Abstract
The mammalian Na+/H+ exchange regulatory factor 1 (NHERF1) is a multidomain scaffolding protein essential for regulating the intracellular trafficking and macromolecular assembly of transmembrane ion channels and receptors. NHERF1 consists of tandem PDZ-1, PDZ-2 domains that interact with the cytoplasmic domains of membrane proteins and a C-terminal (CT) domain that binds the membrane-cytoskeleton linker protein ezrin. NHERF1 is held in an autoinhibited state through intramolecular interactions between PDZ2 and the CT domain that also includes a C-terminal PDZ-binding motif (-SNL). We have determined the structures of the isolated and tandem PDZ2CT domains by high resolution NMR using small angle x-ray scattering as constraints. The PDZ2CT structure shows weak intramolecular interactions between the largely disordered CT domain and the PDZ ligand binding site. The structure reveals a novel helix-turn-helix subdomain that is allosterically coupled to the putative PDZ2 domain by a network of hydrophobic interactions. This helical subdomain increases both the stability and the binding affinity of the extended PDZ structure. Using NMR and small angle neutron scattering for joint structure refinement, we demonstrate the release of intramolecular domain-domain interactions in PDZ2CT upon binding to ezrin. Based on the structural information, we show that human disease-causing mutations in PDZ2, R153Q and E225K, have significantly reduced protein stability. Loss of NHERF1 expressed in cells could result in failure to assemble membrane complexes that are important for normal physiological functions.
Keywords: Methods/NMR, Protein/Allosteric, Protein/Assembly, Protein/Conformation, Protein/Domains, Protein/Structure, Signal Transduction/Adapter Proteins
Introduction
The transduction of biological signals is controlled by the association and disassociation of macromolecular complexes. Dynamic assembly of signaling complexes is, in turn, organized by scaffolding and adapter proteins (1, 2). The mammalian Na+/H+ exchange regulatory factor (NHERF)4 family of scaffolding proteins assembles macromolecular complexes of transmembrane proteins and regulates receptor signaling and ion transport (3–6). More than 50 biological targets have been identified for NHERF1, and some of these proteins are implicated in human diseases, such as cancer, cystic fibrosis, and chronic kidney disease (7–9). Notable functions of NHERF1 include assembling signaling complexes and regulating the endocytic recycling of the cystic fibrosis transmembrane conductance regulator (CFTR), cell surface adhesion and anti-adhesion proteins, G-protein coupled receptors, and tyrosine kinase receptors (10–14). A recent study that correlates NHERF1 mutations with impaired renal phosphate reabsorption in patient underscores the important physiological role of NHERF1 (7).
NHERF1 consists of two modular protein-protein interaction domains, PDZ1 and PDZ2, which bind to the cytoplasmic domains of transmembrane receptors and channels, and a C-terminal domain that binds ezrin (EB) (Fig. 1). A central feature of NHERF1 is its binding to ezrin and to other ezrin-radixin-moesin (ERM) proteins, forming a communication bridge between plasma membrane proteins and the actin cytoskeleton (15, 16). Ezrin and other ERM proteins are membrane-cytoskeleton linkers that participate in a variety of cytoskeletal-related events, such as cell polarity, intracellular trafficking, cell adhesion, cell motility, and cancer metastasis (15, 17–19). The ERM proteins are autoregulated by intramolecular interactions between the N-terminal 4.1-ezrin/radixin/moesin (FERM) domain and the C-terminal actin binding domains (16, 20). In the inactive state, the FERM domain is masked by its own C-terminal domain. Ezrin becomes activated when the intramolecular interactions are disrupted upon phospholipid PIP2 binding and phosphorylation (15). The FERM domain of the activated ezrin binds to target membrane proteins either directly or indirectly through NHERF1 or -2, whereas the C-terminal domain of ezrin interacts with the cytoskeletal actin, completing a linkage between cell membranes and the cytoskeletal network.
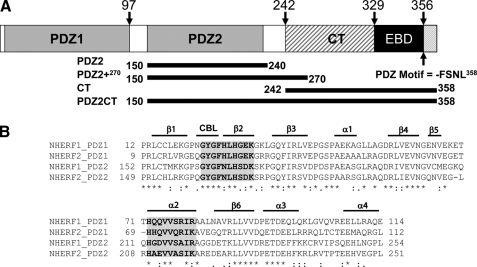
FIGURE 1.
A, schematic representation of the domains of human NHERF1. The C-terminal end of the EB domain contains a canonical PDZ-binding motif. The graph shows the amino acid positions of the differently truncated domains, which include the putative PDZ2240 (residues 150–240), PDZ2+270 (residues 150–270) with the extra C-terminal helical subdomain, and PDZ2CT (residues 150–358). B, sequence alignment of the PDZ domains of human NHERF1 and NHERF2 proteins, annotated with secondary structure elements. CBL, carboxylate binding loop. The residues involved in ligand binding are highlighted in gray and boldface type. The alignment indicates that the sequence in the extra subdomain formed by α3 an α4 is conserved. The alignment was produced in the ClustalX program (66).
Remarkably, ezrin modulates the PDZ domains of NHERF1 to assemble multiprotein complexes in a cooperative fashion. A study by Weinman et al. (21) finds that the association of ezrin with NHERF1 is essential for assembling the transmembrane sodium-hydrogen exchanger isoform 3 (NHE3) complexes and for cAMP-mediated inhibition of NHE3. We have previously shown that when ezrin is bound to the EB domain of NHERF1, the binding affinity of PDZ2 for the C-terminal domain of CFTR increases 24-fold (22). Ezrin binding can also activate NHERF1 to assemble a heterogeneous complex, PTEN, at PDZ1 and β-catenin at PDZ2 (23). A recent study by Mahon (24) finds that ezrin binding to NHERF1 is necessary to promote the co-localization and functional expression of a heterogeneous complex of the transmembrane parathyroid hormone receptor (PTH1R) and the sodium-phosphate cotransporter 2a (NPT2a) at the apical membrane. These studies indicate that ezrin binding to the EB domain enhances the binding of PDZ2 to target proteins and thus positively modulates the full-length NHERF1 to assemble protein complexes. A type I PDZ-binding motif, -SNL358, at the C terminus of NHERF1, which overlaps with the EB domain, is hypothesized to interact with PDZ2, thereby impeding PDZ2 from binding to target proteins. The interaction of PDZ2 with the C-terminal (CT) domain of NHERF1 is proposed to function as an autoregulatory molecular switch (25).
The structures of the isolated PDZ1, PDZ2 domains of NHERF1 and the structures of the NHERF1 PDZ1 domain in complex with the C-terminal peptides of membrane receptors and channels have been determined by x-ray crystallography and NMR (26, 27). Crystal structures of the C-terminal EB peptide of NHERF1 bound to the FERM domains of radixin and moesin have also been reported (28, 29). Recently, a NMR study has characterized the interactions between a PDZ2 domain and the C-terminal fragment of NHERF1 (30), supporting the hypothesis that there are intramolecular interactions between PDZ2 and the C terminus of NHERF1 (23, 25). However, these studies did not reveal the structural mechanism of PDZ2 “inhibition” and “activation” of the intact protein. In particular, the apparent weak binding affinity of the putative PDZ2 for target proteins, as reported in many biochemical studies (22, 31), remains unexplained. The free energy of PDZ2 stability and binding cannot be explained from these previous structural studies. To gain insight into the dynamic control of NHERF1 by ezrin, we have determined the solution structure of the combined PDZ2 and the intact C-terminal domains (PDZ2CT) of NHERF1 as well as the conformational changes in PDZ2CT upon forming a large complex with ezrin, using high resolution NMR spectroscopy.
MATERIALS AND METHODS
Protein Expression and NMR Sample Preparation
The genes for PDZ2240 (residues 150–240), CT (residues 242–358), PDZ2+270 (residues 150–270), PDZ2CT (residues 150–358), and the FERM domain of ezrin (residues 1–300) were inserted in pET151/D-TOPO vector (Invitrogen). The recombinant proteins were expressed in Escherichia coli BL21 (DE3) cells (Novagen, Inc.) and purified according to published methods (22). The epitope with a His6 tag at the N terminus of the protein was cleaved by tobacco etch virus protease. After tobacco etch virus cleavage, all proteins have an extra sequence GIDPFT at their N terminus.
For uniform enrichment of the protein with 15N/13C isotopes, BL21 (DE3) cells were grown in M9-minimal medium containing 15NH4Cl (Cambridge Isotope Laboratories) and [13C6]glucose as sole nitrogen and carbon source, respectively. Deuterated proteins were produced by E. coli grown in M9-minimal medium containing D2O and protonated glucose. For expressing selectively labeled proteins, BL21(DE3) cells were grown in M9 medium enriched with the addition of 1 g/liter each of the 20 unlabeled amino acids except Leu, Phe, and Val (Sigma). Before induction with isopropyl 1-thio-β-d-galactopyranoside, 1 g/liter each of 13C/15N-labeled Leu, Phe, and Val amino acids (Cambridge Isotopes) was added to the cell culture.
