Abstract
Substrates and cofactors of the serine protease thrombin (IIa) employ two anion binding exosites (ABE-I and II) to aid in binding. On the surface of platelets resides the membrane bound receptor complex GpIbα/β-GpIX-GpV. IIa’s ABE-II is proposed to interact with an anionic portion of GpIbα which enhances IIa cleavage of PAR-1 and subsequent activation of platelets. In the current work, 1D and 2D NMR, analytical ultracentrifugation (AUC), and hydrogen deuterium exchange (HDX) coupled with MALDI-TOF MS were performed to further characterize the features of binding to IIa’s ABEs. The described work builds upon investigations performed in a prior study with fibrin(ogen)’s γ′ peptide and IIa (Sabo, T. M., Farrell, D. H., and Maurer, M. C. (2006) Biochemistry 45, 7434–7445). 1D line broadening NMR (1H and 31P) and 2D trNOESY NMR studies indicate that GpIbα D274-E285 interact extensively with the IIa surface in an extended conformation. AUC demonstrates that both GpIbα (269–286) and γ′ (410–427) peptides interact with IIa at 1:1 stoichiometry. Comparing the HDX results to the ABE-I targeting peptide hirudin (54–65), the data implies that GpIbα (269–286), GpIbα (1–290), and γ′ (410–427) are indeed directed to ABE-II. The ABE-II binding fragments reduce HDX for sites distant from the interface suggesting long range conformational effects. These studies illustrate that GpIbα and γ′ target ABE-II with similar consequences on IIa dynamics, albeit with differing structural features.
At the nexus of procoagulation and anticoagulation resides the serine protease thrombin (IIa) (1–3).1 The procoagulant activities of IIa are multifaceted and numerous, including the conversion of fibrinogen to fibrin to form the soft clot (4) and the activation of platelets through cleavage of the protease activated receptors (PAR) (5). A vital anticoagulant function of IIa is the activation of protein C which deactivates several procoagulant enzymes (6). The specificity of IIa is a result of the loops and residues that surround the active site (7). IIa contains two electropositive patches that flank the catalytic site termed anion binding exosites I and II (ABE-I and ABE-II) (8). Exosites guide substrate orientation for catalysis that is more efficient and/or provide a separate cofactor interaction site that either enhances or dampens enzyme reactivity.
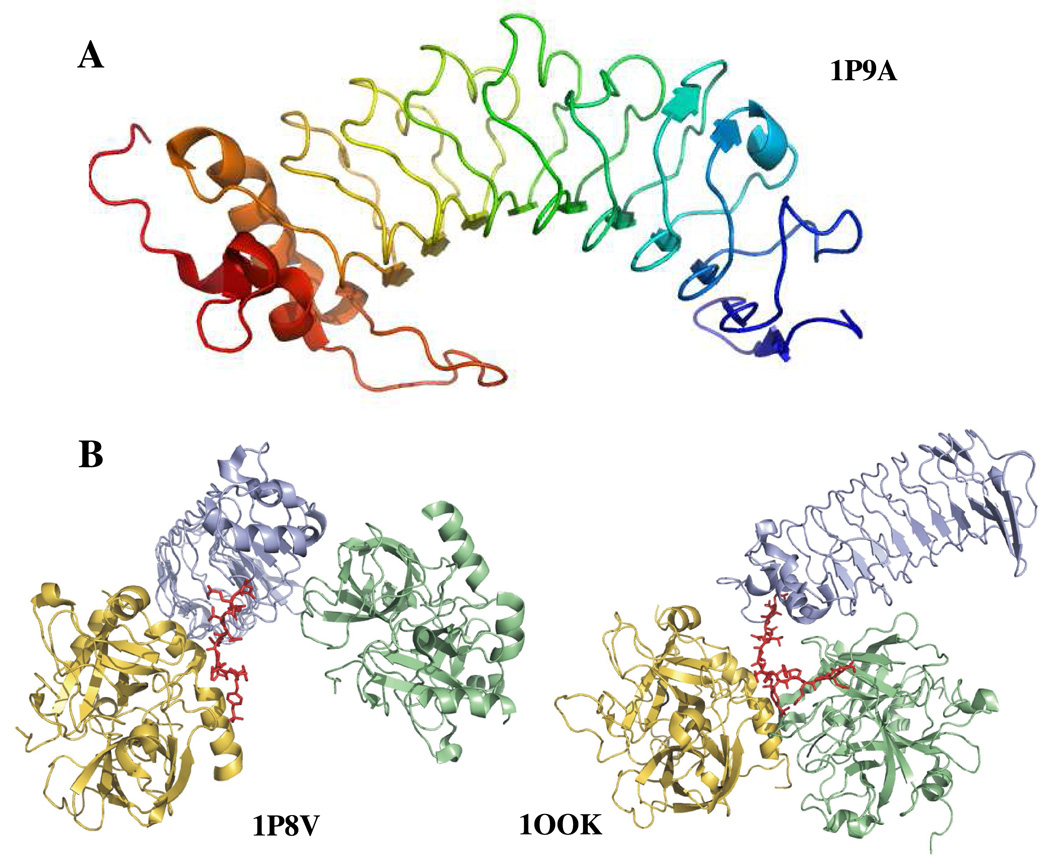
An important event in coagulation is the initial recruitment of IIa by Glycoprotein Ibα (GpIbα) to the surface of platelets. This glycoprotein is part of the highly glycosylated transmembrane platelet receptor GpIbα/β-GpIX-GpV (9, 10). The importance of this receptor is highlighted in Bernard-Soulier syndrome, a bleeding disorder resulting from either dysfunctional or low levels of this complex. GpIbα serves a multitude of functions that include binding coagulation proteins, adhering platelets to the site of damaged tissue, and aiding in leukocyte recruitment to combat infection. Two features of the N-terminus of GpIbα are a leucine-rich-repeat (LRR) domain and an anionic cluster (269DEGDTDLYSDYSYSPEEDTEG286) (Table 1 and Figure 1A).
Table 1.
Peptides Utilized in IIa Binding Studies
| GpIbα YP 276 YP 278 YP 279 | 269DEGDTDLYPDYPYPPEEDTEG286 |
| GpIbα YP 276 Y278 YP 279 | 269DEGDTDLYPDYYPPEEDTEG286 |
| GpIbα Y276 Y278 Y279 | 269DEGDTDLYDYYPEEDTEG286 |
| γ′ YP 418 YP 422 | 410PEHPAETEYPDSLYPPEDDL427 |
| Hirudin YS63 | 54GDFEEIPEEYSLQ65 |
Figure 1.
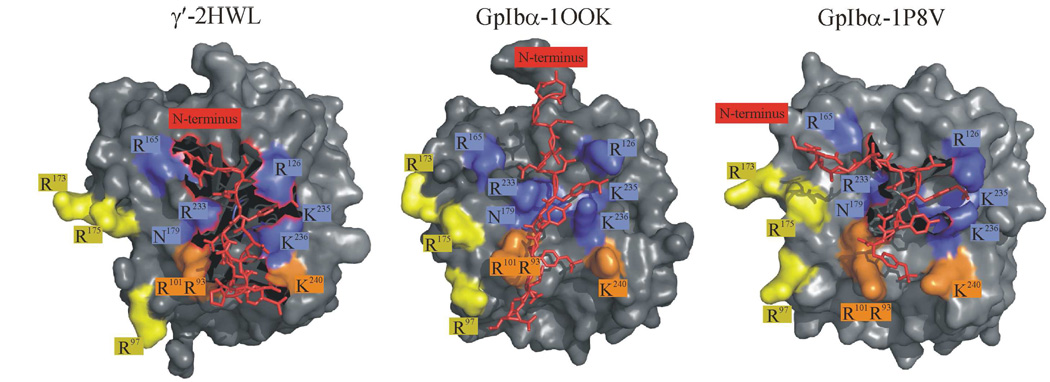
A. Structure of glycoprotein Ibα (1–266) (1P9A) (25). The highly anionic C-terminal tail (269–286) is not modeled in this structure. The most prominent feature of the structure is the leucine rich repeat domain that forms the curvature of the protein. B. Two views of GpIbα binding to thrombin. The crystal structures depict similar ABE-II interactions but differ in the depiction of thrombin dimerization. In blue is GpIbα and the anionic C-terminal tail is in red. Thrombin in gold binds to the C-terminal tail, while thrombin in green binds at different interfaces. The structure on the left is 1P8V (26), the structure on the right 1OOK (27). These figures were created using PyMol (11).
Considerable interest has been directed toward understanding in more detail the nature of the GpIbα-IIa interaction (12, 13). Mutagenesis studies have identified the ABE-II residues R93, R97, R101, R233, K236 and K240 as playing a role in binding, not the ABE-I residues R67, R73, R75, R76, and R77A (14–16).2 Additionally, the ABE-II binding glycosaminoglycan heparin prevents GpIbα interaction with IIa (15). Conversely, others suggest ABE-I as GpIbα’s binding site through competitive inhibition with hirudin (54–65) (17–19). Hirudin is a well-characterized ABE-I targeting IIa inhibitor isolated from the salivary glands of medicinal leeches (20). It should also be noted that GpIbα binding to IIa hinders the release of fibrinopeptide A (FpA) from fibrinogen (14, 21, 22), enhances PAR-1 hydrolysis (23), and decreases catalysis at R372 in FVIII (24).
Two crystal structures of IIa-GpIbα were published concurrently in 2003 (PDB IDs: 1OOK and 1P8V) (25, 26) (Figure 1B). Though both groups report a 1:1 asymmetric unit, symmetry related molecules display unique binding interfaces. Both structures illustrate similar GpIbα interactions with ABE-II. 1OOK shows the C-terminus of GpIbα potentially dimerizing IIa through interactions with both exosites (25). In 1P8V, the second IIa molecule binds to GpIbα at the C-terminal end of the LRR domain (26). Two independent groups have analyzed the available experimental data and have concluded that the stronger interaction likely involves ABE-II and GpIbα residues L275-YS 279, present in both structures (12, 13).
Like GpIbα, fibrin(ogen) has been demonstrated to bind ABE-I (27) and ABE-II (28, 29). The large soluble glycoprotein fibrinogen is a dimer comprised of three domains (AαBβγA)2 (4, 30). During coagulation, IIa conversion of fibrinogen to fibrin (αβγA)2 initiates formation of the non-covalently associated soft clot. The N-termini of the six polypeptides begin in the central E-region, supplying a low affinity ABE-I binding site (27). The C-termini of the polypeptides reach laterally from the E-region to form the flanking D regions. The ABE-II binding γ′ chain (28, 29) is a variant of the γA chain with an insertion of 20 amino acids at the C-terminus (408VRPEHPAETEYSDSLYSPEDDL427) (31, 32) (Table 1).
Between 7–15% of fibrinogen circulates as the γA/γ′ as opposed to γA/γA (33, 34). The potential physiological consequences associated with elevated levels of γA/γ′ remains unclear. Some research has suggested that an abundance of γA/γ′ is linked to increased incidence of cardiovascular diseases (33, 35). Conversely, two independent groups demonstrated that reduced levels of the γ′ chain were indicative of a higher risk for deep vein thrombosis (36, 37).
A previous report utilized nuclear magnetic resonance (NMR) and hydrogen deuterium exchange (HDX) coupled with matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS) to focus on the details of the γ′ peptide (410–427)-IIa ABE-II interaction (38). These experiments localized γ′ peptide binding within ABE-II and detected perturbations in HDX within ABE-I, the autolysis loop, and the A-chain. Many observations reported from the NMR results correlated extremely well with the subsequently reported X-ray structure of γ′ peptide-IIa (39).
In the current investigation, the GpIbα-IIa interaction was examined in further detail. Due to the high degree of similarity between the IIa binding sequences of GpIbα and the γ′ chain (Table 1), perhaps these anionic peptides share conformational features. NMR experiments demonstrate that the GpIbα peptide binds to IIa in an extended conformation, unlike the turn structure reported for the γ′ peptide (38). In addition, these studies were undertaken to better define the IIa exosite(s) responsible for GpIbα interaction, as well as to examine whether GpIbα binding induces the same long-range allostery that is observed when γ′ peptide binds to IIa. The HDX results reveal that binding of both the GpIbα peptide (269–286) and fragment (1–290) to IIa’s ABE-II alters the dynamics of IIa HDX analogous to the γ′ peptide. Finally, analytical ultracentrifugation (AUC) was implemented to observe the behavior of IIa with GpIbα or the γ′ peptide in solution.
Materials and Methods
Synthetic Peptides and GpIbα fragments
Bachem (King of Prussia, PA) synthesized peptides based on residues 269–286 of GpIbα (Table 1). SynPep (Dublin, CA) synthesized a peptide based on residues 410–427 of the human fibrinogen γ′ chain variant (Table 1). For both peptides, phosphotyrosines (YP) were substituted for the naturally occurring sulfotyrosines (YS) without sacrificing binding affinity (28, 40). Sigma-Aldrich and Bachem prepared the sulfonated hirudin fragment 54–65 (Table 1). MALDI-TOF MS measurements on an Applied Biosystems Voyager DE-Pro mass spectrometer were used to verify peptide mass. The concentrations of the peptide were determined by quantitative amino acid analysis (AAA Service Laboratory, Boring, OR).
The GpIbα 1–290 3YS fragment (YS276YS278YS279) and the GpIbα 1–290 2YS fragment (YS276YS279) were generous gifts from the Ruggeri Laboratory at Scripps in La Jolla, CA. The procedure for D. melanogaster expression and purification of the two GpIbα fragments is detailed in a previous report (25). In order to characterize the GpIbα fragments, MALDI-TOF MS was used to observe the mass of the proteins.
On the Use of Bovine Thrombin
Thrombin was purified from bovine plasma barium sulfate eluate (Sigma-Aldrich) as described by Trumbo and Maurer (41). It should be noted that human and bovine IIa exhibit a high degree of sequence conservation (87% identical) (8). Important regions where there are no differences include the active site, the Na+ binding site, the β-insertion (W60D) loop, and both ABEs.
