Abstract
The RecQ family helicases catalyze the DNA unwinding reaction in an ATP hydrolysis-dependent manner. We investigated the mechanism of DNA unwinding by the Escherichia coli RecQ helicase using a new sensitive helicase assay based on fluorescence cross-correlation spectroscopy (FCCS) with two-photon excitation. The FCCS-based assay can be used to measure the unwinding activity under both single and multiple turnover conditions with no limitation related to the size of the DNA strands constituting the DNA substrate. We found that the monomeric helicase was sufficient to perform the unwinding of short DNA substrates. However, a significant increase in the activity was observed using longer DNA substrates, under single turnover conditions, originating from the simultaneous binding of multiple helicase monomers to the same DNA molecule. This functional cooperativity was strongly dependent on several factors, including DNA substrate length, the number and size of single-stranded 3′-tails, and the temperature. Regarding the latter parameter, a strong cooperativity was observed at 37 °C, whereas only modest or no cooperativity was observed at 25 °C regardless of the nature of the DNA substrate. Consistently, the functional cooperativity was found to be tightly associated with a cooperative DNA binding mode. We also showed that the cooperative binding of helicase to the DNA substrate indirectly accounts for the sigmoidal dependence of unwinding activity on ATP concentration, which also occurs only at 37 °C but not at 25 °C. Finally, we further examined the influences of spontaneous DNA rehybridization (after helicase translocation) and the single-stranded DNA binding property of helicase on the unwinding activity as detected in the FCCS assay.
Keywords: Biophysics, DNA/Enzymes, DNA/Helicase, DNA/Protein Interaction, Enzymes/Cooperativity, Methods/Fluorescence, Fluorescence Cross-correlation Spectroscopy, RecQ Helicase
Introduction
Helicases are molecular motor enzymes that unwind and translocate nucleic acids in an ATP hydrolysis-dependent manner (1, 2). RecQ DNA helicases constitute a ubiquitous family of helicases that play a key role in maintaining genome stability in a wide range of organisms from bacteria to higher eukaryotes (for a review, see Ref. 3). These enzymes are involved in many of the processes of DNA metabolism, including recombination, DNA replication, and DNA repair (3–5). In humans, defects in RecQ family helicases, encoded by the blm, wrn, and RecQ4 genes, give rise to the Bloom, Werner, and Rothmund-Thomson syndromes, respectively, characterized by genomic instability and susceptibility to cancer. The Escherichia coli RecQ helicase, the prototype enzyme of this family, is involved in various processes, including the homologous recombination and double strand break repair mediated by the RecF machinery (6) as well as suppression of illegitimate recombination (7).
The structure-function relationship of helicases is difficult to understand at the molecular level, because proteins with different organizations may have similar activities. For example, the need for an oligomeric structure for helicase activity strongly varies between helicase families. Hexameric rings are required for active E. coli DnaB and Rho and bacteriophages T4 gp41 and T7 gp4 (8–14), whereas a dimeric form is required for the Rep helicase (15). Some controversy remains in the literature concerning the oligomeric status of active PcrA and UvrD helicases, which is not well defined. It has been proposed that these helicases are active as monomers (16–19) or dimers (20, 21). Regarding the RecQ family helicase, the quaternary structures of active WRN and BLM proteins are also unclear. Different multimeric states corresponding to catalytically active forms have been identified, including hexamers, tetramers, and monomers (22–25). The exact reasons for these discrepancies are unclear, and the possibility that distinct oligomeric states could be compatible with unwinding activity for a given helicase remains open to debate.
Despite extensive biochemical and structural studies of helicases, the mechanism of DNA unwinding remains obscure, and several enzymatic features of the RecQ family helicases are not clearly understood. As mentioned above for other helicases, the oligomeric status of the active E. coli RecQ helicase is unclear, because enzymologic studies of the unwinding reaction in pre-steady-state conditions have strongly suggested that the active form is monomeric (26), whereas a previous study reported a sigmoidal dependence of unwinding activity on ATP concentration, suggesting a multimeric active form (27). Other difficulties in the interpretation of enzymatic parameters are related to side phenomena that strongly influence the measurement of helicase activity, such as: (i) the DNA rehybridization that occurs after the unwinding process; and (ii) the ability of helicase to bind to the single-stranded DNA product, reducing subsequent turnover. The relative impact of these intrinsic properties of the DNA and helicase molecules, respectively, may be difficult to estimate depending on the method used for monitoring unwinding activity (28).
Fluorescence correlation spectroscopy (FCS)3 measures the translational diffusion of freely diffusing molecules based on the analysis of time-dependent fluorescence intensity fluctuations within a small observation volume (29–31). However, this approach, based on the determination of the diffusion coefficient, is not very sensitive for measuring interactions between molecules or dissociation events when the two interacting entities have similar molecular sizes. This major limitation can be overcome by dual color fluorescence cross-correlation spectroscopy (FCCS), which can be used to monitor the cross-correlation of the fluorescence fluctuations of the two interacting species, labeled by spectrally distinct fluorophores, and then to quantify the amount of fluorescently double-labeled species. A maximal cross-correlation amplitude is obtained when the two molecular species diffuse simultaneously and in a systematic manner through the observed volume, indicating a physical interaction, whereas a decrease in this amplitude accounts for independent diffusion due to a dissociation event. This approach has been shown suitable for monitoring various enzymatic activities or processes, including proteolysis (32), endonucleolytic cleavage by restriction enzymes (33, 34), DNA repair (35), and DNA recombination (36).
We describe here the use of two-photon excitation FCCS to monitor helicase unwinding activity. This assay is sensitive and suitable for studies of unwinding activity, as it can be used for real-time kinetic studies in the low nanomolar range, is compatible with both single and multiple turnover enzymatic conditions, and does not require separation of the single-stranded (ss) DNA product from the double-stranded (ds) DNA substrate by gel electrophoresis. As mentioned above, the cross-correlation of the two fluorescence signals in FCCS is related to the concomitant diffusion of the two fluorescently labeled molecules. Thus, with double-labeled dsDNA substrates, a significant decrease in the amplitude of the cross-correlation function is expected upon DNA unwinding, due to the physical separation of the two labeled DNA strands (the principle of the FCCS-based helicase assay is explained in Fig. 1A). Unlike the fluorescence resonance energy transfer (FRET) approach, FCCS is not sensitive to fluorophore orientation and/or interfluorophore distance. The loss of the cross-correlation signal is sensitive to helicase-mediated strand separation only, with no influence of other phenomena such as unrelated local motions of fluorophores at the DNA ends, which may also affect the donor-acceptor distance. A helicase assay based on fluorescence anisotropy was recently described, but real-time kinetic studies suffer from inherent problems because of the ability of helicase to bind to the ssDNA product (28). Moreover, this assay was compatible with single turnover conditions only (i.e. enzyme concentration exceeding DNA substrate). The FCCS assay has no such limitations because it is not based on a difference in size between the substrate and the reaction product but on the extent of the concomitant diffusion of the two DNA strands.
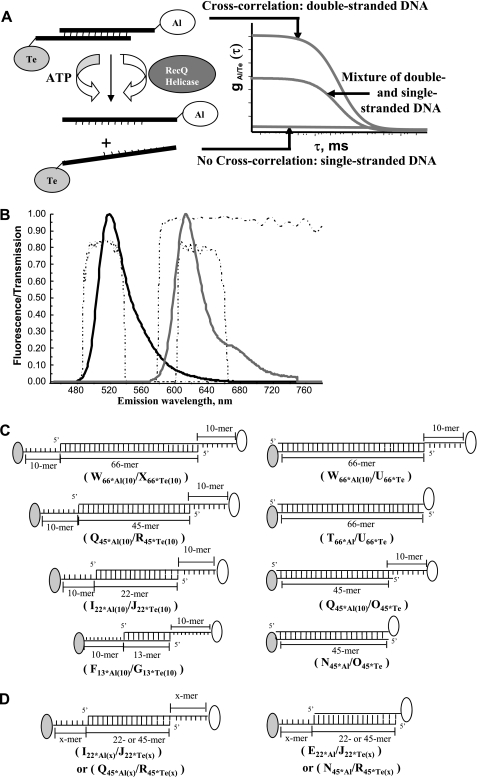
FIGURE 1.
Principle of the FCCS experiment for measuring helicase activity. A, the amplitude of the cross-correlation function gAl/Te(τ) is maximal in the absence of any unwinding activity when using double-stranded DNA substrates double-labeled with Al and Te. A significant decrease in gAl/Te(τ) amplitude with unwinding activity is expected. B, characteristic fluorescence emission spectra of Al-labeled (black line) and Te-labeled (gray line) oligonucleotides upon two-photon excitation (λ = 780 nm). Emission spectra were recorded separately with a home-built setup using a SpectraPro-275 digital triple grating spectrograph coupled to a liquid nitrogen-cooled charge-coupled device detector (Princeton Instruments, Acton, MA). Also shown are transmission profiles (dashed lines) of the dichroic mirror (Chroma 580dcxr) and the two additional filters (Chroma HQ510/50 and HQ630/60m-2p for Al and Te, respectively). C, DNA substrates used in this study. Left, the DNA substrates harbor a duplex region of variable size (13–66 bp) and two ssDNA 3′-tails (10 bases long). Right, two other versions of the 45- and 66-bp DNA substrates were tested: the first contains only one ss-3′-tail, and the other is blunt. Details of sequences are reported in Table 1. The DNA substrate names are indicated in parentheses (see also Table 1 for nomenclature). D, DNA substrates (duplex region of 22 or 45 bp) with one (right) or two (left) ss-3′-tails, designed to study the influence of the ss-3′-tail size on the unwinding rate. An x represents the number of bases in the ss-3′-tail. In the present study, x = 10, 20, or 30 bases (see Table 1).
In this work, we studied the unwinding activity of E. coli RecQ helicase, under both single and multiple turnover conditions, by FCCS. We found that RecQ helicase monomers may function cooperatively or non-cooperatively. Their mode of functioning depends on several parameters, including the length of the DNA substrate (with significant influence from both the duplex and the 3′-ssDNA-flanking regions) and the temperature at which the DNA binding step occurs. Remarkably, the cooperative properties of helicase at the DNA binding level, i.e. helicase binding to the DNA substrate, were found to be predictive, at least qualitatively, of subsequent cooperative effects observed at the catalytic level. Additionally, a post-catalytic event, i.e. helicase binding to newly unwound ssDNA products, was shown also to contribute to the observed functional cooperativity. We also found that the cooperative dependence of the unwinding activity on ATP concentration originated indirectly from the cooperative DNA binding mode of RecQ helicase, reconciling apparently conflicting published results concerning the possibility of an active monomeric form, whereas the activity displays a strong cooperativity on ATP concentration (Hill coefficient ≈ 3), which was interpreted previously as compelling evidence for a multimeric active form (minimally trimeric) (27). Altogether, our data indicate that oligomerization is not a prerequisite for RecQ helicase activity. Nevertheless, in certain conditions, the cooperative assembly of the helicase-DNA complex makes it possible for multiple monomers to align along the same DNA substrate and function in a cooperative manner. Finally, we further examined the influences on the unwinding activity (as measured in our assay) of both spontaneous DNA rehybridization (following helicase translocation) and the ssDNA binding property of helicase. We addressed these questions by evaluating the effects of short single-stranded oligodeoxyribonucleotides (ssODN) and of the single-stranded DNA-binding protein (SSB) on the catalytic rate constant of RecQ helicase.
