Abstract
Schistosome parasites adjust the physiology and behavior of their intermediate molluscan hosts to their own benefit. Previous studies demonstrated effects of the avian-schistosome Trichobilharzia ocellata on peptidergic centers in the brain of the intermediate snail host Lymnaea stagnalis. In particular, electrophysiological properties and peptide release of growth- and reproduction-controlling neuroendocrine neurons were affected. We now have examined the possibility that the expression of genes that control physiology and behavior of the host might be altered during parasitosis. A cDNA library of the brain of parasitized Lymnaea was constructed and differentially screened by using mRNA from the brain of both parasitized and nonparasitized snails. This screening yielded a number of clones, including previously identified cDNAs as well as novel neuronal transcripts, which appear to be differentially regulated. The majority of these transcripts encode neuropeptides. Reverse Northern blot analysis confirmed that neuropeptide gene expression is indeed affected in parasitized animals. Moreover, the expression profiles of 10 transcripts tested showed a differential, parasitic stage-specific regulation. Changes in expression could in many cases already be observed between 1.5 and 5 hr postinfection, suggesting that changes in gene expression are a direct effect of parasitosis. We suggest that direct regulation of neuropeptide gene expression is a strategy of parasites to induce physiological and behavioral changes in the host.
Keywords: growth and reproduction, differential screening of cDNA libraries, avian schistosome parasite, intermediate molluscan host
Endo-parasites have developed different strategies to manipulate vital life processes of their host. They are not only able to circumvent the attacks of the host defense system (1, 2), but at the same time also affect many physiological and behavioral processes in the host to ensure optimal conditions for their own growth and reproduction (3). Schistosome parasites of the phylum Plathyhelminthes, which include various species that cause the disease schistosomiasis (bilharzia), are examples of parasites that interfere with the life processes of both the intermediate (molluscan) and definitive (vertebrate) host (3–6). The avian schistosome Trichobilharzia ocellata and its intermediate host, the freshwater snail Lymnaea stagnalis, form a suitable experimental model system for in-depth studies of parasite-host interactions. Previous studies (7, 8) have demonstrated accelerated body growth in juvenile snails infected with T. ocellata, which is accompanied by a significantly retarded development of both the male and female reproductive systems as well as reproductive activity. It also was shown that parasitic infection of subadult snails causes a strong reduction in female reproductive activity, which becomes manifest at the onset of the production of cercariae, the parasitic stage that leaves the snail to infect the definitive host, i.e., the duck (9).
Lymnaea is a gastropod mollusk, whose nervous system has been extensively used as a model in molecular biological, cellular, neuroendocrine, and behavioral studies (e.g., ref. 10). These studies have shown that growth and metabolism are regulated by the central neuroendocrine light green cells (LGCs), which release several molluscan insulin-related peptides (MIPs) that are encoded by a small family of MIP genes (10–13). Egg-laying and accompanying behaviors are controlled by the neuroendocrine caudodorsal cells (CDCs) (10), which episodically release multiple peptides that are derived from three caudodorsal cell hormone (CDCH) precursors, which also are encoded by a small family of related, yet distinct, genes (14). Evidence that T. ocellata interferes with the activities of the LGCs and CDCs comes from electrophysiological experiments, demonstrating an effect of the peptide schistosomin, which appears in the blood during parasitation (15), on the excitability of the LGCs and CDCs (16). Thus, modifications of various life processes can be induced by alterations of the electrophysiological characteristics of neuroendocrine centers, which in turn cause alterations in the release pattern of biologically active peptides. It is unknown, however, whether parasites are able to interfere with vital life processes at other levels of control. In particular, effects of parasites on the expression of genes that are crucial in the regulation of these life processes have hardly been explored.
We now have examined the effects of T. ocellata on gene expression in the central nervous system (CNS) of L. stagnalis. We screened a broad spectrum of expressed genes and found that parasitosis induces changes in neuronal gene expression. In particular, alterations in the expression patterns of genes encoding neuropeptide precursors were observed. The early onset of many of these changes suggest that they are a direct effect of parasitosis on the host brain. We conclude that the parasite affects neuronal gene expression as part of a strategy to manipulate physiological and behavioral processes in the host for its own benefit.