The interaction of the human derived fibrin(ogen) γ′ and GpIbα fragments with bovine IIa should not be influenced by the minor differences between the species. Supporting this statement is an HDX study, where PPACK inhibition of human and bovine IIa affected HDX in a similar manner for analogous peptides from both types of IIa (42). In addition, 2D trNOESY NMR examining the structural features adopted by fibrinopeptide A (43, 44) and a PAR1 peptide (45) bound to bovine IIa were in agreement with the corresponding human IIa X-ray structures (46, 47). Finally, in the previous study concerning the γ′ peptide and IIa, 2D trNOESY NMR results were similar for γ′ peptide binding to both bovine α-IIa and human γ-IIa (38).
1H and 31P NMR: General Theory
Sample preparation, 1H/31P NMR analysis, and NMR instrumentation were nearly identical to those described in Sabo et al. (38). The only difference is the exclusive use of the 500 MHz NMR spectrometer during the course of this work. A 1:10 ratio of enzyme (150 µM) to peptide (1.5 mM) was maintained for the three complexes studies: IIa-GpIbα (YP 276YP 278YP 279), IIa-GpIbα (YP 276YP 279), and IIa-GpIbα (NoP). All complexes were examined at a pH of 5.6.
When free in solution, a peptide possesses very little secondary structural features due to lack of constraining factors, such as a protein in which to interact (48, 49). This situation results in ωτc ≈ 1 and longer transverse relaxation (T2) times. The 1D spectrum will display very sharp peaks and the 2D trNOESY will lack the presence of many discernible NOEs. Introduction of the binding partner, in this case IIa, leads to the peptide interacting with the protein. Thus, those protons that contact the protein surface adopt the solution characteristics of the larger macromolecule, i.e. ωτc >> 1, shorter T2 times, and large negative NOEs. The peptide now assumes the secondary conformational features associated with binding to the protein. If the koff for the peptide is sufficiently fast, then the peptide will take a “memory” of the bound conformation to the free peptide population. This rejoining with the free peptide population results in 1D line broadening for protons that contact the protein surface and a weighted average of the NOEs of the free and bound ligands in the 2D trNOESY spectrum.
Analytical Ultracentrifugation
IIa was inhibited with a two-fold excess of PPACK for 30 min. An assay involving the chromogenic substrate S2238 was employed to ensure that IIa was inactive. PPACK-IIa was then subjected to overnight dialysis with 150mM NaCl, 25mM phosphate, pH 7.4. The γ′ or GpIbα peptide was lyophilized and reconstituted in the same buffer. A series of sedimentation velocity AUC experiments ranging from 1, 2, 5, 10, and 20 times peptide to IIa (4.5–7.0 µM) in duplicate was performed in a XL-A AUC (Beckman-Coulter). The parameters for each run were 60,000 rpm, 25 °C, and 280 nm. The data was analyzed with the program DC-DT+.
HDX Sample Preparation
IIa was buffer exchanged into the HDX buffer 37.5 mM NaCl, 6.25 mM NaH2PO4, pH 5.6, 6.5, or 7.5 (all pH values are within ± 0.1) with an Amicon-Ultra 4 unit (10 kDa MWCO). The concentration of IIa for all preparations was 25 µM. An aliquot of 24 µL IIa and 24 µL HDX buffer were evaporated to dryness using a SpeedVac unit (Savant). The dry aliquots were stored at −70 °C.
For experiments involving peptides, enough GpIbα, γ′, or hirudin (54–65) was added to 24 µL of buffer exchanged IIa to achieve a 20:1 ratio (40:1 in the case of the γ′ peptide at pH 6.5) during HDX. The peptides were pH adjusted prior to addition. The aliquots of buffer-exchanged IIa and peptide were evaporated to dryness using a SpeedVac unit. For experiments involving the proteins GpIbα-3YS and GpIbα-2YS, the two fragments were exchanged into the pH 6.5 HDX buffer. After the buffer exchange, the GpIbα fragment concentration was 40 µM. An aliquot of 24 µL IIa and 24 µL GpIbα fragment were evaporated to dryness using a SpeedVac unit. The dry aliquots were stored at −70°C.
The increased γ′ (410–427) peptide to IIa ratio raises the possibility of discrepancies between results presented in an earlier report (38) and those described within this paper. Re-analysis of the γ′ (410–427)-IIa HDX pH showed that the value was likely below 6.0 in the prior work and then compared to IIa HDX at pH 6.5 (38). The acidity of the γ′ peptide stock solution surpassed the buffering capabilities of the 25 mM phosphate solution. Thus, both ABE-II binding and pH dependent HDX rate perturbations were being observed (38). When the pH of the γ′ (410–427)-IIa solution was raised to 6.5 in the current project, residence time on the enzyme surface may have decreased. Increasing the peptide to IIa ratio to 40:1 promoted the observation of a single population of ligand-bound IIa. Fortuitously, the carefully monitored pH investigations described in this report validates the conclusions obtained in the previous study for γ′ (410–427)-IIa. Ligand binding to ABE-II decreases HDX at the binding interface, which leads to further decreases in HDX at regions not associated with the interaction site.
HDX Experiments
Dry aliquots were allowed to come to room temperature before beginning the experiment. First, 12 µL of D2O was added to the dry aliquot yielding the final concentrations: 50 µM IIa, 80 µM GpIbα fragment or 1–2 mM peptide, 150 mM NaCl, 25 mM NaH2PO4, pH 5.6, 6.5, or 7.5. The samples were incubated in a desiccator at room temperature for 1 minute or 10 minutes. After incubation, 114 µL of 0.1% TFA pH 2.5 (on ice) was added to quench the solution. An additional 1 µL of 5% TFA was added to insure that the pH was 2.5 in experiments at pH 7.5. Next, 66 µL of the quenched solution was transferred to a tube of activated pepsin bound to agarose. Digestion occurred on ice for 10 minutes. Centrifugation at 4°C separated the digest from the pepsin and 8.2 µL aliquots were immediately frozen in liquid N2. Each frozen HDX aliquot was analyzed as detailed in Sabo et al. with MALDI-TOF MS (38).
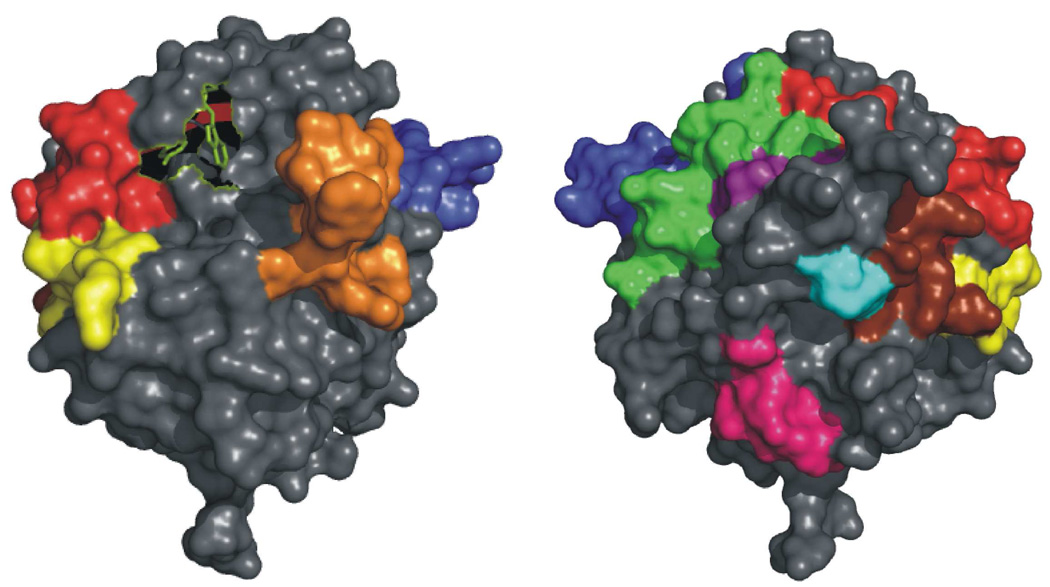
Coverage exists for ten peptides in bovine IIa from a non-reducing pepsin digest (42). Figure 2 depicts the location of the ten peptides within IIa relative to the two ABEs. Residues 65–84 represent a portion of ABE-I, while part of ABE-II is covered with three fragments: 85–99, 173–180, and 173–181. The autolysis loop peptide (135–149D) and 46–52 reside near ABE-I residues 65–84. A β strand within the segment 106–116 is located anti-parallel to a β strand near the N-terminus of the ABE-II fragment 85–99. The A-chain residues −13 to −4 of bovine IIa are highly flexible, but when present in an X-ray structure (such as 2A1D (50)), reach toward the C-terminal helix of the B-chain (233–240), which has several ABE-II residues. Lastly, the segment 202–207 fits into the hinge formed by the boomerang shaped A-chain.
Figure 2.
Depiction of the coverage obtained from a peptic digest of IIa (2AID) relative to Y60A and W60D, which are green. Left: Looking onto the active site of IIa. Right: The structure rotated 180° about the y-axis. Peptides from the peptic digest are colored as follows: the ABE-II regions 85–99 (red) and 173–181 (yellow), the ABE-I peptide 65–84 (blue), the autolysis loop containing peptide 135–149D (orange), 106–116 (green), the A-chain peptide −13 to −4 (cyan), 202–207 (pink), and 46–52 (purple). The additional ABE-II region not observed in the present HDX study, 233–240, is in brown.
The HDX experiments described in the results section cover three topics: 1) the effect of pH on IIa HDX, 2) perturbations in IIa HDX due to peptide binding at pH 5.6 and pH 6.5, and 3) a comparison of GpIbα peptide and GpIbα protein binding to IIa at pH 6.5. The data will be reported in two ways. Raw HDX will be illustrated in a one and ten minute bar graph for the four topics (Figure 6, Figure 8–Figure 10) (38). In addition, the numerical values have been reported in Supplementary Tables 1 and 2. The second method of analysis employs % changes to gauge the significance in fluctuations of HDX (Table 3 and Table 4) (38). This value relates the differences in HDX to the theoretical maximum amount of exchangeable protons present in each observable peptide. These % changes will be classified into three categories: 1) insignificant 0 to ±1.9%, 2) modest change ±2.0% to ±4.4%, and 3) significant change ≥ ±4.5% (38, 51–54).
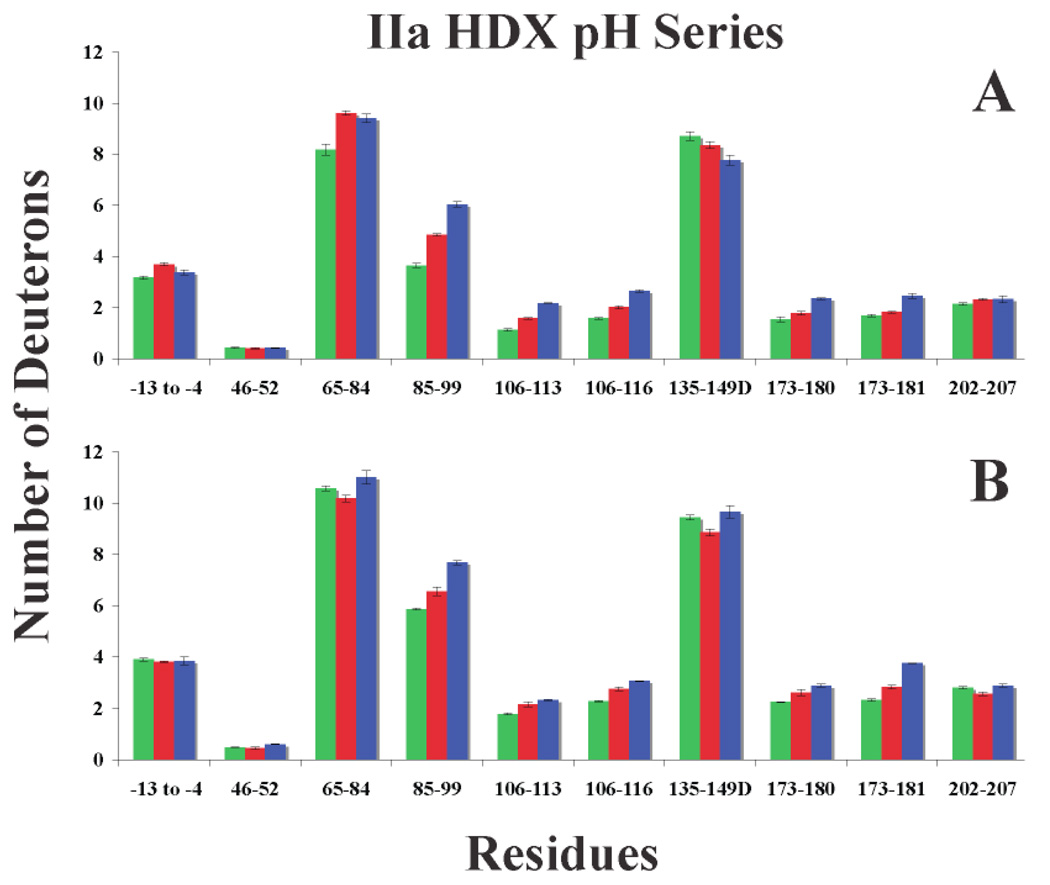
Figure 6.
Graphs of deuterium incorporation for IIa at pH 5.6 (green), pH 6.5 (red), and pH 7.5 (blue). A. One minute of HDX and B. Ten minutes of HDX. Errors correspond to the standard deviation of the mean for three independent experiments.
