Abstract
Background
The genus Aquilegia is an emerging model system in plant evolutionary biology predominantly because of its wide variation in floral traits and associated floral ecology. The anatomy of the Aquilegia flower is also very distinct. There are two whorls of petaloid organs, the outer whorl of sepals and the second whorl of petals that form nectar spurs, as well as a recently evolved fifth whorl of staminodia inserted between stamens and carpels.
Methodology/Principal Findings
We designed an oligonucleotide microarray based on EST sequences from a mixed tissue, normalized cDNA library of an A. formosa x A. pubescens F2 population representing 17,246 unigenes. We then used this array to analyze floral gene expression in late pre-anthesis stage floral organs from a natural A. formosa population. In particular, we tested for gene expression patterns specific to each floral whorl and to combinations of whorls that correspond to traditional and modified ABC model groupings. Similar analyses were performed on gene expression data of Arabidopsis thaliana whorls previously obtained using the Ath1 gene chips (data available through The Arabidopsis Information Resource).
Conclusions/Significance
Our comparative gene expression analyses suggest that 1) petaloid sepals and petals of A. formosa share gene expression patterns more than either have organ-specific patterns, 2) petals of A. formosa and A. thaliana may be independently derived, 3) staminodia express B and C genes similar to stamens but the staminodium genetic program has also converged on aspects of the carpel program and 4) staminodia have unique up-regulation of regulatory genes and genes that have been implicated with defense against microbial infection and herbivory. Our study also highlights the value of comparative gene expression profiling and the Aquilegia microarray in particular for the study of floral evolution and ecology.
Introduction
Flowers intrigue us because of their great diversity of form, colour and smell. This diversity is largely thought to be the result of co-evolution between flowering plants and pollinators, which dates to the Cretaceous when flowering plants first arose [1]. A key aspect of understanding the evolution of floral diversity requires the identification of the underlying genes. For one aspect of floral form, the identity of floral organs, the ABC model has been developed. It states that combinations of three classes of regulatory genes specify the development of sepals (A genes), petals (A + B genes), stamens (B+C genes) and carpels (C genes) [2]. It has been suggested that, once evolved, these regulatory genes could be recruited to other organs and transform them into new floral whorls. For example, B genes are expressed throughout the sterile whorls of monocots and many magnoliid dicots [3], [4], [5] and, as predicted by the ABC model, the entire perianths of these taxa have similar appearances as opposed to clearly distinct sepals and petals. Thus broad expression of B genes in perianth organs has been inferred to be ancestral in flowering plants whereas restriction of B gene expression to an inner whorl of petals in Arabidopsis and other eudicots is considered to be derived [5]. The differential presence of petals is thought to have been driven by the deployment of B gene expression to different positions in the flower after petal identity initially evolved [6], although others have suggested that petals truly evolved multiple times but recruited similar genes to control their development [7].
Many studies that have sought to relate variation in the number and appearance of floral whorls to modifications of the ABC model have examined expression patterns of ABC genes themselves. Recently, expression studies have expanded to include the genes and pathways that the ABC genes regulate both directly and indirectly [8]. Previously such wider analyses of floral gene expression were limited to the eudicot model plant A. thaliana [9], [10]. However, the development of microarrays for emerging model plants has enabled global studies elsewhere in the angiosperm tree, e.g., in the eudicot Gerbera hybrida [11], [12] and, most recently, the basal angiosperm Persea americana [8]. Using these global approaches, it is possible to compare whorl-specific expression patterns with co-expression in the traditionally defined A (sepals+petals), B (petals+stamens) and C (stamens and carpels) domains for many genes and contrast these patterns across different angiosperm lineages.
Apart from drawing attention to many genes simultaneously, global studies of gene expression also provide data that allow novel predictions of biological function. In the context of floral gene expression, this means that not only can expression patterns inform us about genes that are potentially involved in or are markers of the formation of floral organs, but they may also help formulate hypotheses regarding the specific functions of these organs. Such predictions are achieved through gene ontology and gene set enrichment analyses [13]. Expression data are tested for differentially regulated gene sets, which are defined a priori. Gene sets can be based on ontological terms of biological function, molecular function and cellular compartmentalization (www.geneontology.org). Thus the expression patterns for genes likely to underlie floral traits such as colour, scent, defense, nectar production, cell shape and cell size, micro- and macrosporogenesis can be compared within and between angiosperm lineages and provide markers for possible common attributes.
The genus Aquilegia is a member of the Ranunculales, which is phylogenetically positioned as the first diverging branch of the eudicot clade (∼125 mya, [14]). The genus has undergone an adaptive radiation over the last two million years in North America into species that are primarily bee, hummingbird or hawkmoth pollinated and have corresponding morphological floral syndromes [15]. This floral diversity predisposes the genus as a model system for the investigation of pollinator-driven speciation [16]. The anatomy of the Aquilegia flower is also very distinct from many other angiosperm flowers. It has a bipartite perianth with petaloid sepals and petals that possess nectar-producing spurs, followed by four to seven whorls of stamens, one whorl of staminodia and one whorl of free carpels (Figure 1). Spurs and staminodia evolved only recently [17], [18] and while spurs are a modification of petals and produce a nectar reward for pollinators, the underlying developmental program and any specific function of staminodia are still a matter of debate. The sepals of Aquilegia are petaloid owing to their bright coloration and papillated epidermal cells [19]. However, it is not clear to what degree similar organ identity programs are operating in petals and petaloid sepals [19], [20]. Given these morphological features, the Aquilegia flower represents a particularly interesting case to study of the genetic basis of a) petaloidy, b) spur evolution and c) the recent evolution of a novel floral organ, the staminodia [20].
Figure 1. A. formosa pre-anthesis flower and fruit development.
A A. formosa pre-anthesis flower. B Left: A. formosa pre-anthesis flower with stamens removed to expose staminodia. Middle and right: Early and later stages of fruit development, respectively. The sepals, petals and stamens dehisce while the staminodia remain attached to the receptacle and surround the carpels during fruit development.
Growing interest from both the fields of floral genetics and adaptive radiation has prompted the development of a wealth of molecular resources for Aquilegia in the past years including a complete genome sequence [16]. Based on a normalized EST library generated from various tissues of an A. formosa x A. pubescens F2 population [20], a single channel oliognucleotide microarray platform representing more than 17,000 Aquilegia unigenes has been developed. Here we introduce this array and use it to obtain expression profiles from the five floral whorls of wild A. formosa flowers. In particular, we address questions such as: How distinct are the gene expression profiles of petals and petaloid sepals? Do petaloid sepals share expression patterns with petals and stamens (classic B-class organs)? To what extend do staminodia co-express genes with petals, stamens and carpels? Does the staminodia-specific gene expression profile suggest a possible ecological function to this novel organ? Can we identify candidate genes for the identity program for staminodia? To answer these questions, we investigate gene expression in individual whorls and groups of whorls in A. formosa and contrast our findings with those of a similar analysis on a publically available data set on the four floral whorls of A. thaliana. We then apply gene ontology analyses to identify biological processes operating in each whorl.
Results
Whorl-specific gene expression in A. formosa pre-anthesis flowers
Linear model analysis
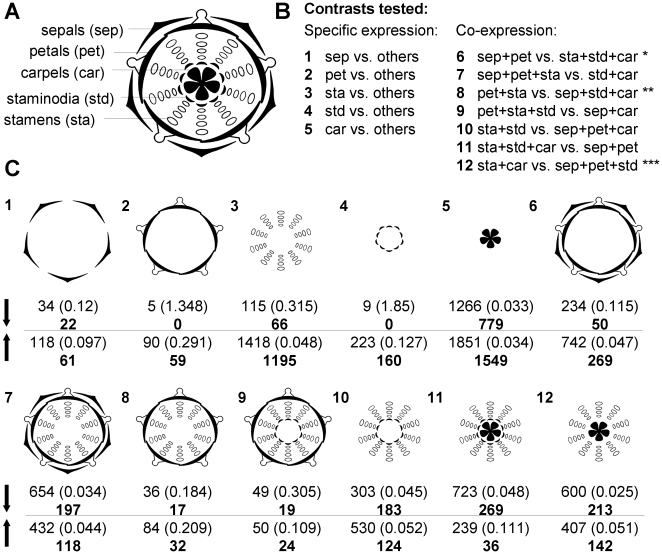
We analysed gene expression in the five floral whorls of Aquilegia formosa late stage pre-anthesis flowers by fitting a linear model to expression data obtained with Aquilegia oligonucleotide arrays. Late stage pre-anthesis flowers are defined as the flower bud has opened, stamens have started to unfurl but anthers have not begun to dehisce [21]. Twelve different models were fit for each gene using different groupings of floral whorls, henceforth called contrasts. First, we tested the extent of whorl-specific expression (contrasts 1–5, Figure 2). Second, we tested for co-expression in sepals and petals, petals and stamens, and stamens and carpels (i.e., for genes expressed in the traditionally defined A, B and C domain, respectively; contrasts 6, 8, 12, Figure 2). Third, we tested groupings pertaining to the specific anatomy of the Aquilegia flower. Particularly, with contrast 7 we tested if the B domain is extended to petaloid sepals; with contrasts 9 and 11 we tested if the B and C domains are extended to staminodia and with contrasts 10 and 7 we tested the extent to which stamens and staminodia or carpels and staminodia were similar in gene expression, respectively. Numbers of up- and down-regulated genes for each contrast are summarized in Figure 2C. We also analysed whorl- and domain-specific gene expression patterns in two publically available datasets of Arabidopsis thaliana. These data are comprised of triplicate measurements of global gene expression in pre- and post-anthesis A. thaliana flowers (stage 12 and 15, [22]) obtained with Affymetrix Ath1 microarrays. The numbers of up- and down-regulated genes for seven A. thaliana contrasts examined in both floral stages and, for comparison, results for the corresponding seven A. formosa contrasts, are given in Figure S1. Lists of differentially expressed genes in A. formosa and A. thaliana can be found in Tables S2, S3, S4. In the following we describe expression patterns referring to specifically expressed genes as defined in Figure 2C.
