Abstract
Parathyroid hormone (PTH) is a hormone regulating bone remodeling through its actions on both bone formation and bone resorption. Previously we reported that PTH induces matrix metalloproteinase-13 (MMP-13) transcription in osteoblastic cells. Here, we show that histone deacetylase 4 (HDAC4) interacts with Runx2, binds the MMP-13 promoter, and suppresses MMP-13 gene transcription in the rat osteoblastic cell line, UMR 106-01. PTH induces the rapid cAMP-dependent protein kinase-dependent release of HDAC4 from the MMP-13 promoter and subsequent transcription of MMP-13. Knock-out of HDAC4 either by siRNA in vitro or by gene deletion in vivo leads to an increase in MMP-13 expression, and overexpression of HDAC4 decreases the PTH induction of MMP-13. All of these observations indicate that HDAC4 represses MMP-13 gene transcription in bone. Moreover, PTH stimulates HDAC4 gene expression and enzymatic activity at times corresponding to the reassociation of HDAC4 with the MMP-13 promoter and a decline in its transcription. Thus, HDAC4 is a basal repressor of MMP-13 transcription, and PTH regulates HDAC4 to control MMP-13 promoter activity. These data identify a novel and discrete mechanism of regulating HDAC4 levels and, subsequently, gene expression.
Keywords: Bone, Gene Transcription, Histone Deacetylase, Hormones, Matrix Metalloproteinase, MMP-13, Osteoblast, Parathyroid Hormone
Introduction
Parathyroid hormone (PTH)2 is an 84-amino acid peptide hormone that functions as an essential regulator of calcium homeostasis and as a mediator of bone remodeling (1). PTH acts via the PTH/PTH-related protein 1 receptor (a G protein-coupled receptor) on osteoblast membranes (2). PTH is a major modulator of bone metabolism via its action on bone formation and resorption. Intermittent administration of PTH increases bone mass by stimulating de novo bone formation (3, 4). The hormone stimulates the expression of matrix metalloproteinase-13 (MMP-13, collagenase-3) (5), RANKL (6), and macrophage colony-stimulating factor (7) among others. MMP-13 is responsible for degrading components of extracellular matrix. Enzyme expression and transcription are strongly induced by PTH in the rat osteoblastic osteosarcoma cell line UMR 106-01 (8). Previously, we showed that Runx2 binding to the runt domain (RD)-binding site and activator protein-1 (AP-1) binding to the AP-1 site are necessary for the PTH-induced MMP-13 promoter activity and that the proteins interact with each other (9). Runx2 (AML-3/Cbfa1) is an important transcription factor in bone cells, and disruption of the Runx2 gene in mice induces skeletal defects (10, 11). Runx2 is essential for osteoblast development and differentiation (12), including MMP-13 expression (13, 14).
Gene expression is regulated by several mechanisms such as DNA methylation, ATP-dependent chromatin remodeling, and post-translational modifications of histones, which include the dynamic acetylation and deacetylation of epsilon-amino groups of lysine residues present in the tails of core histones. Thus, histone deacetylases (HDACs) are crucial regulators of gene expression in transcriptional co-repressor complexes. The class I HDACs (HDAC1, 2, 3, and 8) have homology to the yeast global transcriptional regulator Rpd3 and are widely expressed. In contrast, the class II HDACs (HDAC4, 5, 6, 7, 9, and 10) show homology to yeast Hda1 and are expressed in cell type-restricted patterns. The class IIA histone deacetylases (HDAC4, 5, 7, and 9) can be expressed in a tissue-specific fashion and are regulated by nuclear-cytoplasmic shuttling (15). The 14-3-3 proteins shuttle class II HDACs to the cytoplasm (16, 17). Several class II HDACs appear to have a role in skeletal formation (18). Recently described Hdac6-null mice have a slight increase in bone mineral density (19). Notably, Hdac4-null mice display premature ossification of developing bones caused by constitutive Runx2 expression. Thus, HDAC4 regulates chondrocyte hypertrophy and endochondral bone formation by inhibiting the activity of Runx2 (20). In osteoblasts, HDAC4 and HDAC5 participate in TGFβ signaling pathways that suppress Runx2 activity (21). Moreover, HDAC4 and HDAC5 can deacetylate Runx2 and lead to Smurf-mediated degradation of Runx2 (22). Thus, HDAC4 is considered an important target for osteoblast or chondrocyte differentiation.
Here, we report that the class II histone deacetylase, HDAC4, serves an important role in regulating MMP-13 expression in osteoblastic cells, and PTH controls the function of HDAC4: 1) HDAC4 can suppress MMP-13 gene expression in vivo and in vitro; 2) PTH enhances MMP-13 gene transcription through dissociation of HDAC4 from Runx2 and the promoter of MMP-13; and 3) PTH stimulates HDAC4 gene expression and enzymatic activity, and a feedback system of HDAC4 suppresses PTH-induced MMP-13 gene expression. This is the first observation of the regulation of HDAC4 by PTH in osteoblastic cells, and it may be a clue to understanding chromatin functions for osteoblast differentiation and mineralization.
EXPERIMENTAL PROCEDURES
Materials
Parathyroid hormone (rat PTH 1–34) was purchased from Sigma. HDAC4 siRNA oligonucleotides (TriFECTaTM kit) were purchased from Integrated DNA Technologies. Trichostatin A was purchased from EMD Biosciences.
Antibodies
Anti-HDAC4 (against 10 N-terminal amino acids used for immunoprecipitation and Western blots) and anti-HDAC3 were purchased from Cell Signaling Technology. Anti-HDAC4 (H92, against amino acids 530–631 used for chromatin immunoprecipitation and HDAC4 enzyme assays), anti-HDAC6 (L-18), anti-Runx2 (M-70), and anti-Cdk2 (M2) were purchased from Santa Cruz Biotechnology. Anti-Myc was purchased from Clontech. Anti-FLAG was purchased from Sigma. Anti-MMP-13 was purchased from Millipore.
Cell Culture
The UMR 106-01 cells were cultured in Eagle's minimal essential medium (EMEM) supplemented with 25 mm Hepes, pH 7.4, 1% nonessential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin, and 5% fetal bovine serum. HEK 293 cells were cultured in Dulbecco's modified eagle's medium supplemented with 100 μg/ml streptomycin and 10% fetal bovine serum.
Animal Experiments
HDAC4 knock-out mice were previously described (20). The offspring from mating HDAC4+/− mice were genotyped from DNA obtained by tail clips. Offspring that were HDAC4+/− were subsequently mated, and all of their offspring were genotyped by PCR. Genotyping of the HDAC4 allele was done as follows. Briefly, primers spanning exons 5 and 6 of HDAC4 (reverse primer, 5′-CTTGTTGAGAACAAACTCCTGCAGCT-3′; forward primer, 5′-ATCTGCCCACCAGAGTATGTG-3′) or the nuclear-localized lacZ selection marker (reverse primer, 5′-GATTGACCGTAATGGGATAGGTTACG-3′) were used to identify the wild-type (∼480-bp PCR product) or targeted (∼810-bp PCR product) HDAC4 alleles.
