas.97.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding H. Demple B. In vivo kinetics of a redox-regulated transcriptional switch. Proc Natl Acad Sci U S A. 1997;94:8445–8449. doi: 10.1073/pnas.94.16.8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding H. Hidalgo E. Demple B. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J Biol Chem. 1996;271:33173–33175. doi: 10.1074/jbc.271.52.33173. [DOI] [PubMed] [Google Scholar]
- 37.Dupuy J. Volbeda A. Carpentier P. Darnault C. Moulis JM. Fontecilla-Camps JC. Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure. 2006;14:129–139. doi: 10.1016/j.str.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Eisenstein RS. Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu Rev Nutr. 2000;20:627–662. doi: 10.1146/annurev.nutr.20.1.627. [DOI] [PubMed] [Google Scholar]
- 39.Erlitzki R. Long JC. Theil EC. Multiple, conserved iron-responsive elements in the 3'-untranslated region of transferrin receptor mRNA enhance binding of iron regulatory protein 2. J Biol Chem. 2002;277:42579–42587. doi: 10.1074/jbc.M207918200. [DOI] [PubMed] [Google Scholar]
- 40.Ferreira GC. Franco R. Lloyd SG. Pereira AS. Moura I. Moura JJ. Huynh BH. Mammalian ferrochelatase, a new addition to the metalloenzyme family. J Biol Chem. 1994;269:7062–7065. [PubMed] [Google Scholar]
- 41.Fromme JC. Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furukawa T. Kohno H. Tokunaga R. Taketani S. Nitric oxide-mediated inactivation of mammalian ferrochelatase in vivo and in vitro: possible involvement of the iron-sulphur cluster of the enzyme. Biochem J. 1995;310:533–538. doi: 10.1042/bj3100533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galy B. Ferring D. Minana B. Bell O. Janser HG. Muckenthaler M. Schumann K. Hentze MW. Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2) Blood. 2005;106:2580–2589. doi: 10.1182/blood-2005-04-1365. [DOI] [PubMed] [Google Scholar]
- 44.Gardner AM. Gessner CR. Gardner PR. Regulation of the nitric oxide reduction operon (norRVW) in Escherichia coli: role of NorR and sigma54 in the nitric oxide stress response. J Biol Chem. 2003;278:10081–10086. doi: 10.1074/jbc.M212462200. [DOI] [PubMed] [Google Scholar]
- 45.Gardner AM. Helmick RA. Gardner PR. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J Biol Chem. 2002;277:8172–8177. doi: 10.1074/jbc.M110471200. [DOI] [PubMed] [Google Scholar]
- 46.Gaudu P. Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc Natl Acad Sci U S A. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilles-Gonzalez MA. Ditta GS. Helinski DR. A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature. 1991;350:170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- 48.Gilles-Gonzalez MA. Gonzalez G. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J Inorg Biochem. 2005;99:1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 49.Gilles-Gonzalez Ma. Gonzalez G. Perutz MF. Kiger L. Marden MC. Poyart C. Heme-based sensors, exemplified by the kinase fixl, are a new class of heme protein with distinctive ligand-binding and autoxidation. Biochemistry. 1994;33:8067–8073. doi: 10.1021/bi00192a011. [DOI] [PubMed] [Google Scholar]
- 50.Glaser T. Rose K. Shadle SE. Hedman B. Hodgson KO. Solomon EI. S K-edge X-ray absorption studies of tetranuclear iron-sulfur clusters: mu-sulfide bonding and its contribution to electron delocalization. J Am Chem Soc. 2001;123:442–454. doi: 10.1021/ja002183v. [DOI] [PubMed] [Google Scholar]
- 51.Goessling LS. Mascotti DP. Thach RE. Involvement of heme in the degradation of iron-regulatory protein 2. J Biol Chem. 1998;273:12555–12557. doi: 10.1074/jbc.273.20.12555. [DOI] [PubMed] [Google Scholar]
- 52.Golinelli MP. Chmiel NH. David SS. Site-directed mutagenesis of the cysteine ligands to the [4Fe-4S] cluster of Escherichia coli MutY. Biochemistry. 1999;38:6997–7007. doi: 10.1021/bi982300n. [DOI] [PubMed] [Google Scholar]