EXPERIMENTAL PROCEDURES
RecQ Helicase Purification, SSB, and Oligonucleotides
The E. coli RecQ helicases, wild-type and mutant proteins K53A and D146A, were purified as described previously (28, 37). SSB was purchased from Sigma. Unlabeled and fluorescently labeled oligonucleotides (Table 1) were purchased from Eurogentec (Liege, Belgium) and further purified by electrophoresis in a denaturing 12 or 15% acrylamide/urea gel for long (>10 mer) and short (7 and 10 mer) oligonucleotides, respectively. Double-stranded DNAs were obtained by mixing equimolar amounts of complementary DNA strands in 20 mm Hepes (pH 7.2), 100 mm NaCl. The mixture was heated at 85 °C for 5 min and allowed to anneal by slow cooling to 25 °C.
TABLE 1.
Nomenclature and sequences of oligonucleotides used in this study
| Namea | Length | Sequenceb |
|---|---|---|
| A7*Al | 7 | 5′-GTC AGT G-3′-Al |
| B7 | 7 | 5′-GTC AGT G-3′ |
| B′7 | 7 | 5′-CGT GAT G-3′ |
| C10*Al | 10 | 5′-TTA GTC AGT G-3′-Al |
| D10 | 10 | 5′-TTA GTC AGT G-3′ |
| D′10 | 10 | 5′-TTG CGT GAT A-3′ |
| E22*Al | 22 | 5′-AAT CCG TCG AGC AGA GTT AGG G-3′-Al |
| F13*Al(10) | 23 | 5′-CAG ACT CCC TAG A AGTTAGGGTT-3′-Al |
| G13*Te(10) | 23 | 5′-TCT AGG GAG TCT G GATTGTTATT-3′-Te |
| H13(10) | 23 | 5′-TCT AGG GAG TCT G GATTGTTATT-3′ |
| I22*Al(10)c | 32 | 5′-AAT CCG TCG AGC AGA GTT AGG G AGTTAGGGTT-3′-Al |
| J22*Te(10)d | 32 | 5′-CCC TAA CTC TGC TCG ACG GAT T GATTGTTATT-3′-Te |
| K22(10) | 32 | 5′-AAT CCG TCG AGC AGA GTT AGG G AGTTAGGGTT-3′ |
| L22(10) | 32 | 5′-CCC TAA CTC TGC TCG ACG GAT T GATTGTTATT-3′ |
| M32 | 32 | 5′-AAT CCG TCG AGC AGA GTT AGG AGA TCC CTC AG-3′ |
| N45*Al | 45 | 5′-AGA TCC CTC AGA CCC TTT TAG TCA GTG TGG AAA ATC TCT AGC AGT-3′-Al |
| O45*Te | 45 | 5′-ACT GCT AGA GAT TTT CCA CAC TGA CTA AAA GGG TCT GAG GGA TCT-3′-Te |
| P45 | 45 | 5′-ACT GCT AGA GAT TTT CCA CAC TGA CTA AAA GGG TCT GAG GGA TCT-3′ |
| Q45*Al(10)c | 55 | 5′-AGA TCC CTC AGA CCC TTT TAG TCA GTG TGG AAA ATC TCT AGC AGT AGTTAGGGTT-3′-Al |
| R45*Te(10)d | 55 | 5′-ACT GCT AGA GAT TTT CCA CAC TGA CTA AAA GGG TCT GAG GGA TCT GATTGTTATT-3′-Te |
| S45(10) | 55 | 5′-ACT GCT AGA GAT TTT CCA CAC TGA CTA AAA GGG TCT GAG GGA TCT GATTGTTATT-3′ |
| T66*Al | 66 | 5′-AAT CCG TCG AGC AGA GTT AGG AGA TCC CTC AGA CCC TTT TAG TCA GTG TGG AAA ATC TCT AGC AGT-3′-Al |
| U66*Te | 66 | 5′-ACT GCT AGA GAT TTT CCA CAC TGA CTA AAA GGG TCT GAG GGA TCT CCT AAC TCT GCT CGA CGG ATT-3′-Te |
| V66 | 66 | 5′-ACT GCT AGA GAT TTT CCA CAC TGA CTA AAA GGG TCT GAG GGA TCT CCT AAC TCT GCT CGA CGG ATT-3′ |
| W66*Al(10) | 76 | 5′-AAT CCG TCG AGC AGA GTT AGG AGA TCC CTC AGA CCC TTT TAG TCA GTG TGG AAA ATC TCT AGC AGT AGTTAGGGTT-3′-Al |
| X66*Te(10) | 76 | 5′-ACT GCT AGA GAT TTT CCA CAC TGA CTA AAA GGG TCT GAG GGA TCT CCT AAC TCT GCT CGA CGG ATT GATTGTTATT-3′-Te |
| Y66(10) | 76 | 5′-AAT CCG TCG AGC AGA GTT AGG AGA TCC CTC AGA CCC TTT TAG TCA GTG TGG AAA ATC TCT AGC AGT AGTTAGGGTT-3′ |
| Z66(10) | 76 | 5′-ACT GCT AGA GAT TTT CCA CAC TGA CTA AAA GGG TCT GAG GGA TCT CCT AAC TCT GCT CGA CGG ATT GATTGTTATT-3′ |
a The size of the ss-3′-tail is indicated in parentheses.
b Bases corresponding to non-complementary ss-3′-tails are underlined.
c I22*Al(20), I22*Al(30), Q45*Al(20), and Q45*Al(30) are the equivalent oligonucleotides with a longer ss-3′-tail: 20 nucleotides (AGTTAGGGTTTTTTTTTTTA-3′) for I22*Al(20)and Q45*Al(20) and 30 nucleotides (AGTTAGGGTTTTTTTTTTTAAGTTAGGGTA-3′) for I22*Al(30) and Q45*Al(30).
d J22*Te(20), J22*Te(30), R45*Te(20), and R45*Te(30) are the equivalent oligonucleotides with a longer ss-3′-tail: 20 nucleotides (GATTGTTATTTTTTTTTTTA-3′) for J22*Te(20)and R45*Te(20) and 30 nucleotides (GATTGTTATTTTTTTTTTTAGATTGTTATA-3′) for J22*Te(30) and R45*Te(30).
Fluorescence Cross-correlation Spectroscopy
Dual color FCCS measurements were performed by two-photon excitation using a single laser line on a home-built system (described previously in Ref. 38 for the FCS mode) with a 100-fs pulse, 80-MHz mode-locked Mai Tai Ti:Sapphire tunable laser (Spectra Physics, Mountain View, CA) and a Nikon TE2000 inverted microscope. Briefly, the laser beam was expanded with a two-lens afocal system to overfill the back aperture of the objective (Nikon, Plan Apo, ×100, N.A. 1.4, oil immersion) before its entry via the epifluorescence port of the microscope. The setup was optimized to obtain a diffraction-limited focal spot. Measurements were typically carried out in 50 μl of solution dropped onto a coverslip treated with dimethyldichlorosilane. The fluorescence signals from Alexa Fluor 488 (Alexa488 (Al)) and Texas Red (Te) were collected with the same objective and separated from the excitation by a dichroic mirror (Chroma 700DCSPXR). The output signal from the microscope was further filtered with a Chroma E700SP-2p filter to reject the residual excitation light and split by a dichroic mirror (Chroma 580dcxr). Additional filters (Chroma HQ510/50 and HQ630/60m-2p for Alexa488 and Texas Red, respectively) were used to minimize cross-talk (Fig. 1B), and the split fluorescence signal was focused on two avalanche photodiodes (PerkinElmer Life Sciences, SPCM-AQR-14 single photon counting module with less than 90 dark counts/s) mounted at right angle. The detectors were connected to a digital correlator (ALV 6000, ALV-GmbH, Langen, Germany) for calculation of the normalized cross-correlation function gAl/Te(τ) (or g(τ)) of the two fluorescence intensity fluctuations, according to Equation 1,
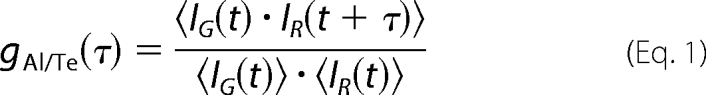 |
where IG(t) and IR(t) are the number of detected fluorescence photons per time unit for the green (Alexa488) and red (Texas Red) channels, respectively. Assuming a three-dimensional Gaussian distribution of excitation intensity, the cross-correlation function for a free Brownian diffusion process is given by Equation 2,
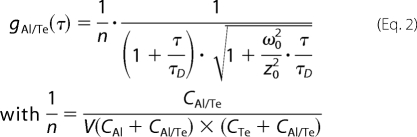 |
where CAl and CTe are the concentrations of singly labeled species, CAl/Te is the concentration of doubly fluorescently labeled species, and τD is the corresponding translational diffusion time. ω0 and z0 are the lateral and axial dimensions of the excitation volume (V), respectively. The excitation volume was calibrated with a 5 nm aqueous solution of Alexa Fluor 488 (succinimidyl ester, Molecular Probes, Eugene, OR; diffusion coefficient, D = 426.3 μm2/s at 21 °C). The excitation wavelength was 780 nm, and the excitation power was 25 milliwatts. Assuming a Gaussian beam shape and according to Equation 2 and τD = ω02/8D, the lateral ω0 and axial z0 dimensions were estimated at 0.380 and 1.40 μm, respectively.
The optimal two-photon excitation wavelength for the FCCS experiments using Alexa488 and Texas Red was found to be 780 nm. In this study, we found that an excitation power of 25 milliwatts was suitable for the two-photon excitation of both Alexa488 and Texas Red (fluorescence intensities displayed quadratic dependences and τD values were constant as a function of incident power below 30 milliwatts. Photobleaching occurred principally above 30 milliwatts). The excitation power was adjusted with a variable attenuator consisting of an achromatic half-wave plate and a polarizing beam splitter (Micro-Controle Spectra-Physics, Evry, France). Recording times were typically between 2.5 and 5 min (average of 5–10 cycles of 30 s each). The cross-correlation curves were fitted with a Levenberg-Marquardt nonlinear least-squares fitting algorithm according to the analytical model (Equation 2) using Igor software (WaveMetrics). The decrease in the amplitude of the cross-correlation, g(0) value, was used to calculate unwinding activity as a function of time, according to Equation 3,
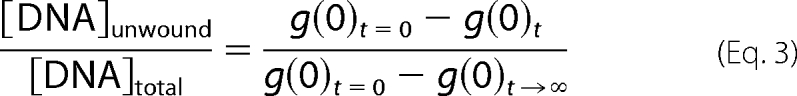 |
with g(0)t=0 and g(0)t→∞ corresponding to the cross-correlation amplitudes at the beginning (zero time) and at the end (infinite time) of the reaction, respectively.