MATERIALS AND METHODS
Construction and Screening of a cDNA Library of the CNS of Parasitized Snails.
Parasitized L. stagnalis were obtained by exposing laboratory-bred snails with a shell length of 8 mm to 4–6 miracidia of the parasite T. ocellata (17). Patently infected snails were used 8 weeks after infection, i.e., when they were shedding cercariae. All snails used in the experiments had a shell length of at least 23 mm, and the control snails of this length were sexually mature. A λZAPII cDNA library was constructed according to the manufacturer’s (Stratagene) instructions. In short, total RNA was extracted, according to Chomczynski and Sacchi (18). Poly(A)+ RNA was prepared, by using oligo(dT)17-linked magnetic Dynabeads (Dynal). Of this poly(A)+ RNA, 4 μg was used to synthesize double-stranded cDNA, to which linkers containing EcoRI restriction sites were ligated. This mixture was size-fractionated to discard cDNAs smaller than 400 nucleotides. The unamplified cDNA library contained 3.2 × 106 independent clones. Of the amplified λZAPII cDNA library, 50,000 individual clones were differentially screened at a density of about 10,000 pfu/400 cm2, by using nylon membranes (Boehringer Mannheim). The screening was performed with α-[32P]dATP radiolabeled cDNA probes (specific activity >1 × 108 dpm/μg), obtained by reverse transcription of oligo(dT)17 primed poly(A)+ RNA of the CNS of parasitized (plus probe) and nonparasitized (minus probe) animals. The membranes were prehybridized at 65°C for 4 hr, and hybridized at 65°C for 16 hr, in a solution of 6× SSC [1× SSC = 150 mM NaCl, 15 mM Na-citrate (pH 7.0)], containing 0.1% Ficoll 400 (wt/vol), 0.1% polyvinylpyrrolidone (wt/vol), 0.1% BSA (wt/vol), and 30 μg/ml denatured nonhomologous DNA. The membranes were washed twice in 1× SSC, 0.1% SDS at 65°C for 20 min, and autoradiographed. Individual clones, corresponding to plaques that gave differential hybridization signals with the plus probe vs. the minus probe, were isolated. The cDNA inserts of these clones were amplified by PCR, by using universal primers on the vector arms, and size-fractionated on a 1.2% agarose gel. PCR products were spotted on nylon membranes and grouped by crosshybridization with randomly labeled cDNA probes, made of PCR products of individual clones. pBluescript plasmid DNA of λZAPII clones was obtained by in vivo excision for sequence analysis.
Reverse Northern Blot Analysis.
Reverse Northern blot analysis (19) was performed by immobilization of 0.5–1 μg of pBluescript plasmid per cDNA clone on nylon membrane (Boehringer Mannheim), by using a Minifold II slotblot apparatus (Schleicher & Schuell). After blotting, plasmid DNA was denatured and neutralized by incubation of the membranes in 0.5 M NaOH/1.5 M NaCl for 3 min and in 0.5 M Tris⋅HCl, pH 7.2/1.5 M NaCl/1 mM EDTA for 3 min, respectively. Brains from 20 infected snails were dissected at 1.5 hr, 5 hr, 6 weeks, and 8 weeks postinfection, respectively, and immediately frozen on dry ice. For each of the four groups, 20 control snails of the same size and age were used. Isolation of poly(A)+ RNA, preparation of radiolabeled cDNA, and hybridization conditions were essentially the same as described above. Autoradiographs were scanned, by using a Hewlett Packard ScanJet IIcx flat-bed scanner and DeskScan IIv2.0 software of Hewlett Packard. Hybridization signal intensities were measured, by using the NIH Image 1.54 program. Data analysis was done, by using the InStat 1.12 program, and performing an unpaired t test on the measured hybridization intensities of corresponding transcripts of parasitized vs. control animals.
RESULTS
Differential Screening of the CNS-Specific cDNA Library of Parasitized Snails Reveales Parasite-Induced Alterations in Neuronal Gene Expression.