- 53.Gong WM. Hao B. Chan MK. New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL. Biochemistry. 2000;39:3955–3962. doi: 10.1021/bi992346w. [DOI] [PubMed] [Google Scholar]
- 54.Gorodetsky AA. Boal AK. Barton JK. Direct electrochemistry of endonuclease III in the presence and absence of DNA. J Am Chem Soc. 2006;128:12082–12083. doi: 10.1021/ja064784d. [DOI] [PubMed] [Google Scholar]
- 55.Gorodetsky AA. Dietrich LE. Lee PE. Demple B. Newman DK. Barton JK. DNA binding shifts the redox potential of the transcription factor SoxR. Proc Natl Acad Sci U S A. 2008;105:3684–3689. doi: 10.1073/pnas.0800093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grant RA. Filman DJ. Finkel SE. Kolter R. Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 57.Greenberg JT. Monach P. Chou JH. Josephy PD. Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci U S A. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gunshin H. Allerson CR. Polycarpou-Schwarz M. Rofts A. Rogers JT. Kishi F. Hentze MW. Rouault TA. Andrews NC. Hediger MA. Iron-dependent regulation of the divalent metal ion transporter. FEBS Lett. 2001;509:309–316. doi: 10.1016/s0014-5793(01)03189-1. [DOI] [PubMed] [Google Scholar]
- 59.Guo B. Phillips JD. Yu Y. Leibold EA. Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J Biol Chem. 1995;270:21645–21651. doi: 10.1074/jbc.270.37.21645. [DOI] [PubMed] [Google Scholar]
- 60.Guo B. Yu Y. Leibold EA. Iron regulates cytoplasmic levels of a novel iron-responsive element-binding protein without aconitase activity. J Biol Chem. 1994;269:24252–24260. [PubMed] [Google Scholar]
- 61.Gurbiel RJ. Batie CJ. Sivaraja M. True AE. Fee JA. Hoffman BM. Ballou DP. Electron-nuclear double resonance spectroscopy of 15N-enriched phthalate dioxygenase from Pseudomonas cepacia proves that two histidines are coordinated to the [2Fe-2S] Rieske-type clusters. Biochemistry. 1989;28:4861–4871. doi: 10.1021/bi00437a051. [DOI] [PubMed] [Google Scholar]
- 62.Hanson ES. Rawlins ML. Leibold EA. Oxygen and iron regulation of iron regulatory protein 2. J Biol Chem. 2003;278:40337–40342. doi: 10.1074/jbc.M302798200. [DOI] [PubMed] [Google Scholar]
- 63.Hao B. Isaza C. Arndt J. Soltis M. Chan MK. Structure-based mechanism of O-2 sensing and ligand discrimination by the FixL heme domain of Bradyrhizobium japonicum. Biochemistry. 2002;41:12952–12958. doi: 10.1021/bi020144l. [DOI] [PubMed] [Google Scholar]
- 64.Hentze MW. Muckenthaler MU. Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 65.Herbig AF. Helmann JD. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol Microbiol. 2001;41:849–859. doi: 10.1046/j.1365-2958.2001.02543.x. [DOI] [PubMed] [Google Scholar]
- 66.Hidalgo E. Bollinger JM., Jr Bradley TM. Walsh CT. Demple B. Binuclear [2Fe-2S] clusters in the Escherichia coli SoxR protein and role of the metal centers in transcription. J Biol Chem. 1995;270:20908–20914. doi: 10.1074/jbc.270.36.20908. [DOI] [PubMed] [Google Scholar]
- 67.Hidalgo E. Demple B. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 1994;13:138–146. doi: 10.1002/j.1460-2075.1994.tb06243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hintze KJ. Katoh Y. Igarashi K. Theil EC. Bach1 repression of ferritin and thioredoxin reductase1 is heme-sensitive in cells and in vitro and coordinates expression with heme oxygenase1, beta-globin, and NADP(H) quinone (oxido) reductase1. J Biol Chem. 2007;282:34365–34371. doi: 10.1074/jbc.M700254200. [DOI] [PubMed] [Google Scholar]
