Abstract
The high glucose consumption of tumor cells even in an oxygen-rich environment, referred to as the Warburg effect, has been noted as a nearly universal biochemical characteristic of cancer cells. Targeting the glycolysis pathway has been explored as an anti-cancer therapeutic strategy to eradicate cancer based on this fundamental biochemical property of cancer cells. Oncoproteins such as Akt and c-Myc regulate cell metabolism. Accumulating studies have uncovered various molecular mechanisms by which oncoproteins affect cellular metabolism, raising a concern as to whether targeting glycolysis will be equally effective in treating cancers arising from different oncogenic activities. Here, we established a dual-regulatable FL5.12 pre-B cell line in which myristoylated Akt is expressed under the control of doxycycline, and c-Myc, fused to the hormone-binding domain of the human estrogen receptor, is activated by 4-hydroxytamoxifen. Using this system, we directly compared the effect of these oncoproteins on cell metabolism in an isogenic background. Activation of either Akt or c-Myc leads to the Warburg effect as indicated by increased cellular glucose uptake, glycolysis, and lactate generation. When cells are treated with glycolysis inhibitors, Akt sensitizes cells to apoptosis, whereas c-Myc does not. In contrast, c-Myc but not Akt sensitizes cells to the inhibition of mitochondrial function. This is correlated with enhanced mitochondrial activities in c-Myc cells. Hence, although both Akt and c-Myc promote aerobic glycolysis, they differentially affect mitochondrial functions and render cells susceptible to the perturbation of cellular metabolic programs.
Keywords: Apoptosis, Bioenergetics, Cancer, Metabolism, Akt/PKB, Glycolysis, Mitochondria, Myc
Introduction
During tumorigenesis, cancer cells rewire their metabolic programs to benefit their growth, proliferation, and survival (1). One of the most drastically altered metabolic pathways in tumor cells is glycolysis. As first documented by Warburg (2), cancerous cells exhibit higher glycolytic activity even when oxygen is abundant as compared with nonmalignant cells. This phenomenon is referred to as the “Warburg effect” or “aerobic glycolysis,” which serves as the foundation for the positron emission tomography that is widely used in clinical cancer diagnosis. The use of positron emission tomography has in turn confirmed the Warburg effect as a universal property of cancer cells (3). An abnormal glucose metabolism fulfills the bioenergetic and biosynthetic needs of cancer cells and is tightly associated with cell death evasion, cell proliferation, angiogenesis, and metastasis of various cancers (1, 4, 5). Targeting the glycolysis pathway thus provides a reasonable therapeutic opportunity.
Recent studies on tumor metabolism have revealed complex mechanisms that underlie the Warburg effect. Besides mitochondrial dysfunction, which was suspected by Warburg himself, other factors, including hypoxia, redox stress, oncogene activation, as well as loss of tumor suppressor genes, can all contribute to the high rate of glycolysis. The connection of genetic alterations in cancers to glucose metabolic regulation suggests that cancer cells not only passively adapt to their environment but can actively manipulate metabolic programs to promote tumorigenesis. This is with particular significance in hematopoietic cancers where oxygen and other nutrient supplies are often abundant (1, 4, 5).
Akt and c-Myc are two frequently dysregulated oncoproteins in various cancers. Both proteins are involved in the regulation of cell growth, proliferation, and death/survival (6, 7). Akt, a serine/threonine kinase, has been found to promote anabolic metabolism, support cell survival, and suppress apoptosis through various mechanisms, including the regulation of glucose metabolism (8–12). Akt promotes the translocation of glucose transporter 1 from the cytosol to the plasma membrane, thus increasing glucose uptake (12). Akt can also promote the association of hexokinase I and II, the most abundant isoforms of glucose kinase in cancer cells, with the mitochondrial outer membrane. This association increases the enzymatic efficiency of the kinase and stabilizes mitochondria, hence rendering the cell resistant to various cell death stimuli (8, 11, 13).
c-Myc, a transcriptional regulator, has also been found to directly regulate the glycolytic pathway (7). In Rat1a fibroblasts and murine livers overexpressing c-Myc, the mRNA levels of the glucose transporter 1, phosphoglucose isomerase, phosphofructokinase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, and enolase are elevated (14). c-Myc also up-regulates the expression of lactate dehydrogenase A, which diverges pyruvate to lactate, and contributes to the Warburg effect (15). Moreover, in addition to these glycolysis-related enzymes, c-Myc has been found to regulate mitochondrial function. Induction of c-Myc in cells results in increased oxygen consumption and mitochondrial mass, whereas Myc-null cells have decreased mitochondrial mass and fewer normal mitochondria (16, 17). cDNA microarray analysis, chromatin immunoprecipitation, and promoter microarray analysis reveal a number of genes involved in mitochondrial DNA replication, transcription, and biogenesis are direct transcriptional targets of c-Myc. These include mitochondrial transcription factor A, DNA polymerase subunit γ-1, and DNA polymerase subunit γ-2, as well as nuclear respiratory factor 1 (16–18). These studies support a direct role for c-Myc on mitochondrial biogenesis, pointing to the possibility that c-Myc not only promotes glycolysis but also enhances the ability of cells to utilize non-glucose substrates to fuel mitochondria. Thus, although both Akt and c-Myc can drive the Warburg effect, they may render cells differentially susceptible to the inhibition of glycolysis. In this work, we establish an isogenic hematopoietic cell culture system to study the effects of Akt and c-Myc on cellular metabolism and to determine how these oncoproteins may influence cell sensitivity to metabolic perturbation.
EXPERIMENTAL PROCEDURES
Establishment of the Dual-regulatable Cell Line
Plasmid tri-cistronically expressing tetracycline-dependent rtTA2-M2 activator, TRSID silencer, and puromycin selection marker (pLIB-rtTAM2iresTRSIDiresPuro, a gift from Dr. M. Ausserlechner of Medical University Innsbruck, Austria) (19) were transfected into FL5.12 cells by Nucleofector transfection (Nucleofector kit V, Program G-016, Amaxa Biosystems). After puromycin selection (0.5 μg/ml for 48 h), stable clones were obtained by limited dilution. The doxycycline (DOX)3 inducibility and stringency were tested, and a single Tet-On cell clone was chosen for further experiments. The DOX-inducible myristoylated Akt (myrAkt)-expressing cells were established by introducing a pRev_TRE-myrAkt-IRES-Hygro plasmid to the Tet-On cells by Nucleofector transfection, and a stable clone was established by hygromycin selection (3 mg/ml for 4 days) and limited dilution. To establish the inducible c-Myc-expressing system, a retroviral c-MycER expressing vector (pBabepuroc- MycERTAM) (20) and a plasmid encoding the ecotropic retroviral envelope proteins were cotransfected into 293T cells. Virus-containing supernatant was used to transduce the DOX-inducible myrAkt cells. The infected cells were selected in 8.0 μg/ml puromycin for 4 days and used as a stable cell pool. This stable cell pool was designated as FL5.12-AM (A for Akt and M for c-Myc). The cells were maintained in FL5.12 complete medium (RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 20 mm HEPES, pH 7.5, 50 μm 2-mercaptoethanol, and 0.4 ng/ml interleukin-3 (IL-3)). Cells were maintained at a density between 5 × 104 and 1 × 106 cells/ml at 37 °C in a humidified incubator with 5% CO2.