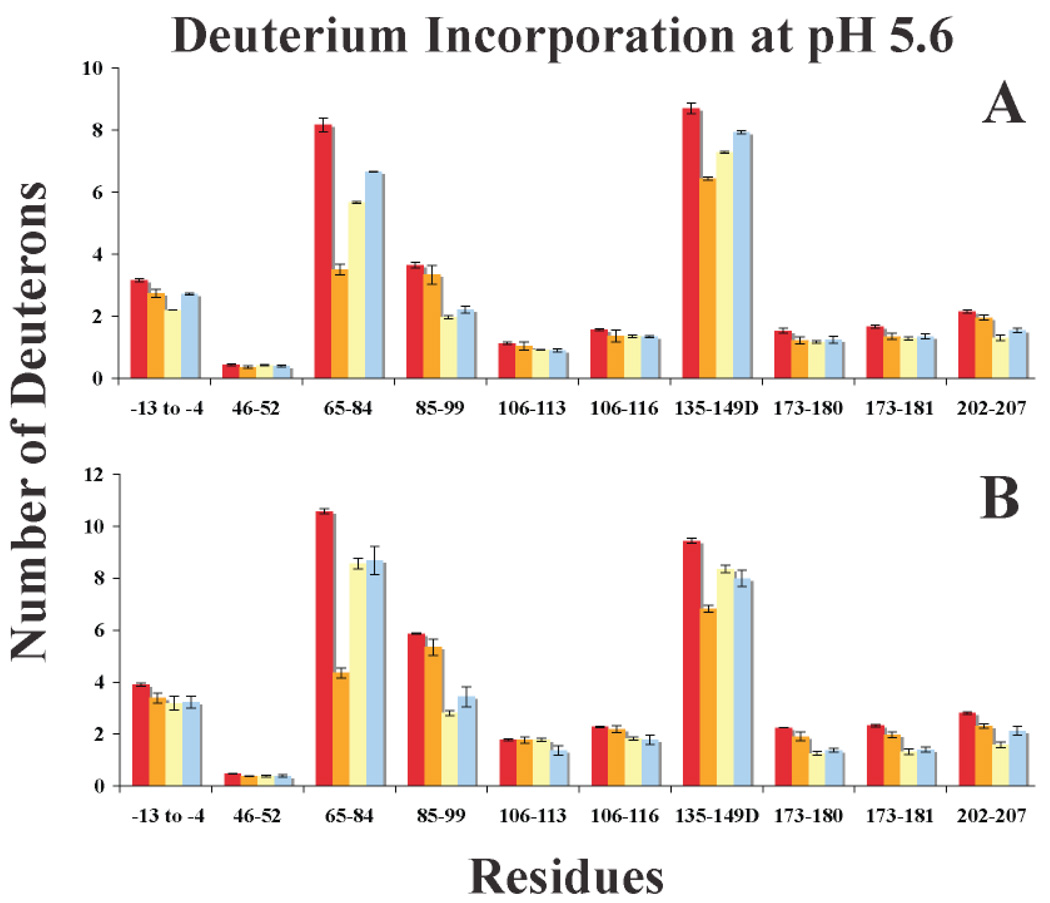
Figure 8.
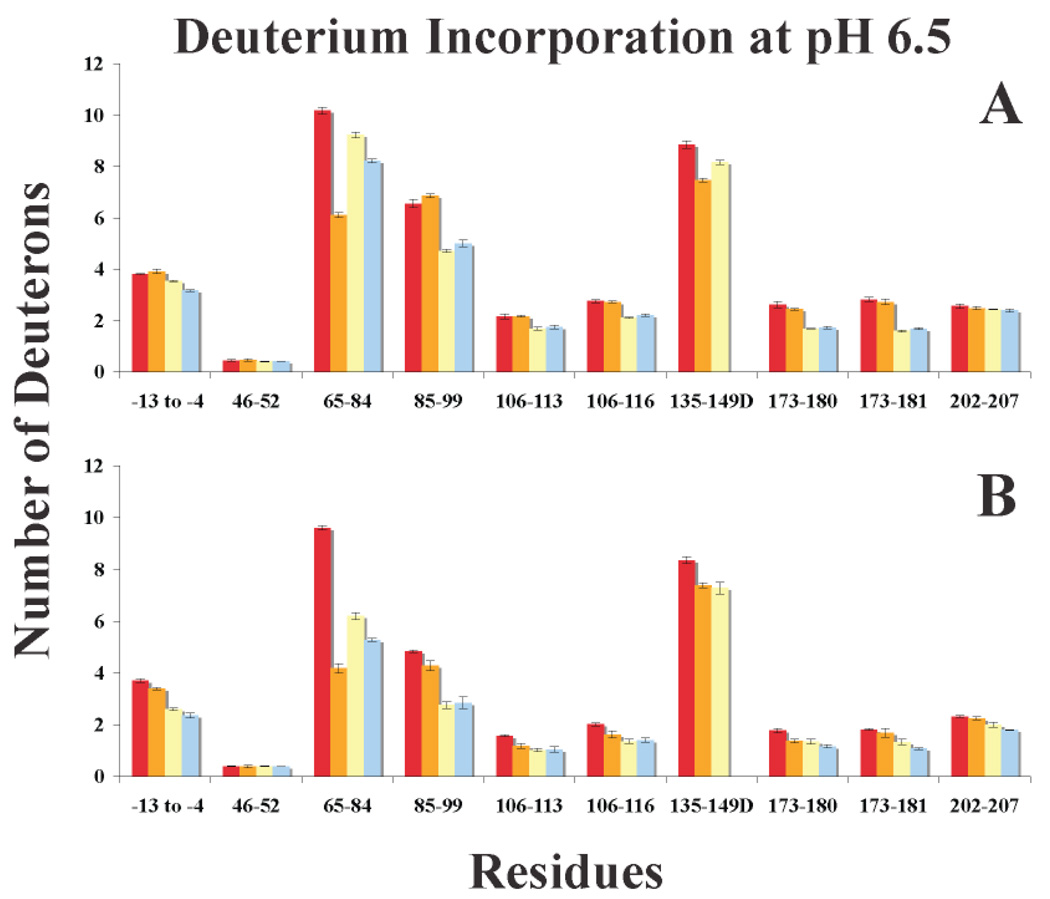
Graphs of deuterium incorporation for IIa and 20:1 peptide-IIa at pH 5.6. A. One Minute of HDX and B. Ten Minutes of HDX. The bars in the graph correspond to IIa alone (red), Hir-IIa (orange), GpIbα-IIa (yellow), and γ′ peptide-IIa (blue). Errors correspond to the standard deviation of the mean for three independent experiments.
Figure 10.
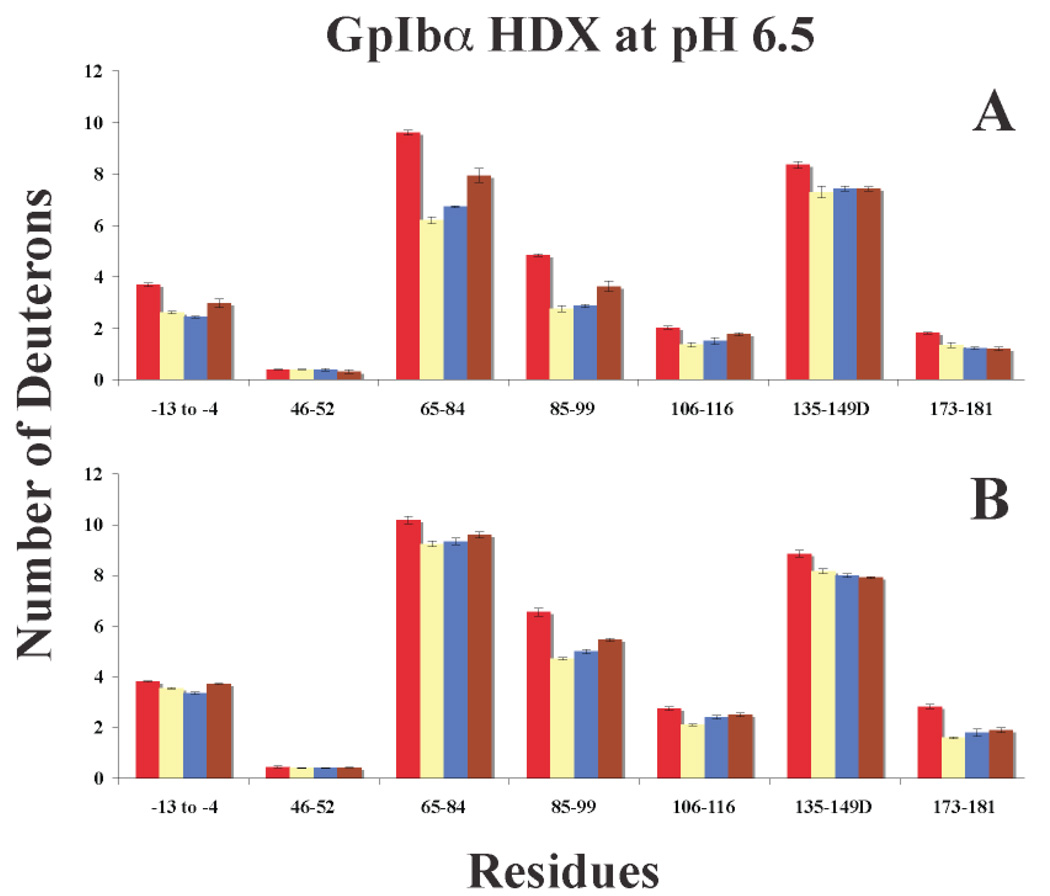
Graphs of deuterium incorporation for IIa, GpIbα peptide-IIa, and GpIbα protein-IIa at pH 6.5. A. One Minute of HDX and B. Ten Minutes of HDX. The bars in the graph correspond to IIa alone (red), 20:1 GpIbα peptide-IIa (yellow), 1.6:1 GpIbα 3S protein-IIa (blue), and 1.6:1 GpIbα 2S protein-IIa (brown). Errors correspond to the standard deviation of the mean for three independent experiments.
Table 3.
Changes in % Deuteration for GpIbα−IIa and γ′ peptide−IIa at 1 and 10 Minutes Relative to IIa pH 5.6a
| 20:1 | ||||||
|---|---|---|---|---|---|---|
| GpIbα | γ′ | Hirudin | ||||
| Residues | 1 m | 10 m | 1 m | 10 m | 1 min | 10 m |
| −13 to −4 | −11.4 b | −8.5 | −5.2 | −8.0 | −4.9 | −6.1 |
| 46–52 | −0.2 | −1.5 | −0.5 | −1.4 | −1.1 | −1.4 |
| 65–84 | −11.5 | −9.3 | −6.9 | −8.7 | −21.4 | −28.5 |
| 85–99 | −11.1 | −20.1 | −9.4 | −16.0 | −2.0 | −3.4 |
| 106–113 | −3.1 | −6.0 | −3.3 | −6.1 | −1.3 | −0.1 |
| 106–116 | −2.2 | −4.6 | −2.4 | −5.1 | −2.1 | −0.9 |
| 135–149D | −7.3 | −5.6 | −4.0 | −7.5 | −11.7 | −13.5 |
| 173–180 | −4.7 | −12.7 | −3.7 | −11.2 | −4.0 | −4.4 |
| 173–181 | −4.4 | −11.5 | −3.8 | −10.6 | −3.8 | −4.1 |
| 202–207 | −12.5 | −17.9 | −8.8 | −10.0 | −2.8 | −7.4 |
The % change for a particular peptide is calculated as described in (38).
The values in bold represent significant changes in deuteration of greater than −4.5%.
Table 4.
Changes in % Deuteration for Peptide and Protein Binding to IIa at 1 and 10 Minutes Relative to Free IIa pH 6.5a
| 20:1 | 40:1 | 1.6:1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hirudin | GpIbα | γ′ | GpIbα 3S | GpIbα 2S | ||||||
| Residues | 1 m | 10 m | 1 m | 10 m | 1 m | 10 m | 1 m | 10 m | 1 m | 10 m |
| −13 to −4 | −3.5 | 1.2 | −12.8 | −3.2 | −15.8 | −7.6 | −14.8 | −5.4 | −8.5 | −1.2 |
| 46–52 | −0.2 | 0.3 | 0.0 | −0.6 | −0.3 | −0.6 | −0.2 | −0.6 | −1.4 | −0.3 |
| 65–84 | −24.9b | −18.6 | −15.6 | −4.3 | −19.9 | −8.9 | −13.2 | −3.9 | −7.7 | −2.7 |
| 85–99 | −3.6 | 2.0 | −13.7 | −12.1 | −13.1 | −10.2 | −13.0 | −10.3 | −8.0 | −7.2 |
| 106–113 | −5.8 | 0.1 | −8.4 | −7.0 | −7.9 | −6.1 | n/a | n/a | n/a | n/a |
| 106–116 | −4.1 | −0.3 | −6.7 | −6.6 | −6.2 | −5.6 | −2.6 | −3.6 | −2.6 | −2.6 |
| 135–149D | −5.0 | −7.1 | −5.5 | −3.6 | n/a | n/a | −4.8 | −4.4 | −4.8 | −4.8 |
| 173–180 | −5.1 | −2.2 | −5.4 | −11.9 | −7.8 | −11.5 | n/a | n/a | n/a | n/a |
| 173–181 | −1.6 | −1.1 | −5.5 | −14.0 | −8.4 | −13.0 | −7.0 | −11.6 | −7.0 | −10.6 |
| 202–207 | −1.0 | −1.2 | −4.7 | −1.9 | −7.6 | −2.5 | n/a | n/a | n/a | n/a |
The % change for a particular peptide is calculated as described in (38)
The values in bold represent significant changes in deuteration of greater than −4.5%.
Results
1H and 31P One-Dimensional Line Broadening NMR
1D NMR experiments were performed to determine which 1H or 31P nuclei within the GpIbα (269–286) peptide interact with IIa. The first set of 1D NMR spectra examined GpIbα (YP 276YP 278YP 279) and displayed the absence of significant line broadening for the peptide in the presence of IIa (data not shown). These findings indicate that the 3YP peptide binds too tightly to IIa, hindering any transfer of information regarding bound structural features to the free peptide population.