DNA Unwinding Assay
The unwinding activity of the RecQ helicase was measured by dual color FCCS (description of the FCCS setup given above) in 20 mm Hepes (pH 7.2), 40 mm NaCl, 1 mm MgCl2, and 0.3 mm dithiothreitol (reaction buffer). The concentration of the double-labeled DNA substrate was typically 5 nm. The DNA binding step (performed at T°binding = 25 or 37 °C) corresponded to the addition of RecQ helicase to the DNA solution. The unwinding reaction was initiated by adding 1 mm ATP and recorded at T°reaction = 25 °C (unless otherwise stated). For multiple turnover experiments ([DNA substrate] > [RecQ helicase]), the various concentrations of total DNA substrate were obtained by mixing double-labeled DNA substrate (constant concentration of 5 nm) with different concentrations of the corresponding unlabeled DNA substrate. ˜nACT, the Hill coefficient characterizing the functional cooperativity, was calculated by fitting the sigmoidal dependence of unwinding activity on the enzyme concentration using the Hill function of Origin 6.0 software.
DNA Binding Assay: Steady-state Fluorescence Anisotropy
The interaction between RecQ helicase and Alexa488-labeled DNA (either double- or single-stranded) was detected by determining steady-state fluorescence anisotropy on a Beacon 2000 instrument (PanVera, Madison, WI) (28, 39–41). The apparent Kd value (Kd,app) was determined by incubating Alexa488-labeled DNA (5 nm) with increasing concentrations of RecQ helicase in 20 mm Hepes (pH 7.2), 40 mm NaCl, 1 mm MgCl2, and 0.3 mm dithiothreitol. Steady-state anisotropy (r) was then recorded. Fractional saturation was calculated as Δr/Δrmax = (r − rfree)/(rbound − rfree), where rbound and rfree represent bound and free DNA anisotropy, respectively. The Hill coefficient, ˜nBIND, was calculated by directly fitting the titration curve using the Hill function of Origin 6.0 software. Kd,app represents the concentration of RecQ helicase required to titrate the DNA to half saturation.
RESULTS
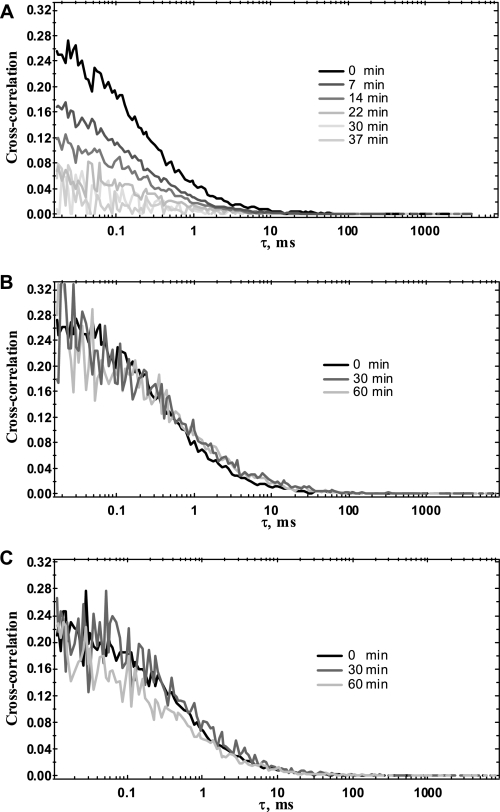
We first assessed whether the unwinding activity of E. coli RecQ helicase would result in a significant and measurable decrease in cross-correlation amplitude, gAl/Te(0) (or g(0)) as measured by FCCS using dsDNA substrates harboring a ssDNA tail (10 bases long) at each 3′-end (Fig. 1C, left. Sequences and nomenclature are as in Table 1). When RecQ helicase and double-labeled DNA substrate were mixed together, a time-dependent decrease in the g(0) value was observed only after the addition of ATP, suggesting that the decrease in cross-correlation amplitude is actually due to RecQ helicase-mediated unwinding of the DNA substrate (Fig. 2A). Indeed, no decrease in the g(0) value was observed either in the absence of ATP (Fig. 2B) or in the presence of ATPγS (a nonhydrolyzable ATP analog) (data not shown). Additionally, two E. coli RecQ helicase ATPase-deficient mutants were tested in the presence of ATP. These protein mutants harbor a single amino acid alteration in helicase conserved motifs I (K53A) and II (D146A) (also known as the walker A and B motif, respectively) and are deficient for both ATP hydrolysis and unwinding activities (37). Again, no decrease in the g(0) value was observed with either the K53A (Fig. 2C) or the D146A mutant (data not shown). Taken together, these results demonstrate that the decrease in the cross-correlation amplitude as measured by FCCS is concomitant to the unwinding activity. However, although the g(0)t→∞ value was low (see Fig. 2A), it was reproducibly different from 0 (about 0.02) and may therefore account for the cross-talk between the detection channels. Indeed, a similar amplitude value was obtained with mixtures of non-complementary Alexa488- and Texas Red-labeled oligonucleotides (data not shown). The g(0)t→∞ value was then further considered in the calculation of the fraction of unwound DNA (see Equation 3). We then studied the unwinding kinetics as a function of DNA substrate size under both single and multiple turnover conditions. In all of the experiments mentioned below, the temperature of the DNA binding step (T°binding) was either 25 or 37 °C, and the reaction temperature (T°reaction) was 25 °C, unless otherwise specified.
FIGURE 2.
Relationship between helicase unwinding activity and the decrease in amplitude of the cross-correlation function. This is an example of a reaction mixture containing 5 nm double-labeled 45-bp DNA substrate (Q45*Al(10)/R45*Te(10)) and 10 nm E. coli RecQ helicase in the reaction buffer (see “Experimental Procedures”). A and B, wild-type helicase. C, K53A mutant. The helicase-DNA complexes were formed at 37 °C and incubated for a further 10 min. The temperature was then lowered to 25 °C, and the unwinding reaction was monitored upon the addition of 1 mm ATP (A and C) or an equivalent volume of reaction buffer without ATP (B).
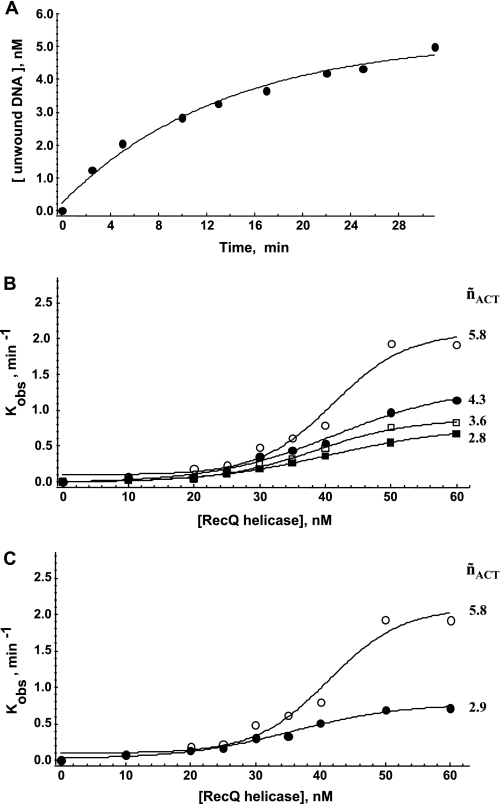
Kinetic Study under Single Turnover Conditions
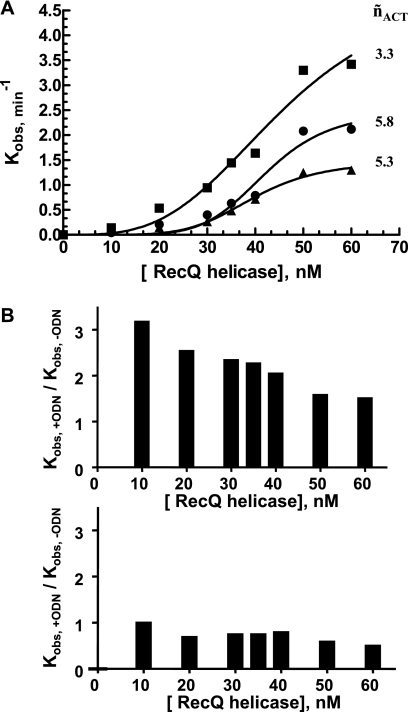
Typically, the time-dependent process described in Fig. 2A was plotted and fitted using a single exponential model (Fig. 3A) corresponding to a single turnover process, with enzyme present in excess over DNA. A similar experimental design was used to assess the influences of (i) enzyme concentration and (ii) DNA substrate size (duplex region) on the first-order kinetic rate constant kobs (T°binding = 37 °C; T°reaction = 25 °C). In the following experiments, all tested DNA substrates harbor two ss-3′-tails (10 bases long) (Fig. 1C, left). Interestingly, the kobs value increased with increasing dsDNA length (13–66 bp) and was also dependent on enzyme concentration (Fig. 3B). Moreover, this response to enzyme concentration strongly depended on dsDNA size and was saturable with, for instance, modest and strong transition effects observed for 13- and 66-bp substrates, respectively (Fig. 3B). The stronger dependence of the first-order kinetic rate constant on helicase concentration for longer DNA substrates strongly suggests that functional cooperativity (˜nACT) between different helicase molecules occurred. Therefore, the stimulation of the unwinding reaction by increasing the size of the DNA substrate probably results from the possibility of multiple RecQ helicase monomers functioning simultaneously on the same DNA substrate.
FIGURE 3.