Activation of Oncoproteins and Induction of Cell Death
To activate the expression of myrAkt and to activate MycER, cells were plated at 5.0 × 104 cells/ml. One μg/ml DOX or 100 nm 4-hydroxytamoxifen (4-OHT) was added to the cell culture for 36 h. For IL-3 deprivation, cells were washed three times with complete medium without IL-3 and resuspended in the IL-3-free medium at 5.0 × 104 cells/ml. For serum deprivation, cells were washed three times and resuspended in FBS-free medium at 5.0 × 104 cells/ml. To induce cell death with bioenergetic inhibitors, 2-deoxyglucose (2-DG, 6 mm) or oligomycin (OG, 1 μg/ml) was used to treat cells. For glutamine deprivation, cells were washed three times and resuspended in glutamine-free medium at 5.0 × 104 cells/ml. Aliquots of the treated cells were taken at various time points for cell viability measurement.
Cell Death Assay
Propidium iodide (PI) was added to each sample at a final concentration of 1 μg/ml. Cell viability was measured by plasma membrane permeability indicated by PI exclusion using a flow cytometer (FACSCalibur, BD Biosciences).
Measurement of Intracellular ATP Levels
Intracellular ATP levels were determined using the CellTiter-GloTM luminescent cell viability assay kit (Promega), following the manufacturer's instruction. Briefly, after treating with 2-DG or OG for certain periods of time, 50 μl of lysis buffer was added to 50 μl of cells (2 × 105 cells/ml), followed by 2 min of agitation on a platform shaker and 10 min of incubation at room temperature. Luminescence was quantified using a SpectraMax M5 microplate reader.
Clonogenic Cell Survival Assay
Clonogenic survival was determined by culturing cells in the presence or absence of 2-DG or OG for 6 h and then performing limited dilution analysis of cell growth following the removal of 2-DG or OG. Percent survival was determined as the number of clones from treated cells normalized to the number of clones from untreated cells (21).
Whole Cell Extraction
Cells were harvested, washed once with ice-cold PBS, and lysed in RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) with freshly added protease inhibitor mixture (ProteCEASETM, G-Biosciences) and phosphatase inhibitors, including 1 mm phenylmethanesulfonyl fluoride, 10 mm sodium fluoride, 10 mm β-glycerophosphate, 10 mm sodium orthovanadate, 10 mm sodium pyrophosphate. After 30 min of incubation on ice and brief sonication, cell lysates were centrifuged at 12,000 × g at 4 °C for 15 min. The supernatant was collected and stored at −80 °C. Protein concentration was determined by the BCA assay (Thermo Scientific).
Subcellular Fractionation
Subcellular components were separated using the ProteoExtract subcellular proteome extraction kit (Calbiochem), following the manufacturer's instruction. 5 × 106 cells were used for subcellular fractionation. Protein concentration was determined by the BCA assay (Thermo Scientific).
Immunoblotting
Equal amounts of whole cell lysates or subcellular fractions were resolved in SDS-PAGE and transferred onto nitrocellulose membrane. After blocking with PBST containing 5% nonfat dry milk, the blots were probed using the following primary antibodies (all from Cell Signaling Technologies unless otherwise stated): phospho-Akt Thr-308 (catalog no. 2965), phospho-Akt Ser-473 (catalog no. 4058), Akt (catalog no. 9272), phospho-GSK3β Ser-9 (catalog no. 9336), GSK3β (catalog no. 9315), c-Myc (catalog no. SC-40, Santa Cruz Biotechnology), PARP (catalog no. SC-7150, Santa Cruz Biotechnology), cleaved caspase-3 (catalog no. 9661), Tom40 (catalog no. SC-11414, Santa Cruz Biotechnology), Cox IV subunit Va (catalog no. 459120, Invitrogen), cytochrome c (catalog no. 556433, Pharmingen), and β-tubulin (catalog no. T4026, Sigma). All primary antibodies were incubated overnight at 4 °C. After washing, the membrane was incubated for 1 h at room temperature with horseradish peroxidase-conjugated or IRDye680-conjugated secondary antibodies. The signals were visualized using SuperSignal West Pico ECL (Thermo Scientific) or using the Odyssey Infrared Imaging System (LI-COR).
Mitochondrial DNA Content
Total cellular DNA was isolated. Ratios of mtDNA to nuclear DNA were determined by quantitative real time PCR using QuantiTect® SYBR Green PCR kit (Qiagen). The primers used for detecting nuclear DNA were as follows: 5′-CCGATTAGGAGTACACACGAAAGGTG-3′ (forward primer) and 5′-ACGCACAAGAGTGGATGCTATTGC-3′ (reverse primer) for poly(A) polymerase; 5′-AGGGGAGAGCGGGTAAGAGA-3′ (forward primer) and 5′-GGACAGGACTAGGCGGAACA-3′ (reverse primer) for 18 S rDNA. The primers used for detecting mtDNA were as follows: 5′-TTATTAACCACTCATTCATTGACC-3′ (forward primer) and 5′-AGCGAAGAATCGGGTCAAGGTGGC-3′ (reverse primer) for cytochrome b; 5′-CCCAATCTCTACCAGCATC-3′ (forward) and 5′-GGCTCATAGTATAGCTGGAG-3′ (reverse) for cytochrome c oxidase subunit 1; 5′-AATCGCCATAGCCTTCCTAACAT-3′ (forward) and 5′-GGCGTCTGCAAATGGTTGTAA-3′ (reverse) for NADH dehydrogenase-1. Sequence detection software version 1.2 (Applied Biosystem) was used to analyze the Ct value of each reaction, and the ΔΔCt method was used to calculate the relative level of mtDNA to nuclear DNA.
Coupled and Uncoupled Endogenous Respiration Rates in Intact Cells
To determine intact cell-coupled and -uncoupled endogenous respiration, 3 × 107 cells were resuspended in 3 ml of fresh medium prewarmed to 37 °C and pre-gassed with 95% air, 5% CO2. The cell suspension was placed in a sealed respiration chamber equipped with temperature control, a microstirring device, and a Clark-type oxygen electrode. The oxygen content in the cell suspension was constantly monitored with an YSI 5300 oxymeter, and oxygen consumption rate was determined as described previously (22). After measuring basal respiration, oligomycin (1 μg/ml final concentration) and carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone (1.5 μm final concentration) were used to acquire the oligomycin-insensitive and carbonyl cyanide p-trifluoromethoxyphenylhydrazone-uncoupled oxygen consumption rate.
Mitochondrial Complex Respiration Rate
Mitochondrial complex respiration was determined by measuring specific substrate oxidation in permeabilized cells (23). 3 × 107 cells were collected by centrifugation and resuspended in 3 ml of mitochondrial respiration buffer (100 mm KCl, 24 mm NaCl, 5 mm MgCl2, 15 mm HEPES, 5 mm KH2PO4, 2 mm EGTA, 0.1 mm ouabain). Cells were placed in the respiratory chamber and permeabilized by incubating with digitonin (50 μg/ml final concentration) for 3 min. Oxygen consumption rates were then measured under the following conditions. (i) Oxidation of glutamate and malate (5 mm each; complex I substrates) was measured in the presence of 2 mm ADP, and the reaction was stopped by rotenone (100 nm final concentration). (ii) Oxidation of succinate (10 mm, complex II substrate) was measured in the presence of rotenone and ADP. The reaction was stopped by antimycin A (20 nm final concentration). (iii) Oxidation of ascorbate (2.5 mm) and N,N,N′,N′-tetramethyl-p-phenylenediamine (0.25 mm) (complex IV substrates) was measured in the presence of rotenone and antimycin A, without ADP.