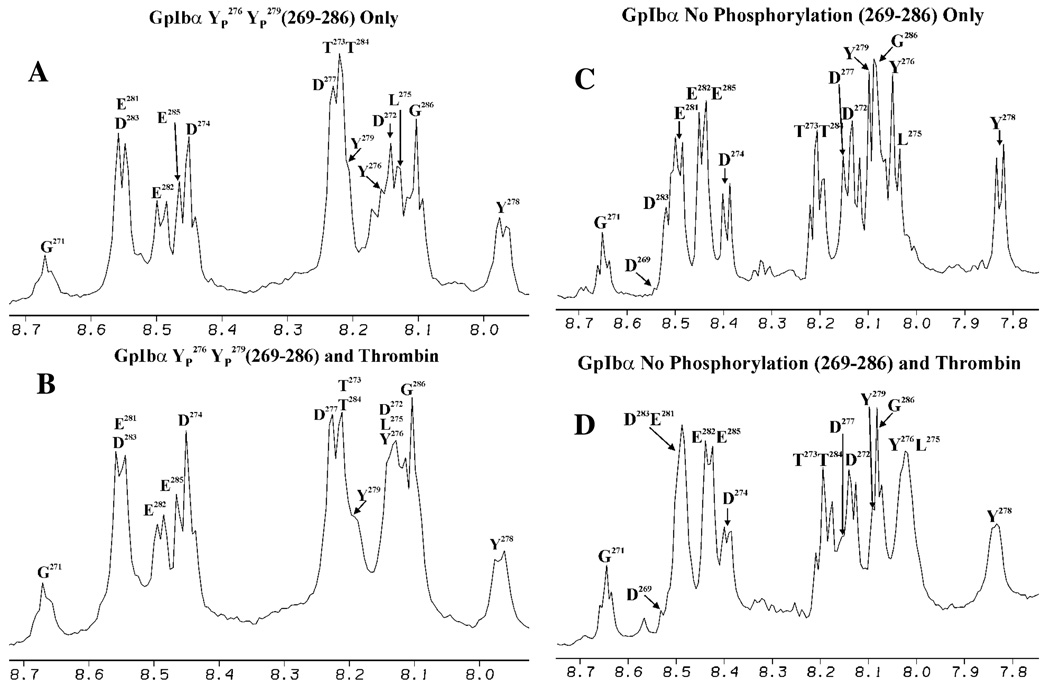
Since the GpIbα (YP 276YP 278YP 279) peptide spectra did not display line broadening, a 2YP variant was synthesized. The GpIbα (YP 276YP 279) peptide still interacts with IIa (demonstrated by the two crystal structures (25, 26)), but the absence of one YP may sufficiently weaken the interaction for the appearance of 1D line broadening. The spectra in Figure 3A and B focus on the amide region and do illustrate some line broadening in the presence of IIa. Most of the broadening encompasses residues D274-YP 279, while a modest amount is observed for residues E281-D283. However, the line broadening is not extensive, with residues D269-D274 and T284-G286 appearing unchanged from the free peptide spectrum. Curiously, the 31P spectra do not present evidence of line broadening (data not shown).
Figure 3.
Line broadening spectra for the GpIbα peptides in the presence of thrombin. All NMR samples are in 25 mM H3PO4, 150 mM NaCl, 200 µM EDTA, and 10% D2O, pH 5.6. A. 1D NMR spectrum for 1.5 mM YP 276YP 279 peptide in solution. B. 1D NMR spectrum for 1.5 mM YP 276YP 279 peptide in the presence of 0.148 mM IIa. The amide protons that display the most line broadening are D274-YP 279. A moderate amount of line broadening is evident for the NHs of E281-D283. Line broadening is not observed for the amide protons of D269-T273 and T284-G286. C. 1D NMR spectrum for 1.5 mM unphosphorylated peptide in solution. D. 1D NMR spectrum for 1.5 mM unphosphorylated peptide in the presence of 0.150 mM IIa. The amide protons that display the most line broadening are D274-YP 279, E281, and D283. A moderate amount of line broadening is evident for the NHs of E282 and E285. Line broadening is not observed for the amide protons of D269-T273, T284, and G286.
A final peptide was synthesized without YP. Much more line broadening is evident with GpIbα (No YP) (Figure 3C and D). The central portion of the peptide spanning D274-Y279, E281, and D283 display a significant amount of line broadening. The residues E282 and E285 show a modest amount of broadening. Finally, the N-terminal residues D269-T273 and the C-terminal residues T284 and G286 do not appear to be essential for the interaction of GpIbα (269–286) with IIa.
Two-Dimensional Transferred Nuclear Overhauser Effect NMR
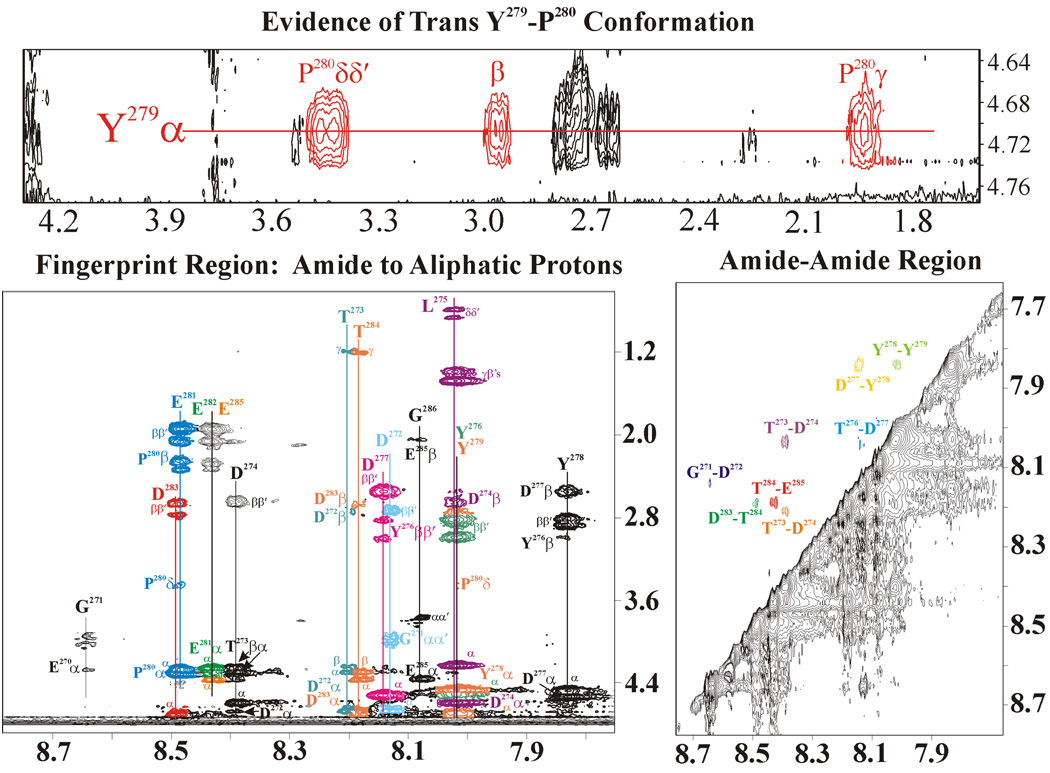
The 2D spectrum of GpIbα (YP 276YP 278YP 279) was indistinguishable from the spectrum with the peptide in the presence of IIa (data not shown). For the YP 276YP 279 and No YP GpIbα peptides, the 2D trNOESY spectra displayed only nearest neighbor NOEs in the presence of IIa. Figure 4 illustrates some of the key features from the GpIbα (No YP)-IIa 2D trNOESY spectra. Detection of only nearest neighbor NOEs suggests that the peptide adopts an extended conformation when binding to IIa. In addition, the Y279-P280 amide bond is in the trans conformation. With the GpIbα (YP 276YP 279) peptide, slightly fewer inter-residue NOEs are observed in the presence of IIa when compared to GpIbα (No YP).
Figure 4.
2D trNOESY spectrum of the unphosphorylated GpIbα (1.5 mM) bound to IIa (0.150 mM). The top panel illustrates the evidence for a trans Y279-P280 peptide bond. The bottom panels demonstrate that GpIbα exists in an extended structure when bound to IIa due to the presence of only nearest neighbor NOEs. The NOEs are color coded according to the corresponding residue.
Analytical Ultracentrifugation
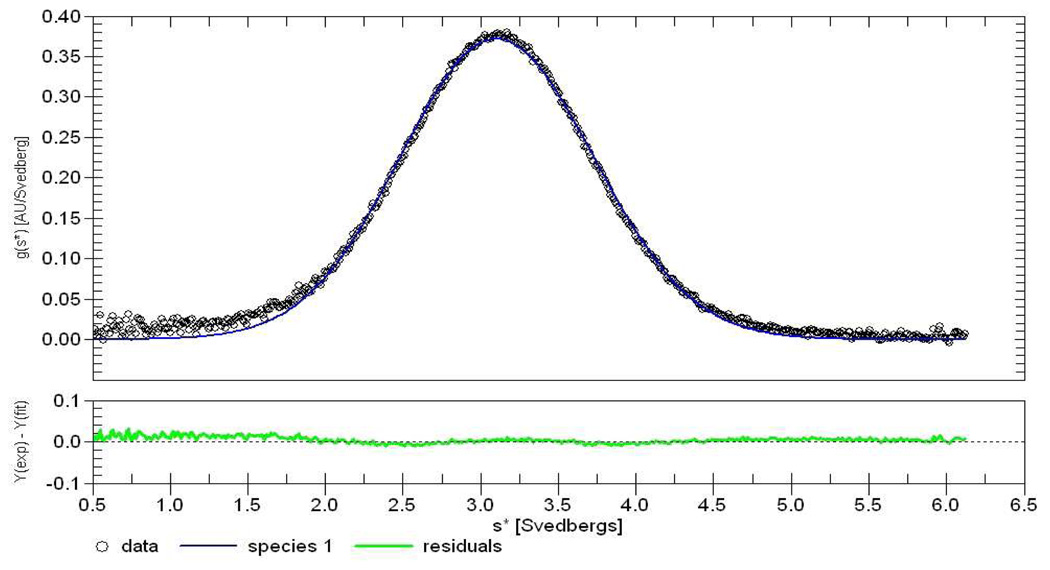
Two X-ray studies offer evidence for potential ABE-II binding ligand-induced IIa dimerization (25, 39). AUC experiments were performed to monitor the behavior of IIa in the presence of the GpIbα or γ′ peptides. In these AUC trials, the concentrations of PPACK inhibited IIa spanned 4.7–7.0 µM. The sedimentation velocity experiments were executed with a range of peptide to IIa molar ratios varying from 0:1 to 20:1. Table 2 presents the data for the average of two runs. Figure 5 displays the distribution of Svedberg constants (S) for the trial examining 2:1 GpIbα peptide to IIa. This figure is representative of all data collected from 1:1 to 20:1 peptide to IIa reported in Table 2.
Table 2.
Analytical Ultracentrifugation Data
| Sedimentation Coefficient (S) | Molecular Weight (kDa) | |
|---|---|---|
| IIa alone | 3.11 ± 0.00 | 36.2 ± 0.3 |
| GpIbα:IIa | ||
| 1:1 | 3.19 ± 0.02 | 37.3 ± 0.0 |
| 2:1 | 3.18 ± 0.01 | 37.5 ± 0.9 |
| 5:1 | 3.27 ± 0.05 | 37.9 ± 0.7 |
| 10:1 | 3.25 ± 0.02 | 37.2 ± 1.1 |
| 20:1 | 3.22 ± 0.01 | 38.1 ± 1.4 |
| γ′:IIa | ||
| 1:1 | 3.19 ± 0.02 | 39.3 ± 1.0 |
| 2:1 | 3.22 ± 0.03 | 38.6 ± 0.4 |
| 5:1 | 3.23 ± 0.03 | 39.9 ± 0.2 |
| 10:1 | 3.27 ± 0.01 | 39.9 ± 0.6 |
| 20:1 | 3.28 ± 0.01 | 40.9 ± 0.7 |
Figure 5.
Sedimentation velocity AUC of GpIbα peptide (14 µM) and IIa (7 µM). Analysis was performed at 280 nm, 20 °C, and 60,000 rpm in 150 mM NaCl, 25 mM phosphate, pH 7.4. The apparent Svedberg constant distribution centered at 3.2 S with the residuals resulting from a global fit of the data using the DCDT+ program.
The Svedberg constant measures the rate of sedimentation, which is related to the size and shape of the molecule (55). When two proteins interact, the surface area is decreased leading to a non-additive increase in the S constant. The calculated Svedberg constants for the peptide binding to the enzyme range from 3.2–3.3, compared to 3.1 for IIa alone. The slight increase in molecular weight and S values with increasing peptide concentration indicate that the peptides bind to IIa but do not instigate protein dimerization under these conditions.
HDX Experiments (I): Observing the Effect of pH on Thrombin’s Backbone Amide Solvent Accessibility
The HDX method exploits differences in the rate of chemical exchange for backbone amide protons in proteins (56). Surface-exposed backbone amide protons are more available for HDX than protons buried within the interior of the protein. Through fluctuations in protein structure (temporary protein unfolding and/or local conformational changes), a buried backbone amide proton may become exposed to solvent (57, 58). Therefore, protein dynamics influence deuterium exchange to a great degree. The rate of chemical exchange is highly dependent on pH and temperature (59).
For the current project, HDX effects at pH values of 5.6, 6.5, and 7.5 were examined. Work at pH 5.6 allows comparisons to be made with the NMR solution conditions. A pH of 7.5 is physiological, but the backbone amides exchange very rapidly. A more favorable working pH is 6.5, a condition that slows the backbone exchange 10-fold relative to pH 7.5 (60) and decreases the rate of IIa autolysis. Figure 2 illustrates the coverage obtained from a peptic digest of IIa (see Materials and Methods for description).