- 69.Hintze KJ. Theil EC. Cellular regulation and molecular interactions of the ferritins. Cell Mol Life Sci. 2006;63:591–600. doi: 10.1007/s00018-005-5285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hintze KJ. Theil EC. DNA and mRNA elements with complementary responses to hemin, antioxidant inducers, and iron control ferritin-L expression. Proc Natl Acad Sci U S A. 2005;102:15048–15052. doi: 10.1073/pnas.0505148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hira S. Tomita T. Matsui T. Igarashi K. Ikeda-Saito M. Bach1, a heme-dependent transcription factor, reveals presence of multiple heme binding sites with distinct coordination structure. IUBMB Life. 2007;59:542–551. doi: 10.1080/15216540701225941. [DOI] [PubMed] [Google Scholar]
- 72.Ho YS. Burden LM. Hurley JH. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 2000;19:5288–5299. doi: 10.1093/emboj/19.20.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hutchings MI. Mandhana N. Spiro S. The NorR protein of Escherichia coli activates expression of the flavorubredoxin gene norV in response to reactive nitrogen species. J Bacteriol. 2002;184:4640–4643. doi: 10.1128/JB.184.16.4640-4643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Igarashi K. Sun J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid Redox Signal. 2006;8:107–118. doi: 10.1089/ars.2006.8.107. [DOI] [PubMed] [Google Scholar]
- 75.Ignarro LJ. Wood KS. Wolin MS. Activation of purified soluble guanylate cyclase by protoporphyrin IX. Proc Natl Acad Sci U S A. 1982;79:2870–2873. doi: 10.1073/pnas.79.9.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ishikawa H. Kato M. Hori H. Ishimori K. Kirisako T. Tokunaga F. Iwai K. Involvement of heme regulatory motif in heme-mediated ubiquitination and degradation of IRP2. Mol Cell. 2005;19:171–181. doi: 10.1016/j.molcel.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 78.Jackson EB., Jr Mukhopadhyay S. Tulis DA. Pharmacologic modulators of soluble guanylate cyclase/cyclic guanosine monophosphate in the vascular system: from bench top to bedside. Curr Vasc Pharmacol. 2007;5:1–14. [PMC free article] [PubMed] [Google Scholar]
- 79.Jeong J. Rouault TA. Levine RL. Identification of a heme-sensing domain in iron regulatory protein 2. J Biol Chem. 2004;279:45450–45454. doi: 10.1074/jbc.M407562200. [DOI] [PubMed] [Google Scholar]
- 80.Kang Y. Weber KD. Qiu Y. Kiley PJ. Blattner FR. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol. 2005;187:1135–1160. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ke Y. Wu J. Leibold EA. Walden WE. Theil EC. Loops and bulge/loops in iron-responsive element isoforms influence iron regulatory protein binding: fine-tuning of mRNA regulation? J Biol Chem. 1998;273:23637–23640. doi: 10.1074/jbc.273.37.23637. [DOI] [PubMed] [Google Scholar]
- 82.Khoroshilova N. Popescu C. Munck E. Beinert H. Kiley PJ. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc Natl Acad Sci U S A. 1997;94:6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kiley PJ. Beinert H. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol Rev. 1998;22:341–352. doi: 10.1111/j.1574-6976.1998.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 84.Kiley PJ. Storz G. Exploiting thiol modifications. PLoS Biol. 2004;2:e400. doi: 10.1371/journal.pbio.0020400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim YM. Bergonia HA. Muller C. Pitt BR. Watkins WD. Lancaster JR., Jr Loss and degradation of enzyme-bound heme induced by cellular nitric oxide synthesis. J Biol Chem. 1995;270:5710–5713. doi: 10.1074/jbc.270.11.5710. [DOI] [PubMed] [Google Scholar]
- 86.Kimata Y. Theil EC. Posttranscriptional regulation of ferritin during nodule development in soybean. Plant Physiol. 1994;104:263–270. doi: 10.1104/pp.104.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kobayashi K. Tagawa S. Isolation of reductase for SoxR that governs an oxidative response regulon from Escherichia coli. FEBS Lett. 1999;451:227–230. doi: 10.1016/s0014-5793(99)00565-7. [DOI] [PubMed] [Google Scholar]