Immunofluorescence Staining and Flow Cytometry
Immunofluorescence staining was performed as described previously (24). Briefly, 1 × 106 cells were fixed for 10 min in 1% paraformaldehyde in PBS. Cells were washed with wash buffer (2% goat serum and 0.03% saponin in PBS) and then incubated sequentially with primary antibodies (mouse anti-human IgG, catalog no. 109-3102, Rockland; Cox IV subunit 1, catalog no. A-6403, Invitrogen) and secondary antibody (fluorescein isothiocyanate-conjugated anti-mouse IgG, catalog no. 710-1232, Rockland) for 30 min at room temperature in wash buffer containing 0.3% saponin. Fluorescence intensity was analyzed by flow cytometry (FACSCalibur, BD Biosciences).
Mitochondrial Mass
1 × 105 cells were resuspended in 0.5 ml of complete medium containing 50 nm mitochondrial fluorescent dye acridine orange 10-nonyl bromide (catalog no. A7847, Sigma). After incubating of 30 min at 37 °C in dark, cells were immediately transferred to a tube on ice for analysis by flow cytometry.
Glucose Uptake
Glucose uptake was measured as described previously with brief modifications (12, 25). 5 × 105 cells were washed and resuspended in 0.5 ml of fresh medium. 2-Deoxy-d-[3H]glucose (5–10 Ci/mmol, 1 μCi per reaction; PerkinElmer Life Sciences) was added for a period of 10 min at 37 °C. The reaction was quenched by addition of ice-cold PBS, and the cells were washed three times with PBS. Cells were collected by centrifugation, and cell pellet was solubilized in 0.1 m NaOH, and radioactivity was measured using a 1900TR liquid scintillation analyzer (Packard).
Glycolytic Rate
Cellular glycolytic rate was determined as described previously (26) with modifications. 2.5 × 105 cells were incubated in 0.25 ml of fresh medium containing 5 μCi of [5-3H]glucose (10–20 Ci/mmol; PerkinElmer Life Sciences) for 1 h at 37 °C. The reaction was stopped by adding an equal volume of 0.2 n HCl. 3H2O was separated from [5-3H]glucose by diffusion in an airtight container for 72 h. Diffused and undiffused tritium was measured using a 1900TR liquid scintillation analyzer (Packard) and compared with controls of [5-3H]glucose only and 3H2O only to determine the rate of glycolysis.
Lactate Concentration
2.5 × 106 cells were collected by centrifugation at 400 × g for 5 min. After three washes with fresh complete media, cells were resuspended in 1 ml of fresh complete media and incubated at 37 °C for 2 h. The media were collected and frozen at −20 °C. Lactate levels were quantified by colorimetric assay following the manufacturer's instructions (EnzyChromTM lactate assay kit, BioAssay Systems).
Glucose Oxidation
Glucose oxidation was determined as described previously (27) with modifications. 1.5 × 106 cells were resuspended in 3 ml of pre-gassed (95% air, 5% CO2) modified Krebs-Henseleit buffer containing 4% fatty acid-free bovine serum albumin and 10 mm glucose. The reaction was carried out in a T25 flask. A total of 0.9 μCi of d-[U-14C]glucose (250–360 mCi/mmol; PerkinElmer Life Sciences) was added to the buffer, and the flask was quickly sealed with a rubber stopper. After a 90-min incubation, the buffer was acidified with 0.10 volume of 70% perchloric acid. 14CO2 was captured over 90 min at 37 °C into a 0.5-ml microcentrifuge tube containing 200 μl of 1 m KOH. The microcentrifuge tube was placed in a scintillation vial and radioactivity measured using a 1900TR liquid scintillation analyzer (Packard).
Fatty Acid β-Oxidation
Fatty acid β-oxidation was measured as described previously (28, 29). 2 × 105 cells were pelleted and resuspended in 200 μl of PBS (with Ca2+ and Mg2+) supplemented with 125 μm [9,10-3H]palmitate/palmitate (30–60 Ci/mmol; PerkinElmer Life Sciences) complexed to 2.5 mg/ml fatty acid-free bovine serum albumin and 1 mm carnitine. A total of 1 μCi of [9,10-3H]palmitate was used for each reaction. Cells were cultured in 24-well plates, and after 2 h, supernatant was recovered and added to 200 μl of 10% trichloroacetic acid. After centrifugation at 10,000 × g for 10 min at 4 °C, 55 μl of 6 m NaOH was added to neutralize trichloroacetic acid. The mixture was applied to ion-exchange columns (Dowex 1X2–400; Sigma), and tritiated water was recovered by eluting with 1.7 ml of H2O. A 1-ml aliquot was used for scintillation counting with a 1900TR liquid scintillation analyzer (Packard).
Statistical Analysis
Data were expressed as average ± S.D. One-way analysis of variance with Tukey's post hoc test was used to determine the differences among three means. For analysis with only two groups, Student's t test was used. Significance was judged when p < 0.05.
RESULTS
Establishment of a Dual-regulatable Cell Line to Activate Akt and c-Myc
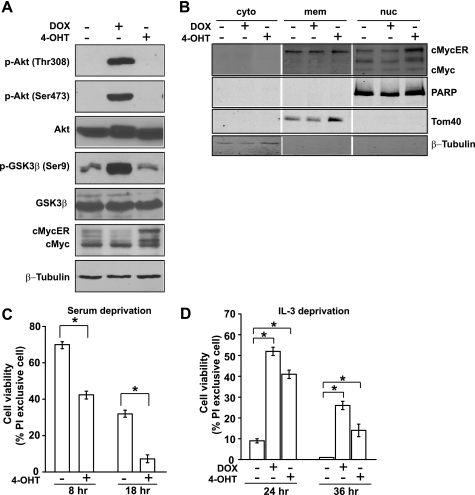
To compare directly the effect of different oncoproteins on cell metabolism, we established a dual-regulatable expression system in the IL-3-dependent mouse pro-B cell line FL5.12. In this system, expression of a constitutively active Akt (myrAkt) is under the control of a tetracycline-responsive promoter (9), whereas c-Myc is fused to the hormone-binding domain of the human estrogen receptor (30). We named the stable cell pool as FL5.12-AM (A for Akt and M for Myc). Addition of DOX to the dual-regulatable cells induced the expression of myrAkt. This is indicated by a marked increase of Akt phosphorylation at threonine 308 and serine 473, as well as by the phosphorylation of the Akt target GSK3β at serine 9 (Fig. 1A). c-MycER fusion protein was constitutively expressed in the dual-regulatable cells. Addition of 4-OHT resulted in the nuclear translocation of c-MycER and enhanced its stability by preventing it from being targeted to protein degradation machinery in the cytosol (Fig. 1A). This is confirmed by subcellular fractionation analysis showing that c-MycER was detected in both the membrane and nuclear fractions and was enriched in the nuclear fraction upon the addition of 4-OHT, whereas endogenous c-Myc stayed steadily in the nuclear fractions (Fig. 1B).
FIGURE 1.
Establishment of the dual-regulatable expression system to activate Akt and c-Myc in FL5.12 cells. A cell pool stably expressing DOX-regulatable myrAkt and 4-OHT-inducible c-MycER was established by retroviral transduction. Cells were treated with DOX (1 μg/ml) or 4-OHT (100 nm) for 36 h to induce the expression of myrAkt and c-MycER, respectively. A, immunoblotting analysis of the expression of Akt and c-MycER, as well as phospho-Akt (Thr-308 and Ser-473) and phospho-GSK3β (Ser-9). B, immunoblotting analysis of the subcellular localization of c-MycER upon addition of 4-OHT. Cells were fractionated. The cytosolic (cyto), membrane (mem), and nuclear (nuc) fractions were resolved by SDS-PAGE and probed for c-Myc, as well as the respective molecular markers for the purity of the fractionation (β-tubulin for the cytosol, Tom40 for the membrane, and PARP for the nucleus). Note that c-MycER fusion protein was constitutively expressed and detected in both the membrane and nuclear fractions. Addition of 4-OHT induced accumulation of c-MycER in the nuclear fraction. The endogenous c-Myc is predominantly localized in the nuclear fraction and does not change levels upon 4-OHT addition. C, activation of c-Myc sensitizes cells to serum withdrawal. Cells were cultured in serum-free media for 8 and 18 h. Cell viability was determined by PI exclusion using flow cytometry. Data shown are average of three experiments ± S.D. (*, p < 0.05). D, activated Akt and c-Myc protect cells from IL-3 withdrawal. After DOX or 4-OHT induction, cells were washed with PBS and cultured in IL-3-free media for the indicated time. Cell viability was determined by PI exclusion using flow cytometry. Data shown are average of three independent experiments ± S.D. (*, p < 0.05).