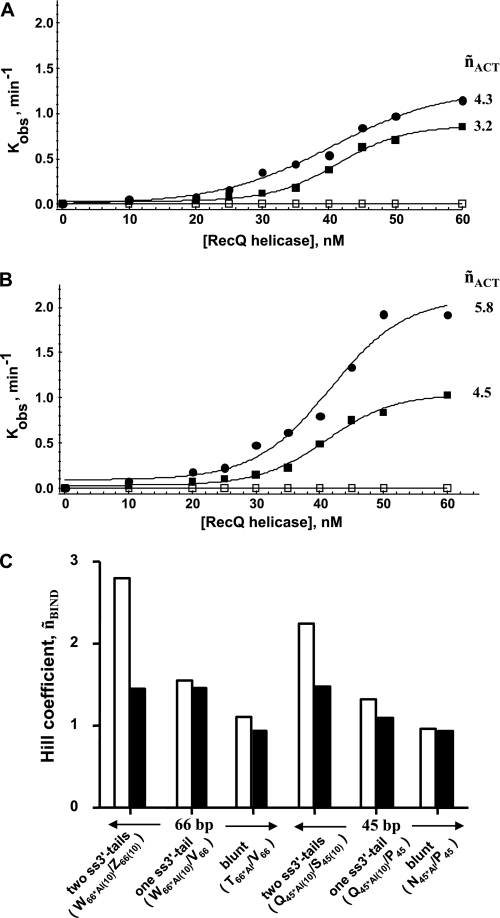
Single turnover study of the unwinding reaction. A, example of a time course of the unwinding reaction under single turnover conditions for the 45-bp DNA substrate (Q45*Al(10)/R45*Te(10)). DNA and E. coli RecQ helicases concentrations were 5 and 10 nm, respectively (T°binding = 37 °C; T°reaction = 25 °C). The amount of unwound DNA was estimated according to the cross-correlation function, gAl/Te(τ), and Equation 3. The single turnover rate constant, kobs, was then calculated from: [DNA]unwound/[DNA]total = 1 − exp(−kobst). B, kobs = f([RecQ helicase]) for different DNA substrate lengths. DNA concentration was 5 nm. The temperature at which DNA binding occurred (before the addition of ATP) was T°binding = 37 °C (T°reaction = 25 °C). White circles, 66 bp (W66*Al(10)/X66*Te(10)); black circles, 45 bp (Q45*Al(10)/R45*Te(10)); white squares, 22 bp (I22*Al(10)/J22*Te(10)); black squares, 13 bp (F13*Al(10)/G13*Te(10)). C, influence of preincubation temperature (corresponding to the DNA binding step: white circles, T°binding = 37 °C; black circles, T°binding = 25 °C) on the subsequent reaction rate (measured at T°reaction = 25 °C). DNA concentration was 5 nm (66-bp substrate (W66*Al(10)/X66*Te(10))). The Hill coefficients characterizing the functional cooperativity (˜nACT) are indicated for each condition.
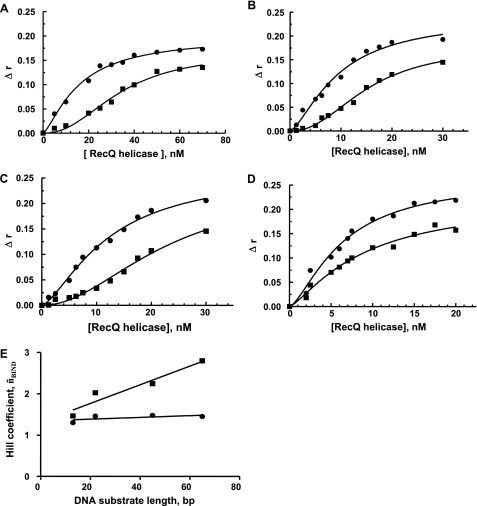
During the time course of our study, we observed that this cooperative behavior was also strongly affected by changes in the temperature of the DNA binding step (before the addition of ATP). As shown in Fig. 3C for the 66-bp substrate, the kobs value (T°reaction = 25 °C) was much less dependent on helicase concentration if the preincubation step was carried out at T°binding = 25 °C (˜nACT = 2.9) than if it was carried out at T°binding = 37 °C (˜nACT = 5.8). We then measured helicase binding to DNA at two temperatures, 25 and 37 °C, by steady-state fluorescence anisotropy (28, 39, 41). DNA binding isotherms for RecQ helicase clearly showed different DNA binding mechanisms at the two different temperatures (Fig. 4, A–D); a cooperative DNA binding mode (˜nBIND) was observed at 37 °C (only for DNA substrates >13 mer), whereas much lower levels of cooperativity were observed at 25 °C. Moreover, the Hill coefficient, ˜nBIND, continuously increased as a function of dsDNA size at 37 °C, whereas DNA binding remained almost non-cooperative at 25 °C regardless of DNA substrate size (Fig. 4E). Our data suggest a direct relationship between the presence of a cooperative DNA binding mode during the preincubation step and the subsequent dependence of unwinding activity on helicase concentration and dsDNA length.
FIGURE 4.
DNA binding properties of the E. coli RecQ helicase. DNA binding was performed in the reaction buffer and measured by monitoring steady-state fluorescence anisotropy (see details under “Experimental Procedures”) at 25 °C (black circles) or 37 °C (black squares). A–D, all DNA substrates were single-labeled with Alexa488: A, 66 bp (W66*Al(10)/Z66(10)); B, 45 bp (Q45*A(10)l/S45(10)); C, 22 bp (I22*Al(10)/L22(10)); D, 13 bp (F13*Al(10)/H13(10)). E, Hill coefficient (related to the DNA binding step, ˜nBIND) as a function of DNA substrate length at two temperatures: 25 °C (black circles) and 37 °C (black squares).
We wondered whether cooperative behavior was influenced by the number of ss/ds junctions (i.e. number of ss-3′-tails (10 bases long)) in the DNA substrate. We assessed unwinding activity with 45- and 66-bp DNA substrates containing zero (blunt), one, or two ss/ds junctions (see Fig. 1C, right). No activity was detected with blunt DNA substrates (Fig. 5, A and B). This result confirms that RecQ helicase requires DNA substrates containing at least one 3′-ssDNA-flanking region for unwinding activity, with little or no unwinding of blunt-ended DNA substrates observed, as reported in previous studies (26, 42, 43). Moreover, with both 45- and 66-bp DNA substrates, unwinding activity was systematically weaker (see kobs values) and less dependent on enzyme concentration (see ˜nACT values) for substrates containing one ss/ds junction than for substrates containing two ss/ds junctions (Fig. 5, A and B). We then studied cooperative DNA binding properties for these DNA substrates. The Hill coefficient, ˜nBIND, varied significantly with the number of ss-3′-tails, with a large difference between substrates containing one ss/ds junction and substrates containing two ss/ds junctions (Fig. 5C). Blunt-ended DNAs displayed almost no cooperativity. Therefore, cooperativity was clearly favored by the presence of ss-3′-tails. Cooperative effects were observed mostly at 37 °C, with weaker effects at 25 °C regardless of the DNA substrate used (consistent with the results shown in Fig. 4E).
FIGURE 5.
Influence of the number of ss-3′-tails on the cooperative DNA binding and reaction modes. The study of kobs = f([RecQ helicase]) (see experimental conditions in the legend for Fig. 3B; T°binding = 37 °C; T°reaction = 25 °C) was repeated, varying the number of ss-3′-tails for two DNA substrate lengths. A, 45 bp: black circles, two ss-3′-tails (Q45*Al(10)/R45*Te(10)); black squares, one ss-3′-tail (Q45*Al(10)/O45*Te); white squares, blunt (N45*Al/O45*Te). B, 66 bp: black circles, two ss-3′-tails (W66*Al(10)/X66*Te(10)); black squares, one ss-3′-tail (W66*Al(10)/U66*Te); white squares, blunt (T66*Al/U66*Te). The Hill coefficients characterizing the functional cooperativity (˜nACT) are indicated for each condition. C, Hill coefficients (DNA binding step, ˜nBIND) for the different DNA substrates at two temperatures: 25 °C (black) and 37 °C (white). DNA binding experiments were carried out as described in the legend for Fig. 4. The DNA substrate names are indicated in parentheses (see also Table 1 for nomenclature).
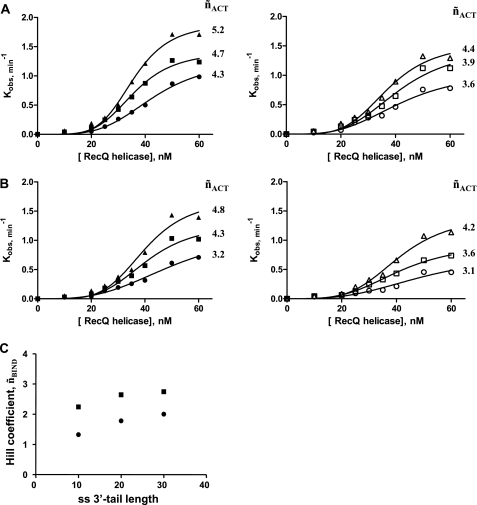
No cooperativity and no significant influence of the ss-3′-tail length on the unwinding rate were evidenced in a recent study by Zhang et al. (26). Nevertheless, the experiments were conducted under conditions that do not favor the cooperative assembly of the helicase-DNA complex, i.e. at 25 °C with rather short DNA substrates (16 bp) harboring one ss-3′-tail. The influence of the ss-3′-tail length for a given size of the duplex region was then further examined under conditions previously shown to be compatible with cooperativity (i.e. T°binding = 37 °C). First, we used either the 22-bp or the 45-bp substrate with two 3′-ssDNA-flanking regions (10, 20, or 30 bases) (see Fig. 1D). Both DNA substrates displayed (i) an unwinding rate and (ii) a functional cooperativity that increased continuously with the size of the ss-3′-tails (Fig. 6A). By varying the size of the ss-3′-tails from 10 to 30 bases, the ˜nACT values increased from 4.3 to 5.2 and from 3.6 to 4.4 for the 45- and 22-bp substrates, respectively. A similar behavior was observed with the corresponding DNA substrates harboring one ss-3′-tail (Fig. 6B). However, as shown previously in Fig. 5, the unwinding rate was systematically less dependent on helicase concentration (see ˜nACT values) for DNA substrates harboring one ss-3′-tail than for substrates harboring two ss-3′-tails for given sizes of the duplex region and ss-3′-tail. Moreover, the influence of the ss-3′-tail length on cooperativity was also observed at the DNA binding level, as shown in Fig. 6C for the 45-bp substrate. Importantly, the influence of the ss-3′-tail length on kobs, ˜nACT, and ˜nBIND values was much less marked when T°binding was 25 °C (data not shown), in accordance with the results obtained by Zhang et al. (26) and confirming the temperature dependence of the cooperative behavior of RecQ helicase.
FIGURE 6.
Influence of the size of the ss-3′-tails on cooperative DNA binding and reaction modes. This is a study of kobs = f([RecQ helicase]) (T°binding = 37 °C; T°reaction = 25 °C) using DNA substrates harboring one or two ss-3′-tails of variable size (10–30 bases long) (see Fig. 1D and Table 1). A, two ss-3′-tails. Left, 45-bp substrate with two 3′-ssDNA-flanking regions of: 10 bases (black circles, Q45*Al(10)/R45*Te(10)), 20 bases (black squares, Q45*Al(20)/R45*Te(20)), or 30 bases (black triangles, Q45*Al(30)/R45*Te(30)). Right, 22-bp substrate with two 3′-ssDNA-flanking regions of: 10 bases (white circles, I22*Al(10)/J22*Te(10)), 20 bases (white squares, I22*Al(20)/J22*Te(20)), or 30 bases (white triangles, I22*Al(30)/J22*Te(30)). B, one ss-3′-tail. Left, 45-bp substrate with one 3′-ssDNA-flanking region of: 10 bases (black circles, N45*Al/R45*Te(10)), 20 bases (black squares, N45*Al/R45*Te(20)), or 30 bases (black triangles, N45*Al/R45*Te(30)). Right, 22-bp substrate with one 3′-ssDNA-flanking region of: 10 bases (white circles, E22*Al/J22*Te(10)), 20 bases (white squares, E22*Al/J22*Te(20)), or 30 bases (white triangles, E22*Al/J22*Te(30)). The Hill coefficients characterizing the functional cooperativity (˜nACT) are indicated for each condition. C, Hill coefficients (DNA binding step, ˜nBIND) for the 45-bp DNA substrate containing either one (circles) or two (squares) ss-3′-tails as a function of the ss-3′-tail length. DNA binding was performed in the reaction buffer and measured by monitoring steady-state fluorescence anisotropy at 37 °C (see details under “Experimental Procedures”).