- 88.Koo MS. Lee JH. Rah SY. Yeo WS. Lee JW. Lee KL. Koh YS. Kang SO. Roe JH. A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J. 2003;22:2614–2622. doi: 10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kulkarni RD. Summers AO. MerR cross-links to the alpha, beta, and sigma 70 subunits of RNA polymerase in the preinitiation complex at the merTPCAD promoter. Biochemistry. 1999;38:3362–3368. doi: 10.1021/bi982814m. [DOI] [PubMed] [Google Scholar]
- 90.Kuo CF. McRee DE. Fisher CL. O'Handley SF. Cunningham RP. Tainer JA. Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III. Science. 1992;258:434–440. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]
- 91.Kurokawa H. Lee DS. Watanabe M. Sagami I. Mikami B. Raman CS. Shimizu T. A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J Biol Chem. 2004;279:20186–20193. doi: 10.1074/jbc.M314199200. [DOI] [PubMed] [Google Scholar]
- 92.Lazazzera BA. Beinert H. Khoroshilova N. Kennedy MC. Kiley PJ. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J Biol Chem. 1996;271:2762–2768. doi: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- 93.Lee JW. Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 94.Leipuviene R. Theil EC. The family of iron responsive RNA structures (IRE) regulated by changes in cellular iron and oxygen. Cell Mol Life Sci. 2007;64:2945–2955. doi: 10.1007/s00018-007-7198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lill R. Muhlenhoff U. Iron-sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu Rev Cell Dev Biol. 2006;22:457–486. doi: 10.1146/annurev.cellbio.22.010305.104538. [DOI] [PubMed] [Google Scholar]
- 96.Lind MI. Missirlis F. Melefors O. Uhrigshardt H. Kirby K. Phillips JP. Soderhall K. Rouault TA. Of two cytosolic aconitases expressed in Drosophila, only one functions as an iron-regulatory protein. J Biol Chem. 2006;281:18707–18714. doi: 10.1074/jbc.M603354200. [DOI] [PubMed] [Google Scholar]
- 97.Liu X. Theil EC. Ferritin: dynamic management of biological iron and oxygen chemistry. Acc Chem Res. 2005;38:167–175. doi: 10.1021/ar0302336. [DOI] [PubMed] [Google Scholar]
- 98.Ludwig ML. Ballou DP. Noodleman L. Phthalate dioxygenase reductase. In: Wieghardt K HR, editor; Poulos T, editor; Messerschmidt A, editor. Handbook of Metalloproteins. Chichester, UK: John Wiley and Sons; 2001. pp. 652–667. [Google Scholar]
- 99.Mascotti DP. Rup D. Thach RE. Regulation of iron metabolism: translational effects mediated by iron, heme, and cytokines. Annu Rev Nutr. 1995;15:239–261. doi: 10.1146/annurev.nu.15.070195.001323. [DOI] [PubMed] [Google Scholar]
- 100.Melville SB. Gunsalus RP. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J Biol Chem. 1990;265:18733–18736. [PubMed] [Google Scholar]
- 101.Mense SM. Zhang L. Heme: a versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 2006;16:681–692. doi: 10.1038/sj.cr.7310086. [DOI] [PubMed] [Google Scholar]
- 102.Merino EJ. Boal AK. Barton JK. Biological contexts for DNA charge transport chemistry. Curr Opin Chem Biol. 2008;12:229–237. doi: 10.1016/j.cbpa.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Metzen E. Ratcliffe PJ. HIF hydroxylation and cellular oxygen sensing. Biol Chem. 2004;385:223–230. doi: 10.1515/BC.2004.016. [DOI] [PubMed] [Google Scholar]
- 104.Meyer J. Iron-sulfur protein folds, iron-sulfur chemistry, and evolution. J Biol Inorg Chem. 2008;13:157–170. doi: 10.1007/s00775-007-0318-7. [DOI] [PubMed] [Google Scholar]
- 105.Monson EK. Weinstein M. Ditta GS. Helinski DR. The fixL protein of Rhizobium meliloti can be separated into a heme-binding oxygen-sensing domain and a functional C-terminal kinase domain. Proc Natl Acad Sci U S A. 1992;89:4280–4284. doi: 10.1073/pnas.89.10.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morrissey JA. Cockayne A. Brummell K. Williams P. The staphylococcal ferritins are differentially regulated in response to iron and manganese and via PerR and Fur. Infect Immun. 2004;72:972–979. doi: 10.1128/IAI.72.2.972-979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mukhopadhyay P. Zheng M. Bedzyk LA. LaRossa RA. Storz G. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc Natl Acad Sci U S A. 2004;101:745–750. doi: 10.1073/pnas.0307741100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nunoshiba T. deRojas-Walker T. Wishnok JS. Tannenbaum SR. Demple B. Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc Natl Acad Sci U S A. 1993;90:9993–9997. doi: 10.1073/pnas.90.21.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ogawa K. Sun J. Taketani S. Nakajima O. Nishitani C. Sassa S. Hayashi N. Yamamoto M. Shibahara S. Fujita H. Igarashi K. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 2001;20:2835–2843. doi: 10.1093/emboj/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Olsen KN. Larsen MH. Gahan CG. Kallipolitis B. Wolf XA. Rea R. Hill C. Ingmer H. The Dps-like protein Fri of Listeria monocytogenes promotes stress tolerance and intracellular multiplication in macrophage-like cells. Microbiology. 2005;151:925–933. doi: 10.1099/mic.0.27552-0. [DOI] [PubMed] [Google Scholar]
- 111.Oyake T. Itoh K. Motohashi H. Hayashi N. Hoshino H. Nishizawa M. Yamamoto M. Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ozer A. Bruick RK. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat Chem Biol. 2007;3:144–153. doi: 10.1038/nchembio863. [DOI] [PubMed] [Google Scholar]
- 113.Park HJ. Suquet C. Satterlee JD. Kang CH. Insights into signal transduction involving PAS domain oxygen-sensing heme proteins from the X-ray crystal structure of Escherichia coli dos heme domain (EcDosH) Biochemistry. 2004;43:2738–2746. doi: 10.1021/bi035980p. [DOI] [PubMed] [Google Scholar]
- 114.Piccinelli P. Samuelsson T. Evolution of the iron-responsive element. RNA. 2007;13:952–966. doi: 10.1261/rna.464807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Porello SL. Cannon MJ. David SS. A substrate recognition role for the [4Fe-4S]2+ cluster of the DNA repair glycosylase MutY. Biochemistry. 1998;37:6465–6475. doi: 10.1021/bi972433t. [DOI] [PubMed] [Google Scholar]
- 116.Poulos TL. Soluble guanylate cyclase. Curr Opin Struct Biol. 2006;16:736–743. doi: 10.1016/j.sbi.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 117.Proudhon D. Wei J. Briat J. Theil EC. Ferritin gene organization: differences between plants and animals suggest possible kingdom-specific selective constraints. Mol Evol. 1996;42:325–336. doi: 10.1007/BF02337543. [DOI] [PubMed] [Google Scholar]
- 118.Ray P. Smith KJ. Parslow RA. Dixon R. Hyde EI. Secondary structure and DNA binding by the C-terminal domain of the transcriptional activator NifA from Klebsiella pneumoniae. Nucleic Acids Res. 2002;30:3972–3980. doi: 10.1093/nar/gkf528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reinhart F. Achebach S. Koch T. Unden G. Reduced apo-fumarate nitrate reductase regulator (apoFNR) as the major form of FNR in aerobically growing Escherichia coli. J Bacteriol. 2008;190:879–886. doi: 10.1128/JB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rothenberger S. Mullner EW. Kuhn LC. The mRNA-binding protein which controls ferritin and transferrin receptor expression is conserved during evolution. Nucleic Acids Res. 1990;18:1175–1179. doi: 10.1093/nar/18.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 122.Rouault TA. Tang CK. Kaptain S. Burgess WH. Haile DJ. Samaniego F. McBride OW. Harford JB. Klausner RD. Cloning of the cDNA encoding an RNA regulatory protein: the human iron-responsive element-binding protein. Proc Natl Acad Sci U S A. 1990;87:7958–7962. doi: 10.1073/pnas.87.20.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Russwurm M. Koesling D. NO activation of guanylyl cyclase. EMBO J. 2004;23:4443–4450. doi: 10.1038/sj.emboj.7600422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sanchez M. Galy B. Dandekar T. Bengert P. Vainshtein Y. Stolte J. Muckenthaler MU. Hentze MW. Iron regulation and the cell cycle: identification of an iron-responsive element in the 3'-untranslated region of human cell division cycle 14A mRNA by a refined microarray-based screening strategy. J Biol Chem. 2006;281:22865–22874. doi: 10.1074/jbc.M603876200. [DOI] [PubMed] [Google Scholar]
