Abstract
In recent years, the biological sciences have seen a surge in the development of methods, including high-throughput global methods, for the quantitative measurement of biomolecule levels (i.e., RNA, proteins, metabolites) from cells and tissues. Just as important as quantitation of biomolecules has been the creation of approaches that uncover the regulatory and signaling connections between biomolecules. Our specific interest is in understanding peptide metabolism in a physiological setting, and this has led us to develop a multidisciplinary approach that integrates genetics, analytical chemistry, synthetic chemistry, biochemistry, and chemical biology to identify the substrates of peptidases in vivo. To accomplish this we utilize a liquid chromatography-mass spectrometry (LC-MS)-based peptidomics platform to measure changes in the peptidome–all peptides in a cell, tissue, or organism–as a function of peptidase activity. Previous analysis of mice lacking the enzyme dipeptidyl peptidase 4 (DPP4−/− mice), a biomedically relevant peptidase, using this approach identified a handful of novel endogenous DPP4 substrates. Here, we utilize these substrates and tissues from DPP4−/− mice to improve the coverage of the peptidomics platform by optimizing the key steps in the workflow, and in doing so, discover a total of 70 renal DPP4 substrates (up from 7 at the beginning of our optimization), a 10-fold improvement in our coverage. The sequences of these DPP4 peptide substrates support a broad role for DPP4 in proline-containing peptide catabolism and strengthen a biochemical model that interlinks aminopeptidase and DPP4 activities. Moreover, the improved peptidome coverage also led to the detection of greater numbers of known bioactive peptides (e.g., peptide hormones) during the analysis of gut samples suggesting additional uses for this optimized workflow. Together these results strengthen our ability to identify endogenous peptide substrates through improved peptidome coverage and demonstrate a broader potential of this peptidomics platform.
Introduction
Peptides play many important roles in cellular and physiological processes ranging from glucose regulation (insulin)1 to immune recognition (MHC peptides).2 In cells and tissues, peptide metabolism controls the composition, as well as the concentrations, of endogenous peptides.3–6 Of the many enzymatic activities that are involved in peptide metabolism, proteolysis is one of the most important as peptidases and proteases are intimately involved in the production,4–5 degradation,7 and signaling of peptides, including peptide hormones that control many physiological processes. The determination of peptidase-substrate interactions is also an important step in the understanding of the cellular and physiological roles of the enzyme,4,7–8 and can also reveal metabolic pathways involved in endogenous peptide metabolism. Additionally, in cases where the peptidase/protease regulates a bioactive peptide, these experiments can reveal new targets for regulating bioactive peptide levels and signaling in vivo.7–8 For example, the knowledge that the angiotensin II peptide—a ligand for the angiotensin receptor and a potent vasoconstrictor that causes hypertension—is produced by angiotensin converting enzyme (ACE) led to the development of ACE inhibitors as anti-hypertensive drugs.8
Typically, peptidase-substrate interactions are defined through in vitro experiments using recombinant enzymes.9–10 While powerful, especially in biochemical studies, in vitro assays are less reliable for discovering endogenous substrates of peptidases. In some cases, in vitro measurements neglect important aspects of in vivo biology (e.g., protein localization, co-activators, etc.) that are necessary to truly understand whether an enzyme and a substrate interact in vivo. Additionally, the lack of knowledge of all potential substrates of peptidases is a significant problem. In our experience, apart from a handful of known bioactive peptides, the composition of the peptidome is largely uncharted and extremely complex.11 To overcome these challenges, a number of new approaches have been applied that use unbiased mass spectrometry (MS)-based methods to measure changes in endogenous peptide levels as a function of peptidase activity.4–6 For example, Fricker and colleagues have pioneered the use of an isotope labeling mass spectrometry approach for quantifying differences in tissue peptide levels as a function of carboxypeptidase E (Cpe) activity to better understand the molecular basis of the extreme obesity associated with Cpe null mice.4 These experiments identified a number of neuropeptides and neuropeptide families regulated by Cpe, and just as importantly serve as a general demonstration of the utility of unbiased peptidome analysis to determine peptides regulated by proteolytic activity. Building off of this example, we recently integrated a label-free MS-based approach with genetics, synthetic chemistry, and biochemistry to identify endogenous substrates of the anti-diabetic drug target dipeptidyl peptidase 4 (DPP4) in the kidney.12
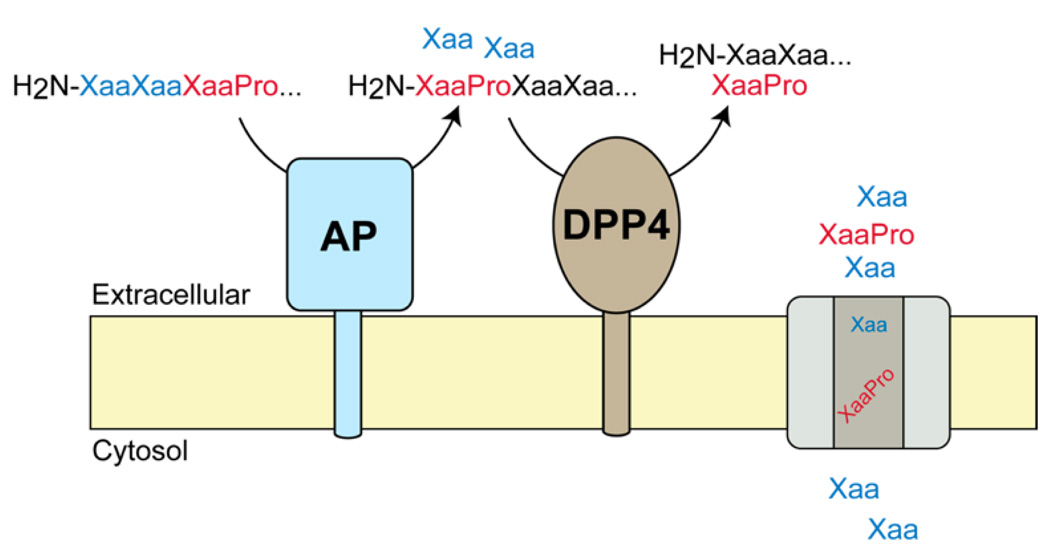
DPP4 is a serine peptidase that is part of the prolyl peptidase superfamily, an enzyme family defined by a preference for cleavage on the c-terminal side of proline and to a lesser extent alanine within peptides.7,13 This enzyme is found in two forms, an extracellular membrane bound peptidase and a secreted protein that is found at high levels in plasma.13 The development of DPP4 inhibitors as drugs was driven by the desire to regulate endogenous levels of the known DPP4 substrate glucagon-like peptide 1 (GLP-1), an insulinotropic peptide.3 In plasma, GLP-1 is found in two predominant forms: the bioactive GLP-1(7–36) amide and the inactive GLP-1(9–36) amide.14 In vivo, DPP4 inactivates GLP-1(7–36) by cleaving this peptide to form the GLP-1(9–36) amide to regulate GLP-1 signaling and indirectly regulate insulin levels.3 As a result of this biochemical connection between GLP-1 and DPP4, inhibitors of DPP4 represent a new class of anti-diabetic medicines because these inhibitors can raise insulin levels through GLP-1 regulation.
DPP4 is also found at high levels in many tissues (gut, kidney, liver) in its extracellular membrane bound form,12 but its role in these tissues is less defined. Our peptidomics analysis of DPP4 was aimed at understanding the biochemical, cellular, and physiological functions of this enzyme in the kidney by comparing the global peptide levels in DPP4 null (DPP4−/−) mice3 to wild type (DPP4+/+) C57BL/6 mice. These initial studies identified a handful (five) of new DPP4 substrates, all of which were fragments of kidney proteins, such as meprin β (Mepβ) and diazepam binding inhibitor (DBI). These substrates contained the canonical DPP4 cleavage site with a penultimate proline at the N-terminal position of the peptide (i.e., H2N-XaaPro). Furthermore, the presence of multiple fragments from these proteins, including peptides that were not regulated by DPP4, indicated that DPP4 is part of the catabolic pathway in the kidney that converts proteins into amino acids, dipeptides, and tripeptides for recovery prior to excretion in the urine.15 Importantly, we confirmed that many of the peptides elevated in DPP4−/− mice were indeed substrates for the enzyme through in vitro biochemical experiments with synthetic substrates and recombinant DPP4. Because all of the DPP4 substrates we discovered contained a proline residue, it follows that DPP4 is responsible for the catabolic degradation of proline-containing peptides in the kidney. This result supported earlier work that showed differences in proline and proline-containing peptide levels in the urine of rats lacking DPP4 activity.15 More generally, this result highlights the value of peptidomics approaches in understanding the cellular and physiological function of a peptidase, in this case DPP4.
Additionally, while a number of penultimate proline-containing (H2N-XaaPro) DPP4 substrates were elevated in DPP4−/− mice we did not detect higher levels of N-terminal proline terminated peptides (H2N-Pro) in these samples. These data indicate that penultimate proline-containing peptides are not converted to proline-terminated peptides in the kidney and, therefore, are not substrates for kidney aminopeptidases.16 This insight led us to develop a new biochemical model for proline-containing peptide catabolism in the kidney that interlinks aminopeptidase and DPP4 activities in the catabolism of proline-containing peptides. In this model, the N-terminus of a proline-containing peptide is processed by an aminopeptidase until a penultimate proline is encountered. At this point, the peptide is released from the aminopeptidase and can then be cleaved by DPP4 to remove the N-terminal H2N-XaaPro dipeptide. We gained support for this biochemical pathway by looking at the processing of proline-containing peptides using tissue lysates from mice lacking the two most abundant aminopeptidases in the kidney, aminopeptidase A and N.16 Lysates from aminopeptidase null mice had impaired ability to generate penultimate proline-containing peptides (i.e., H2N-XaaPro) from proline-containing peptides, indicating that aminopeptidase activity is important in generating DPP4 substrates, and establishing a metabolic pathway where aminopeptidases feed penultimate proline-containing substrates into DPP4. In total, these studies highlight the value of our integrated substrate discovery approach in identifying substrates, biochemical pathways, and physiological functions of DPP4.