To test the hypothesis that the parasite T. ocellata changes gene expression in the CNS of its intermediate host L. stagnalis, we used a differential screening technique, involving 50,000 individual clones of a λZAP II cDNA library of the CNS of parasitized snails. The screening was performed with cDNA probes of equal specific activity, made by reverse transcription of poly(A)+ RNA isolated from the CNS of parasitized (shedding) snails and nonparasitized snails. This screening yielded 148 clones, which displayed differential hybridization signals to either probe, thus corresponding to potentially up- or down-regulated genes in the CNS. Of these 148 cDNA clones, 97 were grouped on the basis of nucleotide sequence identity by crosshybridization, which led to the characterization of 22 groups that ranged in size from 63 clones to one clone (groups A–V; Table 1). From groups consisting of more than one clone (groups A–F), the length of the cDNA inserts was estimated, and the longest cDNA was selected for sequence analysis. In addition, one clone (clone G, see Table 1) of the 16 individual clones was chosen for further characterization.
Table 1.
The 23 groups of cDNA clones characterized from 148 isolated clones representing differentially regulated transcripts
| Group | Group size | Encoded precursor | Length of cDNA length (nucleotides)* |
|---|---|---|---|
| A | 63 | 16S mt rRNA† | 1,000 |
| B | 5 | FMRFa‡ | 1,800 |
| C | 4 | Pedal peptide§ | 1,750 |
| D | 4 | MDM¶ | 1,900 |
| E | 3 | LyNPY§ | 950 |
| F | 2 | Cytochrome c† | 1,500 |
| G | 1 | −LFRFa§ | 1,400 |
| H–V | 15 i.c. | n.i. | 800–2,500 |
| W | 51 n.g. | n.i. | 800–3,500 |
n.i., not identified. i.c. individual clones. n.g., not grouped. 16S mt rRNA, mitochondrial rRNA. LyNPY, Lymnaea NPY. cDNAs encoding neuropeptides are in italics.
Length of longest cDNA insert in the group as estimated by PCR (see Results).
Mitochondrial products.
See ref. 20.
Unpublished data.
See ref. 21.
In total, four cDNAs were completely sequenced and three cDNAs were partially sequenced. Because the full-length sequence data was not relevant for the conclusions of our experiment, only partial sequence information of the encoded proteins is given in Fig. 1; the details of the sequence analyses have been published (21) or concern as-yet-unpublished data. The longest representative cDNAs of group A and F were partially sequenced. On the basis of sequence identity analysis, by using a database search (22), they were shown to encode the Lymnaea homologs of 16S mitochondrial rRNA and the electron carrier cytochrome c, respectively (data not shown). The partial sequence of a representative cDNA of group B was identical to the previously characterized Phe-Met-Arg-Phe-amide (FMRFa) transcript (20), which encodes nine copies of FMRFa together with several related neuropeptides. The representative cDNA of group C encodes a precursor protein, containing 16 copies of a Lymnaea neuropeptide homologous to pedal peptide of Aplysia (23) (Fig. 1A). The representative cDNA of group D encodes a protein, called molluscan defense molecule (MDM) (21) (Fig. 1B), which has a similar overall organization as hemolin, an insect immunoprotein belonging to the Ig superfamily (24). The Lymnaea prohormone encoded by the representative cDNA of group E is homologous to the previously identified human neuropeptide Y (NPY) precursor (25), and, therefore, called LyNPY (Lymnaea NPY) (Fig. 1C). The sequence information of clone G predicts a prohormone, encompassing a set of novel neuropeptides containing the C-terminal sequence −Arg-Phe-amide, and, therefore, belongs to the superfamily of FMRFa-related neuropeptides (27) (Fig. 1D).
Figure 1.