Comparing the levels of HDX across this pH spectrum reveals three distinct groups. See Figure 6, Supplementary Tables 1 and 2. The first grouping concerns those fragments that steadily increase in deuterium incorporation as the pH increases, suggesting a significant degree of solvent exposure for some of the amide protons. These peptides include the ABE-II peptides 85–99 and 173–181, as well as 106–116. The second grouping involves −13 to −4, 46–52, and 202–207. These three peptides seem impervious to pH change. Two probable explanations are that most of the amide protons are solvent inaccessible and/or the amide protons exchange too quickly for any apparent pH effect to be noticed. Finally, both the ABE-I peptide 65–84 and the autolysis loop 135–149D show stagnant levels of HDX at ten minutes. At one minute, an increase in pH appears to slow HDX for the autolysis loop peptide, perhaps indicating a more stable configuration for 135–149D at higher pH. As for 65–84, the rate of deuterium incorporation may slightly increase with higher pH at one minute of HDX.
HDX Experiments (II): Observing the Effect of the GpIbα (YP 276YP 278YP 279) Peptide, the γ′ Peptide (YP 418YP 422), and Hirudin (54–65) on Thrombin’s Backbone Amide Solvent Accessibility at pH 5.6 and pH 6.5
The first reports describing HDX coupled with MALDI-TOF MS to study solvent accessibility at a ligand-protein interface appeared in 1998 (61, 62). With such an HDX experiment, the ligand should be bound to the protein, ideally, for the entire course of the deuterium exchange (61, 63). Thus, the KD describing the strength of the ligand-protein interaction, as well as, the concentration of the binding partners, will determine the lifetime of the complex (63).
The GpIbα, γ′, and hirudin (54–65) peptides were incubated with IIa at room temperature and pH 5.6 or pH 6.5 for one or ten minutes in the presence of 99.99% D2O. A previous study found that GpIbα residues 265–285 bound to IIa with a KD of 5.9 nM (21). In addition, the larger GpIbα fragment 1–282 possesses a KD of 149 nM (64). The weaker KD may stem from the truncated C-terminus. For comparison purposes, an ABE-I targeting peptide was also examined. Sulfonated hirudin (54–65) has a KD of 48 nM recently determined through fluorescence studies (65). Thus, 1 mM GpIbα and 1mM hirudin (54–65) should provide 99.99% occupancy of 50 µM IIa. As for the γ′ peptide, the interaction is weaker (KD = 680 nM) (28), resulting in 99.93% and 99.97% occupancy at 1 and 2 mM peptide, respectively.
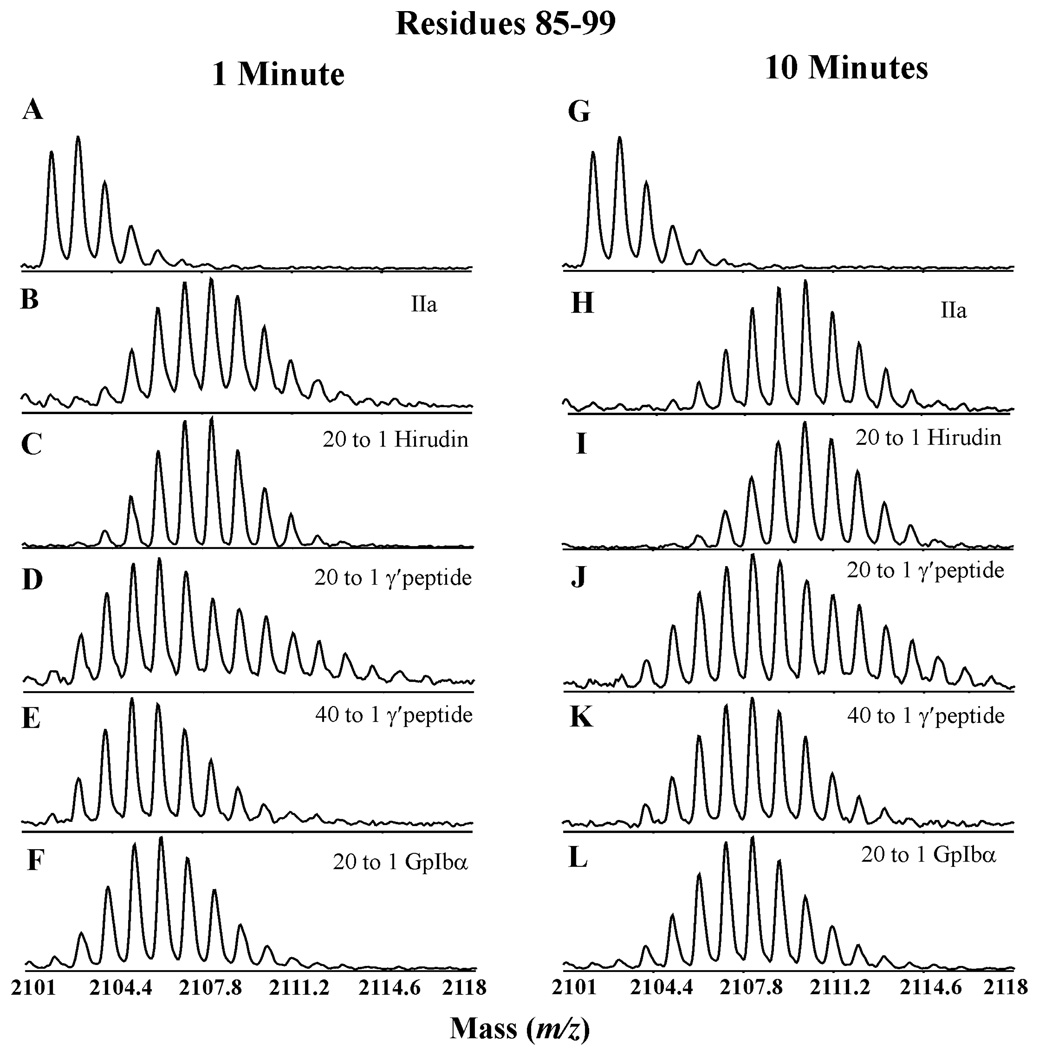
In Figure 7, the isotopic cluster for residues 85–99 of 20:1 γ′ peptide to IIa at pH 6.5 has an interesting feature not evident at pH 5.6. The cluster appears to be expanded with a slight shoulder ending near 2115 m/z (not shown is similar shoulder for 173–181). The results suggest the presence of two different IIa populations at pH 6.5 exposed to deuterium. HDX performed at pH 6.5 on a higher ratio (40:1) of the γ′ peptide (2 mM) to IIa (50 µM) led to the reappearance of a single isotopic cluster (see Materials and Methods for explanation). Therefore, pH 6.5 HDX comparisons will be made between 20:1 hirudin (54–65)-IIa, 20:1 GpIbα-IIa, and 40:1 γ′ peptide-IIa.
Figure 7.
Mass spectra of residues 85–99 after one and ten minutes of HDX. All of the depicted HDX experiments are with 50 µM IIa in 150 mM NaCl, 25 mM NaH2PO3, pH 6.5. A and G: the undeuterated peak cluster, B and H: IIa spectra in the absence of ligands, C and I: 20:1 hirudin (54–65) bound to IIa, D and J: 20:1 γ′ peptide bound to IIa, E and K: 40:1 γ′ peptide bound to IIa, and F and L: 20:1 GpIbα peptide bound to IIa. At the ratio of 20:1, both the γ′ and GpIbα peptides exhibit a similar degree of protection for these ABE-II residues; however in spectra D and J a small shoulder is present suggesting the presence of two IIa populations. Increasing the amount of γ′ peptide completely abrogates the shoulder suggesting fully occupied ABE-II.
The HDX results for peptide binding to IIa are compiled in Table 3 and Table 4, Supplementary Tables 1 and 2, and Figure 8 and Figure 9. At one and ten minutes of HDX, both the GpIbα and γ′ peptides significantly protect the ABE-II fragment 85–99 from deuterium. A unique feature of the data at pH 5.6 is the observed increase in HDX protection at ten minutes for 85–99, either indicating a tighter complex at the lower pH, or representing the slower rate of HDX at this pH. Another segment within ABE-II, 173–181, increases in protection from one to ten minutes of HDX at both pH values in a similar manner. These results suggest that the GpIbα and γ′ peptides are interacting with IIa’s ABE-II. Conversely, hirudin (54–65) exclusively targets ABE-I as evident from the dramatic HDX protection at 65–84.
Figure 9.
Graphs of deuterium incorporation for IIa and peptide-IIa at pH 6.5. A. One Minute of HDX and B. Ten Minutes of HDX. The bars in the graph correspond to IIa alone (red), 20:1 Hir-IIa (orange), 20:1 GpIbα-IIa (yellow) and 40:1 γ′ peptide-IIa (blue). Errors correspond to the standard deviation of the mean for three independent experiments.
Binding to ABE-I and ABE-II appears to protect regions within IIa unaffiliated with their interactive sites. Hirudin (54–65) interactions with IIa lead to mostly modest decreases in HDX for ABE-II peptides 85–99 and 173–181 over the course of ten minutes. The ABE-I fragment 65–84 is affected by GpIbα and γ′ binding to ABE-II to greater degree than hirudin (54–65) ABE-I binding imparts over to ABE-II. From one to ten minutes of HDX at pH 5.6, fragment 65–84 displays slightly less deuterium incorporation with the GpIbα peptide than the γ′ peptide. At pH 6.5, peptide interactions at ABE-II show a large degree of protection at one minute for 65–84 that becomes significantly reduced within ten minutes of HDX.
Located parallel to a portion of the ABE-II fragment 85–99 are the residues 106–116 represented by two overlapping fragments. Hirudin (54–65) binding has very little effect on HDX for 106–116 at pH 5.6, but at pH 6.5 noteworthy protection can be localized to residues 106–113. Both the GpIbα and γ′ peptides display modest protection at one minute, increasing to a level of greater significance at ten minutes for experiments conducted at pH 5.6. As for pH 6.5, the levels of significant HDX protection remain steady from one to ten minutes for 106–116. These data imply that ABE-II binding is transmitted to this neighboring β-strand.
Near ABE-I is the segment 135–149D, which also encompasses most of the autolysis loop. All three peptides significantly slow HDX for this region of IIa. Hirudin (54–65) imparts a larger effect on the dynamics of this region from ABE-I than the GpIbα and γ′ peptides from ABE-II. Another long-range consequence of ABE-I and ABE-II occupation is evident from HDX within the A-chain residues −13 to −4 and the nearby B-chain fragment 202–207. The degree of protection is of a larger magnitude for the γ′ and GpIbα, equating to mostly significant deviations from the HDX of IIa alone. Hirudin (54–65) results indicate that pH plays a role in HDX for −13 to −4 and 202–207. A higher pH of 6.5 results in these two peptides remaining largely unaffected by hirudin (54–65) binding to ABE-1, whereas at pH 5.6 significant protection is observed. All three peptides do not change the dynamics of deuterium exchange for the fragment 46–52.
HDX Experiments (III): Observing the Effect of the GpIbα (YP 276YP 278YP 279) Peptide and the GpIbα fragment (1–290) on Thrombin’s Backbone Amide Solvent Accessibility at pH 6.5
The Ruggeri laboratory at Scripps generously donated expressed GpIbα (1–290) with 3YS (YS 276YS 278YS 279) and 2YS (YS 276YS 279) with the goal of comparing the HDX of the peptide to a larger protein fragment. The GpIbα (1–290)-IIa HDX data recorded at pH 6.5 is quite similar for the 3YS protein and the 3YP peptide (Figure 10, Table 4, and Supplementary Table 1). The 2YS protein displayed similar trends in HDX, however, the degrees of % change are not as large as with the 3YS protein. As with the two ABE-II binding peptides, the GpIbα (1–290) fragment appears to be interacting at ABE-II. Also, long rang HDX perturbations are evident toward ABE-I, the A-chain, and the autolysis loop. It must be noted that the HDX results for hirudin (54–65) and GpIbα (1–290) are not similar. These findings further support the notion that GpIbα’s primary destination on IIa is ABE-II.
Discussion
On the surface of platelets, the receptor GpIbα forms a complex with IIa, however, the literature presents evidence for interactions at both ABEs. Though mutagenesis data strongly suggest that GpIbα binds exclusively at ABE-II (14–16), binding studies display interactions at both exosites (15, 17, 19, 21, 64). Adding to the ambiguity, two crystal structures of GpIbα-IIa illustrate similar interactions at ABE-II for one IIa, while symmetry related IIa binds to GpIbα through ABE-I at different GpIbα interfaces (25, 26) (Figure 1B). The studies described in this report use solution NMR, AUC, and HDX coupled with MALDI-TOF MS to investigate the interaction of GpIbα with IIa.
Analysis of the NMR Data: the GpIbα Peptide (269–286) Binds to Thrombin in an Extended Conformation
To sufficiently weaken the interaction and thus observe the trNOESY effect, the GpIbα peptide (269–286) was dephosphorylated to varying degrees. Conflicting accounts exist regarding the interaction of No YP variants of GpIbα and IIa. One report describes GpIbα (No YP) as unable to bind to IIa (66), while another group demonstrated competitive inhibition of FVIII hydrolysis by IIa in the presence of GpIbα (No YP) (268–282) (24). The present investigation supports the observation that GpIbα (No YP) can interact with IIa.
The lack of line broadening observed for GpIbα residues D269-T273 in the presence of IIa can potentially help resolve the two different conformations depicted for GpIbα 269–277 in the X-ray structures (Figure 11A) (25, 26). Also, a previous study de-emphasizes the importance of GpIbα D269-G271 for binding IIa (64). It can be concluded that the GpIbα residues D269-T273 probably exist as a flexible portion of the glycoprotein, able to adopt multiple conformations, including those observed in the X-ray structures.