Figure 2. Differentially expressed genes in Aquilegia floral whorls and whorl combinations.
A Floral diagram of an Aquilegia flower showing one whorl of five petaloid sepals, one whorl of five petals, four whorls of 10 stamens, one whorl of 10 staminodia and one whorl of five carpels. B Twelve contrasts were tested for differential expression, comparing each whorl against all others (1–5) and combinations of whorls against the remaining whorls (6–12). See text for details. C Numbers of down (downward arrow) and up (upward arrow) regulated genes for each of the 12 contrasts. First line shows number of differentially expressed genes with the corresponding permutation-based false discovery rate in brackets. Bold numbers below state the number of genes that have their highest absolute D statistic under that contrast and are being considered specifically expressed in the context of this study. Note that contrast 11 is reverse to contrast 6 and therefore yields similar numbers but with opposite regulation patterns.
During pre-anthesis in A. formosa, the majority of organ-specific gene expression was found in stamens and carpels whereas specific gene expression in sepals, petals and staminodia was comparably small (Figure 2C, Figure S1). In contrast, in A. thaliana pre-anthesis flowers (stage 12), stamens exhibited the largest extent of organ-specific gene expression followed by sepals, carpels and petals (Figure S1).
When considering sepals and petals combined, A. formosa had more genes specifically co-regulated (319) as compared to organ-specific gene expression (83 and 59 in sepals and petals respectively, Figure 2C). There were over four times as many genes co-up-regulated than specifically up-regulated in either of these organs (Figure 2C). In contrast, A. thaliana had fewer genes co-regulated (188 total) than those with organ-specific expression patterns (854 and 240 in sepals and petals respectively, Figure S1). These data reflect the petaloid nature of sepals in A. formosa compared to their distinct nature in A. thaliana. As expected due to the similar coloured nature of sepals and petals in A. formosa, homologues of four major genes of the anthocyanin biosynthetic pathway, namely chalcone synthase (TC14734), flavanone-3-hydroxylase (TC8210), dihydroflavonol-4-reductase (TC9974) and anthocyanidin synthase (TC18571), were all significantly co-up-regulated (Table 1) corroborating earlier findings [21]. Other genes co-regulated in sepals and petals may reflect other aspects of ‘petaloidy’.
Table 1. Expression statistics for a selected set of genes.
| D-statistics | Log intensities | |||||||||||||||||||||||
| B+ | B+ | STA+ | C+ | SEP | PET | STA | STD | CAR | ||||||||||||||||
| SEQ_ID | Annotation | SEP | PET | STA | STD | CAR | A | B | C | SEP | STD | STD | STD | # probes | m | se | m | se | m | se | m | se | m | se |
| Anthocyanin Biosynthetic Pathway | ||||||||||||||||||||||||
| TC16794 | Chalcone synthase | 4.3 | 13.1 | −3.9 | 0.7 | −12.4 | 14.8 | 4.5 | −17.5 | 6.4 | 5.6 | −1.7 | −13.7 | 24 | 7.7 | 0.1 | 8.5 | 0.2 | 6.7 | 0.1 | 7.3 | 0.1 | 5.9 | 0.0 |
| TC14734 | Chalcone synthase | 9.1 | 5.3 | −1.1 | −9.7 | −4.2 | 13.8 | 3.2 | −2.0 | 10.3 | −3.0 | −6.3 | −12.9 | 25 | 10.0 | 0.2 | 9.7 | 0.2 | 9.2 | 0.2 | 8.5 | 0.2 | 8.9 | 0.2 |
| TC9292 | Chalcone isomerase | 5.2 | 4.2 | −16.3 | 0.5 | 0.4 | 7.9 | −4.6 | −9.0 | −0.3 | −5.1 | −8.6 | −5.5 | 19 | 9.3 | 0.2 | 9.2 | 0.2 | 7.7 | 0.2 | 8.8 | 0.2 | 8.8 | 0.3 |
| TC8210 | Flavanone-3-hydroxylase | 11.7 | 5.9 | −8.3 | −3.6 | −4.7 | 20.0 | −0.9 | −10.7 | 6.8 | −3.7 | −7.8 | −20.1 | 24 | 10.3 | 0.1 | 9.9 | 0.2 | 8.4 | 0.2 | 8.8 | 0.2 | 8.7 | 0.2 |
| TC9974 | Dihydroflavonol-4-reductase | 12.8 | 9.1 | −14.7 | −2.7 | −4.9 | 29.1 | −2.0 | −18.4 | 6.6 | −4.0 | −13.3 | −19.6 | 25 | 9.4 | 0.1 | 9.1 | 0.2 | 5.8 | 0.1 | 7.2 | 0.2 | 6.9 | 0.2 |
| TC18571 | Anthocyanidin synthase | 13.4 | 9.3 | −11.2 | −1.1 | −7.7 | 26.7 | −1.7 | −20.2 | 7.5 | −1.7 | −10.0 | −32.2 | 24 | 10.3 | 0.1 | 9.8 | 0.1 | 6.4 | 0.1 | 8.1 | 0.1 | 6.9 | 0.1 |
| B Genes | ||||||||||||||||||||||||
| TC17477 | PISTILLATA-like protein | 1.5 | 9.5 | 1.6 | 0.8 | −19.6 | 8.3 | 8.8 | −7.0 | 9.8 | 9.3 | 2.1 | −6.6 | 21 | 7.8 | 0.3 | 8.8 | 0.3 | 7.9 | 0.3 | 7.7 | 0.3 | 5.8 | 0.1 |
| TC11683 | PISTILLATA-like protein | 1.4 | 8.8 | 1.6 | 0.7 | −16.1 | 5.8 | 8.0 | −9.5 | 7.8 | 7.4 | 1.2 | −7.1 | 22 | 7.6 | 0.3 | 8.5 | 0.3 | 7.6 | 0.3 | 7.5 | 0.3 | 5.8 | 0.1 |
| TC11684 | PISTILLATA-like protein | 1.3 | 8.9 | 1.1 | 1.0 | −13.9 | 7.5 | 7.4 | −7.6 | 8.4 | 8.6 | 1.7 | −6.7 | 23 | 7.5 | 0.3 | 8.3 | 0.4 | 7.5 | 0.3 | 7.4 | 0.3 | 5.8 | 0.1 |
| TC19085 | APETALA3-3 | −4.5 | 19.7 | 1.2 | −3.0 | −5.1 | 6.0 | 13.3 | −3.4 | 7.4 | 9.3 | −1.3 | −6.6 | 14 | 5.7 | 0.1 | 8.9 | 0.2 | 6.8 | 0.1 | 6.0 | 0.1 | 5.5 | 0.0 |
| TC19725 | APETALA3-2 | −5.6 | 4.4 | 6.1 | 5.1 | −12.5 | −0.7 | 9.6 | −3.2 | 3.8 | 30.0 | 8.3 | 0.7 | 15 | 6.6 | 0.1 | 8.7 | 0.2 | 8.8 | 0.2 | 8.6 | 0.2 | 5.8 | 0.1 |
| TC16289 | APETALA3-1 | 1.1 | −3.5 | 2.3 | 10.3 | −5.9 | −1.6 | −0.6 | −5.2 | 0.2 | 6.3 | 9.8 | 1.6 | 19 | 7.4 | 0.2 | 6.8 | 0.2 | 7.6 | 0.3 | 8.4 | 0.3 | 6.0 | 0.1 |
| C gene | ||||||||||||||||||||||||
| TC8667 | AGAMOUS-like protein | −8.7 | −10.8 | 4.4 | 5.7 | 6.6 | −26.2 | −2.7 | 9.8 | −9.2 | 1.2 | 7.9 | 17.8 | 23 | 6.3 | 0.2 | 6.1 | 0.2 | 9.0 | 0.2 | 9.0 | 0.3 | 9.2 | 0.3 |
| Transcription factors specifically expressed in staminodia | ||||||||||||||||||||||||
| TC13707 | Myb | −4.9 | −3.7 | −4.8 | 27.9 | −1.3 | −6.0 | −5.6 | −4.3 | −12.3 | 4.4 | 8.6 | 7.5 | 24 | 5.9 | 0.1 | 6.0 | 0.1 | 5.9 | 0.0 | 8.8 | 0.1 | 6.4 | 0.1 |
| TC15349 | Bzip | −2.5 | −4.5 | −3.4 | 10.9 | 0.1 | −5.0 | −6.9 | −2.4 | −9.7 | 1.6 | 4.3 | 4.9 | 26 | 6.6 | 0.2 | 6.4 | 0.2 | 6.5 | 0.2 | 7.6 | 0.3 | 6.8 | 0.2 |
| TC15665 | Heat shock | 2.0 | −1.2 | −3.8 | 11.2 | −5.6 | 0.4 | −3.7 | −7.4 | −1.9 | 2.3 | 3.9 | −0.4 | 19 | 6.4 | 0.2 | 6.1 | 0.1 | 5.8 | 0.1 | 7.0 | 0.3 | 5.6 | 0.1 |
| TC16035 | Homeobox 2 protein | 0.3 | 1.1 | −0.6 | 11.5 | −11.3 | 1.1 | 0.0 | −4.4 | 0.3 | 6.7 | 7.4 | −1.2 | 26 | 7.3 | 0.1 | 7.4 | 0.1 | 7.2 | 0.1 | 8.1 | 0.2 | 6.5 | 0.1 |