Transient Transfection and Chloramphenicol Acetyltransferase (CAT) or Luciferase Activities
The −148 rat MMP-13 promoter construct was subcloned upstream of a CAT reporter gene in pSV0 (Promega) as previously described (9). The plasmid DNAs were transiently transfected into cells using GeneJammer (Stratagene). Briefly, the cells were plated at 4 × 105 cells/well in 6-well plates in EMEM containing 5% fetal bovine serum. The following day, the cells were transfected with 1 μg of DNA/well in 1 ml of serum-free EMEM. After 3 h, 1 ml of EMEM containing 10% fetal bovine serum was added and incubated for 24 h. At this time, the agents were added, and the cells were incubated for 24 h prior to lysis for CAT assays. CAT activity was measured by reacting 50 μl of cell lysate in duplicate in a 100-μl reaction volume consisting of final concentrations of 250 μm n-butyryl-coenzyme A and 23 mm [14C]chloramphenicol (0.125 μCi/assay). The values were normalized to protein as determined by the Bradford dye binding (Bio-Rad) method. A standard curve using purified CAT was performed in every experiment to determine the linear range of the enzyme assay. For luciferase assays, UMR 106-01 cells were seeded in 12-well plates overnight and then transfected with indicated plasmids using GeneJammer (Stratagene) according to the manufacturer's protocol. After 2 days, the cells were treated with PTH for 6 h. The lysates were analyzed immediately for luciferase activity using the luciferase assay reagent (Promega) and an OptiCompII luminometer (MGM Instruments, Inc., Hamden, CT). To make the mutation construct of pGL2-MMP-13 promoter, a site-directed mutagenesis kit was used, and detailed procedures were in accord with the manufacturer's instructions.
Real Time Quantitative RT-PCR
Harvested femurs were stored in RNAlater. Total RNA from UMR 106-01 cells or homogenized tissues were isolated with TRIzol reagent following the manufacturer's instructions. Total RNA (0.1 μg) was reverse-transcribed to cDNA with the Invitrogen Superscript kit according to the manufacturer's instructions. PCR was performed on cDNA using primers, and the sequences used are listed in Table 1. The sequences were amplified by adding 2.5 μl of cDNA to the PCR mixture (22.5 μl) containing each primer (0.2 μm) and 12.5 μl of the Platinum SYBR Green qPCR SuperMix UDG (Invitrogen). The reactions were preincubated at 50 °C for 2 min for decontamination of dU-containing DNA by UDG and then incubated at 95 °C for 2 min to inactivate UDG and activate Taq. The PCR program continued 49 cycles of denaturation at 95 °C for 15 s, annealing, and elongation of the primers at 60 °C for 30 s. The relative gene expression for in vivo RNA analysis was determined using the formula 2(−ΔCt). For in vitro RNA analysis, fold changes in gene expression relative to control samples were calculated using the formula 2(ΔCtctrl − ΔCtPTH). All of the samples were normalized to β-actin.
TABLE 1.
Primer sequences
| Gene | Primers |
|---|---|
| Rat HDAC4 | 5′-GCAGAGGTTGAATGTGAGCA-3′ (sense) |
| 5′-GGAAGAAGTTCCCATCGTCA-3′ (antisense) | |
| Rat MMP-13 | 5′-GCCCTATCCCTTGATGCCATT-3′ (sense) |
| 5′-ACAGTTCAGGCTCAACCTGCTG-3′ (antisense) | |
| Rat β-actin | 5′-AGCCATGTACGTAGCCATCC-3′ (sense) |
| 5′-ACCCTCATAGATGGGCACAG-3′ (antisense) |
Immunoprecipitation and Western Blot
To examine the interaction between HDAC4 and Runx2, the FLAG-tagged HDAC4 expression plasmid was co-transfected into HEK 293 cells with Myc-tagged Runx2. A 10-μg portion of each plasmid was transfected into HEK 293 cells (in a 100-mm dish) with 30 μl of GeneJammer (Stratagene) transfection reagent. To test the interaction with endogenous HDAC4 and Runx2, we used UMR 106-01 cells with or without PTH stimulation. UMR 106-01 cells were washed twice in PBS, pH 7.4, and pelleted by centrifugation at 2000 rpm for 5 min at 4 °C. The pellets were resuspended in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 1 mm EDTA, 1% sodium deoxycholate, 0.1% SDS, and protease inhibitors) and incubated for 15 min at 4 °C. Equal amounts of total protein were determined by the Bradford dye binding (Bio-Rad) method.
The preparation of cytoplasmic and nuclear extracts from cells was by the NE-PER nuclear and cytoplasmic extraction reagents (Pierce). The soluble fraction collected by centrifugation was precleared by incubating with protein A/G-agarose beads (Santa Cruz). After the cleared supernatant had been incubated overnight with 2 μg/ml antibody at 4 °C, the agarose beads were washed three times with PBS. The bound proteins were separated using SDS-PAGE and were transferred to polyvinylidene difluoride membranes. The proteins were detected by ECL (Amersham Biosciences) according to the manufacturer's instructions. The quantitative results were obtained using the ImageQuant measurement software, standardized to each loading control.
Plasmids and siRNA Transfection
pCMV-FLAG HDAC4 was kindly provided by Dr. Xiang-Jiao Yang (Molecular Oncology Group, Department of Medicine, McGill University Health Centre, Montreal, Canada). UMR 106-01 cells were seeded in 6-well plates and transiently transfected with 60 nm of rat HDAC4 siRNA oligonucleotides and scrambled oligonucleotides using X-tremeGENE (Roche Applied Science) and incubated for 48 h. These samples were examined by Western blot and real time RT-PCR.