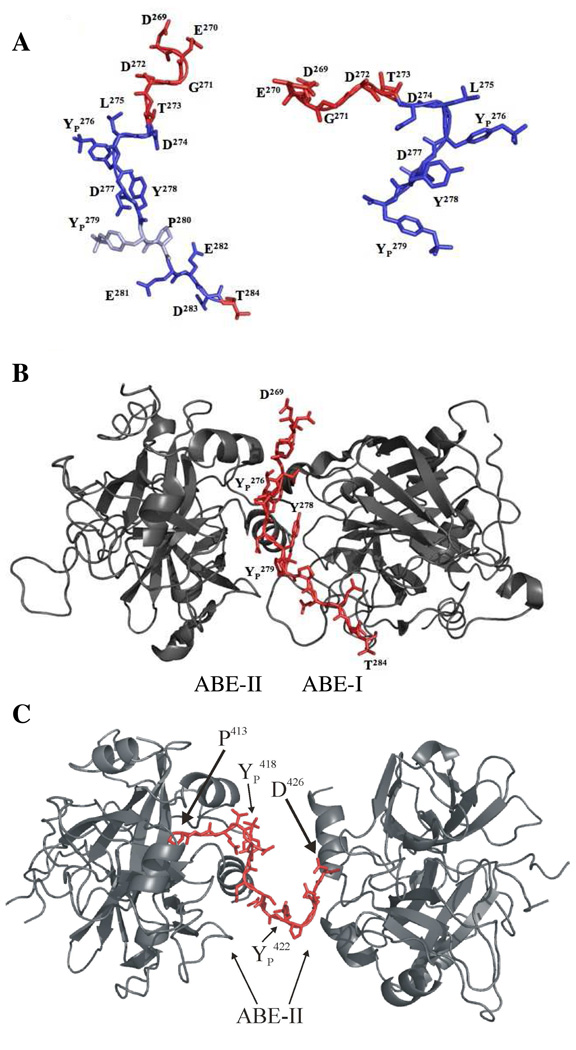
Figure 11.
A. Representation of the GpIbα peptide isolated from thrombin. The peptide on the left is from 1OOK and the structure on the right is from 1P8V. The individual amino acids are labeled. The trans bond between YP 279-P280 in 1OOK is in light blue. Residues experiencing line-broadening in the 1D NMR spectra are in blue. B. Illustration of GpIbα peptide induced dimerization of thrombin in 1OOK. The data suggests this structure more accurately represents the NMR results. The C-terminus of GpIbα is in an extended conformation and residues D274-T284 interact with both ABE-II and ABE-I of opposing thrombin monomers. C. Illustration of γ′ peptide induced dimerization of IIa depicted in the structure 2HWL. The turn conformation is situated between the two symmetry related IIa monomers, both at ABE-II. These figures were created using PyMol (11).
Overall, the GpIbα-IIa NMR work is more consistent with the 1OOK X-ray structure (25), which depicts GpIbα 269–284 in an extended conformation with a trans configuration for the YP 279-P280 bond (Figure 11A) (25). Electrostatic interactions are observed in 1OOK for residues D274-T284 with both ABE-I and ABE-II (Figure 11B) (25). Presumably, residues P280-G286 are too disordered in 1P8V to be effectively modeled (26); yet the NMR data displays significant interactions with IIa for several of the residues within this sequence. The current NMR data and the X-ray structure 1OOK allude to the possibility of GpIbα binding to both exosites of IIa in an extended conformation through dimerization of the protease (Figure 11B).
An analogous situation is apparent when analyzing the NMR and X-ray data for the interaction of the γ′ peptide and IIa (38, 39) (Figure 11C). In this case, the γ′ peptide appears to dimerize IIa in the X-ray structure through utilization of ABE-II of both IIa molecules. The turn structure first described by the NMR work (38) is depicted as being sandwiched between the IIa dimer, perhaps stabilizing this configuration (39). Such interpretations should, however, be viewed with caution since crystal packing effects can influence the structural features being observed.
Analysis of the Analytical Ultracentrifugation Data: GpIbα Peptide (269–286) and γ′ Peptide (410–427) binding to Thrombin
The NMR and X-ray work were the motivations to use AUC to further examine how IIa behaves with increasing concentrations of peptide. The S values calculated for PPACK-IIa (3.1) and PPACK-IIa in the presence of the peptides (3.2–3.3) match those previously determined for PPACK-IIa (3.3) (67). The same study shows a fragment of thrombomodulin with a similar molecular weight to IIa binding to the serine protease and increasing S from 3.3 to 4.2 (67). Another group utilized sedimentation equilibrium AUC to demonstrate that human IIa does not dimerize at 4 µM, matching our results with bovine IIa (68). Taken together, IIa dimerization is not occurring under these conditions.
Analysis of the HDX Data (I): the Influence of pH on HDX dynamics
A series of IIa HDX experiments were performed at pH 5.6, 6.5, and 7.5. The present HDX studies demonstrate that ligand binding to IIa affects HDX in a similar manner at pH 5.6 and 6.5. This finding is important relative to NMR studies performed at pH 5.6 since it suggests that ligand binding effects associated with IIa binding are maintained at lower pH. The highest pH selected was 7.5, a more physiological pH, however, backbone amides exchange at a 10-fold faster rate than at pH 6.5 (60). The main impetus for focusing on pH 6.5 for the current and previous HDX studies (38) are the slower rate of backbone amide exchange coupled with the decreased rate of IIa autolysis.
It is interesting to compare these results with another HDX study with human IIa at pH 6.6 and 7.9 (63). In agreement with the pH studies of Mandell et al., the ABE-II 85–99 region can be described as completely solvent accessible and the 106–116 segment as fairly solvent accessible. Also, the ABE-I region 65–84 is defined by both groups as undergoing a fast rate of HDX. By contrast, the current HDX results for 65–84 at one minute slows significantly with the lower pH of 5.6, a condition not studied by the Komives group. Some differences in HDX are observed when comparing results for 135–149D from this study and 136–149A from Mandell et al. (63). The discrepancies are probably due to the inclusion of the highly solvent accessible autolysis loop (149A–149D) in the current study.
Analysis of the HDX Data (II): GpIbα (269–286), GpIbα (1–290), and the γ′ Peptide (410–427) Target ABE-II
Both peptides appear to bind to IIa’s ABE-II based on HDX protection for fragments 85–99 and 173–181 over the course of ten minutes. Figure 12 presents the γ′ peptide-IIa and GpIbα-IIa X-ray structures viewed from ABE-II. Two residues from the GpIbα mutagenesis studies are present within 85–99 that reduce binding when mutated to A: R93 and R97 (14, 15). While R97 does not appear to interact with GpIbα in the crystal structures, R93 is depicted as being a prominent residue in binding to YP 279 of both GpIbα crystals (25, 26). Also, the GpIbα structures display the importance of IIa N179 in binding to GpIbα D277, which is within fragment 173–181 (25, 26). Similarly, the γ′ HDX data supports the observed electrostatic interactions between IIa R93 and γ′ S420, as well as IIa N179 and γ′ D419 (39).
Figure 12.
Crystallographic views of the γ′ peptide (2HWL) and GpIbα (1OOK and 1P8V) bound to thrombin’s ABE-II. A prominent feature involves GpIbα YP 276 and γ′ YP 418 engulfed by a electropositive pocket formed by IIa R126, K235, and K236. In addition, GpIbα D277 and γ′ D419 reside near IIa N179. Finally, YP 279 and γ′ YP 422 point toward IIa K236. These structures illustrate similar bound characteristics for the central regions of the bound peptide, however differ in regard to the termini. These figures were created using PyMol (11).
Hirudin (54–65) possesses a unique HDX profile when compared to the ABE-II targeting peptides. Numerous X-ray and NMR structures have been reported detailing IIa bound hirudin (69–74). In addition, Myles et al. provide an excellent analysis regarding the electrostatic and hydrophobic nature of hirudin binding to IIa (75). Hirudin (54–65) affects IIa HDX in a similar manner as the larger thrombomodulin fragment TMEGF45 (63). The dramatic protection observed for IIa residues 65–84 can be attributed to hirudin (54–65) electrostatic interactions with R73, R75, and R77a, hydrophobic contacts with L65, T74, Y76, and I82, and the energetically linked residues R67 and K81.
The evidence for a similar destination on IIa for γ′ and GpIbα becomes more compelling when considering recent research. In some of these investigations, the γ′ peptide competitively inhibits IIa binding to GpIbα, consequently hindering platelet aggregation (76, 77). Similarly, Weeterings et al. have shown that fibrin bound to IIa’s ABE-I can promote platelet aggregation through GpIbα interaction with ABE-II (78). Taken together with the HDX data in this report, GpIbα residues 269–286 and 1–290 appear to preferentially target ABE-II.
Finally, it must be pointed out that others have declared ABE-I as GpIbα’s primary binding site on IIa (17–19). Perhaps the concentration of GpIbα and IIa is a factor to consider when examining GpIbα interactions with IIa. In one of these reports, the investigators used IIa at 0.42 nM and GpIbα at 50 µM to monitor the effect of the peptide on catalytic activity, 100,000 times the amount of IIa (17). Conversely, a study proclaiming ABE-II as GpIbα’s binding site implemented a much lower 80:1 ratio of IIa (100 nM) to GpIbα (8 µM) (15). As for the GpIbα protein fragment (1–290), the ratio was much lower at 1.6:1, yielding similar results to the GpIbα peptide (269–286) at 20:1. These observations suggest that protein/peptide concentration plays an important role in determining the destination of GpIbα on IIa.
Analysis of the HDX Data (III): GpIbα (269–286), GpIbα (1–290), and the γ′ Peptide (410–427) Affect Thrombin HDX Dynamics in a Similar Manner
The evidence for exosite binding linkage to the active site is well established (15, 23, 79–81). It has been shown that occupation of ABE-II by GpIbα enhances PAR-1 hydrolysis by IIa (23), yet may non-competitively inhibit FpA release from fibrinogen (15). More recently, research has demonstrated that competitive binding of γ′ peptide to ABE-II reduces the amount of PAR-1 cleaved on the platelet surface (76, 77). As for energetically linked exosites, conflicting reports exist in the literature. Though work has shown a strong negative thermodynamic linkage between the exosites (29, 82), others have found simultaneous exosite occupation without a significant energetic coupling (79, 83).
In this context, questions arise about the nature of the reduced HDX observed when these peptides bind to IIa through ABE-II. Three distinct possibilities can be envisioned. Observed decreases in HDX could be the product of a restricted ensemble of conformations, a conformational change, or as previously discussed, a ligand-protein interface. The last scenario implies that these peptides also bind to other sites on the surface of IIa, such as ABE-I. Evidence against this situation has been provided in the earlier discussion.
The second scenario is that ABE binding could trigger a series of conformational changes that emanate from the binding interface. For example, within the autolysis loop peptide (135–149D), residue W141 has been postulated to connect ABE-I to the active site (42, 63, 84). This line of communication from ABE-I to the active site via W141 has been observed with thrombomodulin binding to ABE-I (63, 84) and PPACK inhibition at the active site of IIa (42). The strong HDX perturbations occurring to the loop residues 135–149D when hirudin (54–65), and to a lesser extent γ′ and GpIbα, bind may be reflective of linkage between the IIa active site, W141, and the exosites ABE I and/or ABE-II.
An alternative explanation centers on a seminal publication from the Griesinger group which presents strong evidence that proteins (in this case ubiquitin) sample most conformations associated with the observed bound conformation before interacting with a ligand (85). Through evolutionary means, the authors suggest that ligands have exploited specific conformations that are sampled by the target protein (i.e. the conformational ensemble). In the context of ligand binding to IIIa, perhaps binding to ABE-I and ABE-II favors one group of IIa conformers over another. This preference would be reflected in the altered HDX dynamics at sites unaffiliated with ligand binding. Regardless of the explanation, the physiological significance of reduced HDX profiles for ABE-occupied IIa remains an intriguing question. Results collected thus far strongly suggest a role for linkage between the two ABE sites.
Correlating the NMR, AUC, and HDX Results: the Influence of Thrombin Concentration
An important variable exists between the three primary investigations reported in this paper: IIa concentration. The IIa required for the NMR, HDX, and AUC studies amounted to 150 µM, 50 µM, and 4 µM, respectively. Early studies attempting to determine the molecular weight of bovine IIa found that as the concentration of IIa increased, the apparent MW doubled (86, 87). Another group reported IIa dimerization in gel-filtration runs (88), which has been corroborated in the presence of GpIbα by Celikel et al (25). Physiologically, research has shown that low levels of IIa activate platelets in a GpIbα dependent manner, whereas higher levels of IIa perform this function independent of GpIbα (18, 89). Clearly, IIa concentration has direct consequences on the mechanism of platelet activation.
Remaining to be determined is whether GpIbα induced IIa dimerization has physiological relevance in regard to bridging neighboring platelets (26) or clustering GpIbα together on the surface of a platelet (25). Perhaps IIa dimerization observed in crystallography and implied in the NMR work is related to the higher amount of material required for these investigations. Recently, serine protease dimerization was reported in IIa-suramin complexes and the effect attributed to crystal packing (90). It is quite intriguing that a potential physiologically relevant dimeric form of prothrombin has been described that retains catalytic activity (91). As several groups have suggested, the KD and concentrations of all IIa interacting partners must be considered at the site of thrombus formation (76, 92, 93).
Conclusions
These studies further characterize peptide binding to IIa’s anion binding exosites. The NMR results suggest that the GpIbα peptide binds to IIa through residues D274-E285 in an extended conformation. HDX data reveal significant similarities between GpIbα and γ′ peptide binding to ABE-II, while hirudin (54–65) demonstrates the HDX profile for an ABE-I targeting peptide. A significant finding from this work concerns the similarity in HDX profiles for the GpIbα peptide and protein fragments, providing strong evidence for the effectiveness of peptide mimics of proteins. By examining γ′ peptide (410–427) and GpIbα (269–286) binding to IIa, a clearer understanding emerges concerning the importance of ABE-II in regulating blood coagulation and platelet activation.