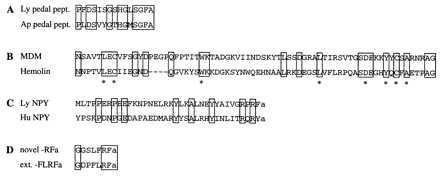
Amino acid sequence alignments of Lymnaea partial proteins and peptides, as predicted from selected cDNAs isolated in the differential screening, with their structural counterparts from other species. Identical amino acid residues are boxed. (A) Lymnaea pedal peptide (Ly pedal pept.) compared with its counterpart in the mollusk Aplysia californica (Ap pedal pept.; ref. 23). (B) A C2-like Ig-like domain of Lymnaea MDM (21) is aligned to that of hemolin of the insect Hyalophora cecropia (24). ∗ indicate residues that are conserved in all C2-like Ig domains. (C) Lymnaea NPY (Ly NPY) compared with human NPY (Hu NPY) (25). (D) Amino acid sequence of a member of a novel class of Lymnaea −Arg-Phe-amide (RFa) neuropeptides compared with one of the extended forms of Lymnaea-Phe-Leu-Arg-Phe-amide (FLRFa) (26). Notice the difference in the N-terminal sequence of the novel −RFa, i.e., LFRFa, instead of FLRFa in the FMRFa family of neuropeptides.
Reverse Northern Blot Analysis of Gene Expression During Parasitosis.
To analyze the parasite-induced changes in gene expression in the brain of Lymnaea in more detail, a reverse Northern blot analysis was performed for 10 transcripts. Six of these transcripts [cytochrome c, MDM, FMRFa, pedal peptide, Leu-Phe-Arg-Phe-amide (LFRFa), and LyNPY] were isolated in the differential screening procedure, described above. Four transcripts (conopressin, CDCH-I, MIP-III, and cytochrome P450) were previously characterized (see Discussion). Transcript levels were measured in the CNS of parasitized animals at 1.5 hr, 5 hr, 6 weeks, and 8 weeks postinfection, and in nonparasitized animals of the same size and age. In Fig. 2, the amount of up- and down-regulation of the various transcripts is expressed as the percentage of control values in nonparasitized animals. Furthermore, to correct for unequal labeling of cDNA probes, these values were divided by the relative expression levels of Lymnaea cytochrome P450, which remained stable during parasitosis.
Figure 2.
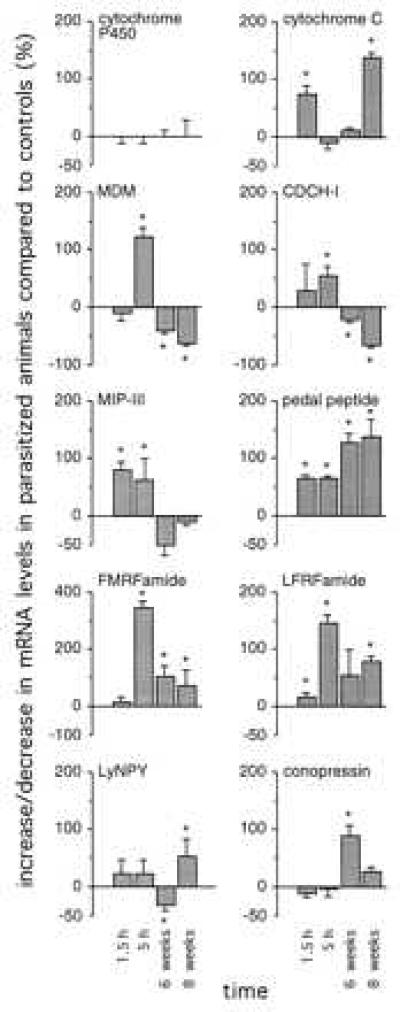
Parasite-induced changes in gene expression in the Lymnaea CNS. The percentage change in transcript levels in parasitized animals at 1.5 hr, 5 hr, 6 weeks, and 8 weeks postinfection was measured by reverse Northern blot analysis, compared with transcript levels in control animals of the same size and age (mean ± SD; n = 3). Changes in cytochrome P450 levels are set at 0% to correct for unequal labeling of cDNA probes. * indicate significant changes (P < 0.05 unpaired t test).