More generally, these results support the use of mass spectrometry based methods, which reveal insights not accessible by other methods, in peptidase and peptide metabolism research. Because our peptidomics platform is the key to our substrate discovery approach, we decided to improve our peptidome coverage through the systematic optimization of the various steps in our peptidomics workflow. In this respect, our previous identification of DPP4 renal peptide substrates provided a significant advantage during this optimization, because these bona fide substrates could be used to assess whether changes made to the current platform improved or worsened our peptidome coverage. The key parameters of the peptidomics workflow that we evaluated include peptide isolation, sample processing, detection (LC-MS) and data analysis methods (Figure 1). We quantified improvements in these parameters by the number of new DPP4 substrates (i.e., penultimate proline-containing peptides) that were identified under different conditions when comparing tissue samples from DPP4+/+ and DPP4−/− mice. The optimization of our peptidomics platform greatly improved our peptidome coverage as evidenced by a large increase in the number of additional DPP4 substrates identified. Lastly, we extended this optimized peptidomics platform in the analysis of the gut, another tissue with high levels of DPP4,17 and found a number of intestinal DPP4 substrates. The broadened coverage of the peptidome coupled with the ability to perform quantitative comparative analysis, facilitate deeper insight into the biochemical connections between the peptidases, such as DPP4, and their substrates. Moreover, these studies have revealed the possibilities, as well as the current limitations, of peptidomics approaches in biology, biochemistry, and chemical biology.
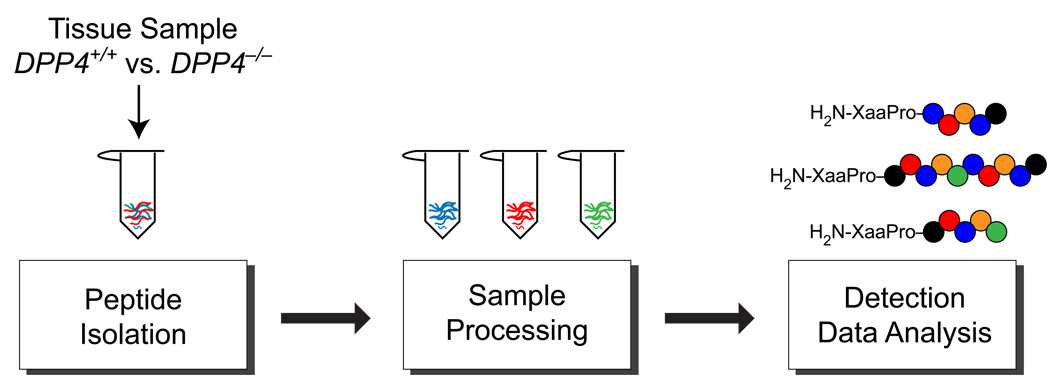
Figure 1.
Key steps in peptidomics workflow. Peptide isolation, sample preparation, peptide detection and data analysis were optimized using tissues from DPP4−/− and DPP+/+ mice. Changes were quantified by the detection of new DPP4 substrates, peptides containing penultimate proline residues (far right). The biggest improvement in peptidome coverage came from improving fractionation of the peptidome through a second offline chromatography step leading to a new workflow and the discovery of many more DPP4 substrates.
Experimental Section
Animal Studies
Wild type (DPP4+/+, C57BL/6) mice used in this study were either purchased (Jackson Labs, Bar Harbor, ME) or taken from a breeding colony. The DPP4−/− mice used in this study (a generous gift from Dr. Didier Marguet) have previously been described3 and are on a C57BL/6 background. Mice in these studies were not littermates from het x het crosses, but were obtained from separate colonies of DPP4−/− and DPP4+/+ (i.e., C57BL/6) mice. Animals were kept on a 12-h light, 12-h dark schedule and fed ad libitum. For kidney and gut tissue collection, animals were euthanized with CO2, their tissue dissected, flash frozen with liquid N2, and stored at −80 °C. All animal care and use procedures were in strict accordance with the standing committee on the use of animals in research and teaching at Harvard University and the National Institute of Health guidelines for the humane treatment of laboratory animals.
Peptidomics Workflow Optimization
All of the following steps toward the optimization of the peptidomics workflow were performed with kidney samples prepared from DPP4+/+ and DPP4−/− mice. Ions corresponding to previously identified DPP4 substrates were used to optimize the workflow parameters. Essentially, steps that improved the ability to detect these bona fide substrates, or discover new substrates were considered useful. The ion intensities of these known renal DPP4 substrates or the discovery of new DPP4 substrates were used as the metric to assess the utility of each of the optimization steps.
Peptide Isolation Step
Frozen pairs of kidney from the same mouse were either boiled for 15 min or microwaved for 2 min in 500 µL of boiling or microwaved water, respectively, to inactivate proteolytic activity. The aqueous fraction was separated and saved, and the tissue was dounce-homogenized in ice-cold 0.25% acetic acid(aq), 6 M GndHCl, or 8 M urea. The aqueous fraction and the homogenate were combined and centrifuged at 20,000 × g for 20 min at 4 °C. The supernatant was sent through a 10 kD molecular weight cutoff filter (Microcon YM-10, Millipore). The filtrate was then sent through a C18 Sep Pak cartridge (HLB 1cc; 30 mg, Oasis). Bound peptides were washed thoroughly with water and then eluted with 50:50 H2O/ACN. The eluted peptides were concentrated in a speed vacuum concentrator and then dissolved in 0.1% formic acid(aq) at 200 mg tissue/40 µL prior to analysis by LC-MS. Only DPP4−/− kidneys (N = 4) were studied in this experiment. Peptide yields were quantified using the Bradford assay. The extraction efficiency of the three dounce-homogenization methods (n = 3) were determined using the peptide standard, RPGL*L*DL*KGKAKWD, synthesized with three d10-leucines (asterisks).
Peptidome Processing Step
To identify thiol-containing peptides or long peptides (>30 amino acids) that are difficult to detect by our standard MS methods, extracted peptides were subjected to reduction/alkylation or trypsin digest, respectively. To perform reduction/alkylation, the peptide extract solution in 0.25% acetic acid(aq) was adjusted to pH 8.0 with NH4OH. A 20 mM TCEP solution in 25 mM NH4HCO3 was added to 10% by volume. The peptide solution was incubated at 37 °C for 1 h. It was then cooled to room temperature for 10 min. A 40 mM iodoacetamide solution in 25 mM NH4HCO3 was added to 10% by volume. The peptide solution was incubated in the dark for 1 h at room temperature. The solution was sent through a 10 kD molecular weight cutoff filter (Microcon YM-10, Millipore), followed by fractionation through a C18 Sep Pak cartridge (HLB 1cc; 30 mg, Oasis). For the trypsin digestion, the peptide sample that results from the “peptide isolation step” was dissolved in 0.02 µg/µL trypsin (Promega) containing 50 mM NH4HCO3 at a ratio of 50:1 (peptide:trypsin) and incubated at 37 °C for 16 h. The reaction was quenched with neat formic acid (final pH ∼3). The solution was then diluted to 200 mg tissue/40 µL in 0.1% formic acid(aq) prior to analysis by LC-MS. Lastly, for each of these experiments DPP4+/+ and DPP4−/− samples were compared (N = 4 to 6 to provide the necessary statistical power to identify true differences between samples) under standard, reduction/alkylation, and trypsin digest conditions.
Peptidome Fractionation
In a previous study, all samples were subjected to a single reverse-phase (C18) high performance liquid chromatography (RP-HPLC; see below) step to fractionate the peptidome.12 To expand coverage of the peptidome, an offline fractionation step was introduced into the workflow prior to the online RP-HPLC step to improve our separation. Two different types of offline separation were tested, strong cation exchange (SCX) and OFFGEL electrophoresis.
SCX was performed using a PolySULFOETHYL A™ column (200 × 2.1mm, 5 µm, 300 Å; PolyLC INC.) connected to an Agilent Technologies 1200 series LC equipped with a degasser. The pump was coupled to a LC-10ATVP pump manual injection set (Shimadzu) with a 1 mL loop. All runs were operated at 0.3 mL/min with a SPD-10A UV-vis detector (Shimadzu) set at λ = 220 nm. The SCX buffers consisted of: A) 7 mM KH2PO4, pH 2.6, 2% ACN (vol/vol); B) 40 mM KCl, 7 mM KH2PO4, pH 2.6, 2% ACN (vol/vol); C) 100 mM KCl, 7 mM KH2PO4, pH 2.6, 2% ACN (vol/vol); D) 400 mM KCl, 7 mM KH2PO4, pH 2.6, 2% ACN (vol/vol).
To obtain reproducibility in peptide elution, an appropriate step-gradient was established using peptide standards with distinct charge states at pH <3.0: LPLFDRVLVE (+2), LPAPEKFVKDIDGGIDQDIFD (+3), GLLDLKGKAKWD (+4), and RPGLLDLKGKAKWD (+5). The step-gradient that was developed includes a 60 min Buffer A wash, a 40 min Buffer B wash, a 40 min Buffer C wash, and a 40 min Buffer D wash, with 20 min transitions between each wash step. Fractions were collected for each wash step. This method was applied to peptidomics analysis of DPP4+/+ vs DPP4−/− kidneys (N = 4) with fractionation of the samples by SCX followed by RP-HPLC. Prior to the SCX runs, all samples were dissolved in 200 µL buffer A. All four salt fractions collected were desalted by a C18 Sep Pak cartridge, concentrated using a speed vac, and dissolved as indicated previously (200 mg tissue/40 µL 0.1% formic acid(aq), normalized according to the weight of the tissue they were extracted from).
OFFGEL electrophoresis (OGE) was performed using a 3100 OFFGEL Fractionator (Agilent Technologies) according to the manufacturer’s protocol. Two focusing buffers were prepared, one containing 5% glycerol and 2% OFFGEL buffer (as supplied) and one simply consisting of LC-MS grade water (J.T. Baker). The 12-lane gel strips with a linear pH gradient ranging at 3–10 were rehydrated with 25 µL focusing buffer per well. All samples were dissolved in 1.8 mL focusing buffer. To each well was added 150 µL of sample. A standard peptide protocol (OG12PE00) was applied for the fractionation. To determine whether ampholytes (present in the OFFGEL buffer; final 1/50 dilution) are important for fractionation, initial runs were performed on human serum album (HSA) trypsin digest standards (200 pmol). Gel lanes 1–2, 3–4, 5–6, and 7–12 were collected, desalted using a C18 Sep Pak cartridge and concentrated using a speed vac. The fractions were dissolved in 20 µL of 0.1% formic acid(aq) per lane. A 5 µL injection of the final samples was performed on the LTQ (see below). Due to superior fractionation with ampholytes, they were included during the analysis of DPP4+/+ vs DPP4−/− samples (N = 4). These samples were desalted using a C18 sep pak cartridge, concentrated using a speed vac, and dissolved as indicated previously (200 mg tissue/40 µL of 0.1% formic acid(aq), normalized according to the weight of the tissue they were extracted from).