All NMR samples were prepared in a buffer containing 20 mm HEPES, 150 mm NaCl, 0.5 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride, pH 7.5, and 10% D2O, 90% H2O. Protein concentration was determined from the molar extinction coefficient at 280 nm and typically ranged from 200 to 600 μm. Chemical shift assignments and structure determination was carried out on a 600 μm sample of PDZ2CT and a 550 μm sample of PDZ2+270.
For analyzing PDZ2CT in complex with the FERM domain of ezrin, 13C/15N-labeled PDZ2CT was mixed with unlabeled FERM at a 1:1 ratio, and the complex was purified by gel filtration and concentrated to 500 μm.
NMR Spectroscopy
The NMR data were acquired on Bruker AVANCE spectrometers equipped with z-axis gradient triple axis CryoProbes at 30 °C. The data were collected at three different field strengths, 700, 800, and 900 MHz, to maximize resolution and sensitivity. Due to the lower stability of isolated PDZ2240 and CT domains, the NMR data for these two samples were acquired at lower temperature (15 °C).
A standard suite of backbone and side-chain experiments were employed for chemical shift assignment of PDZ2CT, PDZ2+270, and CT domains (32, 33). In summary, 94% of resonances for PDZ2+270 and CT and 87% of resonances were assigned for PDZ2CT. To alleviate severe overlap of resonances from the unstructured CT domain linker, selectively labeled (13C/15N-labeled Leu, Phe, and Val) PDZ2CT samples were used to confirm NMR assignments. Distance restraints for structure calculations were obtained from 100-ms mixing time 15N-edited and 13C-edited three-dimensional NOESY-HSQC (aliphatic and aromatic) spectra.
Structure Calculations
NMR data were processed in Topspin 1.3 from Bruker Biospin and analyzed using CARA1.5 (34). Automatic NOE assignment for all three structures (PDZ2+270, PDZ2CT, and PDZ2CT·FERM complex) were carried out using the program CYANA 2.1 (35, 36). The final ensemble of PDZ2+270 structures with water refinement was calculated in ARIA 2.2 (37).
The joint NOE refinement of PDZ2CT and PDZ2CT·FERM complex structures against small angle x-ray scattering (SAXS) and small angle neutron scattering (SANS) data was carried out in XPLOR-NIH 2.24. Details of the protocol have been described by Lee et al. (38). The collection and processing of the SAXS and SANS data has been described in detail elsewhere (40). A total of 500 structures were calculated, and the 20 structures with the lowest total energy and NOE violations were analyzed in PROCHECK_NMR by inspecting the Ramchandran plots (39). The structural statistics of the ensemble of the 20 best structures are reported in Table 1. The PDZ2CT structure was calculated with two different models (random coil and helix) for the 13-residue (Met346–Leu358) C-terminal EB peptide. For the helix model, the coordinates of the peptide were extracted from the radixin FERM domain complex (Protein Data Bank code 2D10) structure (29) and docked to the PDZ2CT structure in the program Haddock (69) (details of docking calculations are given in the supplemental material). The restraints between the peptide and protein from the Haddock model were combined with experimental NOE data in a full refinement of PDZ2CT domains. The structure of PDZ2CT bound to the ezrin FERM domain includes helical restraints derived from the x-ray structure of the EB domain peptide complexed with the moesin FERM domain (Protein Data Bank code 1SGH) in the calculations (28). The SAXS and SANS data were from our previous publications (25, 40).
TABLE 1.
Statistics for NMR ensemble of 20 structures
| PDZ2+270 | PDZ2CTcoil | PDZ2CThelix | PDZ2CTFERMa | |
|---|---|---|---|---|
| Constraints | ||||
| Intraresidue | 527 | 703 | 703 | 481 |
| Sequential | 581 | 799 | 799 | 478 |
| Medium range | 318 | 376 | 384 | 208 |
| Long range | 784 | 888 | 1019 | 504 |
| Total | 2210 | 2766 | 2905 | 1671 |
| Dihedral | 116 | 141 | 157 | 150 |
| SAXS/SANS data points | 143 | 143 | 48 | |
| Precisionb | ||||
| Backbone (Å) | 0.41 ± 0.12 (0.66 ± 0.16) | 0.50 ± 0.11 (0.79 ± 0.20) | 0.38 ± 0.11 (0.60 ± 0.09) | 0.54 ± 0.12 (0.75 ± 0.15) |
| Heavy atoms (Å) | 0.83 ± 0.20 (1.13 ± 0.16) | 1.03 ± 0.09 (1.23 ± 0.15) | 0.86 ± 0.09 (1.04 ± 0.10) | 1.06 ± 0.10 (1.24 ± 0.12) |
| Ramachandranb | ||||
| Favored regions | 96.5% | 91.6% | 97.0% | 97.0% |
| Additional allowed regions | 3.5% | 7.2% | 3.0% | 3.0% |
| Generously allowed regions | 0.0% | 1.2% | 0.0% | 0.0% |
| CNS/ARIA PDZ2+270 | XPLOR-NIH |
|||
|---|---|---|---|---|
| PDZ2CTcoil | PDZ2CThelix | PDZ2CTFERM | ||
| Energies | ||||
| 〈Distance〉 (Å) d ∼ 0.5 Å | 0.0 | 0.0 | 0.0 | 0.0 |
| 〈Angle〉 θ > 5° | 0.1 | 0.0 | 0.0 | 0.25 |
| Etotal (kcal mol−1) | −5040.0 ± 124 | −1332.4 ± 70.8 | −1334.6 ± 74.4 | −1481.8 ± 68.8 |
| E(NOE) | 31.0 ± 3 | 39.9 ± 5.3 | 47.4 ± 5.8 | 39.4 ± 5.4 |
| E(repel) | 135.7 ± 16.0 | 150.3 ± 11.7 | 133.1 ± 15.2 | |
| E(SAX/SANS) | 4.3 ± 1.1 | 6.7 ± 3.0 | 10.6 ± 2.9 | |
| SAXS/SANS(r.m.s.) | 0.09 ± 0.01 | 0.11 ± 0.03 | 0.14 ± 0.02 | |
a PDZ2CTFERM: PDZ2CT in complex with the FERM domain of ezrin.
b The backbone r.m.s. deviation and Ramachandran plot calculated for regular secondary structure elements, β1 (residues 153–158), β2 (residues 166–170), β3 (residues 177–182), β4 (residues 198–202), β5 (residues 205–206), and β6 (residues 225–231) and helices α1 (residues 187–191), α2 (residues 212–220), α3 (residues 233–242), α4 (residues 248–251), and α5 (residues 324–332). The r.m.s. deviations of the polypeptide chain (residues 153–251), including the loops, are indicated in parentheses.
CD Experiments
CD experiments were performed with an Aviv 400 Spectropolarimeter (Aviv Biomedical, Inc., Lakewood, NJ). The protein samples were dissolved at 0.1–0.2 mg/ml in buffer containing 20 mm phosphate, pH 7.5, 150 mm NaCl, and 1 mm dithiothreitol. Urea-induced unfolding curves were measured by recording the ellipiticity at 222 nm on protein samples dissolved at 0.1 mg/ml in urea solutions at different concentrations containing 20 mm potassium phosphate and 150 mm NaCl, pH 7.5, and 1 mm dithiothreitol. The unfolding curves were fit to a two-state model to obtain the ΔG values (41). The experiments were performed at 20 °C.
Surface Plasmon Resonance Binding Experiments
Binding experiments were performed using a Biacore 1000 (Biacore Life Sciences, NJ) at 25.0 °C. The Biacore CM5 Biosensor chips were activated by N-hydroxysuccinimide and N-ethyl-N′-(3-(diethylamino)propyl) carbodimide. The ligand is the 70-amino acid residue C-terminal domain of CFTR (C-CFTR) or the C-terminal domain of NHERF1 (CT). About 3 μl of 10 μg/ml of C-CFTR or 5 μl of 10 μg/ml NHERF1-CT domain dissolved in 10 mm sodium acetate, pH 5.2, was injected to coat the activated surface. The unbound ligand was washed away, and uncoated sites were blocked by 1 m ethanolamine at pH 8.5. The analyte (PDZ2240, PDZ2+270, PDZ2CT, or PDZ2CT·FERM) was exchanged to HBS-EP buffer containing 10 mm HEPES buffer, pH 7.4, 150 mm NaCl, 3 mm EDTA, and 0.005% surfactant polysorbate 20. The analyte was injected over the C-CFTR-coated surfaces at the rate of 50 μl/min for 3 min at a series of concentrations. At the end of each injection, the sensor chip was regenerated with 4.0 m MgCl2, 50 mm triethylamine (pH 9.15), and HBS-EP buffer.