Chromatin Immunoprecipitation (ChIP) Assays
UMR 106-01 cells were incubated for 10 min at room temperature with medium containing 0.8% formaldehyde. The cells were then washed in ice-cold PBS containing protease inhibitors and 1 mm phenylmethylsulfonyl fluoride and resuspended in SDS lysis buffer (1% SDS, 10 mm EDTA, pH 8.0, 25 mm Tris-HCl, pH 8.1, containing protease inhibitors and 1 mm phenylmethylsulfonyl fluoride) for 10 min on ice. The samples were sonicated to reduce the DNA length to 0.5–1 kilobase pairs the cellular debris was removed by centrifugation, and the supernatant was diluted 10-fold in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, 167 mm NaCl supplemented with protease inhibitors). For PCR analysis, aliquots (1:100) of total chromatin DNA before immunoprecipitation were saved (input). Prior to chromatin immunoprecipitations, the samples were precleared with 80 μl of a 25% (v/v) suspension of DNA-coated protein A/G-agarose for 30 min at 4 °C. The supernatant was recovered and used directly for immunoprecipitation experiments with appropriate antibody overnight at 4 °C. Immune complexes were mixed with 60 μl of a 25% precoated protein A/G-agarose suspension followed by incubation for 1 h at 4 °C. The beads were collected and sequentially washed with 1 ml of each of the following buffers: low salt wash buffer, high salt wash buffer, and LiCl wash buffer. The beads were then washed twice using 1 ml of TE buffer. The immunocomplexes were eluted two times by adding a 250-μl aliquot of a freshly prepared solution of 1% SDS, 0.1 m NaHCO3. Twenty μl of 5 m NaCl was added to the samples, and the cross-linking reaction was reversed by a 6-h incubation at 65 °C. Further, the samples were digested with proteinase K (10 mg/ml), 40 mm Tris-HCl, pH 6.5, 10 mm EDTA at 42 °C for 1 h, and DNA was recovered by phenol/chloroform extractions. The DNA was precipitated with two volumes of ethanol using 1 μl of 20 mg/ml glycogen as carrier. The input lysates were also processed as above. The DNA was resuspended in water and used for quantitative PCR. The sequences of the oligonucleotides of the rat MMP-13 promoter used in this study are shown in Table 2 and Fig. 2D. The quantitated data represent real time PCR values normalized to input DNA and to the values obtained with normal rabbit IgG, which were set as one unit in each calculation. The data are presented as fold differences relative to IgG and normalized to input with the formula 2[(ΔCtIgG − CtInput) − (CtAb − CtInput)], where Ct is the threshold cycles, IgG is the normal rabbit IgG, Ab is the specific antibody, and Input is the input genomic DNA (23). To ensure specific PCR amplification, every real time PCR run was followed by a dissociation phase analysis.
TABLE 2.
Sequences of the rat MMP-13 promoter oligonucleotides
| Oligonucleotide | Sequence |
|---|---|
| Distal RD (sense) | 5′-AGAGATGCCCTAATTTTCCATTTC-3′ |
| Distal RD (antisense) | 5′-GTGTTCACTTCCTAGTGAGT-3′ |
| Distal RD and proximal AP-1 (sense) | 5′-CAGATGCGTTTTGATATGCC-3′ |
| Distal RD and proximal AP-1 (antisense) | 5′-AATAGTGATGAGTCACCACTT-3′ |
| Exogenous MMP-13 promoter (sense) | 5′-CACTCAGGTTCTGCCACAAA-3′ |
| Exogenous MMP-13 promoter (antisense) | 5′-TACCGGAATGCCAAGCTTAC-3′ |
FIGURE 2.
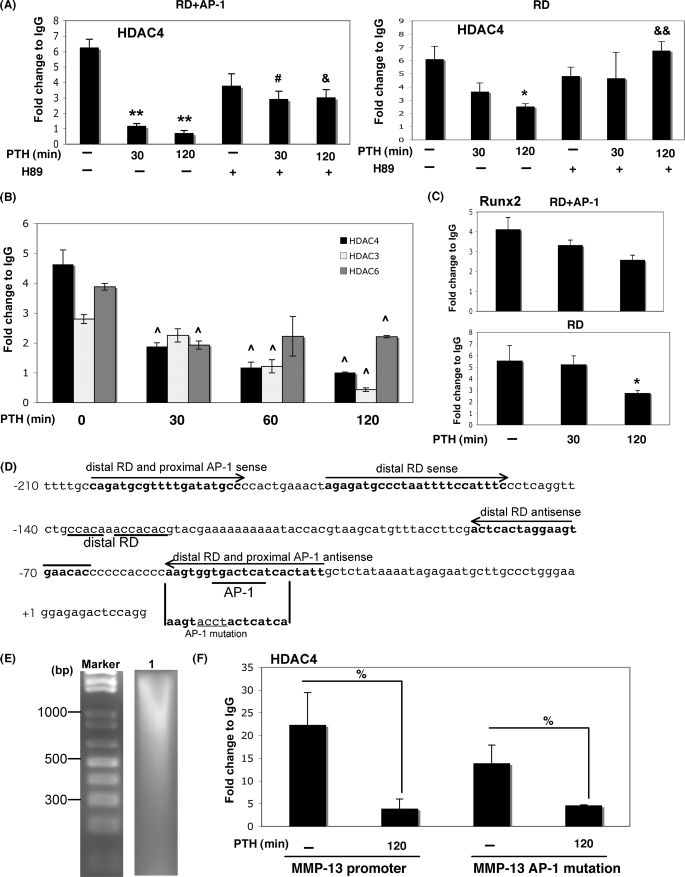
PTH decreases interaction of HDAC4 with Runx2 on the RD site of the MMP-13 promoter. A, UMR 106-01 cells were treated with control or PTH (10−8 m) for 30 and 120 min with or without preincubation with H89 (5 μm) for 30 min. After immunoprecipitation of the cross-linked lysates with HDAC4 antibody or with rabbit IgG as a negative control, the DNA was subjected to PCR with primers that amplify either the distal RD region or distal RD and proximal AP-1 sites of the rat MMP-13 promoter. Input DNA (1:100) is a positive control for the assay. **, p < 0.001 versus control; &&, p < 0.001 versus PTH 120 min; *, p < 0.03 versus control; &, p < 0.03 versus PTH 120 min; #, p < 0.05 versus PTH 30 min. B, UMR 106-01 cells were treated with PTH (10−8 m) for 0, 30, 60, and 120 min. After immunoprecipitation of the cross-linked lysates with anti-HDAC3, HDAC4, or HDAC6 antibodies or with rabbit IgG as a negative control, the DNA was subjected to PCR with primers that amplify the distal RD and proximal AP-1 sites of the rat MMP-13 promoter. Input DNA (1:100) is a positive control for the assay. ^, p < 0.01 versus 0 min. C, UMR 106-01 cells were treated with PTH (10−8 m) for 30 and 120 min. After immunoprecipitation of the cross-linked lysates with anti-Runx2 antibody or with rabbit IgG as a negative control, the DNA was subjected to PCR with primers that amplify either the distal RD region or distal RD and proximal AP-1 sites of the rat MMP-13 promoter. Input DNA (1:100) is a positive control for the assay. *, p < 0.03 versus control. D, DNA sequence of rat MMP-13 promoter (−210/+14). Distal runt domain site and proximal runt domain/activator protein-1 site are indicated as distal RD and RD/AP-1. Primer sequences used for ChIP assay are shown in bold and with arrow. The sequence of AP-1 mutation is indicated by underlining. E, agarose gel electrophoresis after sonication of chromatin. The chromatin length is 500–1000 bp (lane 1). F, UMR 106-01 cells were transfected with −148 rat MMP-13 promoter-Luc or −148 rat MMP-13 mutation (AP-1 mutation). After immunoprecipitation of the cross-linked lysates with anti-HDAC4 antibody or with rabbit IgG as a negative control, the DNA was subjected to PCR with specific primers that amplify distal RD and proximal AP-1 sites of exogenous rat MMP-13 promoter. Input DNA (1:100) is a positive control for the assay. %, p < 0.02 versus control.