The reverse Northern blot analysis shows a differential regulation during parasitosis of all transcripts tested (Fig. 2). The RNA levels of many transcripts were increased already early during parasitosis (between 1.5 and 5 hr postinfection), i.e., CDCH-I (55 ± 16%; mean ± SD, n = 3), MIP-III (80 ± 17%, n = 3), cytochrome c (75 ± 15%, n = 3), MDM (122 ± 16%, n = 3), FMRFa (343 ± 20%, n = 3), pedal peptide (64 ± 5%, n = 3), and LFRFa (144 ± 15%, n = 3). Most of these transcripts had highest expression levels at 5 hr postinfection, except for cytochrome c, which peaked at 1.5 hr postinfection and returned to normal levels between 1.5 and 5 hr postinfection. Interestingly, cytochrome c levels were increased again at 8 weeks postinfection (140 ± 8%, n = 3), when snail were shedding. Some transcripts returned to normal expression levels again at 6–8 weeks postinfection, i.e., MIP-III, or became down-regulated, i.e., CDCH-I and MDM. Others remained up-regulated throughout parasitosis, i.e., FMRFa, pedal peptide, and LFRFa. Conopressin and LyNPY levels were increased (88 ± 18% and 52 ± 30%, respectively, n = 3) only late during parasitosis, at 6 and 8 weeks postinfection, respectively.
DISCUSSION
The primary objective of this study was to examine the possibility of a differential effect of parasitosis on the expression of genes in the CNS of the intermediate host. Here, we present arguments that lead to the conclusion that parasites indeed cause a differential expression pattern of genes, in particular of genes encoding neuropeptide precursors that are involved in the regulation of vital physiological and behavioral processes. This finding may have important implications for theories and models of parasite-host interactions.
We performed two independent experiments: (i) an open differential screening for genes encoding, in principle, both known and unknown proteins, with various functions, and (ii) a reverse Northern blot analysis, focusing on identified genes, encoding primarily neuropeptide precursors. Although the differential screening strategy of the cDNA library of the CNS of parasitized snails could, in principle, detect a large range of genes with altered expression, there are a few limitations. First, the use of 50,000 clones in the screening taken from a library of 3.2 × 106 independent clones reliably covers only the high abundant clones, because in this random sample a confidence limit of >95% occurs only for cDNAs with a frequency of >1 in 17,000. Second, because the screening was performed with a library constructed of mRNA from shedding snails that had been infected for 8 weeks, the cDNA library reflects both long-term changes in gene expression, induced by the prolonged actions of the parasite, and short-term changes, induced by the acute production of large numbers of cercariae, that takes place from approximately week 8 postinfection onward. Effects on gene expression in the CNS of the host that are caused by earlier stages of the life cycle of the parasite, i.e., miracidia and primary sporocysts, very likely cannot be detected by using the present cDNA library. Finally, because the frequency of occurrence of transcripts differs from 1:400 (cytochrome P450) (28) to as little as 1:4 × 106 (conopressin-receptor) (29), highly abundant transcripts might be isolated as false positives (i.e., clones that are picked because of high abundancy rather than parasite-induced changes in abundancy). However, assuming a highly abundant nonregulated cDNA, which occurs in the library with a frequency of 1 in 400, the chance to pick a group larger than 1 in a random sample of 148 of 50,000 (the group size of the differential screening) is less than 5%. Because the frequency of occurrence of neuropeptide transcripts is in the range of 1:1,000 to 1:10,000, this chance is even smaller, e.g., 1% for cDNAs occurring 1:1,000. Thus, the isolation of six groups of cDNAs (sizes ranging from two to 63 clones) indicates true, rather than random, selection.
Interestingly, four of seven groups of differentially regulated mRNAs contain sequences of transcripts encoding neuropeptides. Of the other three groups, we consider the 16S mitochondrial rRNA to be the result of artificial differential labeling in the two cDNA probes because of the lack of a poly(A) tail. This result was repeatedly observed during our reverse Northern blot experiments (data not shown). Thus, our findings suggest that primarily the expression of neuropeptide genes is altered during parasitosis. This finding could be substantiated by quantifying differences in gene expression, by using reverse Northern blot analysis with the six cDNA clones identified from the differential screening and four identified additional cDNA clones, of which three encode neuropeptide precursors involved in the regulation of reproduction, i.e., CDCH-I (14) and conopressin (30), and growth and metabolism, i.e., MIP-III (13). Cytochrome P450 was included, because it remained unchanged throughout parasitation. In all experiments, its expression level was set to 100%, and all other changes in expression were adjusted accordingly. Thus, differences in hybridization intensities because of unequal labeling of the cDNA probes did not influence the outcome of the measurements. As expected, the six genes that were identified as differentially regulated genes in the screening, also showed up- or down-regulation in the reverse Northern blot analysis. In addition, the expression levels of conopressin, CDCH-I, and MIP-III also were affected.