The response curves were obtained by subtracting the background signal generated by injecting the analyte over a control cell without ligand coating to remove the bulk refractive index effects. The nonspecific binding was corrected by subtracting the signal generated by HBS-EP buffer alone. The response unit corresponding to the plateau was taken and plotted as a function of analyte concentrations to obtain the binding curve. The dissociation constant Kd and the S.D. reported was obtained by fitting the binding curve to a monovalent binding model.
RESULTS
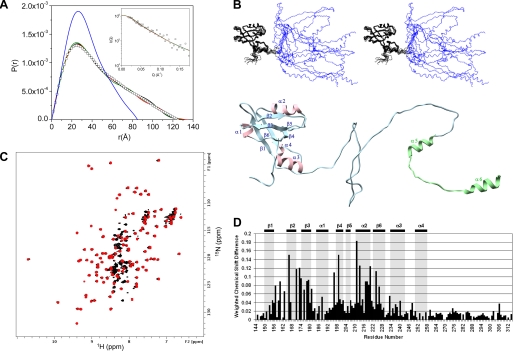
Overall, the 1H-15N HSQC spectrum of PDZ2CT (amino acid residues 150–358) (Fig. 2A) is characteristic of a heterogeneous conformation of a structured PDZ domain, coexisting with a largely disordered C-terminal region (Fig. 2B) that typically yields a high density of sharp peaks between 8 and 8.5 ppm in the proton dimension (supplemental Fig. S3). This observation is supported by our CD results of the isolated domains, which show that the PDZ2 domains are structured, but the CT domain is largely disordered (see Fig. 4). The overlay of NMR spectra offers a glimpse into the extent of the perturbation introduced in the structure by intramolecular domain-domain interactions (Fig. 2, A and B). The shifted resonances of the PDZ domain relative to that of the intact PDZ2CT is characteristic of significant changes of intramolecular interactions between domains that are not structurally independent modules (Fig. 2A).
FIGURE 2.
Comparing 15N-HSQC spectra of different domains in PDZCT. A, overlay of PDZ2CT (black) and PDZ2+270 (red) at 30 °C. The annotated residues in PDZ2+270 are those with severe exchange broadening (blue) and large chemical shift perturbation (>0.14 ppm) (black) in PDZ2CT. B, overlay of PDZ2CT (black) and CT (green) at 15 °C. The extreme C-terminal residues in the isolated CT domain with severe exchange broadening in PDZ2CT have been annotated in magenta.
FIGURE 4.
PDZ2+270 shows increased helicity, stability, and binding affinity for ligands compared with the putative PDZ2240. A, overlay of CD spectra of PDZ2240, PDZ2+270, and PDZ2CT; B, CD spectrum of isolated CT; C, the urea denaturation curves of PDZ2240, PDZ2+270, and PDZ2CT measured by CD; D, comparison of the binding affinities of C-CFTR for PDZ2240, PDZ2+270, and PDZ2CT. The binding curves are from equilibrium SPR measurements.
Redefining the Structural Boundary of a PDZ Domain
Primary sequence analysis, using Blast and pfam (42), predicts the “conventional” PDZ2 domain of NHERF1 starts at Leu154 and ends at Val231. However, by analyzing the backbone chemical shifts of PDZ2CT, we find that the structured region extends to Asn252, well beyond Val231 at the conventionally predicted boundary for a putative PDZ domain and as reported in the x-ray or the NMR structures of PDZ2 (Protein Data Bank codes 2OZF and 2JXO). We find that the structured C-terminal extension is an integral part of the PDZ2 domain as discussed below. This unexpected finding prompted us to clone and to determine the structure of a longer construct (residues 150–270), henceforth referred to as PDZ2+270.
A representative NMR structure of PDZ2+270 is shown in Fig. 3A. The core structure is the conventionally defined PDZ2 fold (residues 150–231) (Fig. 3B), and the backbone superimposes (Cα r.m.s. deviation = 0.9 Å) with that of the putative PDZ2240 structure (Protein Data Bank code 2OZF). The characteristic β-sandwich structure is composed of six strands stacked in an anti-parallel fashion into two β-sheets (Fig. 3A). Strands β1, β6, and β5 are grouped into a single sheet that shares the β4 strand with a second sheet formed by β2 and β3 connected by the two helices α1 and α2. The partially open hydrophobic cavity, enclosed by the β-sandwich, serves as a robust scaffold to recruit peptide-based ligands (Fig. 3B). The conformation of the carboxylate binding (CB) loop (-GYGF-) capable of forming hydrogen bond pairs with the peptide ligand (43) is unaltered in the PDZ2+270 structure. The profile of the heteronuclear 1H-15N NOE values (supplemental Fig. S5) correlates with three flexible regions in the protein (44): the CB loop, the linker connecting strands β2 and β3, and the C-terminal end.
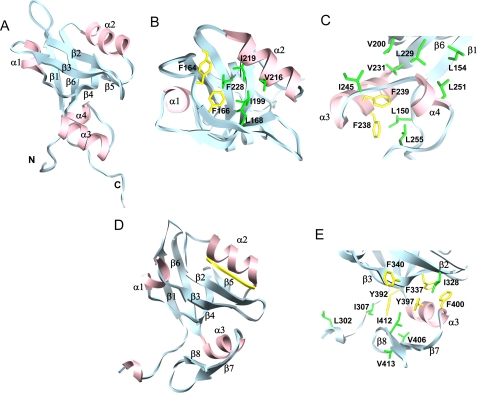
FIGURE 3.
The structure of PDZ2+270 with a novel helix-turn-helix subdomain at the C-terminal end of the putative PDZ fold. A, a single model representing the NMR structure of PDZ2+270 (Protein Data Bank code 2KJD). The secondary structure includes strands β1 (residues 153–158), β2 (residues 166–170), β3 (residues 177–182), β4 (residues 198–202), β5 (residues 205–206), and β6 (residues 225–231) and helices α1 (residues 187–191) and α2 (residues 212–220) that are typical of a putative PDZ fold, together with the extra C-terminal α3 (residues 233–242) and α4 (residues 248–251) helices that are packed against the β1, β4, and β6 strands of PDZ2 by hydrophobic interactions. B, top view of the ligand binding site in PDZ2+270 with important side chains labeled. C, hydrophobic cluster in the C-terminal helical extension of PDZ2+270. D, x-ray structure of the third PDZ domain from PSD-95 complexed with the CRIPT peptide (Protein Data Bank code 1BE9). E, hydrophobic cluster from the C-terminal extension of PDZ3 of PSD-95. The graphics were generated using the University of California San Francisco Chimera package (67).
The novel C-terminal extension (Arg233–Asn252) consists of two helices α3 and α4, forming a closed hydrophobic cluster at the stem of the putative PDZ fold. The pairwise side-chain interactions include residues Phe238, Phe239, Val244, Ile245 (α3), and Leu251 (α4) and those in strands β1 (Leu154), β4 (Val200) and β6 (Leu229 and Val231) (Fig. 3, A and C). Consequently, an exposed hydrophobic patch on the surface of the β-barrel is effectively buried by the packing of the extra helices in the PDZ2+270 domain.
To evaluate the effects of this novel helical extension on the overall thermodynamic stability of the extended PDZ2+270, we have compared the urea unfolding using the CD signals at 222 nm (Fig. 4C and Table 2). The extended PDZ2+270 domain is more stable (ΔΔGn-u0 = 2.4 kcal mol−1) than the canonical structure PDZ2240, but only marginally less stable than PDZ2CT (ΔΔGn-u0 = 3.3 kcal mol−1).
TABLE 2.
Comparison of the stability and binding affinity of PDZ2240, PDZ2270, and PDZ2CT
| PDZ2 variant | Kd binding to C-CFTR70 | ΔΔGbinding | Kd binding to NHERF1-CT | ΔΔGbinding | M (urea denaturation) | Cma | ΔGb | ΔΔGn-u0 |
|---|---|---|---|---|---|---|---|---|
| μm | kcal mol−1 | μm | kcal mol−1 | kcal mol−1m−1 | m | kcal mol−1 | kcal mol−1 | |
| PDZ2240 | 4.800 ± 0.300 | 0 | 803 ± 64 | 0 | 0.42 ± 0.20 | 1.9 ± 0.2 | 1.0 ± 0.1 | |
| PDZ2270 | 0.267 ± 0.011 | −1.71 | 93.4 ± 5.9 | −1.28 | 0.83 ± 0.05 | 3.9 ± 0.1 | 3.4 ± 0.1 | 2.4 |
| PDZ2CT | 5.300 ± 0.200 | 0.06 | 1230 ± 186 | 0.25 | 0.91 ± 0.13 | 4.2 ± 0.1 | 4.3 ± 0.2 | 3.3 |
| PDZ2CT·FERM | 0.202 ± 0.003 | −1.88 | ||||||
| PDZ2270(R153Q) | 0.930 ± 0.045 |
a Cm is the urea concentration at which the protein is 50% unfolded.
b The free energies have been extrapolated to 0 m urea.