We went on to verify the biological consequences of the activation of Akt and c-Myc. As reported previously (31), c-Myc sensitized cells to cell death induced by the deprivation of FBS (Fig. 1C). In the IL-3-dependent FL5.12 cells, a major pro-survival effect of IL-3 comes from the IL-3 receptor-mediated activation of the Akt signaling pathway that promotes cellular glucose uptake and metabolism (9). Under the condition of IL-3 deprivation, constitutively active Akt has been shown to protect cells by promoting glucose transporter 1 localization to the plasma membrane and glycolysis (32). Indeed, in our dual-regulatable FL5.12 cells, activation of myrAkt protected FL5.12 cells from apoptosis induced by IL-3 deprivation (Fig. 1D). Interestingly, activation of c-Myc also protected FL5.12 cells from IL-3 deprivation (Fig. 1D), indicating that c-Myc can also promote cellular bioenergetic activity in the absence of growth factor signaling. Hence, we have established an isogenic cell system in which activation of Akt and c-Myc oncoproteins is tightly regulated.
Activation of Akt and c-Myc Leads to the Warburg Effect
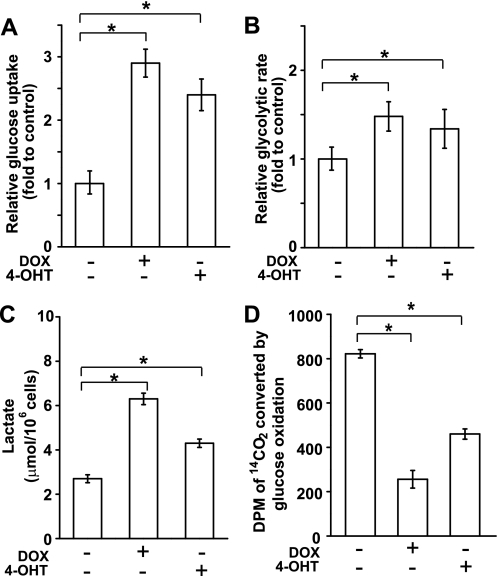
After confirmation of the inducible activation of Akt and c-Myc, the effect of these oncoproteins on the Warburg effect was examined. Several events of glucose metabolism, including glucose uptake, glycolysis rate, and lactate generation, were measured. Glucose uptake was determined by the accumulation of 3H-labeled 2-DG in cells. When Akt or c-Myc was induced, glucose uptake was increased markedly as compared with uninduced cells (Fig. 2A). Addition of DOX or 4-OHT to FL5.12 parental cells slightly reduces the glucose uptake (supplemental Fig. 1A), indicating the increased glucose uptake observed in Akt and c-Myc cells is specifically promoted by these oncoproteins, but not the adverse effect of DOX and 4-OHT. Glycolytic activity was determined by measuring the conversion of [5-3H]glucose to 3H2O. Both Akt and c-Myc enhanced the glycolytic rate (Fig. 2B). Lactate generation increased by 3.0-fold in Akt cells and 1.8-fold in c-Myc cells (Fig. 2C). Moreover, glucose oxidation was decreased in the Akt and c-Myc cells compared with uninduced cells, indicating that glycolytic pyruvate diverged away from mitochondria, consistent with the enhanced lactate generation in Akt and c-Myc cells (compare Fig. 2, C and D). Interestingly, Akt cells constantly showed higher lactate generation and lower glucose oxidation with respect to c-Myc cells (Fig. 2, C and D), suggesting that c-Myc cells have an enhanced ability to shunt pyruvate into mitochondria than Akt. Nevertheless, our findings indicate that both Akt and c-Myc promote the Warburg effect in FL5.12 cells.
FIGURE 2.
Akt and c-Myc cause the Warburg effect. Cells were cultured in the presence of DOX or 4-OHT for 36 h. A, glucose uptake was measured by the accumulation of 2-[1,2-3H]deoxy-d-glucose in cells. B, glycolytic rate was measured by the conversion of d-[5-3H]glucose to 3H2O. C, cells were refed with fresh media for 2 h, and lactate yield was determined by measuring the amount of lactate in the culture medium at the end point. D, glucose oxidation was determined by measuring the conversion of d-[U-14C]glucose into 14CO2. All data shown are average of three experiments ± S.D. (*, p < 0.05).
Activation of Akt and c-Myc Differentially Regulates Mitochondrial Function
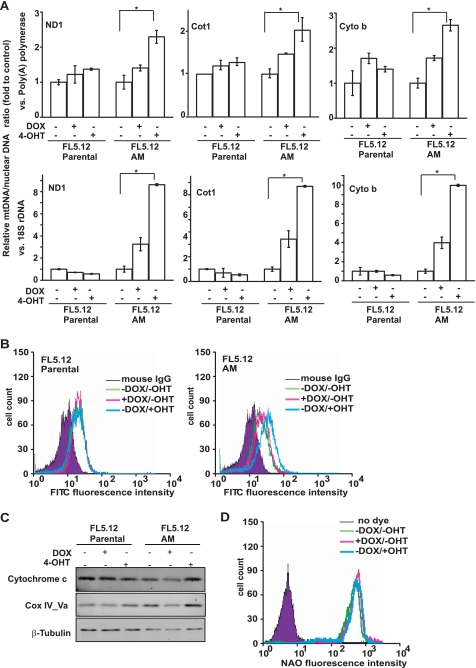
The Warburg effect was initially described to correlate with lower levels of oxidative phosphorylation. However, c-Myc has been implicated in promoting mitochondrial catabolism and biogenesis. Thus, we next examined the effect of Akt and c-Myc on mitochondrial function in the dual-regulatable system. We compared the relative levels of mt-Cot1 (encoding the cytochrome c oxidase subunit 1), mt-cyto b (encoding cytochrome b), and mt-Nd1 (encoding NADH dehydrogenase-1) DNA to nuclear 18 S rDNA or poly(A) polymerase gene. Although DOX, 4-OHT, and Akt activation showed a certain effect on mtDNA copy number, c-Myc activation resulted in the most significant increase in mtDNA in all six combinations (Fig. 3A). In agreement, immunostaining analysis showed that the protein expression level of Cox IV subunit 1 of mitochondrial electron transport chain, which is encoded by mitochondrial DNA, markedly increased when c-Myc was activated (Fig. 3B). These results indicate that c-Myc activation increases the mitochondrial DNA content and thereby enhances the mitochondrial DNA-encoded protein expression and mitochondrial function. Because functional mitochondria need both mitochondrial and nuclear DNA-encoded proteins, we also checked nuclear DNA-encoded mitochondrial proteins. Immunoblotting revealed that the expression levels of cytochrome c and Cox IV subunit Va are both nuclear DNA-encoded mitochondrial proteins and were increased in c-Myc cells, indicating that c-Myc can up-regulate both mitochondrial and nuclear DNA-encoded proteins to increase mitochondrial function. Interestingly, the increased mtDNA and protein expression in c-Myc cells did not seem to correlate with an increased mitochondrial mass, as the mitochondrial membrane potential independent dye acridine orange 10-nonyl bromide failed to detect alterations in mitochondrial mass (Fig. 3D), indicating that in our system the mtDNA copy number and activity can be dissociated from mitochondrial biogenesis, which has been reported previously (33).