| TC17906 | BEL1-related protein | 2.8 | −2.6 | −1.5 | 9.7 | −5.1 | 0.3 | −3.6 | −7.6 | −0.7 | 1.9 | 4.2 | −0.4 | 23 | 8.8 | 0.1 | 8.3 | 0.1 | 8.4 | 0.1 | 9.2 | 0.1 | 7.9 | 0.2 |
| TC17508 | BEL1-related protein | 2.3 | −2.5 | −2.0 | 11.9 | −8.9 | 0.2 | −3.2 | −6.1 | −0.9 | 3.6 | 5.1 | −0.2 | 25 | 8.2 | 0.2 | 7.8 | 0.2 | 7.8 | 0.2 | 8.6 | 0.2 | 7.4 | 0.2 |
| TC15971 | Knotted homeodomain | −0.8 | −2.2 | −1.4 | 12.3 | −5.8 | −2.0 | −1.3 | −6.1 | −2.0 | 5.2 | 5.9 | 1.2 | 24 | 6.5 | 0.2 | 6.4 | 0.1 | 6.5 | 0.1 | 7.4 | 0.2 | 6.2 | 0.1 |
D statistics for anthocyanin biosynthesis genes, B genes, a C gene and transcription factors specifically expressed in staminodia are given under each of the 12 contrasts (figure 1, A = SEP+PET, B = PET+STA, C = STA+CAR) with the D statistic with the highest absolute value highlighted in bold. Also given are average log intensities per probe set and tissue (m) after quantile normalization including their standard errors (se) and numbers of probes in each set. The average log intensities corresponding to the contrast with the highest absolute D-statistic are highlighted.
Interestingly, petals and stamens combined (B domain) had few co-up-regulated genes (32 in A. formosa and 67 in A. thaliana) compared to petals and stamens separately (59 and 1195 in A. formosa and 214 and 1254 in A. thaliana, Figure S1). This pattern perhaps reflects the combination of B and C genes to determine stamen identity and the lack of C-gene expression in petals (at least in A. thaliana, [2]), in addition to the high transcriptional activity in stamens as opposed to petals in pre-anthesis flowers of both species. When we tested for coordinated expression in the B whorls along with either adjacent whorl in Aquilegia, fewer genes were co-up-regulated when staminodia were included (24) but nearly 4-fold more genes were co-up-regulated (118) when sepals were included (Figure 2C) suggesting a significant similarity of expression in sepals, petals and stamens after organ identity is established. These patterns are reflected to some extent by the expression of the B-class identity genes, PISTILLATA (PI) and APETALA3 (AP3), in different ways. The A. formosa homologue of PI is represented by three probe sets (TC17477, TC11683, TC11684) which are all significantly co-up-regulated in sepals, petals, stamens and staminodia compared to carpels (Table 1). The up-regulation of PI in these four tissues has been demonstrated previously in A. vulgaris [19]. The expression of PI is thus extended to both sepals and staminodia in Aquilegia. The three AP3 paralogues are expressed in a whorl-specific manner with AP3-3 (TC19085) being most highly expressed in petals, AP3-2 (TC19725) having the highest expression in petals, stamens and staminodia combined and AP3-1 (TC16289) being most strongly expressed in staminodia (Table 1). Again, these patterns are consistent with the expression of the three AP3 paralogues in A. vulgaris [19]. The expression of AP3 paralogues is thus not extended significantly to sepals. However, the unique expression patterns of AP3 paralogues in Aquilegia in petals, stamens and staminodia is hypothesized to contribute to the identity of these floral tissues [19].
Stamens and carpels (C domain) had far fewer co-up-regulated genes (142 and 50 in A. formosa and A. thaliana respectively) than stamens and carpels individually (1195 and 1549 in A. formosa; 1251 and 456 in A. thaliana, Figure S1). Again, this likely reflects the specific organ identity program of B and C genes for stamens and the lack of B-gene expression in carpels (at least for A. thaliana, [2]). When staminodia were included with the C whorls in A. formosa even fewer genes were co-up-regulated (36) (Figure 2C). Carpels and staminodia together co-up-regulated more genes (197 genes, Figure 2C, contrast 7; these genes are down-regulated in sepals, petals and stamens which is equal to up-regulation in staminodia and carpels) as compared to stamens and staminodia combined (124) (Figure 2C, contrast 10). Thus an interesting gene expression profile for staminodia emerges. In addition to the staminodia-specific up-regulation of 160 genes, there are 197 genes co-up-regulated with carpels and 124 genes co-up-regulated with stamens while stamens and carpels have 142 genes co-up-regulated (Figure 2C). These patterns are interesting given the fact that developmental, morphological and genetic evidence all suggest that staminodia are derived from stamens rather than carpels. However, the transcriptional similarity of staminodia and carpels may be due to shared morphological traits (e.g., staminodia and carpels are both laterally expanded laminar organs while stamens are not) rather than common ancestry. Support for the hypothesis that staminodia evolved from stamens as opposed to being an independently evolved whorl is that the Aquilegia homologue of the C gene AG (TC8667) is most highly up-regulated in stamens, staminodia and carpels combined while, as discussed above, the B genes AP3-2 and PI are detected in both stamens and staminodia (Table 1). As the EST library from which our microarray was designed contained only one AGAMOUS gene (most similar to AGAMOUS1 of Aquilegia alpine, AqAG1), no data are available for the expression of the second previously characterized locus, AqAG2, although studies indicate that this gene is carpel-specific [23]. Regulatory genes specifically up-regulated in staminodia include diverse transcription factors (myb, TC13707; Bzip, TC15349; heat shock, TC15665; homeobox 2, TC16035; BEL1-related, TC17906, TC17508; knotted, TC15971; Table 1).
Correlation analysis
Interestingly, in A. formosa, array-wide expression patterns were significantly negatively correlated between most whorls except for a significant positive correlation between petals and sepals and no significant correlation between sepals and staminodia (Table S1). In A. thaliana, array-wide expression patterns were significantly negatively correlated except for a positive correlation between petals and carpels (Table S1).