Altogether, our results suggest that the cooperative mode of DNA binding and the strong helicase concentration dependence of the single turnover kinetic rate constant are closely related phenomena accounting for the number of active monomers simultaneously present on the DNA substrate. In other words, cooperative DNA binding is predictive, at least qualitatively, of the subsequent cooperative catalytic mode, i.e. with at least two helicase monomers catalyzing the unwinding of a single DNA molecule in a synergistic/cooperative manner. The corresponding assembly mechanism is directly dependent on the number of ss-3′-tails present on the DNA substrate and the total length of the substrate, with both the duplex and the 3′-ssDNA-flanking regions contributing to the cooperative assembly. However, it is important to note that the Hill coefficients characterizing the functional cooperativity and the binding of helicase to the DNA substrate (˜nACT and ˜nBIND, respectively; the values are summarized in Table 2) are not identical. The ˜nACT value was found to be systematically higher than the ˜nBIND value for a given DNA substrate, suggesting that the cooperative DNA binding mode of helicase alone does not fully explain the functional cooperativity (see below).
TABLE 2.
Hill coefficients characterizing the functional cooperativity (ñACT) and the cooperative DNA binding mode (ñBIND)
| DNA substrate |
ñACTb | ñBINDc | |
|---|---|---|---|
| Duplex region | No. of ss-3′-tailsa | ||
| 66 bp | ×2 (10) | 5.8 ± 0.3 | 2.8 ± 0.2 |
| ×1 (10) | 4.5 ± 0.4 | 1.55 ± 0.3 | |
| 45 bp | ×2 (10) | 4.3 ± 0.4 | 2.25 ± 0.25 |
| ×2 (20) | 4.7 ± 0.3 | 2.7 ± 0.3 | |
| ×2 (30) | 5.2 ± 0.4 | 2.8 ± 0.35 | |
| ×1 (10) | 3.2 ± 0.3 | 1.3 ± 0.15 | |
| ×1 (20) | 4.3 ± 0.3 | 1.85 ± 0.25 | |
| ×1 (30) | 4.8 ± 0.4 | 2.05 ± 0.25 | |
| 22 bp | ×2 (10) | 3.3 ± 0.2 | 2.0 ± 0.3 |
| 13 bp | ×2 (10) | 2.8 ± 0.3 | 1.45 ± 0.25 |
Kinetic Study under Multiple Turnover Conditions
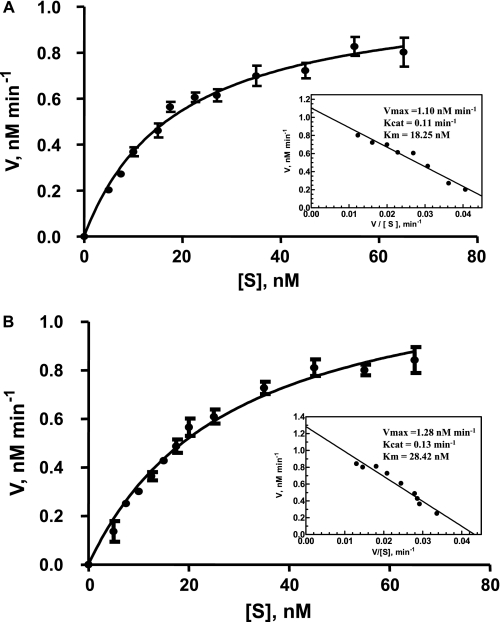
FCCS experiments were also carried out to determine the unwinding activity under Michaelis-Menten conditions in which the DNA substrate is present in excess over enzyme. The concentration of total DNA substrate was varied using mixtures of labeled/unlabeled dsDNA (constant concentration of labeled dsDNA, 5 nm, with increasing concentrations of unlabeled dsDNA), as fluorescence fluctuation spectroscopy and FCS/FCCS analysis are not compatible with high concentrations of fluorescent entities in the excitation volume. The results and corresponding Eadie-Hofstee plots are shown in Fig. 7 for two DNA substrates, a 66-bp (Fig. 7A) and a 22-bp (Fig. 7B) substrate. Unlike the single turnover rate constant kobs (measured under conditions in which enzyme was present in excess over DNA substrate), which was found to be strongly dependent on DNA substrate size, the multiple turnover kcat parameter was not or was only slightly affected by substrate size (kcat,22 mer = 0.128 min−1 and kcat,66 mer = 0.110 min−1), confirming that only a high enzyme:DNA ratio is compatible with the synergistic activity of multiple helicase monomers on the same DNA substrate.
FIGURE 7.
Multiple turnover study of the unwinding reaction. Michaelis-Menten plots for two DNA substrate lengths (inset, corresponding Eadie-Hofstee plots). A, 66-bp substrate. B, 22-bp substrate. 5 nm double-labeled DNA substrate (W66*Al(10)/X66*Te(10) and I22*Al(10)/J22*Te(10) for the 66- and 22-bp substrates, respectively) was mixed with increasing concentrations (0–60 nm) of the unlabeled version of the DNA substrate (Y66(10)/Z66(10) and K22(10)/L22(10) for the 66- and 22-bp substrates, respectively) and preincubated at T°binding = 37 °C with 10 nm E. coli RecQ helicase in the reaction buffer. The temperature was then lowered to 25 °C, and the unwinding reaction was initiated by adding 1 mm ATP (T°reaction = 25 °C). The total concentration of DNA substrate ([S] = labeled + unlabeled) is reported on the x axis. Equilibrium and kinetic parameters are reported in the insets.
Spontaneous Rehybridization after Unwinding Decreases the Apparent Unwinding Rate
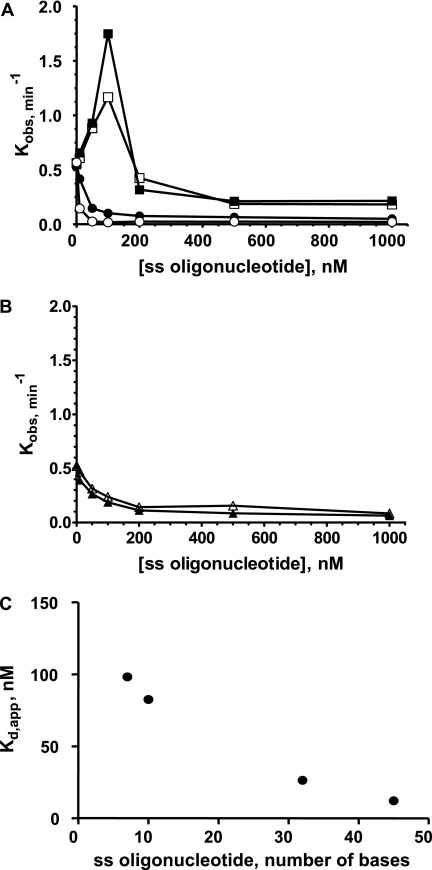
The rehybridization of the two newly separated strands, occurring immediately after DNA unwinding and helicase translocation, may lead to an underestimation of helicase activity as monitored by FCCS, particularly for long DNA substrates. Thus, we characterized quantitatively the effects on the unwinding rate constant of the rehybridization process. To address this question, we measured the unwinding activity using the 66-bp DNA substrate (harboring two ss-3′-tails (10 bases)) under single turnover conditions as described above, except that increasing concentrations of ssODNs (7–45 nucleotides) were added to the preformed helicase-DNA substrate complexes before adding ATP.
All of the ssODNs in Fig. 8A correspond to DNA sequences complementary to one strand of the duplex region of the substrate (see Table 1). Two different effects were observed depending on the size of the ssODN. For short ssODNs (7 and 10 mer), the unwinding activity was first stimulated at low concentrations of ssODN, with concentrations of about 100– 120 nm required for optimal activity. Beyond this critical concentration, unwinding activity was inhibited. By contrast, only inhibitory effects were observed with longer ssODNs, such as 32 and 45 mer. Moreover, non-complementary 7- and 10-mer sequences displayed no stimulation phase (Fig. 8B). These results indicate that the stimulation phase is due to the competitive hybridization of short ssODNs to the unwound complementary strand, whereas inhibition is probably due to trapping effects, i.e. helicase binding to ssODN. Indeed, mostly inhibitory effects were observed with the longer complementary ssODNs, 32 and 45 mer, with higher affinities for helicase (Kd,app = 26 and 12 nm, respectively, Fig. 8C). The affinities of helicase for shorter ssODNs (7 and 10 mer) were much lower (Kd,app = 95 and 88 nm, respectively, Fig. 8C). Consequently, in the latter case, concentrations up to 100 nm were not sufficient to trap helicase to an extent that substantially counteracts the stimulatory hybridization effect. Therefore, such concentrations of ODN stimulate rather than inhibit unwinding, favoring hybridization between the short ODN and the newly unwound complementary strand over rehybridization of the unwound strands.
FIGURE 8.
Dual effect of complementary single-stranded oligonucleotides on the unwinding rate constant. A, the unwinding experiment was performed with the 66-bp DNA substrate (W66*Al(10)/X66*Te(10)) in the presence of increasing concentrations of complementary ssODNs of variable sizes: white squares, 7 mer (B7); black squares, 10 mer (D10); black circles, 32 mer (M32); white circles, 45 mer (P45). The DNA substrate and E. coli RecQ helicase concentrations were 5 and 35 nm, respectively (T°binding = 37 °C; T°reaction = 25 °C). B, control experiments with random non-complementary sequences. White triangles, 7 mer (B′7); black triangles, 10 mer (D′10). C, apparent Kd values characterizing the interaction between RecQ helicase and ssODNs as a function of ODN length. Kd,app values were determined by steady-state fluorescence anisotropy using the following Alexa488-labeled ODNs: A7*Al, C10*Al, I22*Al(10), and N45*Al for 7, 10, 32, and 45 mer, respectively.