FIGURE 3.
Characterization of the effects of Akt and c-Myc on mitochondria. FL5.12 parental and FL5.12-AM cells were cultured with DOX or 4-OHT for 36 h. A, ratio of mitochondrial DNA to nuclear DNA was determined by quantitative real time PCR and normalized to that of uninduced cells. Three pairs of primers for mitochondrial DNA-encoded genes and two pairs of primers for nuclear DNA-encoded genes were used. ND1, NADH dehydrogenase-1; Cot1, cytochrome c oxidase subunit 1; Cyto b, cytochrome b. The experiments were repeated three times, and real time PCR was performed in duplicate for each reaction. Data shown are averages of three experiments ± S.D. (*, p < 0.05). B, cells were incubated with control IgG or anti-Cox IV subunit 1 and then incubated with fluorescein isothiocyanate-conjugated anti-mouse IgG secondary antibody. The expression level of mitochondrial complex IV subunit 1 was analyzed by flow cytometry. Note that the addition of 4-OHT increased the expression of complex IV subunit 1 in FL5.12-AM but not in the parental cells. C, immunoblotting analysis of the expression of nuclear DNA-encoded mitochondrial protein cytochrome c and Cox IV subunit Va. β-Tubulin was used for equal loading. D, mitochondrial mass was detected in FL5.12-AM cells with mitochondrial fluorescent dye acridine orange 10-nonyl bromide (NAO). The relative acridine orange 10-nonyl bromide intensity was analyzed by flow cytometry. No significant difference was detected between the uninduced and induced cells.
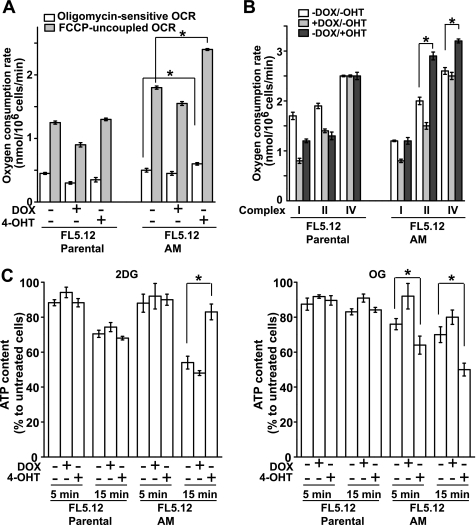
The above data suggest that c-Myc cells may have enhanced mitochondrial activities. To confirm this, cellular oxygen consumption rate (OCR) was measured. DOX slightly decreased both coupled and uncoupled OCR in both parental and FL5.12-AM cells (Fig. 4A), indicating some adverse effect of DOX on mitochondrial function. Nevertheless, both coupled and uncoupled endogenous respiration rates in intact cells were significantly increased in c-Myc cells (Fig. 4A). Similarly, complex-specific measurement of respiratory chain function in c-Myc cells showed increased activity of complexes II and IV (Fig. 4B). This is in agreement with the increased expression levels of Cox IV subunit 1, Cox IV subunit Va, and cytochrome c in c-Myc cells (Fig. 3, B and C). These data indicate that oxidative phosphorylation is enhanced by c-Myc. Consistent with this, the cellular ATP level in c-Myc cell was not sensitive to the glycolysis inhibitor 2-DG but was dramatically affected by mitochondrial F0/F1-ATP synthase inhibitor oligomycin (Fig. 4C). This indicates that the cellular ATP level in c-Myc cell is mainly maintained by mitochondrial oxidative phosphorylation. Taken together, these findings indicate that in FL5.12 cells, c-Myc but not Akt markedly enhances mitochondrial function.
FIGURE 4.
Characterization of the effects of Akt and c-Myc on mitochondrial function. A, oligomycin-sensitive OCR and carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP)-uncoupled OCR were calculated based on the measurement of cellular O2 consumption in complete media using a Clark-type electrode. Data shown are average of three experiments ± S.D. (*, p < 0.05). B, mitochondrial electron transport chain complex-specific ADP-coupled OCR was calculated by measuring O2 consumption of digitonin-permeabilized cells supplied with complex-specific respiratory substrates as follows: glutamate and malate for complex I, succinate for complex II, ascorbate and N,N,N′,N′-tetramethyl-p-phenylenediamine for complex IV. Data shown are average of three experiments ± S.D. (*, p < 0.05). C, cellular ATP level was measured upon the addition of glycolysis inhibitor 2-DG and the mitochondrial F0/F1-ATP synthase inhibitor oligomycin at 5 and 15 min and expressed as percent of ATP level of untreated cells. Data shown are average of three experiments ± S.D. (*, p < 0.05).
Activation of Akt and c-Myc Differentially Sensitizes Cells to Metabolic Perturbation
Currently, cancer therapeutic strategies are being developed targeting glycolysis based on the Warburg effect (5, 34). Our results indicate that activation of Akt and c-Myc can both lead to an altered glucose metabolism, i.e. high glucose uptake and high glycolytic activity, resembling the Warburg effect in most tumor cells (Fig. 2). However, our findings show that activation of Akt and c-Myc can differentially regulate mitochondrial function (Figs. 3 and 4), which may alter the cellular dependence on glycolysis, hence altering cellular sensitivity to the inhibition of glycolysis. To evaluate this possibility, we went on to examine the effect of Akt and c-Myc on cell susceptibility to the inhibition of glycolysis.
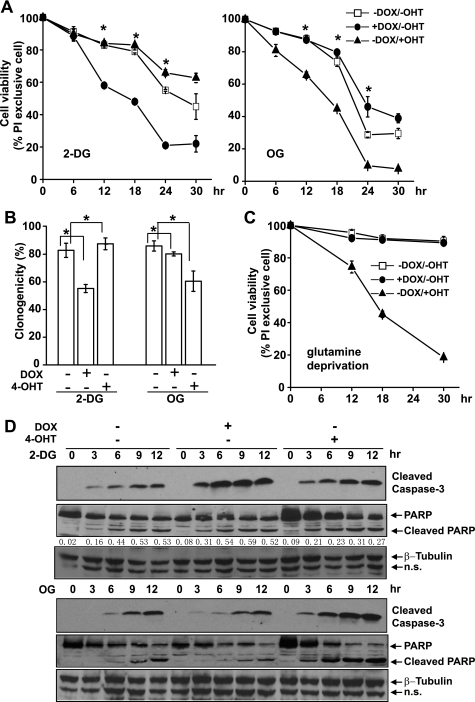
2-DG is an analogue of glucose that enters the cell via glucose transporters. It is phosphorylated by hexokinases but cannot be further catabolized along the glycolysis pathway, and thus serves as a specific competitive inhibitor for glycolysis. To minimize the variation that may arise from the competition between glucose and 2-DG, we first performed a dose titration of the lethal effect of 2-DG on uninduced cells. Below 6 mm, the effect of 2-DG was both dosage- and time-dependent, although above 6 mm the killing effect of 2-DG was only time-dependent (data not shown). Hence, we chose to use 6 mm 2-DG, which is nearly half of the glucose concentration in RPMI 1640 medium, for further experiments. The dual-regulatable FL5.12-AM cells were cultured with DOX or 4-OHT, respectively, to activate Akt or c-Myc. After 36 h, 2-DG was added at a final concentration of 6 mm. Cell death was determined by PI exclusion for plasma membrane permeability. Akt cells were more sensitive to 2-DG treatment, compared with uninduced cells (Fig. 5A). In contrast, although c-Myc cells also displayed enhanced glycolysis, they were slightly resistant to 2-DG compared with uninduced cells (Fig. 5A). These findings indicate the following: 1) Akt enhances cell dependence on aerobic glycolysis and therefore renders cells susceptible to the inhibition of glycolysis, and 2) even though c-Myc also stimulates glycolysis, it does not make cells more vulnerable to glycolysis inhibition.