Our surface plasmon resonance experiments show that, compared with the putative PDZ2240, the extended PDZ2+270 has a 10-fold increase in binding affinities for either the C-terminal domain of CFTR or the C-terminal domain of NHERF1 (Fig. 4D and Table 2). C-CFTR and the CT domain of NHERF1 contain the type I PDZ-binding motifs -DTRL and -FSNL, respectively. By contrast, the affinity of the less stable PDZ2240 for these PDZ-binding motifs is much weaker and similar to that of the autoinhibited PDZ2CT, where the target binding site is obstructed (Table 2). These results corroborate with mutational studies of the PDZ domain of PTB-BL, where a correlation between domain stability and target affinity has been identified (45). Because our results show that the C-terminal subdomain extension in PDZ2+270 is at a distance from the active binding site, the increased binding capability is facilitated through a distal allosteric communication mechanism between the two hydrophobic cores instead of direct engagement.
Side-by-side we have compared the αβ type C-terminal extension observed in the third PDZ domain structure from PSD-95 complexed with the CRIPT peptide (Protein Data Bank code 1BE9) (43). In this example, we see the formation of an entirely different hydrophobic core that involves the distal side of the β2 and β3 strands from the active site (Fig. 3, D and E). The other well known example of an extended PDZ domain is that of the neuronal nitric-oxide synthase with a β-hairpin finger at the C-terminal end. The β-hairpin finger engages the syntrophin PDZ domain directly in an unusual head-to-tail arrangement of the two proteins to form a heterodimer (46, 47). The presence of such extensions adds to the structural diversity of an otherwise homologous family of PDZ domains serving unique roles in each example (43, 46).
The C-terminal helical extension of a PDZ fold appears to be a feature that is shared by many PDZ domains. Based on multiple sequence alignment alone, the C-terminal hydrophobic residues are conserved across all the PDZ domains within the NHERF family of proteins, suggesting similarly functional roles for this extended helical fold (Fig. 1 and supplemental Fig. S1). With this C-terminal extension, the binding affinity of the extended PDZ1 domain of NHERF1 for C-CFTR is also increased 3.7-fold (Table 2).
Our extensive data base searching and multiple alignments suggest that, outside the NHERF family proteins, the amino acid sequence at the C-terminal sections of the PDZ domains is not well conserved (supplemental Fig. S1). Nevertheless, secondary structure analysis predicts α-helical propensity at the C-terminal end in a majority of the PDZ domains analyzed in supplemental Fig. S1. These include many of the PDZ domains that are important in cell signaling, such as those of the human harmonin, protein-tyrosine phosphatase, tamalin, and PARR3, and PDZ1 of PSD-95, as well as the PDZ domains of Drosophila melanogaster INAD (supplemental Fig. S1). We hypothesize that the helical extension at the C-terminal section of these PDZ domains may tune the stability and/or binding affinity of PDZ-mediated interactions. In addition to regulating the target affinity, the C-terminal extensions to PDZ domains are also known to mediate multimerization in harmonin (48).
We point out that there are exceptions in the subset of PDZ domains that we have analyzed. For instance, the PDZ1, PDZ2, and PDZ5 domains of the multiple PDZ domain protein (MUPP1) and PDZ2 of inactivation no afterpotential D lack the helical structure at their respective C-terminal ends (supplemental Fig. S1). Detailed proteomic (49) and mutational analysis (50) can be interpreted as indicating a large distribution through selectivity space, and these additional structural elements are probably of significance.
Disease Mutations Affect PDZ2+270 Stability
A recent study by Karim et al. (7) identifies the correlation of three NHERF1 mutations, L110V, R153Q, and E225K, with impaired renal phosphate reabsorption in patients with chronic kidney disease. In the proximal tubule of the kidney, the transmembrane sodium-phosphate cotransporter 2a (NPT2a) is responsible for the reabsorption of phosphate from urine. Impaired renal phosphate reabsorption leads to kidney stone formation and bone demineralization. The ability of NPT2a to transport phosphate ions depends on the correct localization of NPT2a at the apical membrane of polarized epithelial cells, which is modulated by the parathyroid hormone (PTH). PTH binding to receptor PTH1R triggers a cascade of cellular signaling events that regulate NTP2a endocytosis and thus the capacity of NTP2a to uptake phosphate. NHERF1 interacts with both NPT2a and PTH1R by binding to the PDZ motifs in their respective cytoplasmic tails. NHERF1 is required for correct apical localization of NPT2a and PTH1R (51, 52). The disease mutations of NHERF1, L110V, R153Q, and E225K, are found to inhibit phosphate transport by NPT2a, in a similar fashion as in NHERF1−/− kidney cells (7, 53).
The R153Q and E225K mutations are located in PDZ2 outside the ligand binding sites (Figs. 1 and 5). We have analyzed the effects of R153Q and E225K mutations on the structure and stability of PDZ2+270 using unfolding studies and NMR. The CD unfolding experiments show that R153Q is considerably less stable (Tm = 37 °C) than the wild-type protein (Tm = 55 °C) (Fig. 5D). The higher peak count in the NMR HSQC spectrum of R153Q than that of wild-type PDZ2+270 reflects the small fraction (7%) of unfolded protein in R153Q at 30 °C with characteristic random coil amide proton shifts (Fig. 5C). Notwithstanding the minor population of the unfolded state, the chemical shift mapping and CD spectra of the predominantly folded conformation of R153Q are structurally similar to those of PDZ2+270 (Fig. 5C and supplemental Fig. S4, A and C). The deleterious effects of the R153Q mutation can be attributed to the loss of a potential hydrogen bond/salt bridge between the positively charged bidentate Arg153 Nη group in strand β1 and the negative charge of the COO− groups of Asp197 (β4) and Asp232 at the N terminus of the α2 helix (Fig. 5A). In the wild-type PDZ2+270, Arg153 Nη is involved in hydrogen bond formation, and the attached protons are protected from fast exchange at neutral pH at even 30 °C.
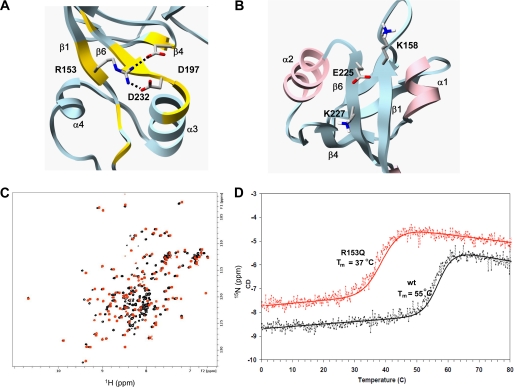
FIGURE 5.
Effects of human disease mutations on PDZ2+270 structure and stability. A, the structure of wild type PDZ2+270 shown with a hydrogen bond formed between positively charged guanidinium group of Arg153 and the negatively charged carboxylate group of Asp232 and Asp197. Residues with weighted backbone chemical shift difference greater than 0.1 ppm of mutant R153Q with reference to wild-type PDZ2+270 (supplemental Fig. S4C) are painted yellow. B, in wild type PDZ2+270, the negatively charged Glu225 is complemented by the positive charge of Lys158 and Lys227. C, overlay of 15N HSQC of PDZ2+270 (red) and mutant R153Q (black) at 30 °C. D, thermal unfolding curves of mutant R153Q (red) and wild-type PDZ2+270 (black) monitored by CD. The molar ellipticity (y axis) as a function of temperature (x axis) was fitted to a standard two-state unfolding equation.
The binding affinity of R153Q for the C-CFTR ligand decreased about 3.5-fold compared with the wild-type PDZ2+270 (Table 2 and supplemental Fig. S4B). Because the critical residues of the PDZ-binding motif (-ATRL) at the NPT2a C terminus are similar to those of CFTR (-DTRL), we expect that the R153Q mutant also has lower binding affinity for NPT2a than does the wild-type PDZ2+270. Due to the significant difference in the sequence of PTH1R PDZ-binding motifs (-ETVM) from that in CFTR, we cannot draw the same conclusion, and this is a subject for future investigation.
The E225K mutation has a dramatic effect on the conformational stability of PDZ2 domain, and this mutant fails to express as an intact protein in E. coli at either 37 or 20 °C. In the wild-type protein, the negatively charged Glu225 is complemented by surrounding positive charge of lysine side chains (Lys158 and Lys227) on the exposed surface of the β-sheet (Fig. 5B). The unfavorable electrostatic energy of the E225K mutant would destabilize the native protein.