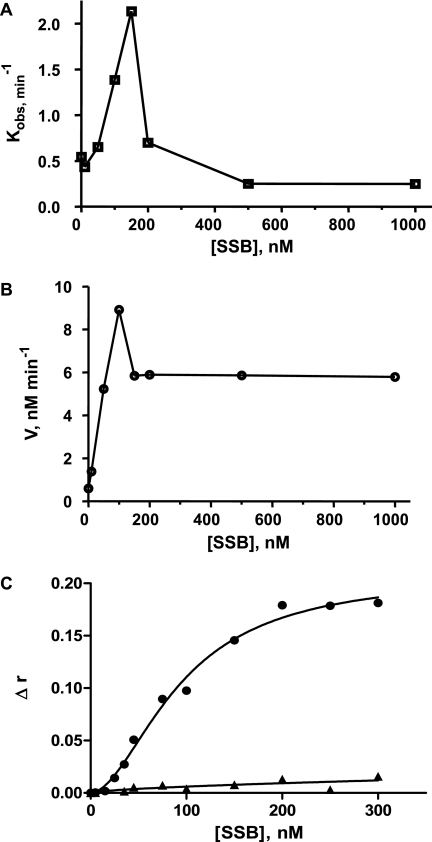
These effects of short ssODNs on unwinding activity were compared with the effects of SSB under similar experimental conditions (Fig. 9A). A similar stimulatory effect on the single turnover rate constant (increase by a factor of up to 4) was observed for concentrations of SSB up to 150 nm. The SSB-dependent stimulation of activity may result from two different mechanisms: (i) SSB may prevent the rehybridization process in a manner similar to that for short ssODNs, because SSB strongly inhibits annealing (44–47); or (ii) SSB may bind to ssDNA products, thereby increasing the concentration of helicase available for catalysis. Under single turnover conditions, the first mechanism alone probably accounts for the stimulatory effect. As shown in Fig. 9B, the stimulatory SSB effect was significantly stronger under multiple turnover conditions (increase by a factor of up to 18). We therefore hypothesized that both mechanisms contribute to the stimulation of helicase activity in multiple turnover conditions. However, high SSB concentrations (>150 nm) decreased helicase activity under both single and multiple turnover conditions, with a more pronounced effect under single turnover conditions. E. coli RecQ helicase has been shown to interact physically with SSB, and a related stimulatory effect of SSB on the unwinding activity has been suggested (48, 49). Nevertheless, the relatively high Kd value characterizing this interaction (6 μm) cannot fully explain the stimulatory effects observed under our experimental conditions (below 150 nm SSB). By contrast, this Kd value is more compatible with a relationship between the formation of helicase-SSB complexes and the observed inhibitory effect for SSB concentrations above 150 nm. This is consistent with the observation that SSB-mediated inhibition is more potent under single turnover conditions, because under multiple turnover conditions (excess of DNA over helicase), ssDNA products may compete with RecQ helicase for binding to SSB.
FIGURE 9.
Comparative study of SSB effects on the unwinding activity of E. coli RecQ helicase under single and multiple turnover conditions. The unwinding experiments were performed with the 66-bp DNA substrate (W66*Al(10)/X66*Te(10)) in the presence of increasing concentrations of SSB. A, single turnover conditions (see experimental conditions in the legend for Fig. 8A). B, multiple turnover conditions (see experimental conditions in the legend for Fig. 7). C, DNA binding isotherms (measured by fluorescence anisotropy) of SSB (at 25 °C) using either ssDNA (circles, W66*Al(10)) or dsDNA (triangles, W66*Al(10)/Z66(10)).
Insight into the Origin of the Functional Cooperativity
The strong dependence of the single turnover kinetic rate constant on the DNA substrate size is compatible with the simultaneous unwinding of an individual DNA substrate by multiple helicase monomers. This functional cooperativity primarily originates from the simultaneous binding of multiple helicase monomers to the same DNA molecule prior to catalysis. However, as mentioned above, the Hill coefficients related to the functional cooperativity were found to be systematically higher than the corresponding coefficients related to the DNA binding step (Table 2). We next assessed whether post-catalytic events may also contribute to the observed positive functional cooperativity. Indeed, under single turnover conditions, the excess of helicase over DNA substrate is compatible with the binding of helicase molecules to the newly unwound DNA strands, leading to an apparent cooperative effect by minimizing the rehybridization process.
We reasoned that if the helicase binding to the ssDNA product significantly contributes to the overall functional cooperativity, this contribution should decrease in the presence of short complementary ssODNs that also minimize the rehybridization process (see previous section). We then performed a comparative study by measuring kobs as a function of helicase concentration (using the 66-bp DNA substrate, W66*Al(10)/X66*Te(10)) in the absence or presence of the short complementary 10-mer ssODN at 120 nm (which prevents the rehybridization process and stimulates unwinding activity, with optimum concentration ≈ 120 nm as shown in Fig. 8). Although an overall stimulatory effect was observed (confirming the result shown in Fig. 8A), the functional cooperativity was significantly reduced (from ˜nACT = 5.8 to ˜nACT = 3.3) upon the addition of the complementary 10-mer ssODN (Fig. 10A). This result indicates that in the presence of the complementary 10-mer ssODN, the response of unwinding activity to helicase concentration is weaker, as confirmed in Fig. 10B (top). It is important to note that only a slight decrease in the ˜nACT value (from 5.8 to 5.3) was observed using a non-complementary 10-mer ssODN (competent for helicase binding but not competent to prevent the rehybridization process) (Fig. 10, A and B (bottom)).
FIGURE 10.
Functional cooperativity is also controlled by a post-catalytic event. A, kobs = f([RecQ helicase]) for the 66-bp DNA substrate (W66*Al(10)/X66*Te(10)) in the absence (circles) or presence of either complementary (squares, D10) or non-complementary (triangles, D′10) single-stranded 10-mer ODN. The experimental conditions were identical to those described in the legend for Fig. 3B (T°binding = 37 °C; T°reaction = 25 °C). The concentration of D10 or D′10 was 120 nm. The Hill coefficients characterizing the functional cooperativity (˜nACT) are indicated for each condition. B, ratio between kobs measured in the presence (+ODN) and absence (−ODN) of single-stranded 10-mer ODN as a function of helicase concentration. Top, ODN = D10; bottom, ODN = D′10.
Taken together, these results show that (i) the specific decrease in the cooperativity coefficient by the short complementary 10-mer ssODN is not explained by the propensity of the ODN to trap helicase, and (ii) the complementary 10-mer ssODN prevents the rehybridization process and then minimizes the impact of the post-catalytic binding of helicase molecules to newly unwound ssDNA products. Consequently, the overall functional cooperativity is significantly reduced, indicating that two mechanisms contribute to the functional cooperativity under standard conditions. The first one is direct, with the possibility of having multiple helicases that cooperate for DNA unwinding. This mechanism appears to be strongly related to the cooperative DNA binding mode of helicase. The second mechanism is indirect and involves the binding of the newly unwound DNA strands by helicase molecules. It is noteworthy that the Hill coefficient characterizing the functional cooperativity as obtained in the presence of the short complementary ssODN (˜nACT = 3.3) is more consistent with the Hill coefficient characterizing the DNA binding step for the same DNA substrate (˜nBIND = 2.8) (see Fig. 4E and Table 2, first line).
DISCUSSION
This study demonstrates that the FCCS approach is particularly suitable for monitoring helicase activity. We used this approach to study the unwinding activity of E. coli RecQ helicase in both single and multiple turnover conditions. Interestingly, in single turnover conditions, strong functional cooperative effects were observed. These effects were dependent on several factors, namely the total length of dsDNA, the number and size of ss-3′-tails, and temperature. The dependence of the single turnover kinetic rate constant on both helicase concentration and DNA substrate size suggests cooperative/synergistic effects compatible with the simultaneous unwinding of an individual DNA substrate by multiple helicase monomers. Consequently, the apparent unwinding activity is strongly stimulated by increasing DNA substrate length and using a high helicase:DNA ratio. Consistent with this finding, no such dependence on DNA size was observed in multiple turnover conditions, i.e. in which the DNA substrate was present in excess over helicase. Furthermore, we observed a direct relationship between the cooperative properties of helicase binding to the DNA substrate and subsequent cooperative effects occurring at the catalytic level. All of the factors listed above (length of duplex region, number and length of ss-3′-tails, and temperature) influence both the DNA binding mode (cooperative or not) and the functional cooperativity. No cooperative DNA binding was evidenced in a recent study by Zhang et al. (26) performed at 25 °C. To date, only a cooperative response of E. coli RecQ helicase activity to ATP has been reported (at 37 °C) (27). We show here that the E. coli RecQ helicase behaves cooperatively only at 37 °C and that the cooperative DNA binding mode of helicase at this temperature fully accounts for the sigmoidal response of helicase activity to ATP concentration (see below). Finally, we show that the functional cooperativity is a composite parameter, taking into account two distinct cooperative effects: the first one corresponds to a cooperative DNA binding mode of helicase prior to unwinding, with the possibility of multiple RecQ helicases functioning simultaneously on the same DNA substrate, as mentioned above; whereas the second mechanism corresponds to a post-catalytic process involving the binding of helicase molecules to newly unwound ssDNA products. The latter mechanism prevents the rehybridization of newly separated strands, leading to an apparent stimulatory effect that is also dependent on enzyme concentration.
FCCS, like other fluorescence-based methods (e.g. fluorescence anisotropy and FRET), is a sensitive method for the direct measurement of helicase activity without the need to separate the ssDNA product from the dsDNA substrate by gel electrophoresis. In steady-state fluorescence anisotropy, the difference in size between the helicase-dsDNA substrate complex and the released fluorescently labeled strand is measured (28). This means that the fluorescence anisotropy approach is compatible with single turnover conditions only (ensuring initial saturation of the DNA substrate). In addition, the decrease in anisotropy accompanying unwinding activity may be very small if helicase remains bound to the labeled strand, complicating analysis. However, this particular problem has been circumvented by Xu et al. (28) using a short labeled strand (<13 mer) with a lower affinity for helicase (confirmed in Fig. 8C). Unlike fluorescence anisotropy, FCCS does not measure a size change; it is sensitive only to the concomitant diffusion of the two fluorescently labeled DNA strands, with high cross-correlation amplitude characterizing the duplex molecule and a significant decrease in this amplitude upon physical separation of the two strands. This method is therefore not limited by the size of the DNA strands initially constituting the DNA substrate.
However, it is important to note that the inherent problem of the ssDNA binding properties of helicase, although not technically limiting in FCCS, results in a strong underestimation of unwinding activity, particularly in multiple turnover conditions. Indeed, the binding of helicase to ssDNA products competes with binding to dsDNA substrates, limiting activity. SSB partly counteracts this effect, thereby increasing apparent catalytic activity (50). Moreover, the other stimulatory effect of SSB on helicase activity, through the inhibition of spontaneous rehybridization of unwound strands, is probably common to both the single and multiple turnover modes of catalysis. The participation of the two mechanisms in the stimulation of helicase activity under multiple turnover conditions, with only one mechanism thought to be involved in single turnover conditions (rehybridization inhibition), probably accounts for the more efficient stimulation of DNA unwinding by SSB in multiple turnover than in single turnover conditions (Fig. 9). However, the precise nature of the mechanism of SSB-mediated stimulation is not clear so far, because SSB was recently reported to also stimulate E. coli RecQ helicase through a direct physical SSB-RecQ helicase interaction (48, 49). However, given its Kd value (6 μm), this interaction is unlikely to be responsible for the SSB-mediated stimulatory effect observed under our experimental conditions. The apparent Kd value characterizing the SSB-ssDNA interaction, as measured by fluorescence anisotropy (Kd,app = 94 nm; Fig. 9C), is more consistent with the observed stimulatory effect below 150 nm SSB, reinforcing the idea that SSB primarily counteracts both trapping and rehybridization effects. By contrast, it seems likely that the inhibition phase, observed for SSB concentrations above 150 nm, may be at least partly due to this helicase-SSB interaction. Alternatively, high SSB concentrations may displace helicase from the DNA substrate ss-3′-tails.