Peptidome Analysis
Samples (10 µL) were injected onto an Eksigent nanoLC-2D HPLC configured with a pre-packaged IntegraFrit trapping column (Proteopep™ II C18, 300 Å, 5 µm) and an in-house packed (C18 AQ, 200 Å, 3 µm silica, 15 cm length, Michrom Bioresources, Inc.) PicoFrit SELF/P (15 µm tip, 25 cm length, New Objective) analytical column. The RP-HPLC gradient proceeded from 3–33% acetonitrile/water (0.1% formic acid) over 180 min. For comparison purposes, all kidney samples were injected at the same concentration (200 mg tissue/40 µL of 0.1% formic acid(aq)).
To improve ion peak integration, mass spectra collected in full MS and tandem MS (Top 3, Top 6, and Top 10, the number indicating the number of most abundant ions targeted for concurrent MS/MS in the linear ion trap with relative collision energy of 30% and 2.5 Da isolation width) modes were compared (N = 4). XCMS, a nonlinear retention time alignment and peak detection software, was used to determine quantitative peptide fold level differences between DPP4+/+ and DPP4−/− samples. The utility of this software was established by comparing data analysis using XCMS and the standard SEQUEST spectral counting based method (N = 3) to identify known DPP4 regulated peptides. A Student’s t-test was used to assess the statistical significance of these differences. DPP4−/− elevated ions established by the XCMS output files were specifically focused on to identify DPP4 substrates. The relative quantitation by ion intensity was also confirmed by using a stable isotope-labeled version of the DBI(92–104) peptide as an internal standard (RPGL*L*DL*KGKAKWD). This standard was added into samples (50 fmol) prior to LC-MS and the peak area of the standard in the LC-MS was used to quantify the amount of natural peptide in the DPP4+/+ and DPP4−/− LC-MS samples (n=4), which enabled the relative quantitation between the two samples to be determined.
Peptide identification was performed with the SEQUEST algorithm with differential modification of methionine to its sulfoxide. The uniprotmus_frc.fasta mouse database, concatenated to a reversed decoy database, served to estimate a false discovery rate (FDR). Peptides were accepted within 1 Da of the expected mass, meeting a series of custom filters on ScoreFinal (Sf), −10 Log P, and charge state that attained an average peptide FDR of <2% across data sets. Manual inspection of spectra, FDR calculation, and protein inference were performed in Proteomics Browser Suite 2.23 (ThermoFisher Scientific). Spectral counting output files generated from SEQUEST were used to identify DPP4 peptide products complementary to the substrates determined by XCMS.
Gut Profiling
Optimal 1D and 2D parameters were applied to profile DPP4+/+ vs. DPP4−/− female gut samples (N = 4; three inches from the connection to the stomach). The final samples were dissolved at 50 mg/40 µL of 0.1% formic acid(aq).
Peptide Standards
The peptide standards that were used for the optimization of SCX fractionation were synthesized and purified as previously described.12 The human serum albumin trypsin digest sample was obtained from Michrom Bio Resources Inc. The heavy-label DBI(92–104) peptide standard, RPGL*L*DL*KGKAKWD, was synthesized manually using solid-phase peptide synthesis by FMOC chemistry starting with an aspartic acid bound Wang resin. L-leucine-d10-N-FMOC was purchased from CDN Isotopes, INC. The crude peptide was purified by RP-HPLC (Shimadzu) using a C18 column (150 mm × 20 mm, 10 µm particle size, Higgins Analytical). Mobile phase A consisted of 99% H2O, 1% acetonitrile, and 0.1% TFA and mobile phase B consisted of 90% acetonitrile, 10% H2O, and 0.07% TFA. The HPLC gradient proceeded from 20–50% B over 40 min. HPLC fractions were analyzed by MALDI-TOF (Waters) to confirm the correct sequence of the peptide using α-cyano-4-hydroxycinnamic acid as the matrix, and the pure fractions were combined and lyophilized.
DPP4 Lysate Activity Assays
DPP4+/+ and DPP4−/− (N = 4) mice were sacrificed and the kidneys and guts were collected and stored at −80 °C prior to use. To test the effect of microwaving on DPP4 activity, one kidney from a mouse was dounce homogenized in ice-cold assay buffer (25 mM Tris-HCl, 140 mM NaCl, 10 mM KCl, pH 7.5, 0.1% BSA), while the other kidney was microwaved at high power in water for 2 min prior to homogenization. A similar experiment was performed to test the heat-induced inactivation of DPP4 activity in gut. Following centrifugation at 20,000 × g for 20 min at 4 °C, DPP4 activity was measured in the lysate using the fluorogenic substrate H-GlyPro-AMC. The assay was performed by addition of H-GlyPro-AMC (22.5 µM final concentration) to lysate (1 mg/mL, 100 µL final) and the fluorescence was monitored at 37 °C using an excitation wavelength of 360 nm and an emission wavelength of 460 nm (Spectramax Gemini XS plate reader, Molecular Devices).
Results and Discussion
Peptidomics Workflow
In our initial efforts toward substrate discovery we integrated a peptidomics platform with genetics, synthetic chemistry, biochemistry, and chemical biology to identify substrates for the biomedically relevant peptidase DPP4.3,14 These studies identified previously uncharacterized DPP4 regulated peptides in the kidney, including a handful of substrates that were elevated in the DPP4−/− mice. The label-free peptidomics workflow used in these experiments consisted of three key steps: peptide isolation, LC-MS analysis, and data analysis–which includes quantitation and peptide identification (Figure 1). This original workflow had been assembled from different literature examples18–19 that in many cases had not been exhaustively tested or optimized. Thus, each step in the peptidomics workflow was evaluated in an attempt to improve the peptidome coverage and by extension increase the number of DPP4 substrates identified. Specifically, we looked at different approaches for peptide isolation, sample processing (reduction/alkylation, trypsin digestion, fractionation), detection, and data analysis (Figure 1). Throughout the remainder of the manuscript we refer to our original conditions as “the standard peptidomics workflow” to distinguish the original setup from the optimized workflows that follow.
The use of the previously identified DPP4 substrates was the key to optimizing the peptidomics workflow because these peptides provided a necessary guide for us to gauge whether any changes to the workflow improved the coverage. Without these peptides it would be impossible to tell if a change to the workflow actually made things worse. Thus, the discovery of new DPP4 substrates helped to characterize the enzyme, but also provided a necessary set of controls to begin a systematic effort to optimize the peptidomics platform itself. Since the DPP4 peptides encountered in our initial study had a distinctive signature sequence, a penultimate proline residue (i.e., H2N-XaaPro), we could ‘quantify’ any improvement in our peptidomics platform by the improved detection of the known DPP4 substrates and/or by the discovery of new DPP4 substrates (i.e., penultimate proline-containing peptides) in the kidney.12
Optimization of the Peptide Isolation Step
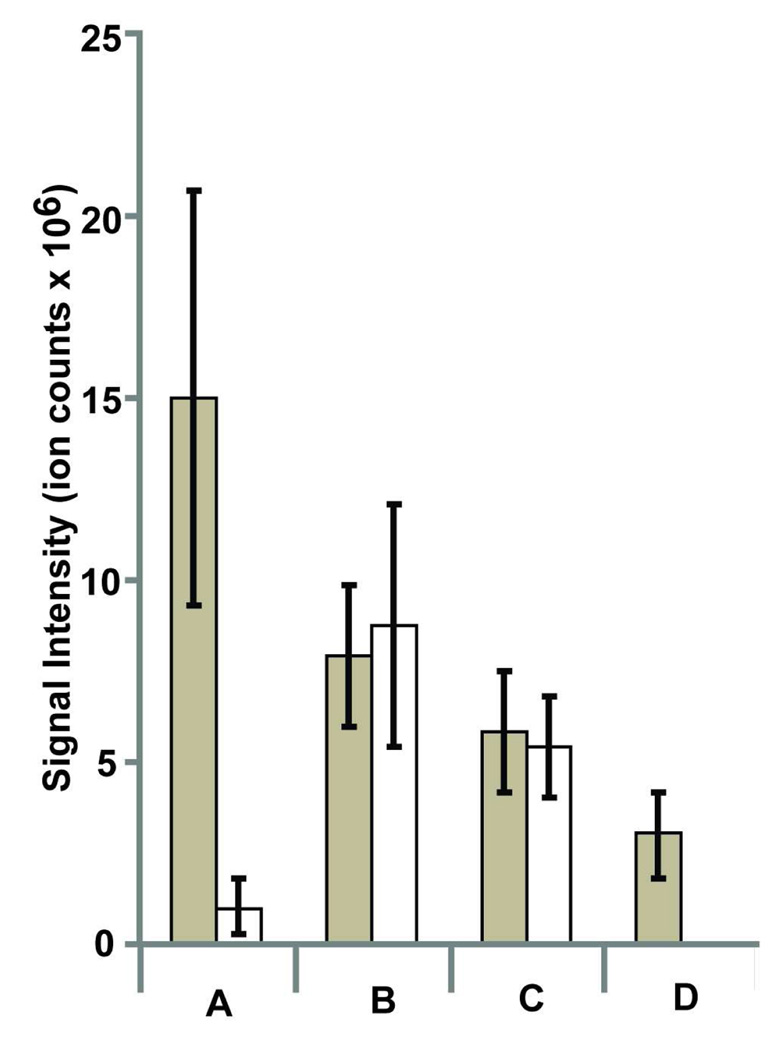
Crucial to the success of a peptidomics workflow is the ability to effectively and efficiently isolate peptides from tissues and to analyze different types of peptides by MS. The first step in this process is the heating of tissues prior to tissue homogenization, which inactivates proteases and prevents the degradation of peptides. Heating methods vary from simple boiling18–19 to microwave irradiation19–20 of the sample prior to homogenization. In the standard peptidomics workflow, kidneys are heated by boiling in hot water and this successfully inactivates proteolysis, including the complete inactivation of DPP4 as measured by a substrate assay.12 To assess a difference, if any, between boiling and microwave irradiation, we compared the two approaches directly by measuring the ion intensities of the known DPP4 substrates, RPGLLDLKGKAKWD (diazepam-binding inhibitor(92–104) [DBI(92–104)]) and LPAPEKFVKDIDGGIDQDIFD (meprinβ(21–41) [Mepβ(21–41)], LPAPEKFVKDIDGGIDQDIFD), in the LC-MS data. Importantly, both heating methods abolished DPP4 activity as measured by a lysate activity assay using the specific DPP4 substrate H-GlyPro-AMC (Supporting Information). Analysis of DPP4−/− regulated peptides revealed that signal intensity of Mepβ(21–41) was more sensitive to the heating conditions than DBI(92–104) (Figure 2). Moreover, the boiled sample gave better overall intensities for the two peptides and since boiling enables more samples to be prepared in parallel we chose to continue to denature samples by boiling of tissues prior to homogenization.