Gene set enrichment analysis
To test which, if any, biological processes were significantly up- or down-regulated in the whorls and whorl combinations of interest, gene ontology categories of biological processes (GOBP) commonly used to annotate A. thaliana loci were assigned to Aquilegia unigenes. A total of 2,571 Aquilegia unigenes were annotated with a total of 842 GOBPs. For the z-test of enrichment, we limited our test to GOBPs that had a minimum of 10 entries, reducing the number of Aquilegia unigenes and GOBPs to 2003 and 163, respectively. We used the z-test to determine if the mean D statistic of a GOBP differed from the mean D statistic of the 2003 genes of a given contrast. The significant GOBPs are listed in Table 2. GOBPs significantly up-regulated in sepals and petals separately and sepals and petals combined (the A domain) included ATP-dependent proteolysis and electron transport. Flavonoid biosynthesis genes were also up-regulated in the perianth, which corroborates earlier findings of up regulation of anthocyanin biosynthetic pathway genes in anthocyanin producing perianths of several Aquilegia species [21]. Another gene set up-regulated in the A domain involves genes responding to auxin stimulus. GOPBs specifically up-regulated in sepals and petals respectively were photosynthesis and aging. In line with expectations, stamens had significant up-regulation of genes involved in pollen development and pollen germination. GOPBs such as mitosis, cytokinesis, microtubule polymerization and movement, and vesicle-mediated transport indicate that A. formosa microspores undergo mitotic divisions involving phragmoblast-mediated cytokinesis. GOBPs significantly up-regulated in the B domain were dominated by those up-regulated in stamens, except for pollen germination and development and microtubule polymerization and movement which were down-regulated in petals. Gene sets enriched in carpels, such as gamete formation, DNA replication and nucleosome assembly indicate that during pre-anthesis, carpels prepare to form megaspores by synthesizing DNA prior to meiosis. Interestingly, stamens and carpels up-regulated different signalling pathways (GTPase mediated signal transduction in stamens, transmembrane receptor protein tyrosin kinase signalling pathway in carpels) and oppositely regulated photosynthesis (down-regulated in stamens and up-regulated in carpels). Both whorls combined (C domain) up-regulated additional categories such as DNA repair and regulation of progression through cell cycle. In staminodia, four GOBPs were significantly up-regulated, namely, lignin biosynthesis, response to wounding, fatty acid beta-oxidation and one carbon compound metabolic process, suggesting an important defence function of staminodia. None of the categories was shared with the four other whorls but similar to stamens, staminodia down-regulated photosynthesis.
Table 2. Significant GO categories.
| GO ID | GO description | GO ID | GO description | GO ID | GO description |
| sepals down | GO.0007018 | microtubule-based movement | GO.0009409 | response to cold | |
| GO.0006730 | one-carbon compound metab.process | GO.0006012 | galactose metabolic process | GO.0000226 | microtubule cytoskeleton organization |
| GO.0045045 | secretory pathway | GO.0007067 | mitosis | GO.0007018 | microtubule-based movement |
| GO.0007067 | mitosis | GO.0007094 | mitotic spindle checkpoint | GO.0051258 | protein polymerization |
| GO.0000910 | cytokinesis | GO.0006887 | exocytosis | GO.0046785 | microtubule polymerization |
| GO.0000074 | regulation cell cycle progression | GO.0000082 | G1/S transition of mitotic cell cycle | A whorls up | |
| GO.0006886 | intracellular protein transport | GO.0007047 | cell wall organization and biogenesis | GO.0006118 | electron transport |
| GO.0007010 | cytoskeleton organization/biogenesis | GO.0000226 | microtubule cytoskeleton organization | GO.0006510 | ATP-dependent proteolysis |
| GO.0006260 | DNA replication | GO.0009826 | unidimensional cell growth | GO.0008152 | metabolic process |
| GO.0006412 | translation | GO.0000160 | two-component signal transduction | GO.0009813 | flavonoid biosynthetic process |
| GO.0007169 | transmembrane receptor protein | GO.0051301 | cell division | GO.0009733 | response to auxin stimulus |
| tyrosine kinase signaling pathway | GO.0006888 | ER to Golgi vesicle-mediated transport | GO.0006629 | lipid metabolic process | |
| GO.0007017 | microtubule-based process | GO.0000162 | tryptophan biosynthetic process | ||
| GO.0000226 | microtubule cytoskeleton organization | staminodia down | |||
| GO.0009826 | unidimensional cell growth | GO.0006457 | protein folding | B whorls down | |
| GO.0051258 | protein polymerization | GO.0006779 | porphyrin biosynthetic process | GO.0009908 | flower development |
| GO.0046785 | microtubule polymerization | GO.0015979 | photosynthesis | GO.0009626 | hypersensitive response |
| GO.0007018 | microtubule-based movement | GO.0000162 | tryptophan biosynthetic process | GO.0006397 | mRNA processing |
| sepals up | GO.0006412 | translation | GO.0000003 | reproduction | |
| GO.0006510 | ATP-dependent proteolysis | staminodia up | GO.0006334 | nucleosome assembly | |
| GO.0009626 | hypersensitive response | GO.0009809 | lignin biosynthetic process | GO.0000398 | nuclear mRNA splicing, spliceosome |
| GO.0015979 | photosynthesis | GO.0009611 | response to wounding | GO.0040007 | growth |
| GO.0009695 | jasmonic acid biosynthetic process | GO.0006635 | fatty acid beta-oxidation | GO.0006412 | translation |
| GO.0006118 | electron transport | GO.0006730 | one-carbon compound metab process | GO.0015979 | photosynthesis |
| GO.0006812 | cation transport | GO.0007568 | aging | B whorls up | |
| GO.0009813 | flavonoid biosynthetic process | GO.0006810 | transport | ||
| GO.0006508 | proteolysis | carpels down | GO.0006886 | intracellular protein transport | |
| GO.0009225 | nucleotide-sugar metabolic process | GO.0006099 | tricarboxylic acid cycle | ||
| petals down | GO.0009738 | abscisic acid mediated signaling | GO.0009225 | nucleotide-sugar metabolic process | |
| GO.0007010 | cytoskeleton organization/biogenesis | GO.0009611 | response to wounding | GO.0015031 | protein transport |
| GO.0007059 | chromosome segregation | GO.0006099 | tricarboxylic acid cycle | GO.0007017 | microtubule-based process |
| GO.0016192 | vesicle-mediated transport | GO.0006118 | electron transport | GO.0007264 | small GTPase mediated signal trans. |
| GO.0009409 | response to cold | GO.0006629 | lipid metabolic process | GO.0009058 | biosynthetic process |
| GO.0000226 | microtubule cytoskeleton organization | GO.0008152 | metabolic process | GO.0000910 | cytokinesis |
| GO.0007018 | microtubule-based movement | GO.0006635 | fatty acid beta-oxidation | GO.0006897 | endocytosis |
| GO.0051258 | protein polymerization | GO.0006810 | transport | GO.0000162 | tryptophan biosynthetic process |
| GO.0046785 | microtubule polymerization | GO.0007568 | aging | GO.0007047 | cell wall organization and biogenesis |
| GO.0009555 | pollen development | carpels up | GO.0006839 | mitochondrial transport | |
| GO.0009846 | pollen germination | GO.0006412 | translation | GO.0006887 | exocytosis |
| petals up | GO.0007169 | transmembrane receptor protein | GO.0000160 | two-component signal transduction | |
| GO.0006118 | electron transport | tyrosine kinase signaling pathway | GO.0006096 | glycolysis | |
| GO.0008152 | metabolic process | GO.0015979 | photosynthesis | GO.0006629 | lipid metabolic process |
| GO.0009733 | response to auxin stimulus | GO.0006260 | DNA replication | ||
| GO.0006510 | ATP-dependent proteolysis | GO.0006334 | nucleosome assembly | C whorls down | |
| GO.0007568 | aging | GO.0000398 | nuclear mRNA splicing, spliceosome | GO.0009809 | lignin biosynthetic process |
| GO.0016575 | histone deacetylation | GO.0040007 | growth | GO.0009813 | flavonoid biosynthetic process |
| GO.0000162 | tryptophan biosynthetic process | GO.0007276 | gamete generation | GO.0006508 | proteolysis |
| GO.0009813 | flavonoid biosynthetic process | GO.0006457 | protein folding | GO.0006979 | response to oxidative stress |
| GO.0006098 | pentose-phosphate shunt | GO.0006364 | rRNA processing | GO.0006510 | ATP-dependent proteolysis |
| GO.0006281 | DNA repair | GO.0008152 | metabolic process | ||
| stamens down | GO.0006414 | translational elongation | GO.0006635 | fatty acid beta-oxidation | |
| GO.0015979 | photosynthesis | GO.0000074 | regulation cell cycle progression | GO.0006118 | electron transport |
| GO.0006412 | translation | GO.0006397 | mRNA processing | GO.0009611 | response to wounding |
| stamens up | GO.0000003 | reproduction | GO.0007568 | aging | |
| GO.0006886 | intracellular protein transport | C whorls down | |||
| GO.0009225 | nucleotide-sugar metabolic process | A whorls down | GO.0006412 | translation | |
| GO.0015031 | protein transport | GO.0000074 | regulation cell cycle progression | GO.0006260 | DNA replication |
| GO.0009058 | biosynthetic process | GO.0009555 | pollen development | GO.0051258 | protein polymerization |
| GO.0006944 | membrane fusion | GO.0007067 | mitosis | GO.0046785 | microtubule polymerization |
| GO.0006810 | transport | GO.0006412 | translation | GO.0007018 | microtubule-based movement |
| GO.0007264 | small GTPase mediated signal trans. | GO.0006260 | DNA replication | GO.0000074 | regulation cell cycle progression |
| GO.0000910 | cytokinesis | GO.0007059 | chromosome segregation | GO.0000226 | microtubule cytoskeleton organization |
| GO.0009846 | pollen germination | GO.0007169 | transmembrane receptor protein | GO.0006281 | DNA repair |
| GO.0007017 | microtubule-based process | tyrosinekinase signaling pathway | GO.0009826 | unidimensional cell growth | |
| GO.0006839 | mitochondrial transport | GO.0007010 | cytoskeleton organization/biogenesis | GO.0006334 | nucleosome assembly |
| GO.0006099 | tricarboxylic acid cycle | GO.0009826 | unidimensional cell growth | GO.0007067 | mitosis |
| GO.0016192 | vesicle-mediated transport | GO.0007017 | microtubule-based process | GO.0007169 | transmembrane receptor protein |
| GO.0051258 | protein polymerization | GO.0009846 | pollen germination | tyrosine kinase signaling pathway | |
| GO.0046785 | microtubule polymerization | GO.0006886 | intracellular protein transport | GO.0006275 | regulation of DNA replication |
| GO.0009555 | pollen development | GO.0000910 | cytokinesis |
Gene categories of biological processes down-regulated or up-regulated in the five floral whorls and traditional A, B and C whorl combinations of Aquilegia formosa pre-anthesis flowers. Significant gene ontologies (GO) were determined by gene set enrichment analysis and had Benjamini-Hochberg-adjusted p values<0.01.