HDAC Enzyme Assay
The HDAC4 assay was carried out using the HDAC assay kit from Enzo Life Sciences, Inc. UMR 106-01 cells were washed with PBS and lysed by sonication in lysis buffer containing 50 mm Tris-HCl, pH 7.5, 120 mm NaCl, 5 mm EDTA, and 0.5% Nonidet P-40. The cleared supernatant using A/G-agarose beads was incubated for 12 h at 4 °C with 10 μg of an anti-HDAC4 antibody; fresh A/G-agarose beads were added and incubated for 1 h. After centrifuging the agarose beads, we used 30 μl of supernatants with 60 μl of 200 μm Fluor de LysTM substrate for the measurement of HDAC4 activity. A/G-agarose beads were washed three times with PBS and 60 μl of 200 μm Fluor de LysTM substrate added and then incubated for 30 min. In both cases, 30 μl were withdrawn, mixed with 20 μl of assay buffer and 50 μl of Fluor de LysTM developer, and fluorescence was measured (CytoFluorTM II, PerSeptive Biosystems; excitation, 360 nm; emission, 460 nm; gain = 70).
Immunohistochemistry
Tibiae were removed from 8-day postnatal mice (wild-type, heterozygous (hetero) and HDAC4-null (knock-out)) and fixed in 4% formaldehyde overnight and decalcified in 10% EDTA, pH 7.5, for 7 days at 4 °C. Tissues were soaked in 10% sucrose for 2 h and in 20% sucrose for 6 h and then in 30% overnight and embedded in optimal cutting temperature compound. The sections (10 μm) were cut longitudinally to the tibia and dried overnight. To evaluate the localization of the MMP-13 protein, immunohistochemical staining was performed by an ABC staining system (Santa Cruz Biotechnology) according to the manufacturer's instructions. The sections were washed in PBS and immersed in methanol containing 1% hydrogen peroxide to block endogenous peroxidase activity. The sections were incubated with 2% blocking serum and then with anti-MMP-13 antibody or normal goat IgG as a negative control at 4 °C overnight and washed in PBS. The sections were incubated with biotin-labeled anti-goat IgG and washed in PBS. Staining was completed by 10 min of incubation with 3,3′-diaminobenzidine.
Statistical Analysis
The data are shown as the means ± S.E. of triplicate measurements with all of the experiments being repeated at least three times. Experimental significance was assessed using Student's t test.
RESULTS
The Binding of HDAC4 to Runx2 Is Decreased after PTH Stimulation
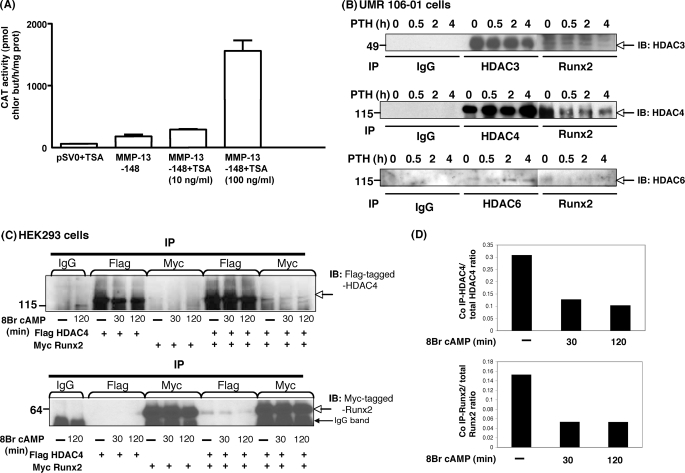
Previously, we identified PTH stimulation of the MMP-13 promoter in the rat osteosarcoma cell line, UMR 106-01 (23), as well as in primary osteoblastic cells (24). In the present study, trichostatin A, an HDAC inhibitor, markedly stimulated basal transcription from the MMP-13 promoter in UMR 106-01 cells (Fig. 1A). These data indicate that HDACs suppress MMP-13 transcription in osteoblastic cells.
FIGURE 1.
HDACs inhibit MMP-13 and HDAC4 binds to Runx2 under basal conditions; after PTH stimulation, HDAC4 is released from Runx2. A, the −148 rat MMP-13 promoter construct was transiently transfected into UMR 106-01 cells, treated with control or trichostatin A (10 ng/ml or 100 ng/ml)-containing medium for 12 h, and then assayed for CAT activity. The data represent the means ± S.E. of three experiments. B, total cell lysates isolated from UMR 106-01 cells with or without PTH (10−8 m) treatment for 0 min, 30 min, 2 h, and 4 h were immunoprecipitated (IP) with anti-Runx2 and HDAC3, HDAC4, or HDAC6 antibodies conjugated with protein A/G-agarose beads. The samples were examined by Western blot with anti-HDAC3, HDAC4, or HDAC6 antibody. C, HEK 293 cells were transfected with FLAG-tagged HDAC4 or Myc-tagged Runx2 constructs. Total cellular lysates isolated from cells treated with 8-Bromo cAMP (10−3 m) for 30 or 120 min or control cells were immunoprecipitated with anti-FLAG and anti-Myc antibodies. The samples were examined by Western blot with anti-FLAG or anti-Myc antibodies. D, the expression level of HDAC4 or Runx2 was expressed as the ratio of co-immunoprecipitated FLAG HDAC4:total FLAG HDAC4 (top panel) or co-immunoprecipitated Myc Runx2:total Myc Runx2 (bottom panel).
Our published work has shown that the proteins binding to the AP-1 and RD-binding sites of the MMP-13 promoter after PTH treatment physically interact (14). Here, we next determined, by co-immunoprecipitation, which endogenous HDACs interact with Runx2 in osteoblastic cells. Other work of ours (data not shown) and published works (25–27) have shown that HDAC3, 4, and 6 are the major HDACs expressed in osteoblastic cells. Precipitated Runx2 interacted predominantly with HDAC3 and 4 but less with HDAC6 under basal conditions. Most notably, the Runx2 interaction with HDAC4 was decreased after PTH stimulation in a time-dependent manner, as were HDAC3 and HDAC6, whereas HDAC4 and HDAC6 protein levels were increased with PTH treatment (Fig. 1B). We also examined whether FLAG-tagged HDAC4 associated with Myc-tagged Runx2 and the effect of 8-bromo cAMP stimulation on this association in transfected HEK 293 cells. The precipitated Myc-tagged Runx2 was found to co-immunoprecipitate with FLAG-tagged HDAC4 under basal conditions. After 8-bromo cAMP treatment for 30 and 120 min, there was a decrease in the amount of HDAC4 bound to precipitated Runx2 (Fig. 1, C and D).