Figure 2.
Optimal conditions for peptide isolation from tissues. Signal intensities were used to quantify the isolation of peptides as a function of changes in the isolation conditions. Two DPP4 substrates, RPGLLDLKGKAKWD (diazepam-binding inhibitor(92–104)) (dark bars) and LPAPEKFVKDIDGGIDQDIFD (meprin-β(21–41)) (white bars) were quantified under different isolation conditions that varied the heating method used to denature proteolytic activity (A and B) or the addition of chaotropic agents to break up any potential peptide aggregates (C and D). Specific conditions were as follows: A. samples were microwaved and homogenized in 0.25% acetic acid(aq); B. samples were boiled and homogenized in 0.25% acetic acid(aq); C. samples were boiled and homogenized in an 8M urea solution; D. samples were boiled and homogenized in a 6M Gnd•HCl solution.
We decided to examine whether aggregation is a factor by looking at different chaotropic agents, which are known to break up peptide and protein aggregates, during the peptide extraction step. Specifically, 6 M GndHCl or 8 M Urea was added to the standard homogenization buffer and the effect of these chaotropes was assessed by looking at the signal intensities of Mepβ(21–41) and DBI(92–104). This experiment did not reveal any improvements in signal intensities for the two peptides with the chaotropic agents, which indicates that aggregation is not likely an issue (Figure 2). The peptide extraction efficiency using these various conditions was also quantified by measuring the recovery of a stable isotope-labeled version of the DBI(92–104) peptide. This peptide was added (1.5 pmoles) to homogenates prepared using 0.25% aqueous acetic acid, 8M Urea, or 6M GndHCl and processed using the standard conditions (MWCO filter, Sep Pak, etc.). The extraction efficiency of these different conditions was determined by comparing the peak area of the peptide standard in the extracted samples to the peak area of the pure peptide standard added to a DPP4−/− LC-MS sample. It was necessary to add the peptide to the LC-MS sample to account for matrix effects (i.e., the presence of other peptides) that influence the ion intensity of any peptide (Supporting Information). The results showed that peptide extraction with 0.25% aqueous acetic acid proved to be the overall superior method with an 84% recovery, versus 36% for 8 M Urea and 50% for 6 M GndHCl. Consequently, all samples were prepared using a 0.25% aqueous acetic acid solution without any additives.
Optimization of the Data Analysis Step
The upcoming optimization steps require a comparison of DPP4+/+ and DPP4−/− samples to identify any additional DPP4 substrates, which is used to assess improved peptidome coverage. As a result, we decided that it would be prudent to improve the data analysis step first, which would help accelerate the downstream experiments. In the “standard peptidomics workflow”, peptides belonging to the DPP4+/+ or DPP4−/− samples were identified by looking for all-or-none differences in the tandem MS data between the samples, typically referred to as spectral counting,21, and then quantifying the difference between the two samples by integration of the corresponding ion in an extracted ion chromatogram (EIC). For example, if a peptide was only identified in the DPP4−/− sample, an EIC of that ion would be generated and quantified in all samples (DPP4+/+ and DPP4−/−, typically an N = 3 or 4). After quantitation, ions (and peptides) are retained if there is a statistically significant increase in the DPP4−/− sample. While this protocol is quite effective, it is tedious due to the large numbers of false positives that must be dealt with during the EIC analysis step. Of course, this limitation is not surprising because spectral counting was developed for proteomics, not peptidomics work.
Instead, we turned to a different approach offered by the program XCMS,22–24 which was developed to determine differences in ion intensities to quantify changes between metabolomics LC-MS datasets. XCMS identifies differences in LC-MS chromatograms by aligning peaks in the LC-MS chromatograms, quantifying these differences using the mass ion intensity (i.e., area under the curve), and providing an output file that statistically ranks changing ions between two datasets. We reasoned that XCMS could also be used to identify changing ions in our LC-MS-based peptidomics data. DPP4+/+ and DPP4−/− kidney samples (N = 3) were analyzed using XCMS and we assessed the performance of XCMS by its ability to detect differences in three ions, corresponding to DPP4 substrates we routinely identify by spectral counting: DBI(92–104); APVNVTTEVKS, elongation factor 1 alpha(281–291) [EF-1α(281–291)]; and Mepβ(21–41).
In our first attempt, XCMS only identified one of these peptides. Upon manual inspection of the EICs and the XCMS output file we realized that XCMS was unable to efficiently identify and integrate peaks in our LC-MS chromatograms due to jagged peak shapes. We found that this jagged peak shape is sensitive to the data collection parameters and if we collect full scan MS data only (i.e. no tandem MS) during the run the peak shapes become smooth and easily identifiable by XCMS (Supporting Information). Indeed, when samples were measured using full scan MS only, XCMS was able to identify all three of the changing peptides between the DPP4+/+ and DPP4−/− samples. In addition, we also confirmed the relative quantitation of DBI(92–104) that we obtain from ion intensity measurements by using the stable isotope-labeled version of the DBI(92–104) peptide, which we spiked into the LC-MS samples before analysis. In this experiment, the area of the natural peptide ion peak was compared to the area of 50 fmol of the stable isotope-labeled DBI(92–104) peptide. As expected, the DBI(92–104) was significantly elevated in the DPP4−/− samples (94.6±31.7 fmol in the DPP4−/− vs 15.5±6.7 fmol in DPP4+/+ LC-MS samples). Similar results were obtained with or without the standard indicating the ion intensity measurements are a reliable means of quantiation.
Thus, the LC-MS analysis now consists of a two-step process, where full scan MS data is initially collected to enable the quantification of ions between samples, followed by a tandem MS experiment to identify the peptide sequences that corresponded to these ions. This two-step process enabled us to replace spectral counting with a faster and more reliable data analysis procedure that uses XCMS for the quantitation of ions, and SEQUEST25 for the subsequent identification of peptides.
Comparison of DPP4 Samples Using the Optimized Workflow
Before moving ahead we needed to establish a baseline number of changes we detect between DPP4+/+ vs DPP4−/− kidneys using the optimized workflow and the current LC-MS setup, which differed from our previous analysis. As mentioned, we could already detect many of the previously identified peptides but we needed to assess the impact of the two-step data analysis approach. Comparison of DPP4+/+ to DPP4−/− kidney samples using XCMS revealed twelve statistically significant (p < 0.05) DPP4-regulated peptides in the samples. The peptides ranged from 9 to 22 amino acids in length, including seven peptides elevated in the DPP4−/− samples and five peptides elevated in the DPP4+/+ samples (Table 1). In these experiments we identified a total of seven DPP4 substrates (i.e. peptides elevated in the DPP4−/− sample), including three previously identified substrates12 DBI(92–104), EF-1α(281–291), Mepβ(21–41), and four novel substrates. Two previously identified substrates were present but not picked up by XCMS because their ion intensities were too low for accurate quantitation.12 We attribute these differences between the present experiments and our previous work to the fact that the instrument setup was completely different (both the LC and the MS). Not surprisingly the most abundant ions (DBI(92–104), EF-1α(281–291), Mepβ(21–41)) were the most consistent between the two datasets, while less abundant ions that are more sensitive to changes in chromatography conditions are less consistent.12 Regardless, any improvements we make in our peptidome coverage will be independent of the instrumentation used, even though some peptides will always differ as a function of different instruments.
Table 1.
Absolute fold changes of peptides identified during global peptide profiling experiments (N = 6) with DPP4+/+ and DPP4−/− mice using the 1D optimized peptidomics workflow. The preferred DPP4 truncation sites in the DPP4−/− elevated peptide sequences are highlighted in bold. The precursor amino acid for the DPP4+/+ elevated peptides is shown in bold and parentheses.
| Protein (peptide region) | Peptide Sequence | Fold Change |
|---|---|---|
| DPP4−/− Elevated Peptides | DPP4−/−/ DPP4+/+ | |
| ATPase, H+ transporting, lysosomal V1 subunit G1(107–118) (Atp6v1g1(107–118)) |
RPEIHENYRING | 8.9** |
| Cathepsin B(74–86) (CtsB(74–86)) | LPETFDAREQWSN | 3.34** |
| Diazepam-binding inhibitor(92–104) (DBI (92–105)) |
RPGLLDLKGKAKWD | 22.7** |
| Elongation factor 1 alpha(281–291) (EF-1α(281–291)) |
APVNVTTEVKS | 14.8** |
| Meprin-β(21–41) (Mepβ(21–41)) | LPAPEKFVKDIDGGIDQDIFD | 28.9** |
| Solute carrier family 9, member 3, regulator 1 (275–296) (Slc9a3r1(275–296)) |
SPRPALARSASSDTSEELNSQD | 2.2** |
| Solute carrier family 22 member 12(3–17) (Slc22a12(3–17)) |
FPELLDRVGGLGRFQ | 7.1** |
| DPP4+/+ Elevated Peptides | DPP4+/+/DPP4−/− | |
| Cathepsin B(76–86) (CtsB(76–86)) | (P)ETFDAREQWSN | 3.0** |
| Cytochrome C oxidase subunit 5A(129–146) (Cox5a(129–146)) |
(P)TLNELGISTPEELGLDKV | 2.6** |
| EF-1α(283–291) | (P)VNVTTEVKS | 3.2** |
| Mepβ(25–41) | (P)EKFVKDIDGGIDQDIFD | 4.4** |
| Peroxiredoxin- 5(52–64) (Prdx5(52–64)) | (P)IKVGDAIPSVEVF | 3.2** |
p<0.05
p<0.01
Comparison of DPP4 Samples with the Introduction of a Trypsin or Reduction/Alkylation Step
The fact that we only detect shorter peptides (9–22 amino acids) and do not detect any cysteine-containing peptides suggested that the standard workflow might benefit from the addition of steps to measure longer peptides and/or peptides containing cysteine. In an attempt to better measure thiol-containing peptides and long peptides (<30 amino acids) regulated by DPP4, we applied a reduction/alkylation step or a trypsin digestion step, respectively, to our isolated peptides prior to LC-MS analysis. From this point forward, we assess any improvement in the peptidome coverage by the number of new DPP4 substrates we identify in the DPP4−/− sample after comparison of the DPP4−/− and DPP4+/+ samples by XCMS, and peptide identification by SEQUEST.