Correlation of floral expression regulation in potential A. formosa – A. thaliana homologues
When aligning a six frame translation of the 17,801 uni genes of the Aquilegia gene index with the A. thaliana proteome (TAIR 7), a match was found for 5,918 genes using BLASTx (E≤5E-06). Vice versa, for 13,511 A. thaliana proteins, a matching Aquilegia uni gene was identified using tBLASTn (E≤7E-06). An intersection of both queries resulted in 2,620 reciprocal pairs of A. thaliana proteins and A. formosa uni genes. For 2,000 of these, expression information was available from both the Aquilegia oligonucleotide array and the Ath1 array. To determine the extent of conservation in floral expression regulation between these potential Aquilegia – Arabidopsis homologues, their expression statistics (D statistics) were correlated. Rank-based correlation coefficients did not exceed 0.42 (Table 3). Gene expression in A. formosa sepals, stamens and carpels was most strongly, and significantly, correlated with their counterparts in the Arabidopsis flower. However, no significant correlation was found between the gene expression patterns of petals of both species, recapitulating their morphological differences on the transcription level. Instead, expression in A. formosa petals was significantly correlated with expression in A. thaliana sepals (0.27) and expression of A. thaliana petals was weakly correlated with expression in A. formosa carpels (0.11). Interestingly, expression in A. formosa staminodia was positively correlated with expression in A. thaliana stamens but negatively correlated with expression in A. thaliana carpels, lending support to the hypothesis that staminodia evolved from stamens rather than carpels. Correlations between whorl groupings were highest for A domains, followed by C and B domains (Table 3).
Table 3. Gene expression correlation between potential homologues of A. formosa and A. thaliana.
| A. formosa | Sepals | Petals | Stamens | Staminodia | Carpels | A domain | B domain | C domain |
| A. thaliana | ||||||||
| Sepals | 0.37* | 0.27* | −0.19* | −0.09* | −0.19* | |||
| Petals | −0.11* | 0.03 | −0.02 | 0.03 | 0.11* | |||
| Stamens | −0.06 | −0.18* | 0.33* | 0.16* | −0.23* | |||
| Carpels | −0.25* | −0.07 | −0.16* | −0.08* | 0.42* | |||
| A domain | 0.34* | −0.01 | −0.30* | |||||
| B domain | −0.18* | 0.20* | 0.09* | |||||
| C domain | −0.34* | 0.01 | 0.30* |
Spearman's rank correlation coefficients for correlations of D statistics of 2000 potentially homologous genes in pair-wise comparisons of A. formosa and A. thaliana (stage 12) floral whorls and traditional A, B, and C domains. Highest A. formosa correlation coefficients are given in bold whereas highest A. thaliana coefficients are italicized. Statistically significant coefficients are marked with an asterisk.
Discussion
In this study we examined gene expression in the five floral whorls of A. formosa with a newly designed oligonucleotide microarray. One of our goals was to compare floral gene expression in the basal eudicot A. formosa with that in the core eudicot A. thaliana. Another aim was to identify genes co-expressed with floral identity genes and characterize the transcriptional signature of petaloid sepals. Lastly, we were interested in generating hypotheses regarding the evolution and ecological function of staminodia, a floral organ type recently evolved in Aquilegia and its close relatives Semiaquilegia and Urophysa [17], [18].
Our study demonstrated the utility and reliability of the Aquilegia microarray by validating previously obtained floral expression patterns. Particularly, the specific expression of anthocyanin biosynthetic genes in the A domain (sepals and petals), AqAG1 in the C domain (stamens and carpels) and PI in all whorls except carpels as well as the unique expression of AP3 paralogues in petals, stamens and staminodia corroborated earlier findings [19], [21].
When contrasting co-expression patterns between the lower eudicot A. formosa and the core eudicot A. thaliana we found that in the latter, organ-specific expression invariably exceeded co-expression between whorls (Figure S1, e.g., stage 12, sepals: 854, petals: 240, stamens: 1658, carpels: 558, A whorls: 188, B whorls: 186, C whorls: 188). In Aquilegia however, co-expression in the A domain (319) was considerably greater than in sepals (83) and petals (59) alone (Figure S1). Also, in Aquilegia co-expression between sepals, petals and stamens (305) was higher than expression in sepals (83) and petals (59) whereas in Arabidopsis, co-expression between sepals, petals and stamens (558) only exceeded expression in petals (240) (Figure S1). These patterns of co-expression are consistent with a similar comparative transcriptomics experiment of the basal angiosperm Persea americana and A. thaliana [8]. This study demonstrated domains of elevated floral gene expression extending across floral whorls in Persea as opposed to expression domains that were more constrained to individual whorls in A. thaliana. In particular, expression levels of Persea genes that clustered with APETALA3 and PISTILLATA peaked in stamens but were also high in tepals and detectable in carpels [8]. In the case of Persea, the results could be interpreted in the context of the ‘fading borders’ model, which correlates the presence of morphological grades between floral organs with similar gradients of floral organ identity gene expression [24]. Aquilegia flowers do not have the same kind of morphological grades observed in magnoliid dicots but the presence of petaloid sepals and the stamen-derived staminodia may provide analogous patterns.
The expression patterns we observed suggest that petaloidy of Aquilegia sepals correlates with a high degree of co-expressed genes in petals and sepals (269 genes) as well as in sepals, petals and stamens (118 genes). Some of these co-expressed genes are likely to be involved in mediating aspects of petaloidy. For example, the production of floral pigments in sepals and petals is consistent with co-expression of anthocyanin genes in these organs. The other identified feature of petaloidy in Aquilegia is papillated epidermal cells [19]. However, we did not identify likely genes involved with this character, e.g., a MIXTA homolog [25], perhaps because they are expressed earlier in development than the pre-anthesis flowers we studied or the genes were not represented on our array. Developmental control of this feature of petaloidy is not determined by the expression of the B class gene PISTILLATA as its down regulation does not affect this character [16]. However, the relatively large number of co-expressed genes in petals and petaloid sepals suggest that while the genetic factors controlling organ identity at a higher level may differ between these organs, identity pathways converge on similar downstream effectors to produce similar coloration and cell types. Despite this common set of expressed genes in Aquilegia sepals and petals, expression of potentially homologous genes in the sepals of Aquilegia correlated most strongly with that of the sepals of Arabidopsis (Table 3). Thus, even though Aquilegia sepals are petaloid, they retain significant ‘sepaloid’ gene expression patterns as well.
Interestingly, we found no correlation in homologous gene expression in A. formosa and A. thaliana petals, which may be indicative of independent origins of petals in A. formosa and A. thaliana or may simply reflect their highly divergent morphologies. Aquilegia petals have been suggested to be derived from sterilized stamens [26] whereas an andropetaloid origin for Arabidopsis petals has recently been challenged and an bracteopetaloid origin has been suggested instead [27]. These two possible origins of petals – petaloid bracts vs sterilized stamens – were first discussed by Takhtajan [28]. On the transcriptional level, andropetals may have arisen through a repression of C gene expression in an outer whorl of stamens whereas bractopetals may have evolved by an expansion of B gene expression into pre-existing sterile organs [29]. Apart from independent evolution, the observed lack of correlation in homologous gene expression in A. thaliana and A. formosa petals may have resulted from a strong divergence of petal identity pathways during the divergence of the Aquilegia and the Arabidopsis lineage. The fact that homologous gene expression in A. formosa petals was most strongly correlated with that in A. thaliana sepals is most likely due to the significant convergence of expression patterns of A. formosa petals and sepals (Table S1) and the strong correlation of expression between A. formosa sepals and A. thaliana sepals.