Short complementary ssODNs (typically 7 or 10 mer) have similar stimulatory effects on helicase activity by minimizing spontaneous rehybridization. However, as found with SSB, this stimulation was limited, as it was followed by a marked inhibition phase, due to competitive interactions, that was detrimental to activity. In the case of short ssODNs, this inhibition can be explained by their significant affinities for helicase (Kd,app = 95 and 88 nm for 7 and 10 mer, respectively). Indeed, longer ssODNs (32 or 45 mer) with even higher affinities (Kd,app < 25 nm) displayed inhibition profiles only. Therefore, due to competitive interactions, SSB and short ssODNs most likely only partially increase the apparent unwinding rate, and their stimulatory effects may therefore be considered modest. Indeed, the apparent unwinding rate of 0.01–0.06 s−1 determined in this study for FCCS (depending on the nature of the DNA substrate used and experimental conditions) is globally consistent with rates from other studies, 0.03–0.2 s−1 (28, 37), but is much lower than the rate of 8 s−1 recently determined by FRET/stopped-flow experiments (26). Unlike FRET, the cross-correlation signal in FCCS is independent of interfluorophore distance and the structural dynamics of the attached fluorophores. FCCS is sensitive only to the physical and complete separation of the two DNA strands and is therefore less sensitive to early catalytic events. This suggests that spontaneous rehybridization may be more problematic in FCCS than in the FRET/stopped-flow approach.
The enzyme excess under single turnover conditions also contributes to the prevention of spontaneous rehybridization, because of the ability of helicase to bind to the newly unwound ssDNA product (post-catalytic process). This contribution is reduced in the presence of a short complementary ssODN but not in the presence of a non-complementary ssODN of equivalent length (Fig. 10). Moreover, we found that this post-catalytic process contributes significantly, together with the possibility of multiple helicase molecules functioning simultaneously on the same DNA substrate, to the high functional cooperativity (˜nACT = 5.8 for the 66-bp substrate) observed under standard conditions, i.e. without any addition of either short complementary ssODNs or SSB. The inhibition of the DNA rehybridization upon the addition of short complementary ssODNs significantly reduces the impact of the helicase binding to newly unwound DNA strands. Consequently, the ˜nACT value decreases (3.3 for the 66-bp substrate). This latter value is more consistent with the corresponding ˜nBIND value (2.8), which is independent of post-catalytic events. It is important to note that the post-catalytic binding of helicase to the ssDNA product is most likely detrimental for activity under multiple turnover conditions because it decreases the concentration of available enzyme for catalysis in subsequent turnovers. There is no such effect under single turnover conditions, as only the activity related to preformed complexes is measured.
We found that the binding to DNA of E. coli RecQ helicase was cooperative at 37 °C but was essentially non-cooperative at 25 °C. Moreover, the cooperativity coefficient continuously increased with the duplex region size of the DNA substrate and was also strongly dependent on the number/size of ss-3′-tails. Consequently, a high cooperativity index was obtained at 37 °C for long DNA substrates harboring two ss-3′-tails. It has been shown previously that the binding of RecQ helicase to DNA is non-cooperative at 25 °C with DNA substrates containing one ss-3′-tail (26). Our results obtained under similar conditions confirmed this statement. Furthermore, our results highlight the existence of different modes of DNA binding by E. coli RecQ helicase as a function of temperature, suggesting that only a specific protein conformation is compatible with cooperative assembly. Cooperativity also was found to be strongly related to the DNA substrate size, and the very low level of cooperativity observed for the short DNA substrate (13 bp) even at 37 °C is compatible with the previously determined binding size of 9–10 nucleotides for a monomeric unit (26, 51). As RecQ helicase is active with short DNA substrates, although no cooperativity is associated with such substrates, we concluded that the monomer is sufficient to ensure DNA unwinding. Moreover, the dependence of both ˜nACT and ˜nBIND coefficients on the DNA substrate length argues against the existence of a specific higher order multimeric state and, most likely, reflects the number of loaded helicases on the substrate; increasing the DNA length favors the binding of multiple catalytic entities, thereby increasing the apparent unwinding activity. This functional cooperativity, in association with a cooperative DNA binding mode, suggests that RecQ helicase monomers may function in a concerted manner on an individual DNA molecule, resulting in a substantial increase in the unwinding activity when multiple monomers are simultaneously bound to the DNA substrate. This property appears to be common to several monomeric helicases such as the hepatitis C virus helicase (52, 53), bacteriophage T4 Dda (54), and yeast Pif1p (55). Several models have been proposed to explain the related stimulation of activity, including the possibility of a monomer preventing spontaneous rehybridization behind another monomer after its translocation. It is important to note that no cooperativity in DNA binding has been evidenced for at least two of these monomeric helicases: hepatitis C virus helicase (53) and Dda (54). For the E. coli RecQ helicase, we show that functional cooperativity is correlated with the presence of a cooperative DNA binding mode. Indeed, all factors modulating the Hill coefficient characterizing the helicase-DNA interaction also influence functional cooperativity. Although the physical significance of this correlation remains to be elucidated, one can reasonably hypothesize that, as mentioned above, the cooperativity reflects the number of helicase molecules bound to the same DNA substrate. This number is clearly controlled by the number and length of ss-3′-tails (Figs. 5 and 6) (already shown by others to be an important factor controlling the number of loaded helicases on the DNA substrate (53)). However, cooperativity significantly increases with increasing duplex DNA region size (Figs. 3 and 4). Taking into account that all DNA substrates used in Figs. 3 and 4 contained the same number of ss-3′-tails and had identical single-stranded tail lengths, our results indicate that the internal duplex region, not only the ss-3′-tail, plays an important role in the cooperative DNA binding mode. Interestingly, no cooperativity was evidenced with blunt-ended DNAs (even with long 45- or 66-bp DNAs (Fig. 5C)) with a concomitant loss of activity. Moreover, cooperativity is not strictly required for subsequent unwinding activity, as shown in the case of the short 13-bp DNA substrate or when helicase activity is measured at 25 °C, regardless of the DNA size. Altogether, our results indicate that: (i) the ss/ds junction is essential to initiate the catalytic reaction; (ii) the ss/ds junction also promotes cooperative assembly of DNA-helicases complexes, although this role is not catalytically essential; and (iii) the duplex region may contribute to the cooperative assembly but only if the DNA substrate harbors at least one ss-3′-tail, which is then strictly required to promote cooperative assembly. It has been suggested that RecQ helicase preferentially binds to ss/ds DNA junctions (26). At first sight, such a preference would account for the effect of the number of ss/ds junctions on the cooperativity. These ss/ds junctions with a 3′-ssDNA-flanking region play a critical role in controlling the entry process of RecQ helicase, which unwinds dsDNA in the 3′ → 5′ direction. Consequently, the presence of two ss-3′-tails makes it possible for helicase molecules to unwind and translocate along DNA from the two opposite 3′-ends, increasing apparent unwinding rate. Taking into account that unwinding activity cannot be initiated from the DNA duplex region, although the cooperative binding of helicase to the duplex region contributes positively to the unwinding reaction, we hypothesize that one role of helicase molecules bound to the duplex region could be to substitute for another helicase, in the case of premature dissociation of a poorly processive helicase from the DNA substrate. However, the exact mechanism behind the cooperative effects occurring at each ss/ds junction with significant contributions from both the duplex region and ss-3′-tail remains to be elucidated.
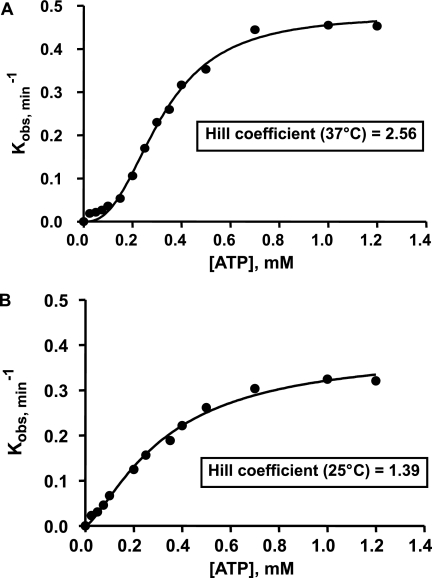
Interestingly, a previous study described a cooperative dependence (sigmoidal response) of E. coli RecQ helicase on ATP concentration (at 37 °C) characterized by a Hill coefficient of 3.3, suggesting that helicase functions as an allosteric multimeric enzyme (at least trimeric and possibly hexameric) (27). This finding apparently conflicts with the finding of others studies showing that RecQ helicase functions as a monomer (26, 37). Moreover, another study states that RecQ helicase binds DNA with no cooperativity (26). It is noteworthy that, in the latter case, the study was carried out at 25 °C, corresponding to the conditions in which no cooperativity was observed in our assays. By contrast, we found that a significant cooperative DNA binding mode exists at 37 °C. We addressed the possibility that the cooperative response of helicase activity to ATP concentration is only apparent and that it originates in the cooperative assembly of helicase-DNA complexes rather than an allosteric transition. Supplemental Equation 11 (see “Appendix”) shows that helicase activity may display a cooperative response to ATP concentration simply because of the indirect cooperative effects at the DNA binding level, in the absence of any allosteric transition and higher order oligomeric organization, which reconciles most of the apparent contradictions in previous reports. To further investigate the relationship between cooperative DNA binding by helicase and its cooperative behavior in terms of ATP dependence, we measured the rate of unwinding in the presence of increasing concentrations of ATP at two temperatures: 25 and 37 °C. In accordance with our model, stronger cooperative behavior was observed at 37 than at 25 °C (Fig. 11). The Hill coefficient at 37 °C, 2.6 (Fig. 11A), is compatible with the Hill coefficient characterizing helicase-DNA complex formation for a similar DNA length, ˜nBIND = 2.8 (Fig. 4B). The corresponding coefficients at 25 °C were also consistent at 1.4 (Fig. 11B) and 1.35 (Fig. 4B), respectively. The slightly lower than previously reported Hill coefficient (2.6 in this study and 3.3 in Ref. 27) characterizing the response to the ATP concentration may be due to the nature of the DNA substrate used, which is shorter in our study. Importantly, the contribution to the overall functional cooperativity of DNA rehybridization inhibition by post-catalytic helicase binding to newly unwound DNA strands probably does not interfere with the cooperative dependence of helicase on ATP concentration, as suggested by the good correlation obtained between ˜nBIND and the Hill coefficient related to the cooperative response to ATP, regardless of the presence or absence of short complementary ssODNs in the helicase assay. The x-ray structure of the DNA-free catalytic core of E. coli RecQ helicase (named RecQΔC; 516 amino acids) shows a monomeric protein (56) in accordance with biochemical and biophysical characterizations of the DNA-free full-length protein (610 amino acids) by time-resolved fluorescence anisotropy and analytical ultracentrifugation (37). Altogether, our results indicate that RecQ helicase monomers bind cooperatively to long DNA substrates and cooperate in DNA unwinding, although the catalytic unit is actually the monomeric form, which is functionally sufficient for short substrates. In conclusion, this cooperative DNA binding mode accounts for the sigmoidal response of helicase unwinding activity to ATP.