Although these mutations are located outside the ligand binding site of the PDZ2 domain, the mutations R153Q and E225K evidently destabilize the native state and would therefore tend to degrade in a cellular context. Such turnover effects may contribute to pathophysiological effects independent of any changes of ligand affinity. Reduced protein stability could translate into the loss of functional NHERF1 expressed in cells and diminish the available number of NHERF1 molecules to assemble transmembrane protein complexes of NPT2a at the cell membrane. Interestingly, based on sequence alignment, the third NHERF1 mutation L110V is located in the α4 helix (Figs. 1 and 3C) of the C-terminal helix-turn-helix subdomain of PDZ1. Homology modeling (54) using the newly determined structure of PDZ2+270 as a template suggests that L110 is involved in forming the extended hydrophobic network with the putative PDZ1. The mutation to a smaller hydrophobic side chain in L110V could potentially disrupt the packing interactions and reduce PDZ1 stability and/or the affinity of PDZ1 binding.
Structural Model of PDZ2CT
In the isolated PDZ2+270, almost all of the NMR resonances could be assigned, and PDZ2+270 has a defined structure. In contrast, the isolated CT is largely disordered with the exception of Phe323–Ala334 (supplemental Fig. S3). In particular, the EB region (Met346–Leu358) possessing the C-terminal type I PDZ-binding motif is a random coil (supplemental Fig. S3).
For the tandem PDZ2CT domains, complete backbone and side-chain chemical shift assignments are challenging. About 87% of the resonances are assigned in the PDZ2CT construct. This is largely due to the unusual relaxation properties at the putative target binding sites of PDZ2 and at the C-terminal end of CT that presumably interacts with PDZ2. Specifically in PDZ2, the amide resonances along the carboxylate binding loop (Tyr164 and Gly165), the β2 strand (Phe166, Asn167, Leu168, His169, and Ser170), Gln177 in the β3 strand, and the α2 helix (His212) (Figs. 2A and 6A) are lost due to exchange broadening in the intermediate time scale regime. Partial side-chain information was recovered from 13C-edited multidimensional experiments. These results are a strong indication that these residues in PDZ2 could be involved in interacting with CT.
FIGURE 6.
Analysis of the interactions of PDZ2+270 with the C-terminal EB. A, top view of the C-terminal EB peptide-binding site in PDZ2CT. Residues that experience severe exchange broadening or large chemical shift perturbation (>0.15 ppm) are shown in orange, and their side chains are marked in yellow. B, a view of the EB peptide (green) with L358 inserted into the hydrophobic pocket (yellow) in the PDZ2 domain. The C-terminal charge of L358 (red) is complemented by the amide groups from the exposed CB loop of PDZ2. Coordinates for this figure are derived from the calculation assuming helix formation in the C-terminal EB domain. Shown are the 15N HSQC spectrum of wild-type PDZ2CT domain (black) overlaid with mutant PDZ2CT(F355A) (red) (C) and overlaid with PDZ2CT(F355P) (red) (D) at 30 °C. Residues with significant chemical shift perturbation are annotated in the overlay plots in C and D.
In the CT region of PDZ2CT, residues Phe323–Ala334 in the N-terminal end of the EB domain form a short helix (α5 in Fig. 7C) based on chemical shift and NOE analysis, whereas line broadening obliterates resonances from the C-terminal residues (Met346–Leu358). Exceptions are the methyl resonances of Leu354 and Leu358 that can be assigned. The disordered EB region in the isolated CT domain does not adopt the well formed α-helical conformation when EB peptide is bound to the FERM domain of radixin, as seen in the crystal structure (29). Collectively, these observations suggest that, through weak intramolecular interactions with PDZ2, helical structures are induced and stabilized in the otherwise largely disordered CT domain.
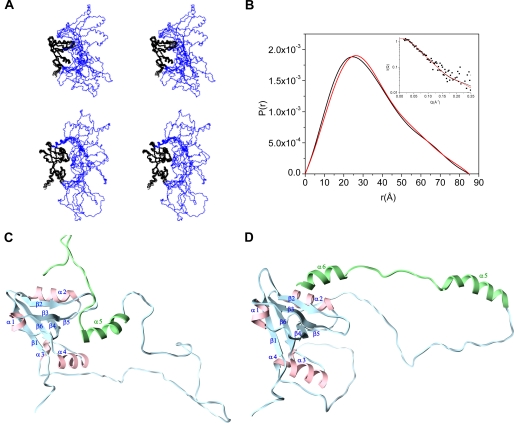
FIGURE 7.
Structural model of the intact PDZ2CT from NMR refined against SAXS. A, stereoview of the ensemble NMR structures of PDZ2CT after refinement against SAXS. The backbone of the PDZ2+270 domain is shown in black, and the CT domain is shown in blue. Top, the C-terminal EB region is shown in random coil conformation. Bottom, the C-terminal EB region is shown in helix conformation. The images were generated in MOLMOL 2.1 (68). B, length distribution functions P(r) calculated from the NMR structure of PDZ2CT with the EB region adopting helix conformation (red) or random coil conformation (black). The inset shows the fits of the calculated scattering profiles from the NMR structures to the experimental SAXS data (filled squares), with the EB region adopting a helix (red) or random coil (black) conformation. C, a representative ribbon structure of the intact NMR structure of PDZ2CT from A. The C-terminal domain includes the single N-terminal helix (residues 323–334) and the EB region that adopts a random coil conformation. D, a representative ribbon structure of PDZ2CT with the C-terminal EB region adopting a helical conformation and docked into the PDZ2 domain via Leu358.
In PDZ2CT, our experiments show that NMR perturbation exists in the extended C-terminal segment Met346–Leu358 of 13 residues, suggesting interactions well beyond the short type 1 PDZ-binding motif (-LFSNL358) in the EB region. The nominal length of the canonical PDZ-binding peptide motifs is only 3–5 residues aligned as an anti-parallel strand with the PDZ β2 strand (Fig. 3B) (26, 43), although longer contacts have been reported (55). By contrast, the binding surface mapped on the PDZ2+270 domain does not increase proportionately in area (Fig. 6A).
Previous biochemical experiments and an NMR study have provided low resolution information about the nature of interactions between the PDZ2 and CT fragments (23, 30). Morales et al. (23) have identified residues Leu358 and Ser356 in the type 1 PDZ-binding motif as crucial to interact with PDZ2 because deletion mutant ΔSNL358 releases autoinhibition in NHERF1. Based on biochemical binding results, Morales et al. (23) have assumed that the length of interaction presented by a helical peptide could match the existing binding pocket in the PDZ2 domain without the necessity of adopting an extended mode of interaction with a β strand. Although direct structural evidence is lacking, helix-breaking mutations (E353P and F355P) are known to release autoinhibitory interactions (23, 30). Additional evidence in favor of a possible helical binding model is the periodic change in the NMR transverse relaxation rates along the length of the putative binding surface of the amphipathic helix measured by Cheng et al. (30).
Because of the largely disordered CT domain, a single “high resolution” structure of the entire PDZ2CT is not an accurate representation of the conformational ensemble. Thus, we have built a suitable model for the coupled domains of PDZ2CT by refining the NMR structure using solution SAXS data as constraints. Several studies have proposed that combining SAXS data with NMR restraints can yield significantly improved accuracy in NMR solution structure determination of multidomain proteins, multimeric assemblies, and tight macromolecular complexes (56–59).
The SAXS data used here are from our recent publication (25). During refinement, the ensemble of PDZ2CT structures was initially calculated assuming the exchange-broadened EB region (Met346–Leu358) to be unstructured because this peptide structure is marginally stable (30). The NOEs between the aromatic ring of Phe166 in PDZ2 and methyl protons of Leu358 in CT provided the sole restraint for docking the C-terminal residue Leu358 to the PDZ2+270 domain.
The NMR and SAXS jointly refined structures of PDZ2CT are shown in Fig. 7A. Fig. 7B shows the fit of the calculated scattering curve generated from a representative NMR structure to the experimental SAXS data. The ensemble averaged χ value of the SAXS fits is quite small (0.09 ± 0.01), indicating that the joint refinement was successful. After refinement, the conformational space sampled by the flexible CT domain is restricted to a more limited conformational space than the unrefined ensemble.
The refined model has an average radius of gyration Rg = 26.7 Å, in agreement with the globular PDZ2+270 domain tethered to the extended loop-like structure from the CT domain (Fig. 7A), which also rules out intermolecular interactions between two different PDZ2CT molecules. In the refined structure, the N-terminal end (residues 323–334) of the EB region is disengaged from the PDZ2+270 domain (Fig. 7C). The superposition of the backbone residues (positions 151–257) improves slightly (1.6 Å) when the binding site residues (β2, β3, α2, and CB loop) are excluded from the r.m.s. deviation calculation (1.1 Å). Hence the β-barrel structure of the PDZ scaffold in the autoinhibited state is intact and well determined despite the conformational flexibility within the binding site.