HDAC4 Interacts with Runx2 on the RD Site of the MMP-13 Promoter
To further explore the potential role of HDAC4 interaction with Runx2, we investigated the presence of HDAC4 on the Runx2 binding site of the MMP-13 promoter by ChIP assay of UMR 106-01 cells with or without PTH stimulation. As shown in Fig. 2 (A and B), the association of HDAC4 with the MMP-13 promoter was decreased after PTH stimulation in a time-dependent manner using primers encompassing the RD and AP-1 sites or only the RD site. This reduction by PTH treatment was maintained at control levels at 30 and 240 min in the presence of the cAMP-dependent protein kinase inhibitor H89, indicating that the hormone is acting via the cAMP-dependent protein kinase pathway to induce the dissociation of HDAC4 from the MMP-13 promoter (Fig. 2A). HDAC3 and HDAC6 were also significantly decreased by PTH treatment (Fig. 2B). As we have previously found (28), Runx2 was bound to the MMP-13 promoter under basal conditions, and the amount bound slightly decreased after 30 and 120 min PTH stimulation on either RD and AP-1 sites or RD site (Fig. 2C). As shown in Fig. 2D, after sonication for ChIP assay, the size of the DNA fragments was 500–1000 kb, which is appropriate for amplification primers of RD and AP-1 sites or RD site. To investigate whether HDAC4 is associated with the AP-1 site on the MMP-13 promoter, we performed ChIP assays using an AP-1 mutation construct of the MMP-13 promoter that is indicated in Fig. 2D. HDAC4 is significantly decreased after PTH stimulation for 2 h on MMP-13 promoter, both wild-type and with AP-1 mutation. These results indicate that HDAC4 is bound to the MMP-13 promoter at the RD, likely with Runx2, under basal conditions, and PTH specifically causes HDAC4 dissociation from the RD of the MMP-13 promoter in osteoblastic cells.
HDAC4 Suppresses MMP-13 Gene Expression
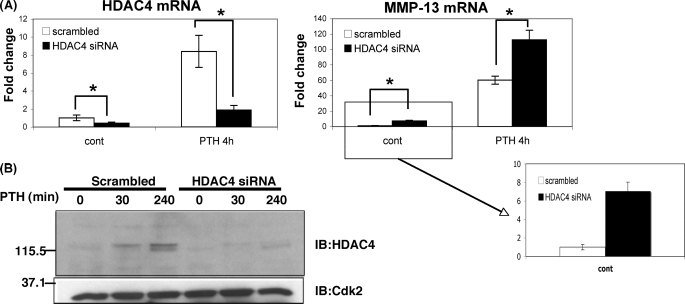
To test whether HDAC4 is involved with the transcriptional regulation of the MMP-13 gene, we suppressed endogenous HDAC4 mRNA and protein levels with HDAC4 siRNA (Fig. 3). MMP-13 mRNA was enhanced after PTH stimulation for 240 min in cells receiving scrambled oligonucleotides. MMP-13 mRNA was even more increased in cells transfected with HDAC4 siRNA and PTH treatment for 240 min (Fig. 3A). Under control conditions, MMP-13 mRNA was also significantly increased in the cells receiving the HDAC4 siRNA. We examined other genes related to the transcriptional regulation of the MMP-13 gene after using HDAC4 siRNA. We investigated the expression of c-Fos, amphiregulin (both highly regulated by PTH), and the PTH receptor, but these genes did not change between scrambled and HDAC4 siRNA (data not shown). We conclude that HDAC4 normally represses MMP-13 expression under basal conditions and after PTH treatment.
FIGURE 3.
HDAC4 suppresses MMP-13 gene expression. A, UMR 106-01 cells were transfected with 60 nm scrambled oligonucleotides or rat HDAC4 siRNA oligonucleotides. The cells were stimulated with PTH (10−8 m) for 30 and 240 min. Total RNA was extracted from the cells transfected with scrambled or siRNA oligonucleotides. RNAs were measured using Real time RT-PCR. The relative levels of mRNAs were normalized to β-actin and then expressed as fold stimulation over control. The error bars represent ± S.E. of three independent experiments. *, p < 0.05 versus scrambled oligonucleotides. B, the nuclear extracts from UMR 106-01 cells with or without PTH treatment were subjected to immunoblotting (IB) with anti-HDAC4 antibody. Anti-Cdk2 was used as a loading control.
PTH Stimulates HDAC4 Gene Expression and Enzymatic Activity
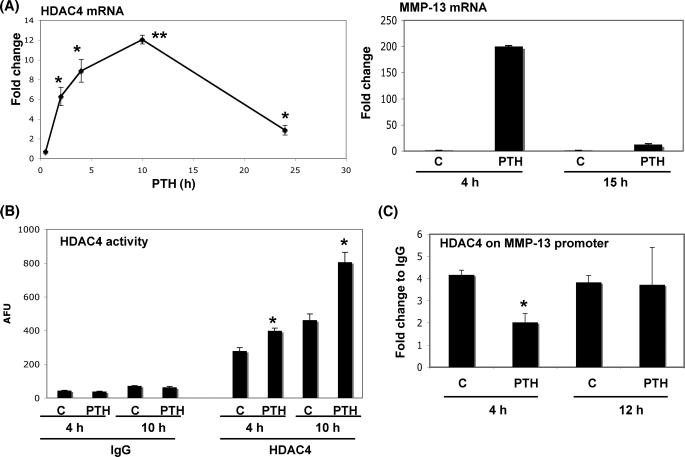
As shown in Fig. 3, PTH induced HDAC4 gene and protein synthesis after 4 h of treatment. To confirm that stimulation of HDAC4 is a response to PTH in osteoblastic cells, we examined HDAC4 mRNA levels over time. PTH stimulated HDAC4 mRNA levels by 12-fold with the peak level around 10 h (Fig. 4A), whereas MMP-13 mRNA levels had decreased after 15 h of PTH stimulation. We have previously identified that PTH induces maximal transcription of MMP-13 at 2 h with peak MMP-13 transcripts at 4 h, declining thereafter (8). To investigate whether PTH affects HDAC activity in osteoblastic cells, we examined endogenous HDAC4 enzymatic activity in UMR 106-01 cells. PTH significantly enhanced HDAC4 enzymatic activity in UMR 106-01 cells at 4 or 10 h (Fig. 4B). The activity of total HDACs (minus HDAC4) were not significantly different after PTH stimulation (data not shown).
FIGURE 4.
PTH stimulates HDAC4 gene expression and enzymatic activity. A, left panel, total RNA was extracted from UMR 106-01 cells treated with PTH (10−8 m) for 2, 4, 10, and 24 h. Right panel, total RNA was extracted from UMR 106-01 cells treated with PTH (10−8 m) for 4 and 15 h. RNAs were measured using Real time RT-PCR. The relative levels of mRNAs were normalized to β-actin and then expressed as fold stimulation over control. The error bars represent ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.001, significant increase compared with control. B, UMR 106-01 cells were stimulated with PTH (10−8 m) for 4 and 10 h. The samples were examined using HDAC enzymatic assay. *, p < 0.01, significant increase compared with respective control. C, UMR 106-01 cells were treated with control or PTH (10−8 m) for 4 and 12 h. After immunoprecipitation of the cross-linked lysates with anti-HDAC4 or with rabbit IgG as a negative control, the DNA was subjected to PCR with primers that amplify the distal RD and proximal AP-1 sites of the rat MMP-13 promoter. Input DNA (1:100) is a positive control for the assay. The error bars represent ± S.E. of three independent experiments. *, p < 0.001 versus respective control. AFU, arbitrary fluorescence units.