Together, the results of the differential screening and the reverse Northern blot analysis show that parasitosis caused specific changes in the expression of several classes of genes, encoding proteins of different functions, i.e., neuropeptide precursors, a mitochondrial enzyme (cytochrome c), and an Ig-like protein (MDM). In time, many differences in the regulation of expression were observed between the various transcripts. Many transcripts were up-regulated early during parasitosis. Some of these transcripts returned to normal levels at 6–8 weeks postinfection (MIP-III) or became down-regulated (CDCH-I and MDM). FMRFa, pedal peptide, and LFRFa remained up-regulated throughout parasitation, and both conopressin and LyNPY were up-regulated only late during parasitation. Interestingly, cytochrome c expression levels peak both early and late during parasitation, immediately on infection, and again when snails were shedding, suggesting alterations in mitochondrial gene expression and cellular metabolic activity, at specified stages of parasitation.
The observation that the transcript encoding the CDCH-I precursor was down-regulated from 6 weeks postinfection onward is in agreement with the inhibition of egg laying and accompanying behavior in parasitized snails (3). In that respect, it is interesting to note that this down-regulation did not occur during the first hours of parasitation. This result makes sense, because at this stage of infection snails were juvenile and not yet sexually active. The expression of the MIP-III gene was up-regulated early during parasitation, which is in agreement with the observation that parasitized snails have an abnormal metabolism (8). Interestingly, MIP-III levels returned to normal again at 6–8 weeks postinfection. At this stage, infected snails start to display abberrant patterns of body growth (8). Possibly, these changes are mediated not by MIP-III, but by any of the other four MIP genes, which have not been studied in the present experiments. The continuing abnormalities in metabolism that are observed also may be mediated by other neuropeptides, such as NPY, which was specifically up-regulated at 8 weeks postinfection, and in vertebrates is involved in the regulation of metabolic processes (31). In parasitized snails, the expression of the gene encoding MDM was up-regulated specifically at 5 hr postinfection, but down-regulated at later stages of parasitation. MDM is expressed in the granular cells that are located in the connective tissue of the brain and other organs, and may play a role in the internal defense system of Lymnaea (21). Thus, early up-regulation of MDM probably reflects an immune response of the snail on infection. Subsequent down-regulation might reflect mechanisms of the parasite to repress the immune response of its host to survive. An initial enhancement of the internal defense system on parasitation, followed by prolonged repression, have been described before (32, 33). Immunosuppressive cellular effects also were reported in the intermediate and definitive hosts of Schistosoma mansoni (i.e., the freshwater snail Biomphalaria glabrata and humans, respectively), through the release of proopiomelanocortin immunoreactive peptides (2).
A considerable amount of research on parasite-host interactions is devoted to the mechanisms parasites have developed to circumvent their elimination by the defense system of the host. To date, however, little is known about how parasites interfere with other regulatory systems in the host to ensure survival and proper development. In insects, parasite-induced effects on neuropeptide expression also may play a crucial role in this respect (34, 35). Here, we demonstrate that parasites are able to adjust vital brain functions through interference at the level of gene expression in the brain, with neuropeptide genes as the prime targets. Because many genes were affected already shortly after infection (1.5–5 hr) strongly suggests that the parasite directly induced the changes that were observed. Furthermore, the differences during parasitation in the expression profiles of the transcripts tested, suggest that the parasite constantly and dynamically adjusts gene expression in the host brain in a parasitic stage-specific manner. We consider it unlikely that the changes are stress-related, because parasitation was achieved by addition of a minimal amount (4–6) miracidia per animal. Moreover, except for not being infected, control animals were handled in the same way as infected animals. Our findings add a dimension to the study of parasite-host interactions and open areas of research. For example, similar data on the interactions of parasites with the brain of vertebrate hosts, including humans, might prove to be of fundamental and practical significance (36).