The molecular mechanisms accompanying the evolution of new floral organs have often been investigated in the framework of the ABC model. For example, the lodicules of monocot grasses are hypothesized to be derived from petals because their identity is controlled homologues of the B genes AP3 and PI (refs in [30]). Thus a novel identity program may have evolved through modifications to an existing identity program in the lodicule. In Aquilegia, following the stamen whorls, there is one whorl of staminodia that have been interpreted as being evolutionarily derived from fertile stamens due to similarities with the development of stamens [17]. The staminodia have a prominent central midrib with ruffled laminae extending to either side and unique epidermal cells. The laminae form an interlocking sheath around the developing ovary. Occasionally, anthers are observed on the tips of the staminodia (pers. obs.). Staminodia express both B and C class identity genes (AG, PI and the AP3-1 paralog) in A. vulgaris [19] and A. formosa (this study), which is consistent with staminodia having evolved from stamens. We interpret our finding that staminodia co-expressed more genes with carpels than with stamens as being the result of convergence of gene expression due to similar morphological features of staminodia and carpels (e.g., their laminar nature) rather than a common evolutionary origin of both organs (Figure 2C).
Staminodia also displayed unique gene expression patterns. In pre-anthesis flowers, staminodia-specific gene expression exceeded that in sepals and petals. A set of approximately 160 genes was specifically up-regulated in staminodia including transcription factors that might be involved in regulating the expression of these genes or even in determining staminodium identity itself. Particularly interesting is the up-regulation of two potential BEL1-related loci (TC17906, TC17508) and one Knotted gene (TC15971) in staminodia. Proteins of both families have been shown to antagonistically interact with AG in the outer floral whorls [31]. However, BEL-Knotted complexes consisting of PENNYWISE (PNY), POUNDFOOLISH (PNF), SHOOT MERISTEMLESS (STM) and KNAT2 have been shown to positively interact with AG in the inner floral whorls [32]. Interestingly, the presence or absence of BEL1 in complexes containing AG-SEP3 is crucial for ovule and carpel identity, respectively [33]. When BEL1 expression is missing, integuments are transformed into carpelloid tissue indicating the need for BEL1 to promote ovule formation in the presence of AG. The fact that BEL1- and Knotted-related proteins are up-regulated in parallel with AG in staminodia of A. formosa leads us to suggest that these three proteins could potentially interact to affect staminodia-specific gene expression or even identity in Aquilegia. Analogously to antagonistic interactions in ovules [33] and outer floral whorls [31], BEL1 and/or Knotted proteins might regulate AG (or complexes thereof) to repress its carpel identity function and enable staminodia identity instead. Clearly, additional experiments are necessary to test this hypothesis. For example, it would be interesting to conduct in situ hybridization experiments in early stages of floral meristem development. In addition, a virus-induced gene silencing system for transient knock-outs of gene expression has recently been established for Aquilegia [34] allowing us to manipulate the expression of all Aquilegia BEL1 and Knotted proteins.
Our gene set enrichment analysis also suggests a possible ecological function of staminodia. Since staminodia remain attached to the receptacle long after the other floral organs have abscised (Figure 1), one hypothesis is that staminodia are impregnated with herbivory defensive compounds that protect the early differentiating fruits [35]. Consistent with this hypothesis, we found lignin biosynthesis genes to be up-regulated in staminodia (e.g., ferulic acid-5-hydroxylase, TC12484, cinnamoyl-CoA reductase, TC8815, TC8816, caffeoyl-CoA-O-methyltransferase, TC11606, TC11605). A strong lignin barrier may protect the developing ovary from microbial and insect enzymes and thus confer protection against predation by pathogens [36] and herbivores. Among the 15 genes with the strongest staminodia-specific expression patterns were two laccases (TC17980, DT727506), two diphenol oxidases (TC12078, TC10815), a peroxidase (TC10188) and two repiratory burst oxidase genes (TC12632, TC13899). Interestingly, these classes of genes have been implicated in other systems for defense against microbial attack and herbivory due to their up-regulation in response to herbivory or mechanical wounding [37], [38], [39] and/or by specific counter defenses in herbivores [40]. Moreover, two phenylpropanoid biosynthesis genes were up-regulated in staminodia (phenylalanine ammonia lyase, TC10503, 4-coumarate-CoA ligase, TC12066, TC11075, TC10642). Phenylpropanoids are precursor not only in lignin biosynthesis but also for isoflavonoid phytoalexins which have been demonstrated to act in microbial defense [41]. Thus metabolomic analyses could augment our expression studies by further testing a defense and protection function for staminodia. Increased lignin production could also enforce the hydrophobic nature of staminodia and prevent excess moisture around the developing seeds. Along these lines, it would be particularly interesting to determine if removal of staminodia affects fruit development or damage.
In summary, our comparative microarray study has enabled a global perspective on floral gene expression in A. formosa. Not only were previous gene expression patterns confirmed but also transcriptional signatures of petaloidy were discerned and candidate genes for the regulation of staminodia-specific genes were identified. Using this newly designed microarray, further questions relating to special features of the Aquilegia flower such as spur formation can be addressed. For example, transcriptional patterns in spur-forming petals of Aquilegia species can be compared with those of spur-less petals in species of Semiaquilegia. Moreover, transcriptional signatures associated with different pollination syndromes may be obtained across the Aquilegia radiation to help characterize the genetics underlying pollinator-driven floral diversification.
Materials and Methods
EST library and microarray construction
An Aquilegia formosa x pubescens normalized cDNA library was constructed from mixed shoot and floral apical meristems, flower buds, leaves and roots from an F2 hybrid population (Invitrogen, USA). The sequencing of 50,000 clones by The Institute of Genomics Research (TIGR, Rockville, USA) led to 85,039 ESTs which assembled into 11,985 contigs (for which a tentative consensus sequence, TC, was obtained) and 5,816 singleton ESTs, resulting in transcribed sequence information for a total of 17,801 Aquilegia unigenes (The Aquilegia Gene Index, version 2.0, http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=aquilegia, for an analysis of release 2.1 refer to [20]). An isothermal set of oligonucleotide probes (3–35 probes per gene depending on length, Tm 76°C) were designed for 17,276 of these genes and used for microarray fabrication (NimbleGen Systems, Reykjavík, Iceland). A total of 17,246 Aquilegia uni genes were represented by more than three probes and therefore included in expression data analysis. The microarray platform specifics have been deposited at the Gene Expression Omnibus genomics data repository hosted by NCBI (GEO accession nr: GPL9791). Sequence variation between A. formosa and A. pubescens is very low [42], thus probes designed from A. pubescens specific alleles are expected to hybridize to A. formosa cDNA. Furthermore, comparisons of gene expression between A. formosa floral whorls will not be affected.
Sampling
Three Aquilegia formosa populations growing in close proximity at Blue Canyon, Sonora Pass (Sierra Nevada mountains, CA) were sampled for this study. Sixty flowers, all from late pre-anthesis stage, were harvested from each population, dissected into the five floral whorls (sepals, petals, stamens, staminodia, carpels) and immediately frozen into liquid nitrogen. Total RNA was isolated (RNeasy kit, Qiagen, USA) from each tissue across the three replicate populations and 40ug of RNA were sent to NimbleGen Systems (Reykjavík, Iceland) for hybridization.
Hybridizations to the Aquilegia oligonucleotide array
Cy3 labeled cDNA from the five tissues and the three biological replicates was singly hybridized to the Aquilegia oligonucleotide array and raw intensities can be found in fifteen files (90004_532.pair, stamens 2; 90005_532.pair, carpels 3; 90013_532.pair, sepals 3; 92391_532.pair, carpels 2; 92477_532.pair, staminodia 2; 92535_532.pair, petals 2; 95084_532.pair, sepals 1; 95191_532.pair, stamens 1; 95192_532.pair, sepals 2; 95195_532.pair, staminodia 1; 95198_532.pair, stamens 3; 98340_532.pair, carpels 1; 98348_532.pair, petals 1; 98350_532.pair, petals 3; 99928_532.pair, staminodia 3, GEO series record: GSE19432). Labelling, hybridization and scanning was performed by NimbleGen Systems Inc., Madison, WI USA, following their standard operating protocols (order number OID5089).