- 125.Sanchez M. Galy B. Muckenthaler MU. Hentze MW. Iron-regulatory proteins limit hypoxia-inducible factor-2alpha expression in iron deficiency. Nat Struct Mol Biol. 2007;14:420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- 126.Sasakura Y. Hirata S. Sugiyama S. Suzuki S. Taguchi S. Watanabe M. Matsui T. Sagami I. Shimizu T. Characterization of a direct oxygen sensor heme protein from Escherichia coli: effects of the heme redox states and mutations at the heme-binding site on catalysis and structure. J Biol Chem. 2002;277:23821–23827. doi: 10.1074/jbc.M202738200. [DOI] [PubMed] [Google Scholar]
- 127.Sasakura Y. Yoshimura-Suzuki T. Kurokawa H. Shimizu T. Structure-function relationships of EcDOS, a heme-regulated phosphodiesterase from Escherichia coli. Acc Chem Res. 2006;39:37–43. doi: 10.1021/ar0501525. [DOI] [PubMed] [Google Scholar]
- 128.Sawers RG. Zehelein E. Bock A. Two-dimensional gel electrophoretic analysis of Escherichia coli proteins: influence of various anaerobic growth conditions and the fnr gene product on cellular protein composition. Arch Microbiol. 1988;149:240–244. doi: 10.1007/BF00422011. [DOI] [PubMed] [Google Scholar]
- 129.Schalinske KL. Anderson SA. Tuazon PT. Chen OS. Kennedy MC. Eisenstein RS. The iron-sulfur cluster of iron regulatory protein 1 modulates the accessibility of RNA binding and phosphorylation sites. Biochemistry. 1997;36:3950–3958. doi: 10.1021/bi9624447. [DOI] [PubMed] [Google Scholar]
- 130.Schalinske KL. Eisenstein RS. Phosphorylation and activation of both iron regulatory proteins 1 and 2 in HL-60 cells. J Biol Chem. 1996;271:7168–7176. doi: 10.1074/jbc.271.12.7168. [DOI] [PubMed] [Google Scholar]
- 131.Schneider-Yin X. Gouya L. Dorsey M. Rufenacht U. Deybach JC. Ferreira GC. Mutations in the iron-sulfur cluster ligands of the human ferrochelatase lead to erythropoietic protoporphyria. Blood. 2000;96:1545–1549. [PubMed] [Google Scholar]
- 132.Schoneich C. Mechanisms of metal-catalyzed oxidation of histidine to 2-oxo-histidine in peptides and proteins. J Pharm Biomed Anal. 2000;21:1093–1097. doi: 10.1016/s0731-7085(99)00182-x. [DOI] [PubMed] [Google Scholar]
- 133.Sellers VM. Johnson MK. Dailey HA. Function of the [2FE-2S] cluster in mammalian ferrochelatase: a possible role as a nitric oxide sensor. Biochemistry. 1996;35:2699–2704. doi: 10.1021/bi952631p. [DOI] [PubMed] [Google Scholar]
- 134.Sellers VM. Wang KF. Johnson MK. Dailey HA. Evidence that the fourth ligand to the [2Fe-2S] cluster in animal ferrochelatase is a cysteine: characterization of the enzyme from Drosophila melanogaster. J Biol Chem. 1998;273:22311–22316. doi: 10.1074/jbc.273.35.22311. [DOI] [PubMed] [Google Scholar]
- 135.Shan Y. Lambrecht RW. Donohue SE. Bonkovsky HL. Role of Bach1 and Nrf2 in up-regulation of the heme oxygenase-1 gene by cobalt protoporphyrin. FASEB J. 2006;20:2651–2653. doi: 10.1096/fj.06-6346fje. [DOI] [PubMed] [Google Scholar]
- 136.Sharrocks AD. Green J. Guest JR. In vivo and in vitro mutants of FNR: the anaerobic transcriptional regulator of E. coli. FEBS Lett. 1990;270:119–122. doi: 10.1016/0014-5793(90)81248-m. [DOI] [PubMed] [Google Scholar]
- 137.Shepherd M. Dailey TA. Dailey HA. A new class of [2Fe-2S]-cluster-containing protoporphyrin (IX) ferrochelatases. Biochem J. 2006;397:47–52. doi: 10.1042/BJ20051967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sheridan RP. Allen LC. Carter CW., Jr Coupling between oxidation state and hydrogen bond conformation in high potential iron-sulfur protein. J Biol Chem. 1981;256:5052–5057. [PubMed] [Google Scholar]