To further elucidate details of side-chain interactions within the binding site, we made use of ambiguous restraints generated from chemical shift perturbation (Fig. 6A) to dock a well folded helical peptide (Met346–Leu358) to PDZ2+270 using the program Haddock (see supplemental material for details). The C-terminal helical peptide is taken from the crystal structure of the EB peptide in complex with the radixin FERM domain (29). Long range restraints between the domains from the docked structure are used to generate a final ensemble of intact PDZ2CT structures and are refined against the SAXS and NOE data. By modeling the C-terminal peptide (Met346–Leu358) as a helix in the NMR calculations, there is no significant change in the χ value from the SAXS fits (0.11 ± 0.03) whereas the Rg value (26.70 Å) is unchanged. The ensemble of the structures, with the C-terminal end adopting a helical conformation, is shown in the lower panel of Fig. 7A. Fig. 7D shows a representative snapshot of the PDZ2CT structure with the C-terminal EB adopting a helical conformation.
The models so derived suggest that the principal driving force for the interaction is the insertion of the C-terminal leucine (Leu358) within a hydrophobic groove enclosed by the aromatic side chain of Tyr164 and Phe166 (Fig. 6B). The anchoring of the methyl groups of Leu358 brings the bidendate terminal carboxylate (COO-) group within bonding distance of the exposed amide proton (NH) in the CB loop (Tyr164, Gly165, Phe166) as seen in Fig. 6B. The pairwise polar interactions involving the backbone of the CB loop and the terminal charge in Leu358 is the molecular basis of recognizing the C-terminal end of the target peptides by PDZ domains, similar to that of NHERF1 PDZ1 binding to ligand peptides, as shown by Karthikeyan (26).
The contribution of other residues in the type 1 PDZ-binding motif, Ser356(−1) and Phe355(−3), can be evaluated by comparing our structural models with the published biochemical data. The structure shows a hydrogen bond pairing between Ser356 backbone O and Asn167 side-chain Nδ2 in the β2 strand whose NMR resonances are selectively broadened in PDZ2CT but are recovered in the PDZ2+270 domain. This particular interaction could explain the much higher specificity of the NHERF1 C terminus for the PDZ2 and not the PDZ1 domain of NHERF1 (23). The amino acid sequence in the PDZ1 binding site is nearly identical to that of PDZ2 domain except for His27 in PDZ1 and Asn167 in PDZ2 (Fig. 1). The imidazole side chain of His27 in PDZ1 could be unfavorable for the specific interaction with the backbone oxygen of Ser356 (Fig. 1). In addition, mutating Ser356 to Ala has a minor impact on binding (23), suggesting that at the −1-position, the interactions of backbone atoms with PDZ2 are more important than the side chain.
The effect of Phe355 on binding to PDZ2 is not well understood, although mutating Phe355 to a Pro or Arg abrogates autoinhibition of NHERF1 (23). In our structural model, the backbone Phe355 carboxylate O is within hydrogen bond distance of Leu168 amide NH, whereas the aromatic ring is partly buried in a hydrophobic cleft between helix α2 and strand β2, contributing to the overall affinity (supplemental Fig. S6B).
To distinguish the role of the side chain from the backbone, we made a third mutation, F355A, assuming that Ala would stabilize the helix but reduce the contribution of the hydrophobic side chain. The mutation failed to eliminate the interaction with the C-terminal peptide as judged by similarities in the 15N HSQC spectra overlay (Fig. 6C) and three-dimensional NOESY data (not shown) of coupled PDZ2CT domains. Relatively small backbone perturbation of residues in strand β2 and helix α2 is in agreement with limited structural change in the mutant (supplemental Fig. S7B). In contrast, the control spectrum of F355P revealed a dramatic shift of resonances with the release of intramolecular interactions (Fig. 6D and supplemental Fig. S7C). The much smaller methyl group of Ala leads to some loss of binding surface and stability of the helix but is not sufficient to disrupt the EB structure in the same manner as the proline. As a consequence, we recover the intensity of several peaks in F355A from the “invisible” parts of the induced EB structure (Trp348-Lys350), whereas the key interactions with the PDZ motif (-SNL358) are intact. Similarly, the mutation E353P has been known to disrupt the peptide structure, whereas the non-conserved mutation E353A has a negligible effect on weakening the intramolecular interaction (30).
In summary, our results suggest that, in intact PDZ2CT, the C-terminal EB region is in dynamic equilibrium between random coil and nascent helical structure (Fig. 7, C and D). The flexible structure facilitates access to the backbone groups analogous to an extended strand in the canonical PDZ ligand. The principal anchor is the terminal residue Leu358, but the correct backbone conformation of the peptide at Ser356 and Phe355 is important for augmenting the polar interactions with strand β2 in the PDZ2 binding site. Further structure is induced along the length of the peptide through contacts between side chains with the protein, but presumably the contribution of these positions to the overall affinity is much smaller than Leu358.
Ezrin Binding Releases PDZ2CT Autoregulation
Using NMR, we have determined the conformational transition induced in 15N labeled PDZ2CT upon binding to unlabeled FERM domain of ezrin. Previous biochemical and biophysical studies have demonstrated that FERM binds to the EB region of NHERF1 with a 1:1 stoichiometry and with high affinity Kd = 19 nm (22, 29, 60). Moreover, binding of FERM to the CT domain of NHERF1 causes a 20-fold increase in the binding affinity of PDZ2 for a target protein (22), suggesting that FERM binding disrupts the intramolecular domain-domain interactions in PDZ2CT. The NMR study on the 15N-PDZ2CT·FERM complex presented here provides a detailed structural mechanism about such functional transitions, which complements our recent low resolution SANS study that shows the overall shape changes in PDZ2CT when forming a complex with FERM (40).
The 15N-labeled PDZ2CT in complex with the unlabeled FERM yields a well behaved amide 1H HSQC spectrum (Fig. 8C), which could be assigned to the N-terminal PDZ2+270 domain (Leu150–Asn252) and a portion of the unstructured CT domain (Gly253–Thr314). The amide resonances of the C-terminal EB domain (Ser315–Leu358) in complex to the FERM domain is no longer observed in the standard 15N HSQC spectrum. The one-to-one correspondence between the backbone amide NH cross-peaks of residues 150–252 in the isolated PDZ2+270 and in the PDZ2CT·FERM complex (Fig. 8C) suggests that the PDZ domain is uncoupled in the modular structure once it is released from autoinhibitory interactions. These results are in agreement with the recent SANS studies that have estimated that the PDZ2 and FERM domains are separated by a distance of 80 Å and hence not in contact with each other (40).
FIGURE 8.
NMR structure of 15N-labeled PDZ2CT in complex with unlabeled FERM domain refined against SANS. A, length distribution function P(r) calculated from the NMR structure of PDZ2CT in solution refined against SAXS (blue line). Also shown are P(r) values calculated from three representative NMR structures of 15N- and 13C-labeled PDZ2CT (black, red, and green lines) in complex with unlabeled FERM after refinement against SANS (open square). SANS data are from deuterium-labeled [U-2H]PDZ2CT in complex with unlabeled FERM in 40% D2O buffer solution, in which FERM is invisible to the neutron and only the structure of the deuterium-labeled PDZ2CT is probed. The inset shows the fit of the three scattering profiles calculated from the NMR structures (black, red, and green lines) to the experimental SANS data (open square). B, the upper panel shows the stereoview of an ensemble PDZ2CT structures in complex with FERM after refinement against SANS, and the lower panel shows a single representative structure. C, overlay of the 15N HSQC spectrum of the labeled PDZ2CT domain in the presence (red) and absence (black) of unlabeled FERM at a 1:1 stoichiometry ratio at 30 °C. D, weighted backbone chemical shift difference of PDZ2CT complexed to FERM (Δ = √((δHN)2 + (δN/5.0)2) with reference to the wild-type PDZ2CT at 30 °C. The last residue observed in the 15N HSQC is Thr314. Secondary structure elements are labeled at the top.
Using the NOE data for residues Arg151–Thr314 and the SANS data as constraints, we are able to calculate a medium resolution structure of the active and open conformation of PDZ2CT in the complex (Fig. 8B). The SANS data are collected on deuterium-labeled PDZ2CT [73%-2H]PDZ2CT) in complex with unlabeled FERM in 1:1 stoichiometry and in 40% D2O buffer solution (40). In 40% D2O, the unlabeled FERM is “invisible” to neutrons; therefore, the SANS data only reflect structural information about the [73%-2H]PDZ2CT in the complex. The C-terminal EB helices are modeled from the x-ray structure of the Moesin FERM domain complex (29). The structure of the PDZ2+270 domain in the open state is superimposible (Cα r.m.s. deviation <1.8 Å) with that of the isolated protein. The flexible linker region (Gly253–Thr314) is found to have all of the characteristics of random coil conformation (chemical shift indexing in supplemental Fig. S8). The joint NMR and SANS refined structures shows that PDZ2CT undergoes significant conformational changes from a compact free protein with a radius of gyration (Rg) = 26.7 Å in solution to an expanded structure in the complex Rg = 37.3 Å.