Acknowledgments
We thank Mr. C. Populier for the maintenance of the snail culture, Mr. R. Van Elk and Ms. M.J.M. Bergamin-Sassen for technical assistance, and Ms. T. Laan for secretarial assistance. This work was supported by the Foundation for Life Sciences (SLW; Grant 805-29-202), which is subsidized by the Netherlands Organization for Scientific Research (NWO).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: CDC, caudodorsal cell; CNS, central nervous system; CDCH, caudodorsal cell hormone; FMRFa, Phe-Met-Arg-Phe-amide; LFRFa, Leu-Phe-Arg-Phe-amide; MDM, molluscan defense molecule; MIP, molluscan insulin-related peptide; NPY, neuropeptide Y.
References
- 1.Maizels R M, Bundy D A P, Selkirk M E, Smith D F, Anderson R M. Nature (London) 1993;365:797–805. doi: 10.1038/365797a0. [DOI] [PubMed] [Google Scholar]
- 2.Duvaux-Miret O, Stefano G B, Smith E M, Dissous G, Capron A. Proc Natl Acad Sci USA. 1992;89:778–781. doi: 10.1073/pnas.89.2.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Jong-Brink M. Adv Neuroimmunol. 1992;2:199–233. [Google Scholar]
- 4.Beckage N E. Receptor. 1993;3:233–245. [PubMed] [Google Scholar]
- 5.Thompson, S. N. & Kavaliers, M. (1994) Parasitology 109, Suppl., S119–S138. [DOI] [PubMed]
- 6.Isseroff H, Sylvester P W, Bessette C L, Jones P L, Fisher W G, Rynkowski T, Gregor K T. Comp Biochem Physiol. 1989;94:41–45. doi: 10.1016/0300-9629(89)90781-0. [DOI] [PubMed] [Google Scholar]
- 7.Sluiters J F. Z Parasitenkd. 1981;64:303–319. doi: 10.1007/BF00927378. [DOI] [PubMed] [Google Scholar]
- 8.Joosse J, Van Elk R. Exp Parasitol. 1986;62:1–13. doi: 10.1016/0014-4894(86)90002-0. [DOI] [PubMed] [Google Scholar]
- 9.Schallig H D F H, Sassen M J M, Hordijk P L, de Jong-Brink M. Parasitology. 1991;102:85–91. doi: 10.1017/s0031182000060376. [DOI] [PubMed] [Google Scholar]
- 10.Geraerts W P M, Smit A B, Li K W, Vreugdenhil E, Van Heerikhuizen H. In: Current Aspects of the Neurosciences. Osborne N N, editor. London: MacMillan; 1991. pp. 255–304. [Google Scholar]
- 11.Smit A B, Vreugdenhil E, Ebberink R H M, Geraerts W P M, Klootwijk J, Joosse J. Nature (London) 1988;331:535–538. doi: 10.1038/331535a0. [DOI] [PubMed] [Google Scholar]
- 12.Geraerts W P M. Gen Comp Endocrinol. 1992;86:433–444. doi: 10.1016/0016-6480(92)90068-u. [DOI] [PubMed] [Google Scholar]
- 13.Smit A B, Van Marle A, Van Elk R, Bogerd J, Van Heerikhuizen H, Geraerts W P M. J Mol Endocrinol. 1993;11:103–113. doi: 10.1677/jme.0.0110103. [DOI] [PubMed] [Google Scholar]
- 14.Vreugdenhil E, Jackson J F, Bouwmeester T, Smit A B, Van Minnen J, Van Heerikhuizen H, Klootwijk J, Joosse J. J Neurosci. 1988;8:4184–4191. doi: 10.1523/JNEUROSCI.08-11-04184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hordijk P L, Schallig H D F H, Ebberink R H M, De Jong-Brink M, Joosse J. Biochem J. 1991;279:837–842. doi: 10.1042/bj2790837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hordijk P L, De Jong-Brink M, Ter Maat A, Pieneman A W, Lodder J C, Kits K S. Neurosci Lett. 1992;136:193–197. doi: 10.1016/0304-3940(92)90047-b. [DOI] [PubMed] [Google Scholar]