Using a reduction/alkylation step we were able to detect all seven DPP4 substrates identified above, but did not identify any additional peptides, including any cysteine-containing peptides. While in this case the reduction/alkylation did not seem to improve our coverage it also did not hinder the detection of any of the bona fide substrates. By contrast, the trypsin digest experiment revealed five additional DPP4−/− elevated peptides (Table 2). On the basis of these results it is clear that the introduction of a trypsin digest step provides a truly orthogonal set of conditions, which in this case increased the total number of DPP4 regulated peptides we identify. Since the peptides we detect are processed by trypsin we cannot determine the structure of the full-length DPP4 substrate but are only identifying a trypsin fragment of the actual substrate. For some of these peptides it appears as if the DPP4 cut site (H2N-XaaPro) comes after the trypsin cut site, which cannot be the case if the peptide is regulated by DPP4 in vivo. For example, the galectin-1(75–88) peptide appears to be cut by trypsin in front of the DPP4 cut site, (R)EPAFPFQPGSSITEV, which would mean that the peptide could not be cut by DPP4 in vivo. The likely explanation for this observation is that the peptide EPAFPFQPGSSITEV was present, but undetected, in our original sample and upon trypsin digestion we cause changes in the peptidome that enable the detection of this peptide in this sample. Additionally, we also find peptides that differ between genotypes but lack a DPP4 cut site. For example, ATP synthase subunit alpha mitochondrial(59–73), ILGADTSVDLEETGR, is substantially elevated in the DPP4−/− sample, but does not contain a DPP4 cut in, or near, the peptide. Of course, some percentage of the DPP4 regulated peptides we identify might not be direct substrates of the enzyme, but are the result of secondary changes associated with the loss of DPP4 activity. For example, DPP4 mice have improved glucose tolerance and this might cause changes in the peptidome as well. In cases, where we do not find a DPP4 cut site we cannot distinguish between direct or indirect regulation.
Table 2.
Absolute fold changes of peptides identified during global peptide profiling experiments (N = 4) with DPP4+/+ and DPP4−/− mice using the 1D optimized peptidomics workflow and trypsin cleavage of the isolated peptidome. The canonical DPP4 cleavage site is highlighted in bold.
| Protein (peptide region) | Peptide Sequence | Fold Change |
|---|---|---|
| DPP4−/−Elevated Peptides | DPP4−/−/ DPP4+/+ | |
| Acyl-coenzyme A synthetase(486–502) | (E)HPAVSETAVISSPDPSR | 14.6* |
| ATP synthase subunit alpha mitochondrial(59–73) | (R)ILGADTSYDLEETGR | 15.5** |
| ATP synthase subunit d, mitochondrial(150–161) | (K)YPYWPHQPIENL | 11.6* |
| Cytochrome b-c1 complex subunit Rieske(62–77) | (A)RPLVATVGLNVPASVR | 3.3** |
| Galectin-1(75–88) | (R)EPAFPFQPGSSITEV | 5.6** |
p<0.05
p<0.01
Additionally, analysis of the tandem MS data from these trypsin digest samples reveals the presence of fragments of Mepβ and DBI, but the signal intensities from these peptides are too low for accurate quantitation by XCMS and, therefore, were not automatically identified as changes. In the future this limitation can be remedied by loading more sample to increase the ion intensity. We did not attempt to load more sample at this point because we wanted to make a direct comparison between our standard peptidomics workflow and these modified conditions, which required an equivalent amount of sample to be used. In total, the results indicate the value of trypsin digestion and, more generally, support our strategy of using DPP4 substrates as a measure of peptidome coverage.
Comparison of DPP4 Samples using Orthogonal Fractionation Strategies
To improve our peptidome coverage further we looked at orthogonal fractionation strategies of our peptidome samples prior to LC-MS analysis. For example, proteome fractionation is often accomplished using a two-dimensional fractionation approach that separates samples using two different (orthogonal) types of chromatography, such as strong cation exchange (SCX) followed by reverse phase chromatography (RP).26 In doing so, each fraction analyzed is less complex, improving the signal-to-noise during MS analysis, which leads to enhanced sensitivity and greater overall coverage. Here, we explore using an offline (i.e., not directly connected to the LC-MS system) strong cation exchange (SCX)27–29 or OFFGEL electrophoresis (OGE) fractionation of the peptidome samples prior to LC-MS analysis as a means to discover additional DPP4 substrates.17,28,30 In the proteomics field, application of these techniques lead to a greater number of proteins detected,31 and we anticipate that introduction of these offline fractionation steps will increase the number of DPP4 substrates we identify. Previous examples of online SCX-RP-peptidomics experiments are known but these attempts did not quantify peptides.32
The decision was made to use offline SCX, instead of the online variants (e.g., MUDPIT26,32) because it would allow us to use the same column for each separation, and give us the most reproducible SCX fractionation. Reproducibility of the SCX fractionation is essential for accurate peak alignment and quantitation during the subsequent LC-MS analysis. The SCX fractionation was optimized using four peptide standards, including three DPP4 regulated peptides, which were selected because they differ in charge at pH 2.6 and, as a result, should separate cleanly by SCX. The peptides and their representative charge states include: heat shock protein 1(10–19), LPLFDRVLVE, +2; Mepβ(21–41), +3; DBI(94–104), +4; and DBI(92–104), +5. A step gradient using no KCl, 40 mM KCl, 100 mM KCl, 200 mM KCl, and 400 mM KCl buffers was chosen, because this format, as opposed to a linear gradient, would provide the most reproducible separation. The four standard peptides eluted according to charge and in distinct salt fractions (Supporting Information); the +2 peptide in 40 mM KCl, the +3 peptide in 100 mM KCl, the +4 peptide in 200 mM KCl, and the +5 peptide in 400 mM KCl.
Initial experiments using DPP4+/+ vs DPP4−/− kidney samples (N = 2) revealed that a majority of the peptides elute into the 40 mM KCl fraction, as expected from peptides in the +2 and +3 charge state, which predominate our sample. Moreover, comparison of the charge state of a peptide with the salt fraction for other salt concentrations indicated that the fractionation was predictable, even though there were some exceptions (Supporting Information). After these initial studies, the experiment was repeated with an increased sample size (N = 4, DPP4+/+ vs. DPP4−/−) to enable the identification of novel substrates. We found a dramatic increase in the number of DPP4 substrates identified using the SCX-RP-peptidomics approach (Table 3 and Supporting Information). We identified 58 substrates using SCX-RP, an 8-fold improvement over the standard peptidomics workflow, which indicates that we had only scratched the surface of the DPP4-regulated peptidome in our initial studies, and that the majority of the differences were not identified in those studies. In addition to the large numbers of DPP4 substrates identified we were also able to detect peptides with DPP4 cleavage sites (H2N-XaaPro) that were unaffected between genotypes (Table 4). This is not surprising since it is not expected that every potential DPP4 substrate and DPP4 will come into contact with each other in the context of a cell or a tissue. Furthermore, unchanged peptides were also seen in every set of conditions examined to support the idea that we are looking at endogenous differences in DPP4 substrates and provide additional confidence in the differences we do find. The identification of additional DPP4 substrates support our original rationale for wanting to improve our peptidomics platform, since a majority of the DPP4 regulated substrates in the kidney are only evident after additional fractionation of the peptidome.
Table 3.
Absolute fold changes of peptides identified during global peptide profiling experiments (N = 4) with DPP4+/+ and DPP4−/− mice using either the SCX-RP-peptidomics workflow or the OGE-RP-peptidomics workflow. The preferred DPP4 truncation sites in the DPP4−/− elevated peptide sequences are highlighted in bold.
| Protein (peptide region) | Peptide Sequence | Fold Change |
|---|---|---|
| DPP4−/− Elevated Peptides | DPP4−/−/ DPP4+/+ | |
| SCX-RP workflow | ||
| 40 S ribosomal protein s2(264–275) | SPYQEFTDHLVK | 13.9* |
| Catalase(23–40) | RPDVLTTGGGNPIGDKLN | 35.6* |
| Histidine triad nucleotide-binding protein(27–34) | IPAKIIFE | 17.96* |
| Lysosomal protective protein(346–354) | IPESLPRWD | 17.63** |
| Major Urinary Protein 6(110–118) | IPKTDYDNF | 28.67** |
| Tripeptidyl-peptidase 1(497–505) | PPLGFLNPR | 10.41** |
| OGE-RP workflow | ||
| Alpha-globin transcription factor CP2(108–115) | LPELNGKL | 5.49** |
| ATP synthase-coupling factor 6, mitochondrial (90–96) |
FPTFKFD | 7.74* |
| Fructose-bisphosphate aldolase B(5–13) | FPALTPEQK | 26.5** |
| Microtubule-associated protein tau(538–546) | APVPMPDLK | 6.69** |
| PDZK1-interacting protein 1(90–113) | FRSSEHKNAYENVLEEEGRVRSTP | 7.92** |
| Protein kinase c and kinase substrate in neurons protein 2(340–358) |
KPGSNLSVPSNPAQSTQLQ | 10.9** |
p<0.05
p<0.01
Table 4.