Supplementary Material
Acknowledgment
We are grateful for the generous gift of GpIbα (1–290) provided by Z. Ruggeri and lab members from the Scripps Institute in La Jolla, California. We are thankful for the guidance and assistance of N.J. Stolowich in performing the NMR experiments. We also appreciate the expertise provided by J. B. Chaires, W. Dean, and N. Garbett in carrying out and analyzing the AUC experiments. Finally, we extend much thanks to P. Doiphode, M. Jadhav, M. Malovichko, and R. Woofter for helpful discussions and critical evaluation of the present work.
Footnotes
Funding for this project was supported by NIH Grant R01HL68440 (to M.C.M.).
Abbreviations: IIa, thrombin; PAR-1, protease-activated receptor 1; ABE-I, anion binding exosite I; ABE-II, anion binding exosite II; GpIbα, glycoprotein Ibα; LRR, leucine rich repeat; FpA, fibrinopeptide A; FVIII, blood clotting factor VIII; PPACK, D-Phe-Pro-Arg chloromethyl ketone; YS, sulfotyrosine; YP, phosphotyrosine; NMR, nuclear magnetic resonance; 1D, one dimensional; 2D, two dimensional; TOCSY, total correlation spectroscopy; tr-NOESY, transferred nuclear Overhauser effect spectroscopy; MALDI-TOF MS, matrix-assisted laser desorption-ionization time-of-flight mass spectrometry; HDX, hydrogen-deuterium exchange; AUC, analytical ultracentrifugation; S, Svedberg constant; KD, equilibrium binding constant.
Throughout the paper, thrombin residues have been referenced according to the chymotrypsin numbering scheme.
Supporting Information Available
Table S1 is Deuterium Incorporation for IIa with the peptides GpIbα, γ′ and Hirudin (54–65) and the protein fragment GpIbα (1–290) 3YS and 2YS at pH 6.5. Table S2 is Deuterium Incorporation for IIa (pH 7.5 and 5.6) with the peptides GpIbα, γ′ and Hirudin (54–65) at pH 5.6 This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Di Cera E. Thrombin interactions. Chest. 2003;124:11S–17S. doi: 10.1378/chest.124.3_suppl.11s. [DOI] [PubMed] [Google Scholar]
- 2.Bode W. The structure of thrombin, a chameleon-like proteinase. J Thromb Haemost. 2005;3:2379–2388. doi: 10.1111/j.1538-7836.2005.01356.x. [DOI] [PubMed] [Google Scholar]
- 3.Huntington JA. Molecular recognition mechanisms of thrombin. J Thromb Haemost. 2005;3:1861–1872. doi: 10.1111/j.1538-7836.2005.01363.x. [DOI] [PubMed] [Google Scholar]
- 4.Weisel JW. Fibrinogen and fibrin. Adv Protein Chem. 2005;70:247–299. doi: 10.1016/S0065-3233(05)70008-5. [DOI] [PubMed] [Google Scholar]
- 5.Coughlin SR. Protease-activated receptors in vascular biology. Thromb Haemost. 2001;86:298–307. [PubMed] [Google Scholar]
- 6.Esmon CT. The protein C pathway. Chest. 2003;124:26S–32S. doi: 10.1378/chest.124.3_suppl.26s. [DOI] [PubMed] [Google Scholar]
- 7.Bode W. The structure of thrombin: a janus-headed proteinase. Semin Thromb Hemost. 2006;32 Suppl 1:16–31. doi: 10.1055/s-2006-939551. [DOI] [PubMed] [Google Scholar]
- 8.Bode W, Turk D, Karshikov A. The refined 1.9-A X-ray crystal structure of D-Phe-Pro-Arg chloromethylketone-inhibited human alpha-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships. Protein Sci. 1992;1:426–471. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrews RK, Gardiner EE, Shen Y, Whisstock JC, Berndt MC. Glycoprotein Ib-IX-V. Int J Biochem Cell Biol. 2003;35:1170–1174. doi: 10.1016/s1357-2725(02)00280-7. [DOI] [PubMed] [Google Scholar]
- 10.Clemetson KJ. A short history of platelet glycoprotein Ib complex. Thromb Haemost. 2007;98:63–68. [PubMed] [Google Scholar]
- 11.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 12.Kobe B, Guncar G, Buchholz R, Huber T, Maco B, Cowieson N, Martin JL, Marfori M, Forwood JK. Crystallography and protein-protein interactions: biological interfaces and crystal contacts. Biochem Soc Trans. 2008;36:1438–1441. doi: 10.1042/BST0361438. [DOI] [PubMed] [Google Scholar]
- 13.Vanhoorelbeke K, Ulrichts H, Romijn RA, Huizinga EG, Deckmyn H. The GPIbalpha-thrombin interaction: far from crystal clear. Trends Mol Med. 2004;10:33–39. doi: 10.1016/j.molmed.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 14.De Cristofaro R, De Candia E, Landolfi R, Rutella S, Hall SW. Structural and functional mapping of the thrombin domain involved in the binding to the platelet glycoprotein Ib. Biochemistry. 2001;40:13268–13273. doi: 10.1021/bi010491f. [DOI] [PubMed] [Google Scholar]
- 15.Li CQ, Vindigni A, Sadler JE, Wardell MR. Platelet glycoprotein Ib alpha binds to thrombin anion-binding exosite II inducing allosteric changes in the activity of thrombin. J Biol Chem. 2001;276:6161–6168. doi: 10.1074/jbc.M004164200. [DOI] [PubMed] [Google Scholar]
- 16.Yun TH, Baglia FA, Myles T, Navaneetham D, Lopez JA, Walsh PN, Leung LL. Thrombin activation of factor XI on activated platelets requires the interaction of factor XI and platelet glycoprotein Ib alpha with thrombin anion-binding exosites I and II, respectively. J Biol Chem. 2003;278:48112–48119. doi: 10.1074/jbc.M306925200. [DOI] [PubMed] [Google Scholar]
- 17.Bouton MC, Thurieau C, Guillin MC, Jandrot-Perrus M. Characteristics of the interaction between thrombin exosite 1 and the sequence 269–287 [correction of 269–297] of platelet glycoprotein Ibalpha. Thromb Haemost. 1998;80:310–315. [PubMed] [Google Scholar]
- 18.Jandrot-Perrus M, Bouton MC, Lanza F, Guillin MC. Thrombin interaction with platelet membrane glycoprotein Ib. Semin Thromb Hemost. 1996;22:151–156. doi: 10.1055/s-2007-999003. [DOI] [PubMed] [Google Scholar]
- 19.Jandrot-Perrus M, Huisse MG, Krstenansky JL, Bezeaud A, Guillin MC. Effect of the hirudin carboxy-terminal peptide 54–65 on the interaction of thrombin with platelets. Thromb Haemost. 1991;66:300–305. [PubMed] [Google Scholar]
- 20.Karshikov A, Bode W, Tulinsky A, Stone SR. Electrostatic interactions in the association of proteins: an analysis of the thrombin-hirudin complex. Protein Sci. 1992;1:727–735. doi: 10.1002/pro.5560010605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Marco L, Mazzucato M, Masotti A, Ruggeri ZM. Localization and characterization of an alpha-thrombin-binding site on platelet glycoprotein Ib alpha. J Biol Chem. 1994;269:6478–6484. [PubMed] [Google Scholar]
- 22.Jandrot-Perrus M, Clemetson KJ, Huisse MG, Guillin MC. Thrombin interaction with platelet glycoprotein Ib: effect of glycocalicin on thrombin specificity. Blood. 1992;80:2781–2786. [PubMed] [Google Scholar]
- 23.De Candia E, Hall SW, Rutella S, Landolfi R, Andrews RK, De Cristofaro R. Binding of thrombin to glycoprotein Ib accelerates the hydrolysis of Par-1 on intact platelets. J Biol Chem. 2001;276:4692–4698. doi: 10.1074/jbc.M008160200. [DOI] [PubMed] [Google Scholar]
- 24.De Cristofaro R, De Filippis V. Interaction of the 268–282 region of glycoprotein Ibalpha with the heparin-binding site of thrombin inhibits the enzyme activation of factor VIII. Biochem J. 2003;373:593–601. doi: 10.1042/BJ20030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Celikel R, McClintock RA, Roberts JR, Mendolicchio GL, Ware J, Varughese KI, Ruggeri ZM. Modulation of alpha-thrombin function by distinct interactions with platelet glycoprotein Ibalpha. Science. 2003;301:218–221. doi: 10.1126/science.1084183. [DOI] [PubMed] [Google Scholar]
- 26.Dumas JJ, Kumar R, Seehra J, Somers WS, Mosyak L. Crystal structure of the GpIbalpha-thrombin complex essential for platelet aggregation. Science. 2003;301:222–226. doi: 10.1126/science.1083917. [DOI] [PubMed] [Google Scholar]
- 27.Vali Z, Scheraga HA. Localization of the binding site on fibrin for the secondary binding site of thrombin. Biochemistry. 1988;27:1956–1963. doi: 10.1021/bi00406a023. [DOI] [PubMed] [Google Scholar]
- 28.Lovely RS, Moaddel M, Farrell DH. Fibrinogen gamma' chain binds thrombin exosite II. J Thromb Haemost. 2003;1:124–131. doi: 10.1046/j.1538-7836.2003.00027.x. [DOI] [PubMed] [Google Scholar]
- 29.Pospisil CH, Stafford AR, Fredenburgh JC, Weitz JI. Evidence that both exosites on thrombin participate in its high affinity interaction with fibrin. J Biol Chem. 2003;278:21584–21591. doi: 10.1074/jbc.M300545200. [DOI] [PubMed] [Google Scholar]
- 30.Mosesson MW, Siebenlist KR, Meh DA. The structure and biological features of fibrinogen and fibrin. Ann N Y Acad Sci. 2001;936:11–30. doi: 10.1111/j.1749-6632.2001.tb03491.x. [DOI] [PubMed] [Google Scholar]
- 31.Chung DW, Davie EW. gamma and gamma' chains of human fibrinogen are produced by alternative mRNA processing. Biochemistry. 1984;23:4232–4236. doi: 10.1021/bi00313a033. [DOI] [PubMed] [Google Scholar]
- 32.Fornace AJ, Jr, Cummings DE, Comeau CM, Kant JA, Crabtree GR. Structure of the human gamma-fibrinogen gene. Alternate mRNA splicing near the 3' end of the gene produces gamma A and gamma B forms of gamma-fibrinogen. J Biol Chem. 1984;259:12826–12830. [PubMed] [Google Scholar]
- 33.Lovely RS, Falls LA, Al-Mondhiry HA, Chambers CE, Sexton GJ, Ni H, Farrell DH. Association of gammaA/gamma' fibrinogen levels and coronary artery disease. Thromb Haemost. 2002;88:26–31. [PubMed] [Google Scholar]
- 34.Mosesson MW, Finlayson JS. Biochemical and chromatographic studies of certain activities associated with human fibrinogen preparations. J Clin Invest. 1963;42:747–755. doi: 10.1172/JCI104767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mannila MN, Lovely RS, Kazmierczak SC, Eriksson P, Samnegard A, Farrell DH, Hamsten A, Silveira A. Elevated plasma fibrinogen gamma' concentration is associated with myocardial infarction: effects of variation in fibrinogen genes and environmental factors. J Thromb Haemost. 2007;5:766–773. doi: 10.1111/j.1538-7836.2007.02406.x. [DOI] [PubMed] [Google Scholar]
- 36.Grunbacher G, Weger W, Marx-Neuhold E, Pilger E, Koppel H, Wascher T, Marz W, Renner W. The fibrinogen gamma (FGG) 10034C>T polymorphism is associated with venous thrombosis. Thromb Res. 2007;121:33–36. doi: 10.1016/j.thromres.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Uitte de Willige S, de Visser MC, Houwing-Duistermaat JJ, Rosendaal FR, Vos HL, Bertina RM. Genetic variation in the fibrinogen gamma gene increases the risk for deep venous thrombosis by reducing plasma fibrinogen gamma' levels. Blood. 2005;106:4176–4183. doi: 10.1182/blood-2005-05-2180. [DOI] [PubMed] [Google Scholar]
- 38.Sabo TM, Farrell DH, Maurer MC. Conformational analysis of gamma' peptide (410–427) interactions with thrombin anion binding exosite II. Biochemistry. 2006;45:7434–7445. doi: 10.1021/bi060360k. [DOI] [PubMed] [Google Scholar]
- 39.Pineda AO, Chen ZW, Marino F, Mathews FS, Mosesson MW, Di Cera E. Crystal structure of thrombin in complex with fibrinogen gamma' peptide. Biophys Chem. 2006;125:556–559. doi: 10.1016/j.bpc.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Stone SR, Hofsteenge J. Kinetics of the inhibition of thrombin by hirudin. Biochemistry. 1986;25:4622–4628. doi: 10.1021/bi00364a025. [DOI] [PubMed] [Google Scholar]
- 41.Trumbo TA, Maurer MC. Examining thrombin hydrolysis of the factor XIII activation peptide segment leads to a proposal for explaining the cardioprotective effects observed with the factor XIII V34L mutation. J Biol Chem. 2000;275:20627–20631. doi: 10.1074/jbc.M000209200. [DOI] [PubMed] [Google Scholar]
- 42.Croy CH, Koeppe JR, Bergqvist S, Komives EA. Allosteric changes in solvent accessibility observed in thrombin upon active site occupation. Biochemistry. 2004;43:5246–5255. doi: 10.1021/bi0499718. [DOI] [PubMed] [Google Scholar]
- 43.Ni F, Zhu Y, Scheraga HA. Thrombin-Bound Structures of Designed Analogs of Human Fibrinopeptide-a Determined by Quantitative Transferred Noe Spectroscopy - a New Structural Basis for Thrombin Specificity. Journal of Molecular Biology. 1995;252:656–671. doi: 10.1006/jmbi.1995.0527. [DOI] [PubMed] [Google Scholar]