The SANS results indicate that, at the resolution of the scattering experiments, the overall structural changes in the FERM domain is small when compared with the crystal structure (40). Based on the joint NMR and SANS refinement studies, we conclude that, upon binding to the FERM domain, the conformational changes in PDZ2CT essentially involve the release of the C-terminal EB domain from interactions with PDZ2 accompanied by small rearrangement in the target binding site of the PDZ2 domain. When complexed to FERM, the largely disordered CT domain becomes more extended. There is no additional structure formation in the extended CT domain beyond the EB region.
DISCUSSION
Using high resolution NMR, we have determined the structure of tandem PDZ2 plus the C-terminal domains of NHERF1. With joint refinement against SAXS data, the NMR structure provides a more complete view of the interactions between PDZ2 and the largely disordered CT domain than any previous studies on the PDZ fragments of NHERF1. By determining the functionally important PDZ2CT construct, our NMR study reveals new structural features associated with the PDZ fold. Our study thus expands the understanding of the relationship between PDZ stability and ligand binding affinity. Further, we have determined the structural transition in PDZ2CT upon forming a complex with the FERM domain of ezrin.
The α3 helix-turn-α4 helix subdomain at the C-terminal end of PDZ2 is an integral part of the extended PDZ2 structure of NHERF1. This subdomain is allosterically connected to the putative PDZ fold through hydrophobic side-chain interactions and increases both the thermodynamic stability and the binding affinity of the extended PDZ structure for target proteins. From sequence and secondary structure analyses, we find that the helical C-terminal extension of the PDZ fold is conserved in the PDZ domains of NHERF family proteins (Fig. 1 and supplemental Fig. S1) and is probably a general feature frequently shared by many PDZ domains that are important in cell signaling.
So far, this important structural feature of PDZ domains may have been overlooked due to canonical definitions. The PDZ domains were discovered based on the identity of the conserved GLGF repeat. The region outside the assumed PDZ fold has been generally ignored in structural studies, because the linker region lacks homology in amino acid sequence and is usually assumed to be of low complexity or disordered. This oversight can lead to loss of crucial insight into the source of weak binding affinities, such as those reported for the abridged PDZ2240 domain of NHERF1 (22, 31) and the dependence of PDZ binding affinities on the lengths of truncation (61, 62). Our results suggest that amino acid sequence variation within a PDZ domain or in the PDZ-binding motif is not the only means to render specificity for PDZ-target protein interactions. The structure flanking the core PDZ fold can also influence target peptide binding. The roles of the extended structure(s) may include modification of “dynamic allostery” as suggested for the PDZ3 domain of PSD-95/SAP90 (63).
The difference in the stability and binding affinities between PDZ2+270 and the putative PDZ2240 supports the view that increased stability of PDZ domains may also translate into higher binding affinity for their cellular target proteins (45). The origin of the higher binding affinity of the extended PDZ+270 can be explained by invoking an allosteric mechanism where the energetically coupled hydrophobic network (64), which extends beyond the putative PDZ fold, provides a more structurally sound framework for PDZ to anchor a ligand. The marginally stable PDZ2240 domain pays an unfavorable entropic cost for any structural reorganization during ligand binding and hence has much lower affinity for the target.
We show that the dynamic intramolecular domain-domain interactions in PDZ2CT regulate the binding capability of the PDZ2 domain. Our NMR results indicate that the relatively weak association is characterized by transient “on-off” contacts at the binding sites. Our results do not exclude the possibility that part of the PDZ-binding motif has a propensity for helix formation as postulated in previous studies (23, 30), but strongly suggest that the FSNL PDZ-binding motif at the EB C terminus is only weakly tethered to the carboxylate binding loop in PDZ2. Any induced structures are transient and marginally stable. These structural results are supported by our thermodynamic data that PDZ2CT is only slightly more stable than PDZ2+270 (Fig. 4 and Table 2). The origin of the weak affinity can be traced to equilibrium between ordered and disordered states of the EB region that destabilizes the contacts at the interface.
Further, we provide a structural framework for understanding the mechanism that regulates the binding affinity of PDZ2 by the distal interactions between EB and the FERM domain of ezrin. The FERM domain of ezrin binds with high affinity for the C-terminal EB domain of NHERF1 (22, 29, 60). Upon FERM binding to the EB domain, the affinity of PDZ2 for the target proteins increases dramatically, indicating long range domain-domain couplings (22, 23). The published structural studies of the isolated FERM and PDZ domains offer no explanation for such cooperative binding behaviors in assembling protein complexes.
By combining NMR and small angle scattering experiments, we reveal the details of the conformational transition in PDZ2CT and the corresponding release of the intramolecular domain-domain couplings between the PDZ2 and CT domains upon binding to FERM. The nanomolar binding affinity of the CT domain for the FERM domain drives the competitive displacement of the weak intramolecular PDZ2-EB interactions. The strong binding between FERM and EB also induces a disorder-to-ordered helix conformational transition in the EB region (22, 29, 60). We expect that the large conformational changes in PDZ2CT associated with the release of autoinhibition are compensated for or overcome by the enthalpic effect of the binding reaction.
Both NMR and SANS indicate that PDZ2+270 and the FERM domain are far apart in the complex. The largely disordered linker region between PDZ2+270 and the EB region thus serves as a flexible spacer between PDZ2+270 and FERM in the complex. The exposed PDZ binding site is poised for docking other targets to form a ternary complex.
Our studies provide mechanistic insight into the allosteric regulation of the scaffolding protein NHERF1 by the membrane-cytoskeleton adapter protein ezrin to assemble large macromolecular complexes. Future efforts will determine how the long range forces are propagated (65) across the full-length NHERF, driving the assembly of the multimeric complexes.
Supplementary Material
Acknowledgments
This work utilized facilities supported in part by the National Science Foundation under Agreement DMR-0454672. We acknowledge the support of the National Institute of Standards and Technology, United States Department of Commerce, in providing the neutron research facilities used in this work.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL086496 (to Z. B.), R01 GM047021 (to D. C.). This work was also supported by a grant from the W. W. Smith Charitable Trust (to Z. B.). The 900-MHz NMR system was purchased with funds from National Institutes of Health Grant GM-66354, the Keck Foundation, and the member institutions of the New York Structural Biology Center.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1–S8.
- NHERF
- Na+/H+ exchange regulatory factor
- CFTR
- cystic fibrosis transmembrane conductance regulator
- EB
- ezrin-binding
- ERM
- ezrin-radixin-moesin
- FERM
- 4.1-ezrin/radixin/moesin
- CT
- C-terminal
- NOE
- nuclear Overhauser effect
- SAXS
- small angle x-ray scattering
- SANS
- small angle neutron scattering
- C-CFTR
- C-terminal domain of CFTR
- r.m.s.
- root mean square
- CB loop
- carboxylate binding loop
- PTH
- parathyroid hormone.