Unaffected XP motif renal peptides observed under different experimental conditions during DPP4+/+ and DPP4−/− peptidomics profiling.
| Protein (peptide sequence/region) | Experimental Condition |
Fold Change |
|---|---|---|
| DPP4−/− Elevated Peptides | DPP4−/−/ DPP4+/+ | |
| Acid sphingomyelinase-like phosphodiesterase (31–43) APAVGQFWHVTDL |
1D Non-reduction/alkylation; non-tryptic |
1.2 |
| Fructose-bisphosphate aldolase A(398–416) TPSGQSGAAASESLFISNH |
1D Reduction/alkylation | 1.1 |
| Apolipoprotein A-I(124–135) APLGAELQESAR | 1D Trypsin digest | 1.1 |
| Sorbitol dehydrogenase(16–25) GPGDIRLENY | 2D (OFFGEL) | 1.2 |
| Thymosin beta-4(193–208) LPSKETIEQEKQAGES |
2D (SCX) | 1.1 |
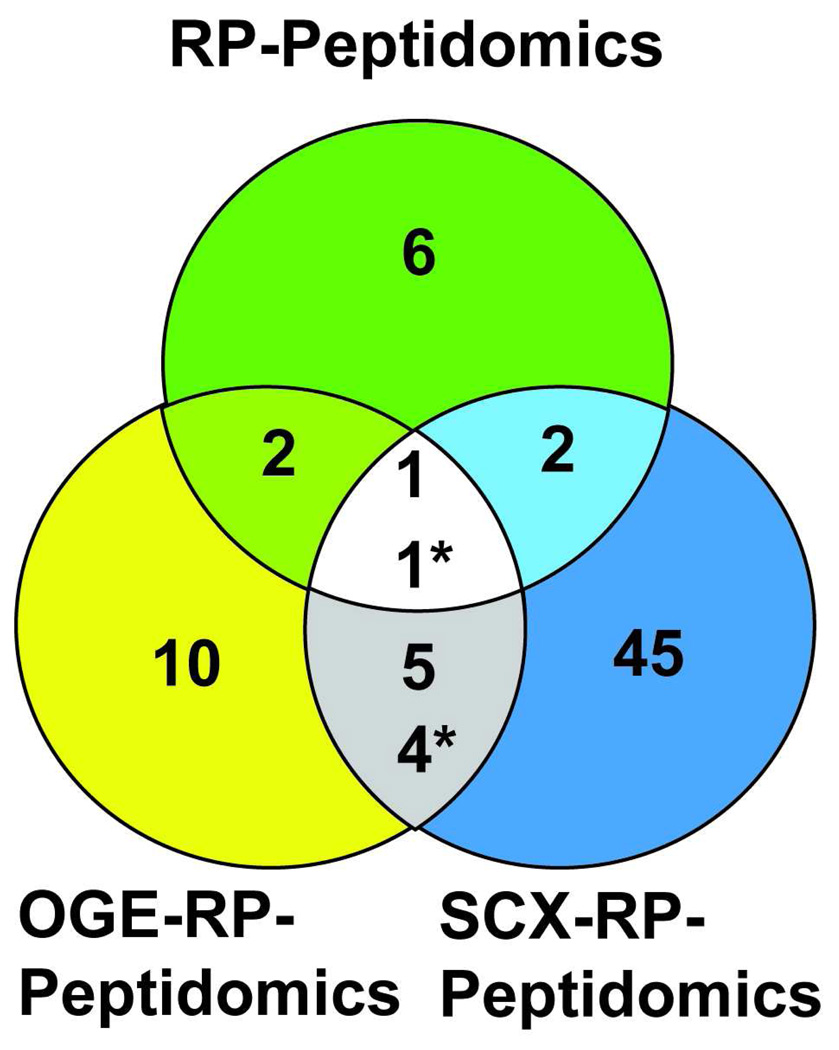
In addition to the SCX-2D workflow, OGE was also applied to the fractionation of the peptidome, which is referred to as the OGE-RP-peptidomics workflow. Again, a small scale test run (N = 2, DPP4+/+ vs. DPP4−/−) profiling of the kidney peptidome was used to identify fractions that contained the most peptides and qualitatively check the reproducibility of the fractionation. Four total fractions were analyzed by LC-MS, which were obtained by grouping lanes 1 and 2, lanes 3 and 4, lanes 5 and 6, and lanes 7 through 12 from the OGE apparatus. The MS data revealed that the peptides separated in the gel, for the most part, according to their isoelectric points (Supporting Information). By examining a larger sample set (N = 4, DPP4+/+ vs. DPP4−/−) using the OGE-RP-peptidomics workflow, a number of new DPP4 regulated peptides were identified (Table 3 and Supporting Information). Importantly, many of these peptides are specific to the OGE-RP-peptidomics workflow, indicating that the OGE and SCX can be run in parallel to increase the peptidome coverage even further (Figure 3).
Figure 3.
Venn diagram showing the number of DPP4 regulated peptides identified under different conditions. The conditions compared include the standard peptidomics workflow, OGE-RP-peptidomics workflow, and the SCX-RP-peptidomics workflow. The SCX-RP-peptidomics workflow provides the most peptidome coverage, but the minimal overlap between methods suggests that the greatest coverage will only come from using different approaches.
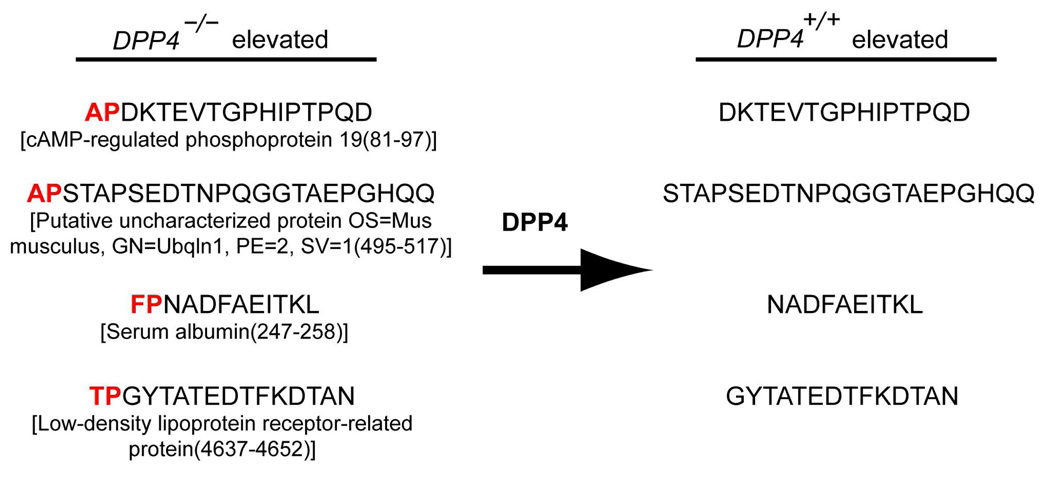
Overall, peptidome fractionation results are very encouraging because of the dramatic increase in the number of DPP4 regulated peptides identified, which indicates an improvement in our peptidome coverage. Indeed, a majority of the peptides are clearly DPP4 substrates due to the presence of the canonical DPP4 cleavage site (H2N-XaaPro) in the sequence and, in some cases, we were able to detect both the substrate (elevated in DPP4−/−) and product (elevated in DPP4+/+) in our samples. For example, we found the novel DPP4 substrate APDKTEVTGPHIPTPQD (cAMP-regulated phosphoprotein 19(81–97)) elevated in the DPP4−/− sample and the expected DPP4 cleavage product, DKTEVTGPHIPTPQD, elevated in the DPP4+/+ sample (Figure 4 and Supporting Information). We refer to these peptides that are linked between KO and WT samples as “substrate-product pairs”,12 and they provide the best evidence that an enzyme is regulating a specific peptide. In our hands the SCX-RP-peptidomics workflow was superior to the OGE-RP-peptidomics approach, but for maximal coverage both approaches can be applied in parallel.
Figure 4.
Newly identified substrate-product pairs during global peptide profiling experiments (n=4) with DPP4+/+ and DPP4−/− mice kidneys applying the SCX-RP peptidomics workflowfractionation.
Overall analysis of the kidney data
In total, the results obtained from these offline fractionation experiments were encouraging for a number of reasons. First, from a technical standpoint these experiments demonstrate the value of fractionation and the utility of DPP4 substrates in guiding the optimization strategy. Next, these results provide additional evidence for the suspected physiological and biochemical functions associated with DPP4 in the kidney. Leibach and colleagues had originally postulated that DPP4 is involved in renal peptide catabolism, through measurement of urine proline-containing peptide levels in rats possessing a natural mutation in DPP4.15 Our original experiments in mice supported this idea through the identification of a handful of DPP4 substrates, all of which are fragments of kidney proteins. These studies greatly strengthen the evidence for a catabolic role for DPP4 due to the significantly larger numbers of DPP4 substrates identified that are derived from a variety of different proteins. For example, we see examples of fragments of membrane (i.e. low-density lipoprotein receptor-related protein 2), mitochondrial (i.e. ATP synthase-coupling factor 6), and cytosolic (i.e. cytoplasmic dynein 1 light intermediate chain 1) proteins as DPP4 substrates, which supports a general role for DPP4 in peptide and protein catabolism in the kidney.
Interestingly, many of the substrates we discover are derived from intracellular proteins. In tissues, DPP4 is an extracellular membrane protein, which necessitates that DPP4 substrates must be present outside the cell. We cannot provide a physiological mechanism for the ability of DPP4 to cut peptides derived from intracellular proteins, but we suspect that some of the precursor proteins and peptides are in the glomerular filtrate as it passes through the kidney, or are released from cells in the kidney as they lyse. Furthermore, even with our improved coverage we are probably still not detecting all potential DPP4 substrates due to the normalization of tissue by weight, instead of histological composition. Normalization to kidney weight provided a practical means to assess total amount of sample extracted, but the kidney is a complex organ with many histological regions that might also differ slightly between samples. Therefore, our results represent the most robust changes that can be picked up by looking at the whole kidney but there are probably other changes that are being overlooked because we are not controlling for these histological differences.
A peculiarity in our initial studies was the lack of any proline-terminated peptides elevated in DPP4−/− samples, which indicated that kidney aminopeptidases do not cut adjacent to proline residues. This aminopeptidase selectivity suggested a model where aminopeptidases and DPP4 are interlinked in a metabolic pathway for processing peptides with internal proline residues. In this model, peptides with internal prolines are processed initially by aminopeptidase activity until a penultimate proline is reached and the peptide is no longer a substrate. At this point, aminopeptidase activity ceases and the peptide is released and DPP4 is able to cleave this substrate (Figure 5). We found strong evidence for this in a series of in vitro experiments using tissues from aminopeptidase null mice. However, our previous studies did not provide enough DPP4 substrates in vivo to unambiguously support this model. Now that we have greatly increased the number of DPP4 substrates we can state that an interlinked aminopeptidase and DPP4 pathway is operative in vivo. Indeed, out of a total of 70 DPP4−/− elevated peptides, we find only 4 peptides do not have an n-terminal H2N-XaaPro motif, and only one of these peptides terminates in a proline. Thus, the improved peptidome coverage has helped strengthen a role for DPP4 in renal peptide catabolism, and provides additional evidence for a biochemical pathway comprised of aminopeptidase and DPP4 activities.
Figure 5.
Model for the regulation of proline-containing peptides through the combined action of aminopeptidase and DPP4 activities. On the basis of in vitro experiments in brush border membrane lysates we determined that aminopeptidase activity is necessary for the production of DPP4 substrates. In this model, aminopeptidase activity begins to process proline-containing peptides until a penultimate proline is reached. At this point, the peptide is no longer a substrate for aminopeptidases and it is then cleaved by DPP4. These enzyme activities generate amino acids and dipeptides that can be recovered through amino acid and dipeptide transporters, respectively, to provide an overall pathway for protein catabolism in the kidney. This model would predict that in the absence of DPP4 there would be an accumulation of H2N–XaaPro-peptides, but no accumulation of H2N–Pro-peptides, in tissues from DPP4−/− mice, which is strongly supported by the current peptidomics studies.