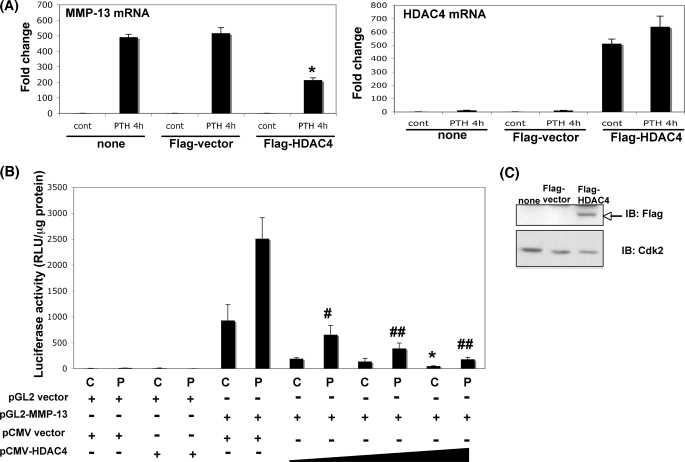
We hypothesized that PTH regulates HDAC4 such that increased HDAC4 switches off MMP-13 transcription. As shown in Fig. 4C, the association of HDAC4 with the MMP-13 promoter is decreased compared with the control after 4 h of PTH treatment, but by 12 h the enzyme had reassociated with the promoter to levels similar to the control (Fig. 4C). Although the binding had returned to the same as control levels, PTH also stimulated HDAC4 activity to greater than control, and we hypothesized that both of these suppressed MMP-13 gene transcription. To further investigate this hypothesis, we examined MMP-13 gene expression when HDAC4 was overexpressed in the osteoblastic cells. Under these conditions, we found that overexpression of HDAC4 significantly suppressed PTH-induced MMP-13 gene expression compared with nontransfected or FLAG vector-transfected cells (Fig. 5A). The increase in exogenous mRNA levels of FLAG-tagged HDAC4 in UMR 106-01 cells are shown. This effect is at the promoter, because overexpression of HDAC4 significantly suppressed the transcriptional activity of the MMP-13 promoter (Fig. 5B). The expression level of FLAG-tagged HDAC4 transfected into UMR 106-01 cells is indicated in Fig. 5C. These results indicate that PTH induces negative feedback of HDAC4 to the MMP-13 promoter in osteoblastic cells, which suppresses and down-regulates MMP-13 transcription.
FIGURE 5.
Transfected HDAC4 represses MMP-13 transcription. A, UMR 106-01 cells were transfected with FLAG vector or rat FLAG-tagged HDAC4 construct or no vector. The cells were stimulated with PTH (10−8 m) for 4 h. Total RNA was extracted from the cells transfected with none (no transfection), FLAG vector, or FLAG-tagged HDAC4. RNAs were measured using Real time RT-PCR. The relative levels of mRNAs were normalized to β-actin and then expressed as fold stimulation over control (cont). The error bars represent ± S.E. of three independent experiments. *, p < 0.05 versus no vector or FLAG vector and treated with PTH for 4 h. B, UMR 106-01 cells were transiently transfected with various vectors (100 ng of pGL2 vector, 100 ng of −148 rat MMP-13 promoter-Luc, 100 ng of FLAG vector, and 100, 200, or 500 ng of FLAG-tagged rat HDAC4), and the luciferase activities were measured with (P) or without (C) 6 h of 10−8 m PTH treatment. The luciferase activities were normalized to the amount of total protein. The error bars represent ± S.E. of three independent experiments. *, p < 0.05 versus FLAG control. #, p < 0.02; ##, p < 0.01 versus FLAG PTH 6 h. C, UMR 106-01 cells were transfected with FLAG vector or rat FLAG-tagged HDAC4 construct or no vector (none). The nuclear extracts from cells were subjected to immunoblotting with anti-FLAG antibody. Anti-Cdk2 was used as a loading control.
HDAC4 Suppresses MMP-13 Gene Expression in Vivo
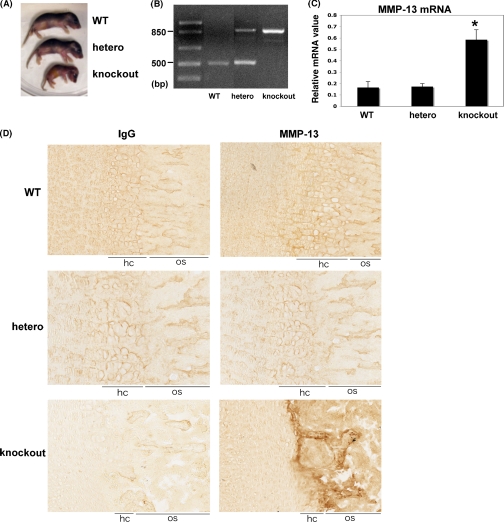
Vega et al. (20) showed that Hdac4-deficient mice display premature ossification of developing bones caused by ectopic and early onset of chondrocyte hypertrophy. MMP-13 is normally expressed by the mineralizing hypertrophic chondrocytes as well as osteoblasts (29). To investigate whether HDAC4 affects MMP-13 gene expression in vivo, we examined the femurs of postnatal day 5 wild-type, heterozygous, and Hdac4-deficient pups. As shown in Fig. 6A, Hdac4 knock-out mice were significantly smaller than wild-type or heterozygous littermates. Genotyping was confirmed by PCR analysis (Fig. 6B). We found that MMP-13 mRNA abundance was greatly enhanced in Hdac4-deficient mice compared with wild-type or heterozygous littermates (Fig. 6C). In contrast, osteocalcin and bone sialoprotein genes were slightly increased in Hdac4-deficient pups, but there was no significant difference (data not shown). As shown in Fig. 6D, MMP-13 staining in the tibiae of knock-out mice was strongly detected in late hypertrophic chondrocytes and bone regions compared with wild-type or heterozygous mice. Moreover, the hypertrophic chondrocyte zone in knock-out mice was notably smaller than their littermates. These results suggest HDAC4 suppresses MMP-13 gene and protein expression in vivo.
FIGURE 6.
HDAC4 suppresses MMP-13 gene expression in vivo. A, photograph of postnatal day 5 (P5) wild-type (WT), heterozygous (hetero), and HDAC4-null (knockout) pups. B, genotypes of 4-day-old pups from matings of heterozygous mice. C, total RNA was extracted from femurs of 5-day postnatal mice (n = 4). RNAs were measured using real time RT-PCR. The relative levels of mRNAs were normalized to β-actin and then expressed as relative mRNA value. The error bars represent ± S.E. of four animals. *, p < 0.01, significant increase compared with wild-type or heterozygous animals. D, immunohistochemical staining of MMP-13 and negative control in the tibiae of day 8 mice (×200). MMP-13 was strongly detected in hypertrophic and ossification zones (arrow). hc, hypertrophic chondrocyte zone; os, ossification zone.