- 44.Ni F, Meinwald YC, Vasquez M, Scheraga HA. High-resolution NMR studies of fibrinogen-like peptides in solution: structure of a thrombin-bound peptide corresponding to residues 7–16 of the A alpha chain of human fibrinogen. Biochemistry. 1989;28:3094–3105. doi: 10.1021/bi00433a053. [DOI] [PubMed] [Google Scholar]
- 45.Ni F, Ripoll DR, Martin PD, Edwards BF. Solution structure of a platelet receptor peptide bound to bovine alpha-thrombin. Biochemistry. 1992;31:11551–11557. doi: 10.1021/bi00161a037. [DOI] [PubMed] [Google Scholar]
- 46.Martin PD, Robertson W, Turk D, Huber R, Bode W, Edwards BF. The structure of residues 7–16 of the A alpha-chain of human fibrinogen bound to bovine thrombin at 2.3-A resolution. J Biol Chem. 1992;267:7911–7920. [PubMed] [Google Scholar]
- 47.Mathews II, Padmanabhan KP, Ganesh V, Tulinsky A, Ishii M, Chen J, Turck CW, Coughlin SR, Fenton JW., 2nd Crystallographic structures of thrombin complexed with thrombin receptor peptides: existence of expected and novel binding modes. Biochemistry. 1994;33:3266–3279. doi: 10.1021/bi00177a018. [DOI] [PubMed] [Google Scholar]
- 48.Ni F. 2-Dimensional Transferred Nuclear-Overhauser-Effects with Incomplete Averaging of Free-Ligand and Bound-Ligand Resonances. Journal of Magnetic Resonance Series B. 1995;106:147–155. [Google Scholar]
- 49.Ni F, Scheraga HA. Use of the Transferred Nuclear Overhauser Effect to Determine the Conformations of Ligands Bound to Proteins. Accounts of Chemical Research. 1994;27:257–264. [Google Scholar]
- 50.Friedrich R, Panizzi P, Kawabata S, Bode W, Bock PE, Fuentes-Prior P. Structural basis for reduced staphylocoagulase-mediated bovine prothrombin activation. J Biol Chem. 2006;281:1188–1195. doi: 10.1074/jbc.M507957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sabo TM, Brasher PB, Maurer MC. Perturbations in Factor XIII Resulting from Activation and Inhibition Examined by Solution Based Methods and Detected by MALDI-TOF MS. Biochemistry. 2007;46:10089–10101. doi: 10.1021/bi700579z. [DOI] [PubMed] [Google Scholar]
- 52.Chen J, Smith DL. Amide hydrogen exchange shows that malate dehydrogenase is a folded monomer at pH 5. Protein Sci. 2001;10:1079–1083. doi: 10.1110/ps.53201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Lane LC, Smith DL. Detecting structural changes in viral capsids by hydrogen exchange and mass spectrometry. Protein Sci. 2001;10:1234–1243. doi: 10.1110/ps.100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner BT, Jr, Maurer MC. Evaluating the roles of thrombin and calcium in the activation of coagulation factor XIII using H/D exchange and MALDI-TOF MS. Biochemistry. 2002;41:7947–7954. doi: 10.1021/bi025630n. [DOI] [PubMed] [Google Scholar]
- 55.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Resing KA, Hoofnagle AN, Ahn NG. Modeling deuterium exchange behavior of ERK2 using pepsin mapping to probe secondary structure. J Am Soc Mass Spectrom. 1999;10:685–702. doi: 10.1016/S1044-0305(99)00037-9. [DOI] [PubMed] [Google Scholar]
- 57.Arrington CB, Robertson AD. Correlated motions in native proteins from MS analysis of NH exchange: evidence for a manifold of unfolding reactions in ovomucoid third domain. J Mol Biol. 2000;300:221–232. doi: 10.1006/jmbi.2000.3859. [DOI] [PubMed] [Google Scholar]
- 58.Clarke J, Itzhaki LS. Hydrogen exchange and protein folding. Curr Opin Struct Biol. 1998;8:112–118. doi: 10.1016/s0959-440x(98)80018-3. [DOI] [PubMed] [Google Scholar]
- 59.Swint-Kruse L, Robertson AD. Temperature and pH dependences of hydrogen exchange and global stability for ovomucoid third domain. Biochemistry. 1996;35:171–180. doi: 10.1021/bi9517603. [DOI] [PubMed] [Google Scholar]
- 60.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mandell JG, Falick AM, Komives EA. Identification of protein-protein interfaces by decreased amide proton solvent accessibility. Proc Natl Acad Sci U S A. 1998;95:14705–14710. doi: 10.1073/pnas.95.25.14705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mandell JG, Falick AM, Komives EA. Measurement of amide hydrogen exchange by MALDI-TOF mass spectrometry. Anal Chem. 1998;70:3987–3995. doi: 10.1021/ac980553g. [DOI] [PubMed] [Google Scholar]
- 63.Mandell JG, Baerga-Ortiz A, Akashi S, Takio K, Komives EA. Solvent accessibility of the thrombin-thrombomodulin interface. J Mol Biol. 2001;306:575–589. doi: 10.1006/jmbi.2000.4416. [DOI] [PubMed] [Google Scholar]
- 64.De Cristofaro R, De Candia E, Rutella S, Weitz JI. The Asp(272)-Glu(282) region of platelet glycoprotein Ibalpha interacts with the heparin-binding site of alpha-thrombin and protects the enzyme from the heparin-catalyzed inhibition by antithrombin III. J Biol Chem. 2000;275:3887–3895. doi: 10.1074/jbc.275.6.3887. [DOI] [PubMed] [Google Scholar]
- 65.Kroh HK, Tans G, Nicolaes GA, Rosing J, Bock PE. Expression of allosteric linkage between the sodium ion binding site and exosite I of thrombin during prothrombin activation. J Biol Chem. 2007;282:16095–16104. doi: 10.1074/jbc.M610577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marchese P, Murata M, Mazzucato M, Pradella P, De Marco L, Ware J, Ruggeri ZM. Identification of three tyrosine residues of glycoprotein Ib alpha with distinct roles in von Willebrand factor and alpha-thrombin binding. J Biol Chem. 1995;270:9571–9578. doi: 10.1074/jbc.270.16.9571. [DOI] [PubMed] [Google Scholar]
- 67.Olsen PH, Esmon NL, Esmon CT, Laue TM. Ca2+ dependence of the interactions between protein C, thrombin, and the elastase fragment of thrombomodulin. Analysis by ultracentrifugation. Biochemistry. 1992;31:746–754. doi: 10.1021/bi00118a016. [DOI] [PubMed] [Google Scholar]
- 68.Lonhienne TG, Jackson CM, Winzor DJ. Thermodynamic non-ideality as an alternative source of the effect of sucrose on the thrombin-catalyzed hydrolysis of peptide p-nitroanilide substrates. Biophys Chem. 2003;103:259–269. doi: 10.1016/s0301-4622(02)00322-8. [DOI] [PubMed] [Google Scholar]
- 69.Grutter MG, Priestle JP, Rahuel J, Grossenbacher H, Bode W, Hofsteenge J, Stone SR. Crystal structure of the thrombin-hirudin complex: a novel mode of serine protease inhibition. EMBO J. 1990;9:2361–2365. doi: 10.1002/j.1460-2075.1990.tb07410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rydel TJ, Ravichandran KG, Tulinsky A, Bode W, Huber R, Roitsch C, Fenton JW., 2nd The structure of a complex of recombinant hirudin and human alpha-thrombin. Science. 1990;249:277–280. doi: 10.1126/science.2374926. [DOI] [PubMed] [Google Scholar]
- 71.Rydel TJ, Tulinsky A, Bode W, Huber R. Refined structure of the hirudin-thrombin complex. J Mol Biol. 1991;221:583–601. doi: 10.1016/0022-2836(91)80074-5. [DOI] [PubMed] [Google Scholar]
- 72.Vitali J, Martin PD, Malkowski MG, Robertson WD, Lazar JB, Winant RC, Johnson PH, Edwards BF. The structure of a complex of bovine alpha-thrombin and recombinant hirudin at 2.8-A resolution. J Biol Chem. 1992;267:17670–17678. [PubMed] [Google Scholar]
- 73.Ni F, Konishi Y, Scheraga HA. Thrombin-bound conformation of the C-terminal fragments of hirudin determined by transferred nuclear Overhauser effects. Biochemistry. 1990;29:4479–4489. doi: 10.1021/bi00470a030. [DOI] [PubMed] [Google Scholar]
- 74.Ni F, Ripoll DR, Purisima EO. Conformational stability of a thrombin-binding peptide derived from the hirudin C-terminus. Biochemistry. 1992;31:2545–2554. doi: 10.1021/bi00124a015. [DOI] [PubMed] [Google Scholar]
- 75.Myles T, Le Bonniec BF, Betz A, Stone SR. Electrostatic steering and ionic tethering in the formation of thrombin-hirudin complexes: the role of the thrombin anion-binding exosite-I. Biochemistry. 2001;40:4972–4979. doi: 10.1021/bi0023549. [DOI] [PubMed] [Google Scholar]
- 76.Lovely RS, Rein CM, White TC, Jouihan SA, Boshkov LK, Bakke AC, McCarty OJ, Farrell DH. gammaA/gamma' fibrinogen inhibits thrombin-induced platelet aggregation. Thromb Haemost. 2008;100:837–846. [PubMed] [Google Scholar]
- 77.Lancellotti S, Rutella S, De Filippis V, Pozzi N, Rocca B, De Cristofaro R. Fibrinogen-elongated gamma chain inhibits thrombin-induced platelet response, hindering the interaction with different receptors. J Biol Chem. 2008;283:30193–30204. doi: 10.1074/jbc.M803659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weeterings C, Adelmeijer J, Myles T, de Groot PG, Lisman T. Glycoprotein Ibalpha-mediated platelet adhesion and aggregation to immobilized thrombin under conditions of flow. Arterioscler Thromb Vasc Biol. 2006;26:670–675. doi: 10.1161/01.ATV.0000200391.70818.a9. [DOI] [PubMed] [Google Scholar]
- 79.Verhamme IM, Olson ST, Tollefsen DM, Bock PE. Binding of exosite ligands to human thrombin. Re-evaluation of allosteric linkage between thrombin exosites I and II. J Biol Chem. 2002;277:6788–6798. doi: 10.1074/jbc.M110257200. [DOI] [PubMed] [Google Scholar]
- 80.Siebenlist KR, Mosesson MW, Hernandez I, Bush LA, Di Cera E, Shainoff JR, Di Orio JP, Stojanovic L. Studies on the basis for the properties of fibrin produced from fibrinogen-containing gamma' chains. Blood. 2005;106:2730–2736. doi: 10.1182/blood-2005-01-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lane DA, Philippou H, Huntington JA. Directing thrombin. Blood. 2005;106:2605–2612. doi: 10.1182/blood-2005-04-1710. [DOI] [PubMed] [Google Scholar]
- 82.Fredenburgh JC, Stafford AR, Weitz JI. Evidence for allosteric linkage between exosites 1 and 2 of thrombin. J Biol Chem. 1997;272:25493–25499. doi: 10.1074/jbc.272.41.25493. [DOI] [PubMed] [Google Scholar]
- 83.Hogg PJ, Jackson CM, Labanowski JK, Bock PE. Binding of fibrin monomer and heparin to thrombin in a ternary complex alters the environment of the thrombin catalytic site, reduces affinity for hirudin, and inhibits cleavage of fibrinogen. J Biol Chem. 1996;271:26088–26095. doi: 10.1074/jbc.271.42.26088. [DOI] [PubMed] [Google Scholar]
- 84.Koeppe JR, Seitova A, Mather T, Komives EA. Thrombomodulin tightens the thrombin active site loops to promote protein C activation. Biochemistry. 2005;44:14784–14791. doi: 10.1021/bi0510577. [DOI] [PubMed] [Google Scholar]
- 85.Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 86.Cohly MA, Scheraga HA. Aggregation of thrombin. Arch Biochem Biophys. 1961;95:428–434. doi: 10.1016/0003-9861(61)90172-2. [DOI] [PubMed] [Google Scholar]
- 87.Winzor DJ, Scheraga HA. Studies of Chemically Reacting Systems on Sephadex. II. Molecular Weights of Monomers in Rapid Association Equilibrium. The Journal of Physical Chemistry. 1964;68:338–343. [Google Scholar]
- 88.Batt CW, Mikulka TW, Mann KG, Guarracino CL, Altiere RJ, Graham RG, Quigley JP, Wolf JW, Zafonte CW. The purification and properties of bovine thrombin. J Biol Chem. 1970;245:4857–4862.. [PubMed] [Google Scholar]
- 89.De Marco L, Mazzucato M, Masotti A, Fenton JW, 2nd, Ruggeri ZM. Function of glycoprotein Ib alpha in platelet activation induced by alpha-thrombin. J Biol Chem. 1991;266:23776–23783. [PubMed] [Google Scholar]
- 90.Lima LM, Becker CF, Giesel GM, Marques AF, Cargnelutti MT, de Oliveira Neto M, Queiroz Monteiro R, Verli H, Polikarpov I. Structural and thermodynamic analysis of thrombin:suramin interaction in solution and crystal phases. Biochim Biophys Acta. 2009;1794:873–881. doi: 10.1016/j.bbapap.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 91.Anderson PJ. A dimeric form of prothrombin on membrane surfaces. Biochem J. 1998;336(Pt 3):631–638. doi: 10.1042/bj3360631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Adam F, Bouton MC, Huisse MG, Jandrot-Perrus M. Thrombin interaction with platelet membrane glycoprotein Ib alpha. Trends Mol Med. 2003;9:461–464. doi: 10.1016/j.molmed.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 93.Mann KG, Brummel-Ziedins K, Orfeo T, Butenas S. Models of blood coagulation. Blood Cells Mol Dis. 2006;36:108–117. doi: 10.1016/j.bcmd.2005.12.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.