Comparison of Gut Samples using the SCX-RP-peptidomics platform
After using the kidney peptidome, and known DPP4 substrates, to successfully optimize the peptidomics workflow and acquire deeper insight into DPP4 renal activity, we sought to apply this approach to another tissue. The goals of these experiments were twofold: 1) to test the generality of this approach and 2) characterize DPP4 regulated peptides, and substrates, in another tissue. As mentioned, the gut has high levels of DPP4 and is of great interest because many important bioactive peptides are located there which exhibit strong interactions with the endocrine, nervous, and immune systems.33 While DPP4 regulation of the GLP-1 and consequently insulin levels is well documented in the blood, a general characterization of DPP4 activity in the gut is lacking, making these experiments an interesting endeavor due to the potential of making a new discovery on the role of DPP4 in the gut.
Gut samples were boiled to quench proteolytic activity and a DPP4 lysate activity assay showed that boiling the gut inactivated DPP4 (data not shown). A comparative profiling experiment (N = 4, DPP4+/+ vs. DPP4−/−) of the gut was attempted twice using the standard peptidomics workflow but the LC-MS data were extremely noisy, making it difficult to align the samples by XCMS. We believe that certain factors in the gut, like residual food and bile salts, were interfering with our LC-MS experiments. Instead of using XCMS, we reverted to determining differences by looking for all-or-none changes in the tandem MS files to identify differences between the DPP4+/+ and DPP4−/− samples from the standard peptidomics workflow. For the offline fractionation experiments, we chose to only use the SCX-RP-peptidomics workflow due to the improved coverage of this approach. Comparative profiling of the DPP4+/+ vs. DPP4−/− gut samples (N = 4) revealed that the LC-MS data were still somewhat noisy, especially when compared to the kidney data, but the fractionated LC-MS chromatograms could be aligned and analyzed by XCMS. The additional steps involved in the SCX fractionation must have resulted in better purification of samples and minimized the contaminants that were interfering with the standard peptidomics workflow.
Several DPP4−/− elevated peptides were identified in the gut and many of these contained the canonical DPP4 cleavage site (e.g., H2N-XaaPro) indicating the likelihood that these peptides are substrates (Table 5 and Supporting Information). As in the kidney, we found that a majority of these DPP4 substrates are derived from proteins, such as glyceraldehyde-3-phosphate dehydrogenase and histone H2B type 1-M (Table 5). Interestingly, the histone H2B type 1-M peptide contains a penultimate alanine residue (i.e., H2N-XaaAla), a sequence that can also be cleaved by DPP4 in vitro,7 which indicates that our in vivo analysis provides the same biochemical information about DPP4 that can be gained from in vitro experiments. More generally, the discovery of these protein fragments as DPP4 substrates suggests that DPP4 is likely playing a role in protein catabolism in the gut, a similar function to what it was doing in the kidney. Moreover, the same aminopeptidases that work in concert with DPP4 in the kidney brush border membranes are also found in gut membranes.16 The co-expression of these proteins in the gut means that the same pathway that was operative in the kidney, where aminopeptidase activity generated DPP4 substrates, is likely operating in the gut as well and that the combined proteolytic activities of these enzymes has been co-opted to perform the same biochemistry, protein catabolism, in two different tissues.
Table 5.
Absolute fold changes of peptides identified during global peptide profiling experiments (N = 4) with gut tissue from DPP4+/+ and DPP4−/− mice using the SCX-RP-peptidomics workflow. The preferred DPP4 truncation sites in the DPP4−/− elevated peptide sequences are highlighted in bold.
| Protein (peptide region) | Peptide Sequence | Fold Change |
|---|---|---|
| DPP4−/− Elevated Peptides | DPP4−/−/ DPP4+/+ | |
| Chymotrypsinogen B(21–28) | VPAIQPVLTG | 9.83* |
| Glyceraldehyde-3-phosphate dehydrogenase (219–228) |
IPELNGKTG | 14.95* |
| Histone H2B type 1-M(110–122) | HAVSEGTKAVTKY | 10.55** |
| Junction plakoglobin(709–717) | VPLDPLDMH | 11.13** |
| Putative uncharacterized protein GN=Tf, PE=2, SV=1(94–114) |
GREEKPAASDSSGKQSTQVMA | 18.86** |
| Secretogranin-1(21–35) | APVDNRDHNEEMVTR | 29.85* |
p<0.05
p<0.01
In the gut, we also find evidence for DPP4 regulation of endogenous peptides linked to specific biological processes. A fragment of a defensin peptide (defensin-related cryptdin-5(20–33) DPIHKTDEETNTEE; Supporting Information) was found to be a substrate of DPP4. Defensins are known for their antibacterial properties34–35 and they are initially processed as 73 amino acid pro-bioactive peptides. Studies have shown that one or two peptidases may be involved in the formation of the mature bioactive peptide,36–37 with metalloproteinase matrilysin proposed to be the main peptidase responsible for the cleavage forming the mature peptide. While the defensin-related cryptdin-5(20–33) is thought to be an inactive fragment of the defensin, it is possible that DPP4 regulates longer fragments of the defensin-related cryptdin-5(20–33), which might not be detected under our current methods. In addition to the defensins, we also identify a secretogranin peptide as a DPP4 substrate (Table 5). Secretogranins and secretogranin-derived peptides are components of secretory vesicles, and while the functions of these proteins/peptides are still being elucidated, recent experiments have revealed a potential role for secretogranin peptides in hormonal signaling pathways.38 In the future, it will be interesting to see whether this particular secretogranin has any bioactivity, which would link DPP4 to additional signaling pathways and biology.
In addition to increasing the number of DPP4 regulated peptides identified, the SCX-RP-peptidomics platform was able to detect a number fragments of known bioactive peptides–glucagon, neuropeptide Y, and somatostatin–that were not detected using the standard peptidomics platform (Table 6). The ability to detect these peptides suggests that in addition to using this approach to identify peptidase substrates, these optimized peptidomics workflows will also find use as a general profiling tool to better understand the regulation of some of the body’s most important molecules, peptide hormones. These promising results also point to the need for more targeted approaches for the quantitation of bioactive peptides, since most of the instrument bandwidth is being spent collecting data on peptides that are not bioactive. In the future, we will look to develop protocols for the targeted analysis of bioactive peptides39 by using our optimized peptidomics workflow in combination with triple quadrupole (QQQ) mass spectrometer, which provide the most sensitive detection scheme. As a result, this work provides an important tool for peptidase substrate discovery and looking forward will enable us to build a platform for bioactive peptide analysis.
Table 6.
Increase in the number of bioactive peptide fragments detected in datasets corresponding to global peptide profiling experiments (N = 4) upon application of the SCX-RP-peptidomics workflow.
| Peptide |
|---|
| Standard Peptidomics Workflow |
| Defensin-related cryptdin 3, 5, 9, 20, 23, 24 |
| Defensin-related cryptdin-related sequence 1, 2 |
| Somatostatin |
| VIP peptides |
| SCX-RP-Peptidomics Workflow |
| Defensin-related cryptdin 3, 5, 9, 11, 20, 23, 24 |
| Defensin-related cryptdin-related sequence 1, 2, 10 |
| Glucagon (Including a GLP 1 fragment) |
| Insulin-1 |
| Insulin-2 |
| Neuropeptide Y |
| Somatostatin |
| VIP peptides (Including PHI 2741) |
Conclusion
We set out to improve our peptidomics platform by varying key parameters in the workflow while using the number of newly identified DPP4 substrates as a way to quantify any improvements. These studies led to an ∼10-fold improvement in the number of total DPP4 substrates identified in the kidney (7 vs. 70) confirming the idea that the platform could be improved. Big improvements in the peptidome coverage resulted from the application of XCMS to accelerate the discovery of changing ions (and eventually peptides) and, most importantly, the improved fractionation of the peptidome by off-line fractionation methods (SCX and OGE). While additional fractionation steps increase the time it takes to analyze a sample, the dramatically increased peptidome coverage using offline fractionation is well worth the additional time (Table 7). Furthermore, the gains we see in our optimized method should be independent of the mass spectrometer used, but the total number of peptides correctly identified could improve by using a state-of-the-art mass spectrometer capable of making measurements with high resolution and improved mass accuracy.
Table 7.
Total list of peptides identified during global peptide profiling experiments (N = 4) with DPP4+/+ and DPP4−/− samples using the standard, SCX-RP, and OGE-RP workflows. The canonical DPP4 cleavage site is highlighted in bold.