FIGURE 11.
Differential cooperative effects of ATP on helicase unwinding activity as a function of temperature. The unwinding experiments were performed with the 66-bp DNA substrate (W66*Al(10)/X66*Te(10)) (5 nm) in the presence of 35 nm E. coli RecQ helicase and increasing concentrations of ATP. A, T°binding = T°reaction = 37 °C. B, T°binding = T°reaction = 25 °C.
Supplementary Material
Acknowledgment
We thank Prof. Zhao Yulong from East China Normal University, Shanghai, for interest in this work.
This work was supported by grants from the CNRS, Ecole Normale Supérieure de Cachan, East China Normal University (ECNU)-Shanghai, and the Institut d'Alembert.

The on-line version of this article (available at http://www.jbc.org) contains a supplemental “Appendix.”
- FCS
- fluorescence correlation spectroscopy
- FCCS
- fluorescence cross-correlation spectroscopy
- Al
- Alexa Fluor 488 (Alexa488)
- ds
- double-stranded
- FRET
- fluorescence resonance energy transfer
- ODN
- oligodeoxyribonucleotide
- ss
- single-stranded
- SSB
- single-stranded DNA-binding protein
- Te
- Texas Red
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1.Patel S. S., Donmez I. (2006) J. Biol. Chem. 281, 18265–18268 [DOI] [PubMed] [Google Scholar]
- 2.Singleton M. R., Dillingham M. S., Wigley D. B. (2007) Annu. Rev. Biochem. 76, 23–50 [DOI] [PubMed] [Google Scholar]
- 3.Killoran M. P., Keck J. L. (2006) Nucleic Acids Res. 34, 4098–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosh R. M., Jr., Bohr V. A. (2007) Nucleic Acids Res. 35, 7527–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma S., Brosh R. M., Jr. (2008) Cell Cycle 7, 989–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakayama H., Nakayama K., Nakayama R., Irino N., Nakayama Y., Hanawalt P. C. (1984) Mol. Gen. Genet. 195, 474–480 [DOI] [PubMed] [Google Scholar]
- 7.Hanada K., Ukita T., Kohno Y., Saito K., Kato J., Ikeda H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3860–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galletto R., Jezewska M. J., Bujalowski W. (2004) J. Mol. Biol. 343, 101–114 [DOI] [PubMed] [Google Scholar]
- 9.Galletto R., Jezewska M. J., Bujalowski W. (2004) J. Mol. Biol. 343, 83–99 [DOI] [PubMed] [Google Scholar]
- 10.Yu X., Jezewska M. J., Bujalowski W., Egelman E. H. (1996) J. Mol. Biol. 259, 7–14 [DOI] [PubMed] [Google Scholar]
- 11.Yu X., Horiguchi T., Shigesada K., Egelman E. H. (2000) J. Mol. Biol. 299, 1279–1287 [DOI] [PubMed] [Google Scholar]
- 12.Norcum M. T., Warrington J. A., Spiering M. M., Ishmael F. T., Trakselis M. A., Benkovic S. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3623–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singleton M. R., Sawaya M. R., Ellenberger T., Wigley D. B. (2000) Cell 101, 589–600 [DOI] [PubMed] [Google Scholar]
- 14.Jeong Y. J., Levin M. K., Patel S. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7264–7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng W., Hsieh J., Brendza K. M., Lohman T. M. (2001) J. Mol. Biol. 310, 327–350 [DOI] [PubMed] [Google Scholar]
- 16.Soultanas P., Dillingham M. S., Wiley P., Webb M. R., Wigley D. B. (2000) EMBO J. 19, 3799–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soultanas P., Wigley D. B. (2000) Curr. Opin. Struct. Biol. 10, 124–128 [DOI] [PubMed] [Google Scholar]
- 18.Velankar S. S., Soultanas P., Dillingham M. S., Subramanya H. S., Wigley D. B. (1999) Cell 97, 75–84 [DOI] [PubMed] [Google Scholar]
- 19.Mechanic L. E., Hall M. C., Matson S. W. (1999) J. Biol. Chem. 274, 12488–12498 [DOI] [PubMed] [Google Scholar]
- 20.Maluf N. K., Fischer C. J., Lohman T. M. (2003) J. Mol. Biol. 325, 913–935 [DOI] [PubMed] [Google Scholar]
- 21.Yang Y., Dou S. X., Ren H., Wang P. Y., Zhang X. D., Qian M., Pan B. Y., Xi X. G. (2008) Nucleic Acids Res. 36, 1976–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Compton S. A., Tolun G., Kamath-Loeb A. S., Loeb L. A., Griffith J. D. (2008) J. Biol. Chem. 283, 24478–24483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karow J. K., Newman R. H., Freemont P. S., Hickson I. D. (1999) Curr. Biol. 9, 597–600 [DOI] [PubMed] [Google Scholar]
- 24.Choudhary S., Sommers J. A., Brosh R. M., Jr. (2004) J. Biol. Chem. 279, 34603–34613 [DOI] [PubMed] [Google Scholar]
- 25.Janscak P., Garcia P. L., Hamburger F., Makuta Y., Shiraishi K., Imai Y., Ikeda H., Bickle T. A. (2003) J. Mol. Biol. 330, 29–42 [DOI] [PubMed] [Google Scholar]
- 26.Zhang X. D., Dou S. X., Xie P., Hu J. S., Wang P. Y., Xi X. G. (2006) J. Biol. Chem. 281, 12655–12663 [DOI] [PubMed] [Google Scholar]
- 27.Harmon F. G., Kowalczykowski S. C. (2001) J. Biol. Chem. 276, 232–243 [DOI] [PubMed] [Google Scholar]
- 28.Xu H. Q., Zhang A. H., Auclair C., Xi X. G. (2003) Nucleic Acids Res. 31, e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haustein E., Schwille P. (2007) Annu. Rev. Biophys. Biomol. Struct. 36, 151–169 [DOI] [PubMed] [Google Scholar]
- 30.Clamme J. P., Azoulay J., Mély Y. (2003) Biophys. J. 84, 1960–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., Ercelen S., Duportail G., Schaub E., Tikhonov V., Slita A., Zarubaev V., Babak V., Mély Y. (2008) J. Gene Med. 10, 527–539 [DOI] [PubMed] [Google Scholar]
- 32.Kohl T., Haustein E., Schwille P. (2005) Biophys. J. 89, 2770–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rarbach M., Kettling U., Koltermann A., Eigen M. (2001) Methods 24, 104–116 [DOI] [PubMed] [Google Scholar]
- 34.Kettling U., Koltermann A., Schwille P., Eigen M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1416–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collini M., Caccia M., Chirico G., Barone F., Dogliotti E., Mazzei F. (2005) Nucleic Acids Res. 33, e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jahnz M., Schwille P. (2005) Nucleic Acids Res. 33, e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu H. Q., Deprez E., Zhang A. H., Tauc P., Ladjimi M. M., Brochon J. C., Auclair C., Xi X. G. (2003) J. Biol. Chem. 278, 34925–34933 [DOI] [PubMed] [Google Scholar]
- 38.Delelis O., Carayon K., Guiot E., Leh H., Tauc P., Brochon J. C., Mouscadet J. F., Deprez E. (2008) J. Biol. Chem. 283, 27838–27849 [DOI] [PubMed] [Google Scholar]
- 39.Guiot E., Carayon K., Delelis O., Simon F., Tauc P., Zubin E., Gottikh M., Mouscadet J. F., Brochon J. C., Deprez E. (2006) J. Biol. Chem. 281, 22707–22719 [DOI] [PubMed] [Google Scholar]
- 40.Pinskaya M., Romanova E., Volkov E., Deprez E., Leh H., Brochon J. C., Mouscadet J. F., Gottikh M. (2004) Biochemistry 43, 8735–8743 [DOI] [PubMed] [Google Scholar]
- 41.Deprez E., Barbe S., Kolaski M., Leh H., Zouhiri F., Auclair C., Brochon J. C., Le Bret M., Mouscadet J. F. (2004) Mol. Pharmacol. 65, 85–98 [DOI] [PubMed] [Google Scholar]
- 42.Zhang X. D., Dou S. X., Xie P., Wang P. Y., Xi X. G. (2005) Acta Biochim. Biophys. Sin. (Shanghai) 37, 593–600 [DOI] [PubMed] [Google Scholar]
- 43.Hishida T., Han Y. W., Shibata T., Kubota Y., Ishino Y., Iwasaki H., Shinagawa H. (2004) Genes Dev. 18, 1886–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheok C. F., Wu L., Garcia P. L., Janscak P., Hickson I. D. (2005) Nucleic Acids Res. 33, 3932–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mortier-Barrière I., Velten M., Dupaigne P., Mirouze N., Piétrement O., McGovern S., Fichant G., Martin B., Noirot P., Le Cam E., Polard P., Claverys J. P. (2007) Cell 130, 824–836 [DOI] [PubMed] [Google Scholar]
- 46.Martin S. L., Bushman F. D. (2001) Mol. Cell. Biol. 21, 467–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kantake N., Madiraju M. V., Sugiyama T., Kowalczykowski S. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15327–15332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shereda R. D., Bernstein D. A., Keck J. L. (2007) J. Biol. Chem. 282, 19247–19258 [DOI] [PubMed] [Google Scholar]
- 49.Shereda R. D., Reiter N. J., Butcher S. E., Keck J. L. (2009) J. Mol. Biol. 386, 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roman L. J., Dixon D. A., Kowalczykowski S. C. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 3367–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dou S. X., Wang P. Y., Xu H. Q., Xi X. G. (2004) J. Biol. Chem. 279, 6354–6363 [DOI] [PubMed] [Google Scholar]
- 52.Levin M. K., Gurjar M., Patel S. S. (2005) Nat. Struct. Mol. Biol. 12, 429–435 [DOI] [PubMed] [Google Scholar]
- 53.Levin M. K., Wang Y. H., Patel S. S. (2004) J. Biol. Chem. 279, 26005–26012 [DOI] [PubMed] [Google Scholar]
- 54.Byrd A. K., Raney K. D. (2004) Nat. Struct. Mol. Biol. 11, 531–538 [DOI] [PubMed] [Google Scholar]
- 55.Boulé J. B., Zakian V. A. (2007) Nucleic Acids Res. 35, 5809–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernstein D. A., Zittel M. C., Keck J. L. (2003) EMBO J. 22, 4910–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.