DISCUSSION
MMP-13 plays a crucial role in human fetal bone development through endochondral and intramembranous ossification (30). This enzyme is involved with bone remodeling, endochondral bone formation, and bone repair (31). Inada et al. (32) have shown that Mmp-13-deficient mice have a delayed development of primary ossification centers. The absence of MMP-13 results in accumulation of collagen, which is normally broken down by collagenases in growth plates and primary ossification centers. The same researchers showed that PTH-induced bone resorption and calcemic responses were markedly diminished in mice with a homozygous targeted mutation in Col1a1 that is resistant to collagenase cleavage (33). Therefore, MMP-13 is important in bone development and PTH calcemic responses. Our previous data and that of others (9, 34) demonstrated that MMP-13 gene transcription requires Runx2, which binds to the runt domain site of the MMP-13 promoter and PTH stimulates transactivation by Runx2 in rat osteoblastic cells (23).
Recent work by several laboratories showed that HDAC4 is expressed in prehypertrophic chondrocytres in vivo and regulates mineralization of endochondral bones through inhibiting Runx2 (20), and HDAC4 or HDAC5 are required for efficient TGF-β-mediated inhibition of Runx2 function and are involved in osteoblastic differentiation (21). Schroeder et al. (25) showed that HDAC3 and HDAC4 bind to Runx2 in UMR 106-01 cells, and HDAC3 suppresses Runx2-mediated activation of the osteocalcin promoter. Moreover, inhibition of HDAC4 and HDAC5 increases Runx2 acetylation, potentiates BMP-2-stimulated osteoblastic differentiation, and increases bone formation (22). Here, we show that trichostatin A, an HDAC inhibitor, markedly stimulates basal transcription from the MMP-13 promoter, indicating that the MMP-13 gene is under basal repression by HDACs.
Vega et al. (20) have concluded that HDAC4 interferes with the ability of Runx2 to bind its response elements in DNA, thus repressing activation of Runx2 target genes. As a consequence, this phenomenon delays chondrocyte hypertrophy and bone formation. Our experiments indicate that HDAC4 associates with Runx2 bound to the RD site of the MMP-13 promoter under basal conditions in UMR 106-01 cells. We suggest that, through cAMP-dependent protein kinase-dependent pathways, HDAC4 is released from Runx2 on the MMP-13 promoter after PTH treatment, and then histone acetyl transferases, especially p300, associate with Runx2 on the MMP-13 promoter (28). Thus, HDAC4 suppresses MMP-13 gene expression by binding with Runx2. We hypothesize that HDAC4 competes with p300 for Runx2 interaction after PTH treatment.
In addition to its role in suppressing the initiation of MMP-13 transcription, we conclude that PTH stimulates HDAC4 gene expression and activity, which feeds back to repress MMP-13. Under basal conditions, we expect that other HDACs, especially HDAC3 or HDAC6, also bind to the MMP-13 promoter and suppress gene transcription. With PTH treatment, HDAC3 or HDAC6 binding to Runx2 and the MMP-13 promoter are also decreased, and this likely stimulates gene expression. Other HDACs may rebind to the MMP-13 promoter after 12 h as well as HDAC4 leading to decreased MMP-13 transcription and return to basal conditions. However, because PTH stimulates HDAC4 activity at 10 h greater than at 4 h, HDAC4 would notably suppress MMP-13 gene expression even if the amount of HDAC4 bound on the MMP-13 promoter is almost the same at 12 h as the control. In contrast, we have examined the activity of other HDACs and find no significant difference compared with control. In particular, PTH increases HDAC4 enzymatic activity at late times.
Our data demonstrate that MMP-13 expression is enhanced in Hdac4-deficient mice compared with wild-type or heterozygous mice. Therefore, we have shown that HDAC4 suppresses the MMP-13 gene under basal conditions both in vitro and in vivo. Interestingly, we have found that PTH induces HDAC4 gene expression and enzymatic activity in osteoblastic cells. The rat HDAC4 promoter has not been identified yet, but we have conducted homology searches of the human HDAC4 promoter that Liu et al. (35) have reported. The human HDAC4 promoter has GC-rich DNA sequences, which bind Sp-1/Sp-3. We have analyzed for regulatory motifs in the rat promoter and found 86% consensus sequences of the human HDAC4 promoter including Sp-1/Sp-3 or CCAAT elements in upstream HDAC4 sequences. These sequences may be associated with PTH stimulation of HDAC4 gene expression. Moreover, the association of HDAC4 with the MMP-13 promoter returns to control levels after prolonged PTH stimulation, whereas HDAC4 association with the MMP-13 promoter was at its lowest at a time when MMP-13 transcript levels are maximal. Overexpression of HDAC4 suppresses MMP-13 transcription. These results suggest that PTH causes the recruitment of HDAC4, which then suppresses the PTH-induced MMP-13 gene transcription. Thus, PTH induces feedback regulation by HDAC4 in osteoblastic cells.
The class II HDACs control tissue growth and development. HDAC7 is expressed in the vascular endothelium of early stage embryos and represses MMP-10 (stromelysin 2) through an inhibition of the transcription factor myocyte enhancer factor 2 (36). Lack of Hdac7 induces MMP-10 expression in the vascular endothelium, resulting in embryonic lethality caused by cardiovascular defects including dilated and ruptured blood vessels. HDAC7 directly binds to myocyte enhancer factor 2 and inhibits its activation of the MMP-10 promoter. This has remarkable parallels to our work described here, which is the first to show that HDAC4 is a repressor of MMP-13 in osteoblastic cells and bone.
In this report, we demonstrate the effects of PTH on the MMP-13 gene through class II histone deacetylases in rat osteoblastic cells. We focused on HDAC4 function, because HDAC4 plays an important role in suppressing chondrocyte and osteoblast differentiation through association with Runx2. Our results lead to several conclusions: 1) HDAC4 represses MMP-13 expression by binding to Runx2 at the RD site of the MMP-13 promoter; 2) PTH leads to HDAC4 release from the MMP-13 promoter through cAMP-dependent protein kinase-dependent pathways; 3) PTH induces expression of HDAC4, which feeds back to turn off PTH-induced MMP-13 transcription; and 4) both in vitro and in vivo studies demonstrate that HDAC4 suppresses MMP-13 gene and protein expression.
Acknowledgments
We thank Dr. Xiang-Jiao Yang for providing pCMV-FLAG HDAC4. We thank KAZUSA DNA research institute for human HDAC4 cDNA (KIAA0288). We thank Drs. Istvan Sohar and Peter Lobel for use of the CytoFluorTM II.
This work was supported, in whole or in part, by National Institutes of Health Grant DK47420 (to N. C. P.).