FIGURE 5.
Akt and c-Myc render cells differentially susceptible to the inhibition of glycolysis or mitochondrial function. Cells were cultured with DOX or 4-OHT to activate Akt or c-Myc for 36 h. A, cells were treated with 6 mm 2-DG or 1 μg/ml OG. Cell viability was measured by PI exclusion at indicated time points. Data are average of three experiments ± S.D. (*, p < 0.05). B, cells were treated with 2-DG or OG for 6 h and washed with fresh media without the inhibitors. 80 cells were placed in a 96-well plate and cultured for 14 days. Clonogenicity of the cells represents cell survival ability after 6 h of treatment with the metabolic inhibitors. Data are average of three experiments ± S.D. (*, p < 0.05). C, cells were cultured in glutamine-free media for the indicated time. Cell viability was measured by PI exclusion. Data are average of three experiments ± S.D. D, cell lysates were collected at the indicated time points, and immunoblotting was performed with antibodies against cleaved caspase-3, PARP, and β-tubulin. The ratio of cleaved PARP versus full-length PARP is indicated in cells treated with 2-DG. n.s., nonspecific bands.
The killing mechanism of 2-DG is primarily due to its ability to disturb bioenergetics through glycolysis. It is possible that the elevated mitochondrial function in c-Myc cells accounts for their resistance to 2-DG treatment by allowing cells to use other metabolic substrates to support bioenergetic activities when glucose supply is limited. If this is the case, c-Myc cells would be more sensitive to inhibition of mitochondrial function. To test this possibility, we treated the cells with OG to inhibit the mitochondrial F0/F1-ATP synthase. At the concentration of 1 μg/ml, the kinetics of OG-induced cell death is time-dependent (data not shown); therefore, we used this concentration for our following experiments. As shown in Fig. 5A, c-Myc cells were more sensitive to OG treatment, compared with uninduced cells and Akt cells, indicating that mitochondrial function plays a more critical role in c-Myc cells on maintaining cellular bioenergetic homeostasis. The differential cell death rates of Akt cells to 2-DG and c-Myc cells to OG were also revealed by clonogenicity analysis (Fig. 5B), as well as by immunoblotting analysis of apoptotic cleavage of caspase-3 and caspase-3-mediated cleavage of PARP (Fig. 5D). Addition of DOX and 4-OHT had little or virtually no effect on cell susceptibility to 2-DG and OG in the FL5.12 parental cells (supplemental Fig. 1B).
It has been reported that c-Myc confers cells to glutamine addiction by regulating the glutaminolysis pathway (35, 36). c-Myc cells use glutamine as a bioenergetic and biosynthetic substrate through the tricarboxylic acid cycle. Consistent with previous reports (35, 36), glutamine deprivation induced massive cell death of c-Myc cells, although Akt cells and uninduced cells were not affected (Fig. 5C).
Ability to Switch Bioenergetic Substrates Is Essential for c-Myc to Confer Resistance to Glycolysis Inhibition
The above results indicate that differing glucose metabolism and mitochondrial status in Akt and c-Myc cells may determine the cellular response to the perturbations of different metabolic programs. Akt may stimulate glucose metabolism, hence sensitizing cells to glycolytic inhibition while sparing them when mitochondrial function is perturbed. On the other hand, c-Myc may stimulate both glucose metabolism and mitochondrial activity, hence reducing cell dependence on glucose and increasing cell susceptibility to mitochondrial inhibitors. Indeed, it has been reported that c-Myc promotes the use of nutrient sources other than glucose, such as fatty acids and glutamine to fuel mitochondria and to provide substrates for biosynthesis (35–37).
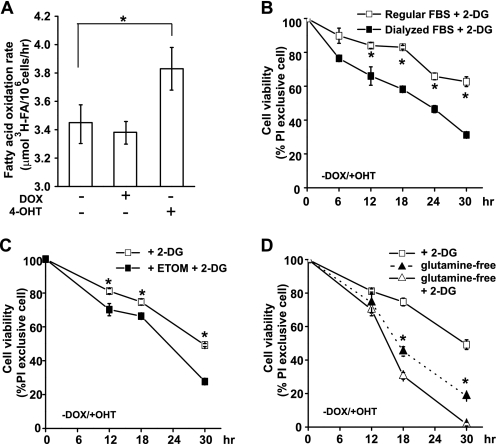
The ability of c-Myc cells to switch to non-glucose nutrient sources may explain their resistance to the direct inhibition of glycolysis. Indeed, fatty acid oxidation was higher in c-Myc cells (Fig. 6A). It can then be predicted that blockage of the access of non-glucose nutrient substrates, such as fatty acids and glutamine, to mitochondria would sensitize c-Myc cells to 2-DG treatment. To test this, we first cultured the cells in culture medium in which regular FBS was substituted with dialyzed FBS, thus depleting a major source of fatty acids. As shown in Fig. 6B, c-Myc cells cultured in medium with dialyzed FBS were more sensitive to 2-DG treatment, compared with cells cultured in medium containing regular FBS. Moreover, etomoxir, an inhibitor of malonyl-CoA-dependent CPT1 (carnitine palmitoyltransferase 1) was used to block fatty acid β-oxidation (28). CPT1 is an enzyme located on the mitochondrial outer membrane that esterifies long chain fatty acids to carnitine and controls the fatty acid entry into mitochondria for β-oxidation. Although the addition of etomoxir itself did not affect cell viability, it sensitized c-Myc cells to 2-DG treatment (Fig. 6C). Similar results were obtained when c-Myc cells were treated with 2-DG in glutamine-free media. Glutamine deprivation enhanced cell death in 2-DG-treated c-Myc cells (Fig. 6D). These results indicate that enhanced fatty acid and glutamine usage may be the reason for c-Myc cells to be protected from glycolysis inhibition.
FIGURE 6.
Fatty acid and glutamine depletion sensitizes c-Myc cells to glycolysis inhibition. A, fatty acid β-oxidation was measured by the conversion of 3H-labeled palmitic acid into 3H2O. Data shown are average of three experiments ± S.D. (*, p < 0.05). B, cells were preconditioned in media in which regular FBS was substituted with dialyzed FBS. Cells were cultured with 4-OHT to activate c-Myc and then treated with 6 mm 2-DG. Cell viability was determined by PI exclusion. Data shown are average of three experiments ± S.D. (*, p < 0.05). C, cells were cultured in media with regular FBS, and c-Myc was activated by the addition of 4-OHT. Cells were treated with 6 mm 2-DG only, or in combination with CPT1 inhibitor etomoxir (ETOM, 0.2 mm) that was added 30 min prior to the treatment of 2-DG. Cell viability was determined by PI exclusion. Data shown are average of three experiments ± S.D. (*, p < 0.05). D, cells were cultured in medium with or without glutamine and treated with 6 mm 2-DG. Cell viability was determined by PI exclusion. The cell death curve induced by glutamine deprivation is also shown (solid triangle). Data shown are average of three experiments ± S.D. (*, p < 0.05 for comparisons of cell viability between glutamine-free and glutamine-free plus 2-DG treatment).