REFERENCES
- 1.Pawson T., Scott J. D. (1997) Science 278, 2075–2080 [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharyya R. P., Remenyi A., Yeh B. J., Lim W. A. (2006) Annu. Rev. Biochem. 75, 655–680 [DOI] [PubMed] [Google Scholar]
- 3.Shenolikar S., Voltz J. W., Cunningham R., Weinman E. J. (2004) Physiology 19, 362–369 [DOI] [PubMed] [Google Scholar]
- 4.Thelin W. R., Hodson C. A., Milgram S. L. (2005) J. Physiol. 567, 13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donowitz M., Cha B., Zachos N. C., Brett C. L., Sharma A., Tse C. M., Li X. (2005) J. Physiol. 567, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamprecht G., Seidler U. (2006) Am. J. Physiol. Gastrointest. Liver Physiol. 291, G766–G777 [DOI] [PubMed] [Google Scholar]
- 7.Karim Z., Gérard B., Bakouh N., Alili R., Leroy C., Beck L., Silve C., Planelles G., Urena-Torres P., Grandchamp B., Friedlander G., Prié D. (2008) N. Engl. J. Med. 359, 1128–1135 [DOI] [PubMed] [Google Scholar]
- 8.Sizemore S., Cicek M., Sizemore N., Ng K. P., Casey G. (2007) Cancer Res. 67, 6183–6191 [DOI] [PubMed] [Google Scholar]
- 9.Kwon S. H., Pollard H., Guggino W. B. (2007) Cell Physiol. Biochem. 20, 763–772 [DOI] [PubMed] [Google Scholar]
- 10.Weinman E. J., Hall R. A., Friedman P. A., Liu-Chen L. Y., Shenolikar S. (2006) Annu. Rev. Physiol. 68, 491–505 [DOI] [PubMed] [Google Scholar]
- 11.Cao T. T., Deacon H. W., Reczek D., Bretscher A., von Zastrow M. (1999) Nature 401, 286–290 [DOI] [PubMed] [Google Scholar]
- 12.Schmieder S., Nagai M., Orlando R. A., Takeda T., Farquhar M. G. (2004) J. Am. Soc. Nephrol. 15, 2289–2298 [DOI] [PubMed] [Google Scholar]
- 13.Ko S. B., Zeng W., Dorwart M. R., Luo X., Kim K. H., Millen L., Goto H., Naruse S., Soyombo A., Thomas P. J., Muallem S. (2004) Nat. Cell Biol. 6, 343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall R. A., Ostedgaard L. S., Premont R. T., Blitzer J. T., Rahman N., Welsh M. J., Lefkowitz R. J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8496–8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bretscher A., Edwards K., Fehon R. G. (2002) Nat. Rev. Mol. Cell Biol. 3, 586–599 [DOI] [PubMed] [Google Scholar]
- 16.Algrain M., Turunen O., Vaheri A., Louvard D., Arpin M. (1993) J. Cell Biol. 120, 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott B. E., Meens J. A., SenGupta S. K., Louvard D., Arpin M. (2005) Breast Cancer Res. 7, R365–R373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khanna C., Wan X., Bose S., Cassaday R., Olomu O., Mendoza A., Yeung C., Gorlick R., Hewitt S. M., Helman L. J. (2004) Nat. Med. 10, 182–186 [DOI] [PubMed] [Google Scholar]
- 19.Curto M., McClatchey A. I. (2004) Cancer Cell 5, 113–114 [DOI] [PubMed] [Google Scholar]
- 20.Gary R., Bretscher A. (1995) Mol. Biol. Cell 6, 1061–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinman E. J., Steplock D., Donowitz M., Shenolikar S. (2000) Biochemistry 39, 6123–6129 [DOI] [PubMed] [Google Scholar]
- 22.Li J., Dai Z., Jana D., Callaway D. J., Bu Z. (2005) J. Biol. Chem. 280, 37634–37643 [DOI] [PubMed] [Google Scholar]
- 23.Morales F. C., Takahashi Y., Momin S., Adams H., Chen X., Georgescu M. M. (2007) Mol. Cell. Biol. 27, 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahon M. J. (2008) Am. J. Physiol. Renal. Physiol. 294, F667–F675 [DOI] [PubMed] [Google Scholar]
- 25.Li J., Poulikakos P. I., Dai Z., Testa J. R., Callaway D. J., Bu Z. (2007) J. Biol. Chem. 282, 27086–27099 [DOI] [PubMed] [Google Scholar]
- 26.Karthikeyan S., Leung T., Birrane G., Webster G., Ladias J. A. (2001) J. Mol. Biol. 308, 963–973 [DOI] [PubMed] [Google Scholar]
- 27.Karthikeyan S., Leung T., Ladias J. A. (2002) J. Biol. Chem. 277, 18973–18978 [DOI] [PubMed] [Google Scholar]
- 28.Finnerty C. M., Chambers D., Ingraffea J., Faber H. R., Karplus P. A., Bretscher A. (2004) J. Cell Sci. 117, 1547–1552 [DOI] [PubMed] [Google Scholar]
- 29.Terawaki S., Maesaki R., Hakoshima T. (2006) Structure 14, 777–789 [DOI] [PubMed] [Google Scholar]
- 30.Cheng H., Li J., Fazlieva R., Dai Z., Bu Z., Roder H. (2009) Structure 17, 660–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voltz J. W., Weinman E. J., Shenolikar S. (2001) Oncogene 20, 6309–6314 [DOI] [PubMed] [Google Scholar]
- 32.Cavanagh J., Fairbrother W. J., Palmer A. G., Skelton N. J. (1996) Protein NMR spectroscopy: Principles and Practice, pp. 411–528, Academic Press, Inc., San Diego [Google Scholar]
- 33.Sattler M., Schleucher J., Griesinger C. (1999) Prog. NMR Spectrosc. 34, 93–158 [Google Scholar]
- 34.Keller R. L. J. (2004) Optimizing the Process of Nuclear Magnetic Resonance Spectrum Analysis and Computer Aided Resonance Assignment, Ph.D. thesis, Swiss Federal Institute of Technology Zurich [Google Scholar]
- 35.Güntert P., Mumenthaler C., Wüthrich K. (1997) J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 36.Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 37.Nilges M., Macias M. J., O'Donoghue S. I., Oschkinat H. (1997) J. Mol. Biol. 269, 408–422 [DOI] [PubMed] [Google Scholar]
- 38.Lee D., Walsh J. D., Yu P., Markus M. A., Choli-Papadopoulou T., Schwieters C. D., Krueger S., Draper D. E., Wang Y. X. (2007) J. Mol. Biol. 367, 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 40.Li J., Callaway D. J., Bu Z. (2009) J. Mol. Biol. 392, 166–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santoro M. M., Bolen D. W. (1988) Biochemistry 27, 8063–8068 [DOI] [PubMed] [Google Scholar]
- 42.Finn R. D., Tate J., Mistry J., Coggill P. C., Sammut S. J., Hotz H. R., Ceric G., Forslund K., Eddy S. R., Sonnhammer E. L., Bateman A. (2008) Nucleic Acids Res. 36, D281–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doyle D. A., Lee A., Lewis J., Kim E., Sheng M., MacKinnon R. (1996) Cell 85, 1067–1076 [DOI] [PubMed] [Google Scholar]
- 44.Kay L. E., Torchia D. A., Bax A. (1989) Biochemistry 28, 8972–8979 [DOI] [PubMed] [Google Scholar]
- 45.Jemth P., Gianni S. (2007) Biochemistry 46, 8701–8708 [DOI] [PubMed] [Google Scholar]
- 46.Hillier B. J., Christopherson K. S., Prehoda K. E., Bredt D. S., Lim W. A. (1999) Science 284, 812–815 [PubMed] [Google Scholar]
- 47.Brenman J. E., Christopherson K. S., Craven S. E., McGee A. W., Bredt D. S. (1996) J. Neurosci. 16, 7407–7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verpy E., Leibovici M., Zwaenepoel I., Liu X. Z., Gal A., Salem N., Mansour A., Blanchard S., Kobayashi I., Keats B. J., Slim R., Petit C. (2000) Nat. Genet. 26, 51–55 [DOI] [PubMed] [Google Scholar]
- 49.Stiffler M. A., Grantcharova V. P., Sevecka M., MacBeath G. (2006) J. Am. Chem. Soc. 128, 5913–5922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J. R., Chang B. H., Allen J. E., Stiffler M. A., MacBeath G. (2008) Nat. Biotechnol. 26, 1041–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahon M. J., Cole J. A., Lederer E. D., Segre G. V. (2003) Mol. Endocrinol. 17, 2355–2364 [DOI] [PubMed] [Google Scholar]
- 52.Hernando N., Déliot N., Gisler S. M., Lederer E., Weinman E. J., Biber J., Murer H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11957–11962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cunningham R., E X., Steplock D., Shenolikar S., Weinman E. J. (2005) Am. J. Physiol. Renal. Physiol. 289, F933–F938 [DOI] [PubMed] [Google Scholar]
- 54.Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 55.Tonikian R., Zhang Y., Sazinsky S. L., Currell B., Yeh J. H., Reva B., Held H. A., Appleton B. A., Evangelista M., Wu Y., Xin X., Chan A. C., Seshagiri S., Lasky L. A., Sander C., Boone C., Bader G. D., Sidhu S. S. (2008) PLoS Biol. 6, e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grishaev A., Wu J., Trewhella J., Bax A. (2005) J. Am. Chem. Soc. 127, 16621–16628 [DOI] [PubMed] [Google Scholar]
- 57.Grishaev A., Tugarinov V., Kay L. E., Trewhella J., Bax A. (2008) J. Biomol. NMR 40, 95–106 [DOI] [PubMed] [Google Scholar]
- 58.Lynn G. W., Heller W. T., Mayasundari A., Minor K. H., Peterson C. B. (2005) Biochemistry 44, 565–574 [DOI] [PubMed] [Google Scholar]
- 59.Mattinen M. L., Pääkkönen K., Ikonen T., Craven J., Drakenberg T., Serimaa R., Waltho J., Annila A. (2002) Biophys. J. 83, 1177–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reczek D., Bretscher A. (1998) J. Biol. Chem. 273, 18452–18458 [DOI] [PubMed] [Google Scholar]
- 61.Cushing P. R., Fellows A., Villone D., Boisguérin P., Madden D. R. (2008) Biochemistry 47, 10084–10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raghuram V., Mak D. O., Foskett J. K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1300–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petit C. M., Zhang J., Sapienza P. J., Fuentes E. J., Lee A. L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18249–18254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lockless S. W., Ranganathan R. (1999) Science 286, 295–299 [DOI] [PubMed] [Google Scholar]
- 65.Bu Z., Biehl R., Monkenbusch M., Richter D., Callaway D. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17646–17651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson J. D., Gibson T. J., Higgins D. G. (2002) Curr. Protocols in Bioinformatics, Chapter 2, Unit 2.3 [DOI] [PubMed] [Google Scholar]
- 67.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 68.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 69.Dominguez C., Boelens R., Bonvin A. M. (2003) J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.