- 17.Sluiters J F, Brussaard-Wust C M, Meuleman E A. Z Parasitenkd. 1980;70:13–26. doi: 10.1007/BF00927722. [DOI] [PubMed] [Google Scholar]
- 18.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 19.Mackler S A, Eberwine J H. Proc Natl Acad Sci USA. 1994;91:385–389. doi: 10.1073/pnas.91.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linacre A, Kellet E, Saunders S, Bright K, Benjamin P R, Burke J F. J Neurosci. 1990;10:412–419. doi: 10.1523/JNEUROSCI.10-02-00412.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoek R M, Smit A B, Frings H, Vink J M, De Jong-Brink M, Geraerts W P M. Eur J Immunol. 1996;26:939–944. doi: 10.1002/eji.1830260433. [DOI] [PubMed] [Google Scholar]
- 22.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lloyd P E, Connolly C M. J Neurosci. 1989;9:312–317. doi: 10.1523/JNEUROSCI.09-01-00312.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun S-C, Lindstöm L, Boman H G, Faye I, Schmidt O. Science. 1990;250:1729–1732. doi: 10.1126/science.2270488. [DOI] [PubMed] [Google Scholar]
- 25.Tatemoto K, Carlquist M, Mutt V. Nature (London) 1982;296:659–662. doi: 10.1038/296659a0. [DOI] [PubMed] [Google Scholar]
- 26.Saunders S E, Bright K, Kellet E, Benjamin P R, Burke J F. J Neurosci. 1991;11:740–745. doi: 10.1523/JNEUROSCI.11-03-00740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg M J, Payza K, Nachman R J, Holman G M, Price D A. Peptides. 1988;9:125–135. doi: 10.1016/0196-9781(88)90236-7. [DOI] [PubMed] [Google Scholar]
- 28.Teunissen Y, Geraerts W P M, Van Heerikhuizen H, Planta R J, Joosse J. J Biochem. 1992;112:249–252. doi: 10.1093/oxfordjournals.jbchem.a123885. [DOI] [PubMed] [Google Scholar]
- 29.Van Kesteren R E, Tensen C P, Smit A B, Van Minnen J, Van Soest P F, Kits K S, Meyerhof W, Richter D, Van Heerikhuizen H, Geraerts W P M. Neuron. 1995;15:897–908. doi: 10.1016/0896-6273(95)90180-9. [DOI] [PubMed] [Google Scholar]
- 30.Van Kesteren R E, Smit A B, De Lange R P J, Kits K S, Van Golen F A, Van Der Schors R C, De With N D, Burke J F, Geraerts W P M. J Neurosci. 1995;15:5989–5998. doi: 10.1523/JNEUROSCI.15-09-05989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leibowitz S F. Brain Res Bull. 1991;27:333–337. doi: 10.1016/0361-9230(91)90121-y. [DOI] [PubMed] [Google Scholar]
- 32.Amen R J, Baggen J M C, Bezemer P D, De Jong-Brink M. Parasitology. 1992;104:33–40. doi: 10.1017/s0031182000060777. [DOI] [PubMed] [Google Scholar]
- 33.Núñez P E, Adema C M, De Jong-Brink M. Parasitology. 1994;109:299–310. doi: 10.1017/s0031182000078331. [DOI] [PubMed] [Google Scholar]
- 34.Zitnan D, Kingan T G, Kramer S J, Beckage N E. J Comp Neurobiol. 1995;356:83–100. doi: 10.1002/cne.903560106. [DOI] [PubMed] [Google Scholar]
- 35.Zitnan D, Kingan T G, Beckage N E. Insect Biochem Mol Biol. 1995;25:669–678. doi: 10.1016/0965-1748(95)00006-h. [DOI] [PubMed] [Google Scholar]
- 36.De Jong-Brink M. Adv Parasit. 1995;35:178–256. doi: 10.1016/s0065-308x(08)60072-x. [DOI] [PubMed] [Google Scholar]