- 139.Shi H. Bencze KZ. Stemmler TL. Philpott CC. A cytosolic iron chaperone that delivers iron to ferritin. Science. 2008;320:1207–1210. doi: 10.1126/science.1157643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Spiro S. Guest JR. Inactivation of the FNR protein of Escherichia coli by targeted mutagenesis in the N-terminal region. Mol Microbiol. 1988;2:701–707. doi: 10.1111/j.1365-2958.1988.tb00080.x. [DOI] [PubMed] [Google Scholar]
- 141.Stehling O. Netz DJ. Niggemeyer B. Rosser R. Eisenstein RS. Puccio H. Pierik AJ. Lill R. The human CIA component huNbp35 is essential for both cytosolic iron-sulfur protein assembly and iron homeostasis. Mol Cell Biol. 2008 Jun 23; doi: 10.1128/MCB.00545-08. E-pub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stephens PJ. Jollie DR. Warshel A. Protein control of redox potentials of iron-sulfur proteins. Chem Rev. 1996;96:2491–2514. doi: 10.1021/cr950045w. [DOI] [PubMed] [Google Scholar]
- 143.Stone JR. Marletta MA. Soluble guanylate cyclase from bovine lung: activation with nitric oxide and carbon monoxide and spectral characterization of the ferrous and ferric states. Biochemistry. 1994;33:5636–5640. doi: 10.1021/bi00184a036. [DOI] [PubMed] [Google Scholar]
- 144.Sun J. Brand M. Zenke Y. Tashiro S. Groudine M. Igarashi K. Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc Natl Acad Sci U S A. 2004;101:1461–1466. doi: 10.1073/pnas.0308083100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sutton VR. Mettert EL. Beinert H. Kiley PJ. Kinetic analysis of the oxidative conversion of the [4Fe-4S]2+ cluster of FNR to a [2Fe-2S]2+ cluster. J Bacteriol. 2004;186:8018–8025. doi: 10.1128/JB.186.23.8018-8025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Suzuki H. Tashiro S. Hira S. Sun J. Yamazaki C. Zenke Y. Ikeda-Saito M. Yoshida M. Igarashi K. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J. 2004;23:2544–2553. doi: 10.1038/sj.emboj.7600248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Suzuki H. Tashiro S. Sun J. Doi H. Satomi S. Igarashi K. Cadmium induces nuclear export of Bach1, a transcriptional repressor of heme oxygenase-1 gene. J Biol Chem. 2003;278:49246–49253. doi: 10.1074/jbc.M306764200. [DOI] [PubMed] [Google Scholar]
- 148.Tahara T. Sun J. Nakanishi K. Yamamoto M. Mori H. Saito T. Fujita H. Igarashi K. Taketani S. Heme positively regulates the expression of beta-globin at the locus control region via the transcriptional factor Bach1 in erythroid cells. J Biol Chem. 2004;279:5480–5487. doi: 10.1074/jbc.M302733200. [DOI] [PubMed] [Google Scholar]
- 149.Tamura K. Nakamura H. Tanaka Y. Oue S. Tsukamoto K. Nomura M. Tsuchiya T. Adachi S. Takahashi S. Iizuka T. Shiro Y. Nature of endogenous ligand binding to heme iron in oxygen sensor FixL. J Am Chem Soc. 1996;118:9434–9435. [Google Scholar]
- 150.Tang Y. Guest JR. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology. 1999;145:3069–3079. doi: 10.1099/00221287-145-11-3069. [DOI] [PubMed] [Google Scholar]
- 151.Tang Y. Guest JR. Artymiuk PJ. Green J. Switching aconitase B between catalytic and regulatory modes involves iron-dependent dimer formation. Mol Microbiol. 2005;56:1149–1158. doi: 10.1111/j.1365-2958.2005.04610.x. [DOI] [PubMed] [Google Scholar]
- 152.Thayer MM. Ahern H. Xing D. Cunningham RP. Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Theil EC. Eisenstein RS. Combinatorial mRNA regulation: iron regulatory proteins and iso-iron-responsive elements (Iso-IREs) J Biol Chem. 2000;275:40659–40662. doi: 10.1074/jbc.R000019200. [DOI] [PubMed] [Google Scholar]