Source of Arabidopsis thaliana microarray data
Gene expression data of stage 12 and 15 A. thaliana flowers were generated by the Arabidopsis gene expression atlas project. These data sets are part of a developmental series of floral expression data generated from experiments with A. thaliana Col-0 plants and they are available from NCBI's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5632, [43]). Triplicate expression data were retrieved for sepals, petals, stamens and carpels from stage 12 and stage 15 flowers [22]. The respective raw data files used in the analysis were: ATGE_34 wild type flowers stage 12, sepals (GSM131585.CEL, GSM131586.CEL, GSM131587.CEL), ATGE_35 wild type flowers stage 12, petals (GSM131588.CEL, GSM131589.CEL, GSM131590.CEL), ATGE_36 wild type flowers stage 12, stamens (GSM131591.CEL, GSM131592.CEL, GSM131593.CEL), ATGE_37 wild type flowers stage 12, carpels (GSM131594.CEL, GSM131595.CEL, GSM131596.CEL), ATGE_41 wild type flowers stage 15, sepals (GSM131603.CEL, GSM131604.CEL, GSM131605.CEL) ATGE_42 wild type flowers stage 15, petals (GSM131606.CEL, GSM131607.CEL, GSM131608.CEL), ATGE_43 wild type flowers stage 15, stamens (GSM131609.CEL, GSM131610.CEL, GSM131611.CEL), ATGE_45 wild type flowers stage 15, carpels (GSM131612.CEL, GSM131613.CEL, GSM131614.CEL). Data were obtained using Affymetrix GeneChip Arabidopsis ATH1 Genome Array (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL198). For the purpose of this study only probes located in exons designed to the sense strand were analysed, which reduced the number of probes to 352002 representing a total of 17,246 genes (genes with less than 4 probes were omitted).
Microarray analysis
A. formosa
Raw intensity data were log-transformed, spatially corrected [44] and quantile-normalized [45]. After correcting for probe effect (by subtracting probe means), the gene means were determined from each probe set. For each of the 17,246 genes, a linear model was fit either using individual tissues or combinations thereof as fixed effects (Figure 2B) and populations as random effects. The goal was to test how many genes were specifically expressed in each tissue (up- or down-regulated with respect to the other four) and how many genes would be co-expressed in 1) sepals and petals (the traditional A domain, contrast 6), 2) petals and stamens (the traditional B domain, contrast 8), 3) stamens and carpels (the traditional C domain, contrast 12), 4) sepals, petals and stamens (contrast 7) to test to what extend the B domain is extended to petaloid sepals, 5) petals, stamens and staminodia (contrast 9) to test the extension of the B domain to staminodia, 6) stamens and staminodia (contrast 10) and stamens and carpels (contrast 7) to test if gene expression in staminodia would be more similar to that in stamens or carpels. A linear model was fit to each gene in both the experimental data as well as 99 sets of permuted data with arrays re-sampled from the experimental dataset. A standardized expression value (D statistic = coefficient/ (error + median error of all genes), [46]) was then calculated for each gene in the experimental and the permuted data sets. Ranked D-statistics of the permuted data sets were averaged for each gene and compared to the D-statistics of the experimental dataset. False discovery rates were calculated using a delta threshold of 4. In Figure 2C, numbers of differentially expressed genes under each contrast are given with their corresponding false discovery rate. Some genes were significantly differentially expressed under more than one contrast. For the purpose of this study we define genes as being specifically expressed under a given contrast when the D statistic for that contrast is the highest absolute D statistic across all contrasts (bold numbers in Figure 2C).
A. thaliana
The dataset with the four floral whorls in stage 12 was separately analysed from the dataset with four floral whorls in stage 15. Normalization was performed as described for the Aquilegia arrays. Similarly to the A. formosa analysis, a linear model was fit for each gene using each of the four tissues and combinations thereof as fixed effects. The goal was to test how many genes were specifically expressed in each tissue (up- or down-regulated with respect to the other three) and how many genes would be co-expressed in 1) sepals and petals (the traditional A domain), 2) petals and stamens (the traditional B domain) and 3) stamens and carpels (the traditional C domain). Permutation based false discovery rates were calculated as described for A. formosa. Numbers of differentially expressed genes for both A. thaliana datasets are contrasted with those from A. formosa for the seven contrasts that were common to all three analyses in Figure S1. All normalization and permutation analyses were carried out using customized R-scripts which can be found at http://naturalvariation.org/aquilegia.
Gene set enrichment analysis
Gene ontology matrices of biological processes were constructed for genes on both arrays. The A. thaliana matrix consisted of 25,111 gene loci annotated with 1,542 GO categories [47]. Only GO categories with at least 10 genes and only genes that were present on the Ath1 array were used in parametric gene set enrichment analysis. Applying these filters led to a final matrix of 16,985 genes annotated with 306 GO categories. In case of Aquilegia, 2132 GO categories were assigned to 5,889 Aquilegia unigenes through the Plant Gene Index project (http://compbio.dfci.harvard.edu/tgi/plant.html). An Aquilegia GO matrix was then designed by eliminating GO terms not related to plants and GO terms with less than ten genes resulting in a final matrix of 2003 genes annotated in 163 GO categories. Parametric analyses of gene set enrichment were performed on D statistics of both A. thaliana datasets and the A. formosa dataset based on statistical procedures described in [48]. Results from A. thaliana are not shown but results from A. formosa are summarized in Table 1.
Aquilegia-Arabidopsis homology assignment
First, a six frame translation of the 17,801 unigenes of the Aquilegia Gene Index (AQGI.release_2) was aligned against the A. thaliana proteome (TAIR7_pep_20070425) using the BLASTx algorithm. Then, the A. thaliana proteome was matched against the six translations of the Aquilegia unigene set using tBLASTn. Good quality hits from both alignments were extracted based on E values. Entries found in both alignments represent reciprocal matches between A. thaliana and Aquilegia and were considered potential homologues [49] for the purpose of this study.
Supporting Information
Differentially expressed genes in A. formosa (pre-anthesis) and A. thaliana (stages 12 and 15) flowers. Each square represents one contrast and reports the number of differentially genes, the corresponding false discovery rate as determined by bootstrap analysis in brackets and the number of differentially expressed genes adjusted for genes with higher D statistics with other contrasts. Upper and lower panel depict numbers for down- and up-regulated genes, respectively.
(5.74 MB TIF)
Spearman's rank correlation coefficients of array-wide expression of all pair wise combinations of whorls (* denotes p<0.001). In Aquilegia formosa (AF), most correlations are negative, except for a positive correlation of sepals and petals and no correlation between staminodia and sepals. In Arabidopsis thaliana (AT, stage 12), most correlations are also negative, except for a positive correlation between carpels and petals.
(0.05 MB DOC)
Differentially expressed genes in late pre-anthesis A. formosa floral whorls.
(5.15 MB XLS)
Differentially expressed genes in stage 12 A. thaliana floral whorls.
(1.57 MB XLS)
Differentially expressed genes in stage 15 A. thaliana floral whorls.