DISCUSSION
The alteration of tumor cell metabolism is signified by two coupled events, enhanced glycolysis and altered mitochondrial function. Evidence exists indicating that in addition to passive adaptation to the tumor microenvironment, i.e. the low oxygen tension often encountered during tumor development, the aberrant tumor cell metabolism can be actively regulated by oncoproteins and tumor suppressors (4, 38). Many if not all prominent oncoproteins, including c-Myc, Ras, Src, and Akt, stimulate aerobic glycolysis in transformed cells (4, 39, 40). This warrants the molecular basis for targeting cell metabolism as an anti-cancer strategy, which is intended to target the fundamental property of cancer cells and bypass the complex proliferation and pro-survival signaling pathways in tumors that are almost always heterogeneous. In this study, we attempt to compare how tumor cells with elevated glycolytic activity, driven by different oncoproteins, respond to metabolic perturbation. To this end, we established a dual-regulatable FL5.12 cell line to control the activation of Akt and c-Myc and to compare their responses to glycolysis and mitochondrial respiration inhibitors. Our results show that although Akt cells are highly dependent on glycolysis for survival and are sensitive to glycolysis inhibition, c-Myc cells, which also display an enhanced glycolytic activity, are more susceptible to perturbation of mitochondrial function.
Akt has been shown to protect cells from numerous causes of death through its ability to up-regulate glucose metabolism. This ironically makes malignant cells with high Akt activity more susceptible to limited glucose supply (40). Inhibition of glucose metabolism has been shown to induce apoptosis via up-regulation of Puma and degradation of Mcl-1 (21, 41). In addition to directly up-regulating glycolysis, Akt has been found to drive synthesis pathways to promote cell growth, which requires a constant supply of substrates for biosynthetic building blocks. Under this condition, amino acids and lipids, instead of being used as alternative bioenergetic fuel, may be channeled into cell growth, and their catabolism is reciprocally suppressed (40). Indeed, constitutive Akt activation blunts fatty acid β-oxidation by suppressing expression of CPT1A (carnitine palmitoyltransferase 1A) that is required for the entry of fatty acid into mitochondria (28). Pharmacological activation of fatty acid β-oxidation can reverse glucose dependence of Akt-transformed cells (42).
The c-Myc proto-oncoprotein is a potent regulator of multiple metabolic pathways essential for cancer cell growth (43). Oncogenic c-Myc promotes increased aerobic glycolysis through the constitutive elevation of glycolytic enzymes such as lactate dehydrogenase A (14, 44). Induction of lactate dehydrogenase A by c-Myc is required for Myc-dependent transformation (15, 44). Lactate dehydrogenase A diverts glucose-derived pyruvate into lactate, thereby depleting the mitochondrial oxidation substrate. Despite this, Myc-transformed cells display increased mitochondrial mass and O2 consumption, consistent with the findings that a number of c-Myc targets are involved in mitochondrial biogenesis (16–18). Our results are consistent with a recent report that c-Myc cells are sensitive to the inhibition of the mitochondrial electron transport chain (18). In c-Myc cells, mitochondrial respiration can be maintained by catabolizing non-glucose substrates, especially when glucose supply is limited. In glioblastoma cells, fatty acid β-oxidation can be quickly switched on to sustain ATP generation during glucose deprivation (45). Moreover, c-Myc can directly or indirectly up-regulate the glutaminolysis pathway and promote catabolism of glutamine (35, 36). Although glutamine may not be used by tumor cells for ATP generation when the glucose supply is abundant, it is critical for biosynthesis and can be used as an energy substrate when glucose supply is limited (37). Indeed, we observed here that c-Myc, but not Akt, sensitized cells to the inhibition of fatty acid and glutamine metabolism (Fig. 5C and Fig. 6).
Inhibition of glycolysis has been evaluated as an anti-cancer approach. The application of glycolysis inhibitors has produced encouraging antineoplastic results in vitro and in vivo (25, 46–49). However, little work has gone into the development of drugs that target mitochondrial function in tumor cells. It was recently reported that in a xenograft tumor model, tumor cells growing in oxygenated regions can use lactate for oxidative energy production, suggesting that tumor cells are bioenergetically heterogeneous. Although some cells have compromised mitochondrial activities, some cells retain their mitochondrial function (50). These tumor cells can form symbiotic relationships with hypoxic cells that can be advantageous for tumor development. Hence, targeting mitochondrial bioenergetic function may also be a reasonable therapeutic strategy (51, 52). Our findings indicate that variable oncogenic activities may dictate cell susceptibility to the perturbation of cellular metabolic programs. This may explain why therapeutic agents that target glycolysis do not always provide satisfying outcomes and points to the importance of understanding cancer cell etiology and the complexity of their metabolic processes when designing strategies to interfere with their bioenergetic supply.
Supplementary Material
Acknowledgments
We thank Dr. Michael J. Ausserlechner for the Tet-On plasmids; Dr. David Plas for the myrAkt expression construct; Dr. Yan Song and Dr. Erich Mackow for assistance on quantitative real time PCR; Patric Mclaughlin for technical assistance; and Drs. Claire C. Bastie and Jeffrey E. Pessin for advice on fatty acid oxidation. We also thank members in the Zong laboratory for critical readings of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants CA129536 and CA098092 from NCI. This work was also supported by Susan Komen for the Cure Grant KG081538 and Long Island League Against Cancer.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- DOX
- doxycycline
- 4-OHT
- 4-hydroxytamoxifen
- 2-DG
- 2-deoxyglucose
- OG
- oligomycin
- IL-3
- interleukin-3
- FBS
- fetal bovine serum
- PBS
- phosphate-buffered saline
- PI
- propidium iodide
- PARP
- poly(ADP-ribose) polymerase
- myrAkt
- myristoylated Akt
- OCR
- oxygen consumption rate
- Cox
- complex.