- PTH
- parathyroid hormone
- HDAC
- histone deacetylase
- MMP
- matrix metalloproteinase
- RD
- runt domain
- AP-1
- activator protein-1
- EMEM
- Eagle's minimal essential medium
- CAT
- chloramphenicol acetyltransferase
- PBS
- phosphate-buffered saline
- siRNA
- small interfering RNA
- RT
- reverse transcription
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.Strewler G. J., Stern P. H., Jacobs J. W., Eveloff J., Klein R. F., Leung S. C., Rosenblatt M., Nissenson R. A. (1987) J. Clin. Invest. 80, 1803–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jüppner H., Abou-Samra A. B., Freeman M., Kong X. F., Schipani E., Richards J., Kolakowski L. F., Jr., Hock J., Potts J. T., Jr., Kronenberg H. M. (1991) Science 254, 1024–1026 [DOI] [PubMed] [Google Scholar]
- 3.Hock J. M., Gera I., Fonseca J., Raisz L. G. (1988) Endocrinology 122, 2899–2904 [DOI] [PubMed] [Google Scholar]
- 4.Hock J. M., Gera I. (1992) J. Bone Miner. Res. 7, 65–72 [DOI] [PubMed] [Google Scholar]
- 5.Partridge N. C., Jeffrey J. J., Ehlich L. S., Teitelbaum S. L., Fliszar C., Welgus H. G., Kahn A. J. (1987) Endocrinology 120, 1956–1962 [DOI] [PubMed] [Google Scholar]
- 6.Lee S. K., Lorenzo J. A. (1999) Endocrinology 140, 3552–3561 [DOI] [PubMed] [Google Scholar]
- 7.Weir E. C., Lowik C. W., Paliwal I., Insogna K. L. (1996) J. Bone Miner. Res. 11, 1474–1481 [DOI] [PubMed] [Google Scholar]
- 8.Scott D. K., Brakenhoff K. D., Clohisy J. C., Quinn C. O., Partridge N. C. (1992) Mol. Endocrinol. 6, 2153–2159 [DOI] [PubMed] [Google Scholar]
- 9.Selvamurugan N., Chou W. Y., Pearman A. T., Pulumati M. R., Partridge N. C. (1998) J. Biol. Chem. 273, 10647–10657 [DOI] [PubMed] [Google Scholar]
- 10.Mundlos S., Otto F., Mundlos C., Mulliken J. B., Aylsworth A. S., Albright S., Lindhout D., Cole W. G., Henn W., Knoll J. H., Owen M. J., Mertelsmann R., Zabel B. U., Olsen B. R. (1997) Cell 89, 773–779 [DOI] [PubMed] [Google Scholar]
- 11.Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., Owen M. J. (1997) Cell 89, 765–771 [DOI] [PubMed] [Google Scholar]
- 12.Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997) Cell 89, 747–754 [DOI] [PubMed] [Google Scholar]
- 13.Inada M., Yasui T., Nomura S., Miyake S., Deguchi K., Himeno M., Sato M., Yamagiwa H., Kimura T., Yasui N., Ochi T., Endo N., Kitamura Y., Kishimoto T., Komori T. (1999) Dev. Dyn. 214, 279–290 [DOI] [PubMed] [Google Scholar]
- 14.D'Alonzo R. C., Selvamurugan N., Karsenty G., Partridge N. C. (2002) J. Biol. Chem. 277, 816–822 [DOI] [PubMed] [Google Scholar]
- 15.Verdin E., Dequiedt F., Kasler H. G. (2003) Trends Genet. 19, 286–293 [DOI] [PubMed] [Google Scholar]
- 16.Grozinger C. M., Schreiber S. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7835–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang A. H., Kruhlak M. J., Wu J., Bertos N. R., Vezmar M., Posner B. I., Bazett-Jones D. P., Yang X. J. (2000) Mol. Cell. Biol. 20, 6904–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westendorf J. J. (2007) J. Cell. Biochem. 102, 332–340 [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Kwon S., Yamaguchi T., Cubizolles F., Rousseaux S., Kneissel M., Cao C., Li N., Cheng H. L., Chua K., Lombard D., Mizeracki A., Matthias G., Alt F. W., Khochbin S., Matthias P. (2008) Mol. Cell. Biol. 28, 1688–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vega R. B., Matsuda K., Oh J., Barbosa A. C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J. M., Richardson J. A., Karsenty G., Olson E. N. (2004) Cell 119, 555–566 [DOI] [PubMed] [Google Scholar]
- 21.Kang J. S., Alliston T., Delston R., Derynck R. (2005) EMBO J. 24, 2543–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeon E. J., Lee K. Y., Choi N. S., Lee M. H., Kim H. N., Jin Y. H., Ryoo H. M., Choi J. Y., Yoshida M., Nishino N., Oh B. C., Lee K. S., Lee Y. H., Bae S. C. (2006) J. Biol. Chem. 281, 16502–16511 [DOI] [PubMed] [Google Scholar]
- 23.Selvamurugan N., Pulumati M. R., Tyson D. R., Partridge N. C. (2000) J. Biol. Chem. 275, 5037–5042 [DOI] [PubMed] [Google Scholar]
- 24.Winchester S. K., Selvamurugan N., D'Alonzo R. C., Partridge N. C. (2000) J. Biol. Chem. 275, 23310–23318 [DOI] [PubMed] [Google Scholar]
- 25.Schroeder T. M., Kahler R. A., Li X., Westendorf J. J. (2004) J. Biol. Chem. 279, 41998–42007 [DOI] [PubMed] [Google Scholar]
- 26.Westendorf J. J., Zaidi S. K., Cascino J. E., Kahler R., van Wijnen A. J., Lian J. B., Yoshida M., Stein G. S., Li X. (2002) Mol. Cell. Biol. 22, 7982–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z., Hassan M. Q., Jafferji M., Aqeilan R. I., Garzon R., Croce C. M., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2009) J. Biol. Chem. 284, 15676–15684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boumah C. E., Lee M., Selvamurugan N., Shimizu E., Partridge N. C. (2009) Mol. Endocrinol. 23, 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuckermann J. P., Pittois K., Partridge N. C., Merregaert J., Angel P. (2000) J. Bone Miner Res. 15, 1257–1265 [DOI] [PubMed] [Google Scholar]
- 30.Johansson N., Saarialho-Kere U., Airola K., Herva R., Nissinen L., Westermarck J., Vuorio E., Heino J., Kähäri V. M. (1997) Dev. Dyn. 208, 387–397 [DOI] [PubMed] [Google Scholar]
- 31.Yamagiwa H., Tokunaga K., Hayami T., Hatano H., Uchida M., Endo N., Takahashi H. E. (1999) Bone 25, 197–203 [DOI] [PubMed] [Google Scholar]
- 32.Inada M., Wang Y., Byrne M. H., Rahman M. U., Miyaura C., López-Otín C., Krane S. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17192–17197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao W., Byrne M. H., Boyce B. F., Krane S. M. (1999) J. Clin. Invest. 103, 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porte D., Tuckermann J., Becker M., Baumann B., Teurich S., Higgins T., Owen M. J., Schorpp-Kistner M., Angel P. (1999) Oncogene 18, 667–678 [DOI] [PubMed] [Google Scholar]
- 35.Liu F., Pore N., Kim M., Voong K. R., Dowling M., Maity A., Kao G. D. (2006) Mol. Biol. Cell 17, 585–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang S., Young B. D., Li S., Qi X., Richardson J. A., Olson E. N. (2006) Cell 126, 321–334 [DOI] [PubMed] [Google Scholar]