| Normal 1D | SCX-2D | OG-2D |
|---|---|---|
| LPETFDAREQWSN | RPEIHENYRING | RPEIHENYRING |
| RPGLLDLKGKAKWD | LPETFDAREQWSN | LPAPEKFVKDIDGGIDQDIFD |
| APVNVTTEVKS | APVNVTTEVKS | SPRPALARSASSDTSEELNSQD |
| SPRPALARSASSDTSEELNSQD | LPAPEKFV | FPELLDRVGGLGRFQ |
| LPAPEKFVKDIDGGIDQDIFD | PPYGQPQPGFG | LPFGDEDALK |
| FPELLDRVGGLGRFQ | DDIANSEENPTPGVV | TPGYTATEDTFKDTAN |
| RPEIHENYRING | PENVENQN | IPKTDYDNF |
| PPNPFGPVSGAQIQ | TPRPTDPIPTSEVN | |
| APAPVGPLVG | KPEFVDIINAKQ | |
| VPQLQGYLR | IPSVEVFEGEPGKKVN | |
| LPHTFTPTTQLS(M)N | KPNPDQLLKELPFPLN | |
| VPKTGVTGPYVLG | LPELNGKL | |
| VPKTGVTGPYVLGTGLS | FPTFKFD | |
| PPVQVSPLIKF | VPAASEPPVLDVKRPFL | |
| FPTFKFD | LPGVGVSML | |
| IPKNWSL | FPALTPEQK | |
| LPETFDARE | APVPMPDLK | |
| LPETFDAREQ | GPGGKEATWVVDVKN | |
| LPETFDAREQW | FRSSEHKNAYENVLEEEGRVRSTP | |
| LPETFDAREQWS | KPQAQEQPPASPEALRG | |
| LPFGDEDALK | HPDAENAFKVRL | |
| LPAETSLPLVFPKPMT | KPFPWGDGNHTL | |
| VPAASEPPVLDVKR | KPGSNLSVPSNPAQSTQLQ | |
| VPEPKIIDA | ||
| KPFPWGDG | ||
| YPLPVAHVTMLS | ||
| APFDSRFPNQNQ | ||
| KPASVSPTTPTSPTEGEAS | ||
| PPVPSPSQPPSSKPVS | ||
| GPLKDIPSDA | ||
| IPELNGKLT | ||
| APSGSFFARDNTANF | ||
| VGGTNDKGVGMGMTVPVSF | ||
| IPAKIIFE | ||
| APLQVSRGLSASTVDLSSSS | ||
| TPGYTATEDTFKDTAN | ||
| IPESLPRWD | ||
| IPKTDYDN | ||
| IPKTDYDNF | ||
| APVPMPDLKN | ||
| TPRPTDPIPTSEVN | ||
| GPITTDIREGQ | ||
| APIKVGDAIPSVE | ||
| VPKLYEQL | ||
| KPGSNLSVPSNPAQSTQLQ | ||
| LPGVLHQF | ||
| TPNSGATGNNAGPKSMEVS | ||
| APSTAPSEDTNPQGGTAEPGHQQ | ||
| YPSSSRTPQAPTPAN | ||
| FPNADFAEITKL | ||
| YPIPELGPNDVLLK | ||
| YPIPELGPNDVLLKMH | ||
| APKSILDQSISPF | ||
| KPEFVDIINA | ||
| PPLGFLNPR | ||
| PPLGFLNPRL | ||
| PPLGFLNPRLY | ||
| KPDIDAWELRKGMNTLVGY |
In this work we also enhanced our understanding of the biochemistry and biology of DPP4. The increased peptidome coverage in the kidney provided additional evidence for a role for DPP4 in proline peptide catabolism in the kidney. Moreover, since we found a much higher number of penultimate proline-containing substrates, but only one N-terminal proline peptide, elevated in DPP4−/− samples these datasets strengthened a model that suggests that N-terminal proline-containing peptide processing is a collaborative effort between aminopeptidase and dipeptidyl peptidase activities. The improved coverage afforded by our optimized platform also suggested that we might have the requisite breadth and depth to measure differences in bioactive peptide levels. In this regard, we focused our efforts in the gut, which is known to contain a number of bioactive peptide hormones, mostly involved in feeding and immunomodulation.17 Comparison of gut samples from DPP4+/+ and DPP4−/− mice identified a number of new DPP4 substrates. Some of these substrates include peptides such as secretogranins, which have been implicated in a number of biological processes,40 and a fragment of a defensin peptide, which are antimicrobial peptides.34–35 However, on the basis of a very broad substrate profile, which includes fragments of histone proteins and chymotrypsinogen, we suspect that membrane bound DPP4 in the gut serves a catabolic role in processing proline-containing peptides so that the corresponding amino acids can be extracted from proteins in the intestine. In total, this work signifies an important step forward in our development of a peptidomics platform for endogenous substrate discovery that can impact research in biochemistry and chemical biology.
Supplementary Material
Acknowledgment
We would like to thank Angela C. Lou, Channing K. Carter, and Tim Maher for technical assistance, and members of the Saghatelian lab in the preparation of this manuscript. This work was supported by the Mary Fieser Postdoctoral Fellowship (A.D.T.), Searle Scholar Award (A.S.), Burroughs Wellcome Fund Career Award in the Biomedical Sciences (A.S.), and National Institutes of Health Grants 1DP2OD002374 (A.S.).
Footnotes
Supporting Information Available. Tables on renal peptide fractionation by SCX and OFFGEL electrophoresis. Tables on DPP4 regulated peptides found in the gut and the kidney. Table on renal DPP4 regulated peptides determined through 1D and 2D methods. Figure of DPP4 activity assay on kidney lysates in the presence and absence of boiling. Figure on the comparison of MS peak shapes and areas in the presence and absence of tandem MS. Figure on peptide elution from PolySULFOETHYL™ A SCX column.
References
- 1.Saltiel AR, Kahn CR. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 2.Sayegh MH, Watschinger B, Carpenter CB. Transplantation. 1994;57:1295–1302. doi: 10.1097/00007890-199405150-00001. [DOI] [PubMed] [Google Scholar]
- 3.Marguet D, Baggio L, Kobayashi T, Bernard AM, Pierres M, Nielsen PF, Ribel U, Watanabe T, Drucker DJ, Wagtmann N. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6874–6879. doi: 10.1073/pnas.120069197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Che FY, Fricker LD. J. Mass Spectrom. 2005;40:238–249. doi: 10.1002/jms.743. [DOI] [PubMed] [Google Scholar]
- 5.Beinfeld MC, Funkelstein L, Foulon T, Cadel S, Kitagawa K, Toneff T, Reinheckel T, Peters C, Hook V. Peptides. 2009;30:1882–1891. doi: 10.1016/j.peptides.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tenorio-Laranga J, Valero ML, Mannisto PT, Sanchez del Pino M, Garcia-Horsman JA. Anal Biochem. 2009;393:80–87. doi: 10.1016/j.ab.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 7.Leiting B, Pryor KD, Wu JK, Marsilio F, Patel RA, Craik CS, Ellman JA, Cummings RT, Thornberry NA. Biochem. J. 2003;371:525–532. doi: 10.1042/BJ20021643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Materson BJ, Preston RA. Arch. Intern. Med. 1994;154:513–523. [PubMed] [Google Scholar]
- 9.Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS. Proc Natl Acad Sci U S A. 2000;97:7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Backes BJ, Harris JL, Leonetti F, Craik CS, Ellman JA. Nat Biotechnol. 2000;18:187–193. doi: 10.1038/72642. [DOI] [PubMed] [Google Scholar]
- 11.Hu LH, Ye ML, Zou HF. Expert Rev. Proteomics. 2009;6:433–447. doi: 10.1586/epr.09.55. [DOI] [PubMed] [Google Scholar]
- 12.Tagore DM, Nolte WM, Neveu JM, Rangel R, Guzman-Rojas L, Pasqualini R, Arap W, Lane WS, Saghatelian A. Nature Chemical Biology. 2009;5:23–25. doi: 10.1038/nchembio.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenblum JS, Kozarich JW. Curr. Opin. Chem. Biol. 2003;7:496–504. doi: 10.1016/s1367-5931(03)00084-x. [DOI] [PubMed] [Google Scholar]
- 14.Weber AE. J. Med Chem. 2004;47:4135–4141. doi: 10.1021/jm030628v. [DOI] [PubMed] [Google Scholar]
- 15.Tiruppathi C, Miyamoto Y, Ganapathy V, Roesel RA, Whitford GM, Leibach FH. J. Biol Chem. 1990;265:1476–1483. [PubMed] [Google Scholar]
- 16.Medeiros MD, Turner AJ. Endocrinology. 1994;134:2088–2094. doi: 10.1210/endo.134.5.7908871. [DOI] [PubMed] [Google Scholar]
- 17.Hubner NC, Ren S, Mann M. Proteomics. 2008;8:4862–4872. doi: 10.1002/pmic.200800351. [DOI] [PubMed] [Google Scholar]
- 18.Dowell JA, Vander Heyden W, Li L. J. Proteome Res. 2006;5:3368–3375. doi: 10.1021/pr0603452. [DOI] [PubMed] [Google Scholar]
- 19.Parkin MC, Wei H, O'Callaghan JP, Kennedy RT. Anal. Chem. 2005;77:6331–6338. doi: 10.1021/ac050712d. [DOI] [PubMed] [Google Scholar]
- 20.Che FY, Lim J, Pan H, Biswas R, Fricker LD. Mol Cell Proteomics. 2005;4:1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Nature Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 22.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nordstrom A, O'Maille G, Qin C, Siuzdak G. Anal. Chem. 2006;78:3289–3295. doi: 10.1021/ac060245f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 25.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, Smith RD. J. Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 26.Washburn MP, Wolters D, Yates JR., 3rd Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 27.Villen J, Gygi SP. Nat. Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slebos RJC, Brock JWC, Winters NF, Stuart SR, Martinez MA, Li M, Chambers MC, Zimmerman LJ, Ham AJ, Tabb DL, Liebler DC. J. Proteome Res. 2008;7:5286–5294. doi: 10.1021/pr8004666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang N, Xie CH, Young JB, Li L. Anal. Chem. 2009;81:1049–1060. doi: 10.1021/ac802106z. [DOI] [PubMed] [Google Scholar]
- 30.Fraterman S, Zeiger U, Khurana TS, Rubinstein NA, Wilm M. Proteomics. 2007;7:3404–3416. doi: 10.1002/pmic.200700382. [DOI] [PubMed] [Google Scholar]
- 31.Motoyama A, Yates JR. Anal. Chem. 2008;80:7187–7193. doi: 10.1021/ac8013669. [DOI] [PubMed] [Google Scholar]
- 32.McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR. Int. J. Mass Spectrom. 2002;219:245–251. [Google Scholar]
- 33.Ahlman H, Nilsson O. Ann. Oncol. 2001;12:S63–S68. doi: 10.1093/annonc/12.suppl_2.s63. [DOI] [PubMed] [Google Scholar]
- 34.Ganz T. Nat. Rev. Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 35.Hadjicharalambous C, Sheynis T, Jelinek R, Shanahan MT, Ouellette AJ, Gizeli E. Biochemistry. 2008;47:12626–12634. doi: 10.1021/bi800335e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 37.Putsep K, Axelsson LG, Boman A, Midtvedt T, Normark S, Boman HG, Andersson M. J. Biol. Chem. 2000;275:40478–40482. doi: 10.1074/jbc.M007816200. [DOI] [PubMed] [Google Scholar]
- 38.Zhao E, Basak A, Wong AOL, Ko W, Chen A, Lopez GC, Grey CL, Canosa LF, Somoza GM, Chang JP, Trudeau VL. Endocrinology. 2009;150:2273–2282. doi: 10.1210/en.2008-1060. [DOI] [PubMed] [Google Scholar]
- 39.Lortie M, Bark S, Blantz R, Hook V. Anal Biochem. 2009;394:164–170. doi: 10.1016/j.ab.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taupenot L, Harper KL, O'Connor DT. N. Engl. J. Med. 2003;348:1134–1149. doi: 10.1056/NEJMra021405. [DOI] [PubMed] [Google Scholar]
- 41.Dejda A, Sokolowska P, Nowak JZ. Pharmacol. Rep. 2005;57:307–320. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.