(1.65 MB XLS)
Acknowledgments
We thank Xu Xhang and Christos Noutsos for help with R and BLAST, the University of California for the use of Owens Valley Laboratory at their White Mountain Research Station, Marilyn Hodges for the photographic contribution and consultation, and Annette Becker and an anonymous reviewer for helpful comments on an earlier draft.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded under a National Science Foundation grant to SA Hodges (http://www.nsf.gov/awardsearch/showAward.do?AwardNumber=0412727). C Voelckel acknowledges funding from the Alexander von Humboldt Foundation through a Feodor Lynen Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ren D, Labandeira CC, Santiago-Blay JA, Rasnitsyn A, Shih C, et al. A probable pollination mode before angiosperms: Eurasian,long-proboscid scorpion flies. Science. 2009;326:840–847. doi: 10.1126/science.1178338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coen ES, Meyerowitz EM. The war of the whorls - genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 3.Kalivas A, Pasentsis K, Polidoros AN, Tsaftaris AS. Heterotopic expression of B-class floral homeotic genes PISTILLATA/GLOBOSA supports a modified model for crocus (Crocus sativus L.) flower formation. DNA Sequence. 2007;18:120–130. doi: 10.1080/10425170601060582. [DOI] [PubMed] [Google Scholar]
- 4.Kanno A, Saeki H, Kameya T, Saedler H, Theissen G. Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Molecular Biology. 2003;52:831–841. doi: 10.1023/a:1025070827979. [DOI] [PubMed] [Google Scholar]
- 5.Kim S, Koh J, Yoo MJ, Kong HZ, Hu Y, et al. Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. Plant Journal. 2005;43:724–744. doi: 10.1111/j.1365-313X.2005.02487.x. [DOI] [PubMed] [Google Scholar]
- 6.Bowman JL. Evolutionary conservation of angiosperm flower development at the molecular and genetic levels. Journal of Biosciences. 1997;22:515–527. [Google Scholar]
- 7.Irish VF. Evolution of petal identity. Journal of Experimental Botany. 2009;60:2517–2527. doi: 10.1093/jxb/erp159. [DOI] [PubMed] [Google Scholar]
- 8.Chanderbali AS, Albert VA, Leebens-Mack J, Altman NS, Soltis DE, et al. Transcriptional signatures of ancient floral developmental genetics in avocado (Persea americana; Lauraceae). Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8929–8934. doi: 10.1073/pnas.0811476106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peiffer JA, Kaushik S, Sakai H, Arteaga-Vazquez M, Sanchez-Leon N, et al. A spatial dissection of the Arabidopsis floral transcriptome by MPSS. BMC Plant Biology. 2008;8 doi: 10.1186/1471-2229-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM. Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell. 2004;16:1314–1326. doi: 10.1105/tpc.021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laitinen RAE, Immanen J, Auvinen P, Rudd S, Alatalo E, et al. Analysis of the floral transcriptome uncovers new regulators of organ determination and gene families related to flower organ differentiation in Gerbera hybrida (Asteraceae). Genome Research. 2005;15:475–486. doi: 10.1101/gr.3043705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laitinen RAE, Pollanen E, Teeri TH, Elomaa P, Kotilainen M. Transcriptional analysis of petal organogenesis in Gerbera hybrida. Planta. 2007;226:347–360. doi: 10.1007/s00425-007-0486-2. [DOI] [PubMed] [Google Scholar]
- 13.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 14.Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whittall JB, Hodges SA. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature. 2007;447:706–U712. doi: 10.1038/nature05857. [DOI] [PubMed] [Google Scholar]
- 16.Hodges SA, Derieg NJ. Adaptive radiations: From field to genomic studies. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9947–9954. doi: 10.1073/pnas.0901594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tucker SC, Hodges SA. Floral ontogeny of Aquilegia, Semiaquilegia, and Enemion (Ranunculaceae). International Journal of Plant Sciences. 2005;166:557–574. [Google Scholar]
- 18.Wang W, Chen ZD. Generic level phylogeny of Thalictroideae (Ranunculaceae) - implications for the taxonomic status of Paropyrum and petal evolution. Taxon. 2007;56:811–821. [Google Scholar]
- 19.Kramer EM, Holappa L, Gould B, Jaramillo MA, Setnikov D, et al. Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. Plant Cell. 2007;19:750–766. doi: 10.1105/tpc.107.050385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer EM, Hodges SA. Aquilegia as a model system for the evolution and ecology of petals. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365:477–490. doi: 10.1098/rstb.2009.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whittall JB, Voelckel C, Kliebenstein DJ, Hodges SA. Convergence, constraint and the role of gene expression during adaptive radiation: floral anthocyanins in Aquilegia. Molecular Ecology. 2006;15:4645–4657. doi: 10.1111/j.1365-294X.2006.03114.x. [DOI] [PubMed] [Google Scholar]
- 22.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramer EM, Jaramillo MA, Di Stilio VS. Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics. 2004;166:1011–1023. doi: 10.1534/genetics.166.2.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soltis DE, Chanderbali AS, Kim S, Buzgo M, Soltis PS. The ABC model and its applicability to basal angiosperms. Annals of Botany. 2007;100:155–163. doi: 10.1093/aob/mcm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noda K, Glover BJ, Linstead P, Martin C. Flower color intensity depends on specialized cell-shape controlled by a myb-related transcription factor. Nature. 1994;369:661–664. doi: 10.1038/369661a0. [DOI] [PubMed] [Google Scholar]
- 26.Erbar C, Kusma S, Leins P. Development and interpretation of nectary organs in Ranunculaceae. Flora. 1999;194:317–332. [Google Scholar]
- 27.Ronse De Craene LP. Are petals sterile stamens or bracts? The origin and evolution of petals in the core eudicots. Ann Bot. 2007;100:621–630. doi: 10.1093/aob/mcm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takhtajan . Evolutionary trends in flowering plants. New York: Columbia University Press; 1991. [Google Scholar]
- 29.Baum DA. The evolution of plant development. Current Opinion in Plant Biology. 1998;1:79–86. doi: 10.1016/s1369-5266(98)80132-5. [DOI] [PubMed] [Google Scholar]
- 30.Kramer EM, Jaramillo MA. Genetic basis for innovations in floral organ identity. Journal of Experimental Zoology Part B-Molecular and Developmental Evolution. 2005;304B:526–535. doi: 10.1002/jez.b.21046. [DOI] [PubMed] [Google Scholar]
- 31.Bao X, Franks RG, Levin JZ, Liu Z. Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell. 2004;16:1478–1489. doi: 10.1105/tpc.021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu LF, Patibanda V, Smith HMS. A novel role of bell1-like homeobox genes, pennywise and pound-foolish, in floral patterning. Planta. 2009;229:693–707. doi: 10.1007/s00425-008-0867-1. [DOI] [PubMed] [Google Scholar]
- 33.Brambilla V, Battaglia R, Colombo M, Masiero S, Bencivenga S, et al. Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell. 2007;19:2544–2556. doi: 10.1105/tpc.107.051797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gould B, Kramer EM. Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia (columbine, Ranunculaceae). Plant Methods. 2007;3 doi: 10.1186/1746-4811-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer EM. Aquilegia: a new model for plant development, ecology, and evolution. Annual Review of Plant Biology. 2009;60:261–277. doi: 10.1146/annurev.arplant.043008.092051. [DOI] [PubMed] [Google Scholar]
- 36.Bhuiyan NH, Selvaraj G, Wei YD, King J. Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. Journal of Experimental Botany. 2009;60:509–521. doi: 10.1093/jxb/ern290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bindschedler LV, Dewdney J, Blee KA, Stone JM, Asai T, et al. Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant Journal. 2006;47:851–863. doi: 10.1111/j.1365-313X.2006.02837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ralph SG, Yueh H, Friedmann M, Aeschliman D, Zeznik JA, et al. Conifer defence against insects: microarray gene expression profiling of Sitka spruce (Picea sitchensis) induced by mechanical wounding or feeding by spruce budworms (Choristoneura occidentalis) or white pine weevils (Pissodes strobi) reveals large-scale changes of the host transcriptome. Plant Cell and Environment. 2006;29:1545–1570. doi: 10.1111/j.1365-3040.2006.01532.x. [DOI] [PubMed] [Google Scholar]
- 39.Torres MA, Dangl JL. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Current Opinion in Plant Biology. 2005;8:397–403. doi: 10.1016/j.pbi.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 40.Weech MH, Chapleau M, Pan L, Ide C, Bede JC. Caterpillar saliva interferes with induced Arabidopsis thaliana defence responses via the systemic acquired resistance pathway. Journal of Experimental Botany. 2008;59:2437–2448. doi: 10.1093/jxb/ern108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zabala G, Zou JJ, Tuteja J, Gonzalez DO, Clough SJ, et al. Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection. BMC Plant Biology. 2006;6 doi: 10.1186/1471-2229-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooper EA, Whittall JB, Hodges SA, Nordborg M. Genetic variation at nuclear loci fail to distinguish two morphologically distinct specis of Aquilegia. PLoS ONE. 2010;5(1):e8655. doi: 10.1371/journal.pone.0008655. doi: 10.1371/journal.pone.0008655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, et al. A gene expression map of Arabidopsis thaliana development. Nature Genetics. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 44.Borevitz JO, Liang D, Plouffe D, Chang HS, Zhu T, et al. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Research. 2003;13:513–523. doi: 10.1101/gr.541303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 46.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X, Shiu S, Cal A, Borevitz JO. Global analysis of genetic, epigenetic and transcriptional polymorphisms in Arabidopsis thaliana using whole genome tiling arrays. Plos Genetics. 2008;4 doi: 10.1371/journal.pgen.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim SY, Volsky DJ. PAGE: Parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6 doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed genes in A. formosa (pre-anthesis) and A. thaliana (stages 12 and 15) flowers. Each square represents one contrast and reports the number of differentially genes, the corresponding false discovery rate as determined by bootstrap analysis in brackets and the number of differentially expressed genes adjusted for genes with higher D statistics with other contrasts. Upper and lower panel depict numbers for down- and up-regulated genes, respectively.
(5.74 MB TIF)
Spearman's rank correlation coefficients of array-wide expression of all pair wise combinations of whorls (* denotes p<0.001). In Aquilegia formosa (AF), most correlations are negative, except for a positive correlation of sepals and petals and no correlation between staminodia and sepals. In Arabidopsis thaliana (AT, stage 12), most correlations are also negative, except for a positive correlation between carpels and petals.
(0.05 MB DOC)
Differentially expressed genes in late pre-anthesis A. formosa floral whorls.
(5.15 MB XLS)
Differentially expressed genes in stage 12 A. thaliana floral whorls.
(1.57 MB XLS)
Differentially expressed genes in stage 15 A. thaliana floral whorls.
(1.65 MB XLS)