- 154.Tsaneva IR. Weiss B. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol. 1990;172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tseng CP. Albrecht J. Gunsalus RP. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J Bacteriol. 1996;178:1094–1098. doi: 10.1128/jb.178.4.1094-1098.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tucker NP. D'Autreaux B. Studholme DJ. Spiro S. Dixon R. DNA binding activity of the Escherichia coli nitric oxide sensor NorR suggests a conserved target sequence in diverse proteobacteria. J Bacteriol. 2004;186:6656–6660. doi: 10.1128/JB.186.19.6656-6660.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tucker NP. D'Autreaux B. Yousafzai FK. Fairhurst SA. Spiro S. Dixon R. Analysis of the nitric oxide-sensing non-heme iron center in the NorR regulatory protein. J Biol Chem. 2008;283:908–918. doi: 10.1074/jbc.M705850200. [DOI] [PubMed] [Google Scholar]
- 158.Varghese S. Tang Y. Imlay JA. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J Bacteriol. 2003;185:221–230. doi: 10.1128/JB.185.1.221-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vicente JB. Teixeira M. Redox and spectroscopic properties of the Escherichia coli nitric oxide-detoxifying system involving flavorubredoxin and its NADH-oxidizing redox partner. J Biol Chem. 2005;280:34599–34608. doi: 10.1074/jbc.M506349200. [DOI] [PubMed] [Google Scholar]
- 160.Walden WE. Selezneva AI. Dupuy J. Volbeda A. Fontecilla-Camps JC. Theil EC. Volz K. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science. 2006;314:1903–1908. doi: 10.1126/science.1133116. [DOI] [PubMed] [Google Scholar]
- 161.Wallander ML. Leibold EA. Eisenstein RS. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim Biophys Acta. 2006;1763:668–689. doi: 10.1016/j.bbamcr.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Watanabe S. Kita A. Kobayashi K. Miki K. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc Natl Acad Sci U S A. 2008;105:4121–4126. doi: 10.1073/pnas.0709188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wedel B. Humbert P. Harteneck C. Foerster J. Malkewitz J. Bohme E. Schultz G. Koesling D. Mutation of His-105 in the beta 1 subunit yields a nitric oxide-insensitive form of soluble guanylyl cyclase. Proc Natl Acad Sci U S A. 1994;91:2592–2596. doi: 10.1073/pnas.91.7.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wiedenheft B. Mosolf J. Willits D. Yeager M. Dryden KA. Young M. Douglas T. An archaeal antioxidant: characterization of a Dps-like protein from Sulfolobus solfataricus. Proc Natl Acad Sci U S A. 2005;102:10551–10556. doi: 10.1073/pnas.0501497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wu CK. Dailey HA. Rose JP. Burden A. Sellers VM. Wang BC. The 2.0 A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat Struct Biol. 2001;8:156–160. doi: 10.1038/84152. [DOI] [PubMed] [Google Scholar]
- 166.Wu J. Dunham WR. Weiss B. Overproduction and physical characterization of SoxR, a [2Fe-2S] protein that governs an oxidative response regulon in Escherichia coli. J Biol Chem. 1995;270:10323–10327. doi: 10.1074/jbc.270.17.10323. [DOI] [PubMed] [Google Scholar]
- 167.Zhang D. Dimopoulos G. Wolf A. Minana B. Kafatos FC. Winzerling JJ. Cloning and molecular characterization of two mosquito iron regulatory proteins. Insect Biochem Mol Biol. 2002;32:579–589. doi: 10.1016/s0965-1748(01)00138-2. [DOI] [PubMed] [Google Scholar]
- 168.Zhang X. Chaney M. Wigneshweraraj SR. Schumacher J. Bordes P. Cannon W. Buck M. Mechanochemical ATPases and transcriptional activation. Mol Microbiol. 2002;45:895–903. doi: 10.1046/j.1365-2958.2002.03065.x. [DOI] [PubMed] [Google Scholar]
- 169.Zhao Y. Schelvis JP. Babcock GT. Marletta MA. Identification of histidine 105 in the beta1 subunit of soluble guanylate cyclase as the heme proximal ligand. Biochemistry. 1998;37:4502–4509. doi: 10.1021/bi972686m. [DOI] [PubMed] [Google Scholar]
- 170.Zheng L. Kennedy MC. Blondin GA. Beinert H. Zalkin H. Binding of cytosolic aconitase to the iron responsive element of porcine mitochondrial aconitase mRNA. Arch Biochem Biophys. 1992;299:356–360. doi: 10.1016/0003-9861(92)90287-7. [DOI] [PubMed] [Google Scholar]