REFERENCES
- 1.DeBerardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B. (2008) Cell Metab. 7, 11–20 [DOI] [PubMed] [Google Scholar]
- 2.Warburg O. (1956) Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 3.Wechalekar K., Sharma B., Cook G. (2005) Clin. Radiol. 60, 1143–1155 [DOI] [PubMed] [Google Scholar]
- 4.Kroemer G., Pouyssegur J. (2008) Cancer Cell 13, 472–482 [DOI] [PubMed] [Google Scholar]
- 5.Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009) Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manning B. D., Cantley L. C. (2007) Cell 129, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang C. V., O'Donnell K. A., Zeller K. I., Nguyen T., Osthus R. C., Li F. (2006) Semin. Cancer Biol. 16, 253–264 [DOI] [PubMed] [Google Scholar]
- 8.Gottlob K., Majewski N., Kennedy S., Kandel E., Robey R. B., Hay N. (2001) Genes Dev. 15, 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plas D. R., Talapatra S., Edinger A. L., Rathmell J. C., Thompson C. B. (2001) J. Biol. Chem. 276, 12041–12048 [DOI] [PubMed] [Google Scholar]
- 10.Birnbaum M. J. (2004) Dev. Cell 7, 781–782 [DOI] [PubMed] [Google Scholar]
- 11.Majewski N., Nogueira V., Bhaskar P., Coy P. E., Skeen J. E., Gottlob K., Chandel N. S., Thompson C. B., Robey R. B., Hay N. (2004) Mol. Cell 16, 819–830 [DOI] [PubMed] [Google Scholar]
- 12.Rathmell J. C., Fox C. J., Plas D. R., Hammerman P. S., Cinalli R. M., Thompson C. B. (2003) Mol. Cell. Biol. 23, 7315–7328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robey R. B., Hay N. (2006) Oncogene 25, 4683–4696 [DOI] [PubMed] [Google Scholar]
- 14.Osthus R. C., Shim H., Kim S., Li Q., Reddy R., Mukherjee M., Xu Y., Wonsey D., Lee L. A., Dang C. V. (2000) J. Biol. Chem. 275, 21797–21800 [DOI] [PubMed] [Google Scholar]
- 15.Lewis B. C., Prescott J. E., Campbell S. E., Shim H., Orlowski R. Z., Dang C. V. (2000) Cancer Res. 60, 6178–6183 [PubMed] [Google Scholar]
- 16.Kim J., Lee J. H., Iyer V. R. (2008) PLoS ONE 3, e1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F., Wang Y., Zeller K. I., Potter J. J., Wonsey D. R., O'Donnell K. A., Kim J. W., Yustein J. T., Lee L. A., Dang C. V. (2005) Mol. Cell. Biol. 25, 6225–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrish F., Neretti N., Sedivy J. M., Hockenbery D. M. (2008) Cell Cycle 7, 1054–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ausserlechner M. J., Obexer P., Deutschmann A., Geiger K., Kofler R. (2006) Mol. Cancer Ther. 5, 1927–1934 [DOI] [PubMed] [Google Scholar]
- 20.Ricci M. S., Jin Z., Dews M., Yu D., Thomas-Tikhonenko A., Dicker D. T., El-Deiry W. S. (2004) Mol. Cell. Biol. 24, 8541–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y., Altman B. J., Coloff J. L., Herman C. E., Jacobs S. R., Wieman H. L., Wofford J. A., Dimascio L. N., Ilkayeva O., Kelekar A., Reya T., Rathmell J. C. (2007) Mol. Cell. Biol. 27, 4328–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickman K. G., Hempson S. J., Anderson J., Lippe S., Zhao L., Burakoff R., Shaw R. D. (2000) Am. J. Physiol. Gastrointest. Liver Physiol. 279, G757–G766 [DOI] [PubMed] [Google Scholar]
- 23.Barrientos A. (2002) Methods 26, 307–316 [DOI] [PubMed] [Google Scholar]
- 24.Edinger A. L., Thompson C. B. (2002) Mol. Biol. Cell 13, 2276–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu R. H., Pelicano H., Zhou Y., Carew J. S., Feng L., Bhalla K. N., Keating M. J., Huang P. (2005) Cancer Res. 65, 613–621 [PubMed] [Google Scholar]
- 26.Zong W. X., Ditsworth D., Bauer D. E., Wang Z. Q., Thompson C. B. (2004) Genes Dev. 18, 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith A. C., Bruce C. R., Dyck D. J. (2005) J. Physiol. 565, 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deberardinis R. J., Lum J. J., Thompson C. B. (2006) J. Biol. Chem. 281, 37372–37380 [DOI] [PubMed] [Google Scholar]
- 29.Djouadi F., Bonnefont J. P., Munnich A., Bastin J. (2003) Mol. Genet. Metab. 78, 112–118 [DOI] [PubMed] [Google Scholar]
- 30.Littlewood T. D., Hancock D. C., Danielian P. S., Parker M. G., Evan G. I. (1995) Nucleic Acids Res. 23, 1686–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evan G. I., Wyllie A. H., Gilbert C. S., Littlewood T. D., Land H., Brooks M., Waters C. M., Penn L. Z., Hancock D. C. (1992) Cell 69, 119–128 [DOI] [PubMed] [Google Scholar]
- 32.Wieman H. L., Wofford J. A., Rathmell J. C. (2007) Mol. Biol. Cell 18, 1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ekstrand M. I., Falkenberg M., Rantanen A., Park C. B., Gaspari M., Hultenby K., Rustin P., Gustafsson C. M., Larsson N. G. (2004) Hum. Mol. Genet. 13, 935–944 [DOI] [PubMed] [Google Scholar]
- 34.Pelicano H., Martin D. S., Xu R. H., Huang P. (2006) Oncogene 25, 4633–4646 [DOI] [PubMed] [Google Scholar]
- 35.Wise D. R., DeBerardinis R. J., Mancuso A., Sayed N., Zhang X. Y., Pfeiffer H. K., Nissim I., Daikhin E., Yudkoff M., McMahon S. B., Thompson C. B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao P., Tchernyshyov I., Chang T. C., Lee Y. S., Kita K., Ochi T., Zeller K. I., De Marzo A. M., Van Eyk J. E., Mendell J. T., Dang C. V. (2009) Nature 458, 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuneva M., Zamboni N., Oefner P., Sachidanandam R., Lazebnik Y. (2007) J. Cell Biol. 178, 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones R. G., Thompson C. B. (2009) Genes Dev. 23, 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dang C. V., Semenza G. L. (1999) Trends Biochem. Sci. 24, 68–72 [DOI] [PubMed] [Google Scholar]
- 40.Elstrom R. L., Bauer D. E., Buzzai M., Karnauskas R., Harris M. H., Plas D. R., Zhuang H., Cinalli R. M., Alavi A., Rudin C. M., Thompson C. B. (2004) Cancer Res. 64, 3892–3899 [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y., Coloff J. L., Ferguson E. C., Jacobs S. R., Cui K., Rathmell J. C. (2008) J. Biol. Chem. 283, 36344–36353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buzzai M., Bauer D. E., Jones R. G., Deberardinis R. J., Hatzivassiliou G., Elstrom R. L., Thompson C. B. (2005) Oncogene 24, 4165–4173 [DOI] [PubMed] [Google Scholar]
- 43.Dang C. V., Kim J. W., Gao P., Yustein J. (2008) Nat. Rev. Cancer 8, 51–56 [DOI] [PubMed] [Google Scholar]
- 44.Shim H., Dolde C., Lewis B. C., Wu C. S., Dang G., Jungmann R. A., Dalla-Favera R., Dang C. V. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jelluma N., Yang X., Stokoe D., Evan G. I., Dansen T. B., Haas-Kogan D. A. (2006) Mol. Cancer Res. 4, 319–330 [DOI] [PubMed] [Google Scholar]
- 46.Geschwind J. F., Georgiades C. S., Ko Y. H., Pedersen P. L. (2004) Expert Rev. Anticancer Ther. 4, 449–457 [DOI] [PubMed] [Google Scholar]
- 47.Maschek G., Savaraj N., Priebe W., Braunschweiger P., Hamilton K., Tidmarsh G. F., De Young L. R., Lampidis T. J. (2004) Cancer Res. 64, 31–34 [DOI] [PubMed] [Google Scholar]
- 48.Maher J. C., Krishan A., Lampidis T. J. (2004) Cancer Chemother. Pharmacol. 53, 116–122 [DOI] [PubMed] [Google Scholar]
- 49.Chen Z., Zhang H., Lu W., Huang P. (2009) Biochim. Biophys. Acta 1787, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonveaux P., Végran F., Schroeder T., Wergin M. C., Verrax J., Rabbani Z. N., De Saedeleer C. J., Kennedy K. M., Diepart C., Jordan B. F., Kelley M. J., Gallez B., Wahl M. L., Feron O., Dewhirst M. W. (2008) J. Clin. Invest. 118, 3930–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gogvadze V., Orrenius S., Zhivotovsky B. (2009) Semin. Cancer Biol. 19, 57–66 [DOI] [PubMed] [Google Scholar]
- 52.Gogvadze V., Orrenius S., Zhivotovsky B. (2009) Apoptosis 14, 624–640 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.