Abstract
In prion diseases, the infectious isoform of the prion protein (PrPSc) may subvert a normal, physiological activity of the cellular isoform (PrPC). A deletion mutant of the prion protein (Δ105–125) that produces a neonatal lethal phenotype when expressed in transgenic mice provides a window into the normal function of PrPC and how it can be corrupted to produce neurotoxic effects. We report here the surprising and unexpected observation that cells expressing Δ105–125 PrP and related mutants are hypersensitive to the toxic effects of two classes of antibiotics (aminoglycosides and bleomycin analogues) that are commonly used for selection of stably transfected cell lines. This unusual phenomenon mimics several essential features of Δ105–125 PrP toxicity seen in transgenic mice, including rescue by co-expression of wild type PrP. Cells expressing Δ105–125 PrP are susceptible to drug toxicity within minutes, suggesting that the mutant protein enhances cellular accumulation of these cationic compounds. Our results establish a screenable cellular phenotype for the activity of neurotoxic forms of PrP, and they suggest possible mechanisms by which these molecules could produce their pathological effects in vivo.
Keywords: Prions, Antibiotics, Cell Death, Drug Action, Mutant, Aminoglycoside, Cell Assay, Drug, Mutant, Neurotoxicity
Introduction
Transmissible spongiform encephalopathies, or prion diseases, are fatal neurodegenerative disorders that affect both humans and animals. The central molecular event in these diseases is the conversion of a normal, cell surface glycoprotein (PrPC) into a conformationally altered isoform (PrPSc) that is capable of propagating itself via a molecular templating mechanism (1, 2).
Although a great deal of progress has been made in elucidating the molecular identity of the infectious agent in prion diseases, the pathogenic mechanisms responsible for prion-induced neurodegeneration remain poorly understood (3). Several pieces of evidence indicate that cell surface PrPC may play an important role in transducing neurotoxic signals elicited by PrPSc, possibly as a result of physical interaction between the two isoforms (4–7). These observations have sparked renewed interest in deciphering the normal, physiological function of PrPC, because a subversion of this function may figure in the pathological process. A variety of functions have been proposed for PrPC, including roles in metal ion homeostasis, cell adhesion, signal transduction, stem cell proliferation, and protection from cellular stress (reviewed in Ref. 8). However, which of these are physiologically relevant is uncertain.
Important insights into the physiological activity of PrPC and how it might be altered in the disease state come from studies of transgenic mice expressing certain deleted forms of PrP. Shmerling et al. (9) originally reported that transgenic mice expressing PrP harboring either of two large, N-terminal deletions (Δ32–121 and Δ32–134) developed a spontaneous neurodegenerative illness characterized by ataxia and massive degeneration of cerebellar granule neurons. Importantly, this phenotype was only observed on the Prn-p0/0 (PrP-null) genetic background: co-expression of endogenous, wild type (WT)4 PrP from a single Prn-p allele completely abrogated clinical symptoms and neuropathology. A subsequent study reported that mice expressing a shorter PrP deletion (Δ94–134) also developed ataxia and neuropathological changes (10). Finally, ectopic central nervous system expression of Doppel (Dpl), a PrP paralog that is structurally equivalent to Δ32–134 PrP, produced a neurodegenerative phenotype in transgenic mice that was suppressed by co-expression of WT PrP (11, 12). Taken together, these mouse models demonstrate that deletion of critical residues within the flexible, N-terminal tail of PrP endow the protein with a powerful neurotoxic activity that is antagonized by the presence of WT PrP.
To map more precisely the region of PrP responsible for this phenomenon, we created Tg(ΔCR) mice expressing PrP with a much smaller deletion, comprising residues 105–125 within the central region of the molecule (13). The deleted segment encompasses a cluster of three positively charged amino acids (residues 105, 109, and 110) followed by a stretch of 15 hydrophobic residues (residues 111–125) that are highly conserved in PrP from fish to humans (14). Tg(ΔCR) mice display a neonatal lethal phenotype characterized by granule cell degeneration and vacuolar degeneration of white matter areas of the brain and spinal cord (13, 15). This phenotype is reversed in a dose-dependent fashion by co-expression of WT PrP, with 5-fold overexpression of the WT protein from a second transgene, allowing the mice to live for over 1 year. The biochemical and cell biological properties of ΔCR PrP are similar to those of WT PrP (16), suggesting that the neurotoxicity of the ΔCR molecule results from an alteration of a normal activity of PrPC rather than from accumulation of misfolded protein aggregates or cellular mislocalization.
To understand the mechanisms underlying the powerful toxicity of ΔCR PrP and other deleted forms of PrP and Dpl, it is essential to develop cell culture models. Strikingly, it has proven difficult to reproduce in vitro the toxic effects of deleted PrP and Dpl seen in vivo. For example, we have found that cerebellar granule neurons from neonatal Tg(ΔCR) mice, which massively degenerate in vivo, display normal viability when maintained in dissociated cultures for many weeks.5
In the course of testing an array of drugs for their effects on the viability of cells expressing ΔCR PrP, we made a surprising and unexpected observation; these cells are hypersensitive to the toxic effects of two classes of antibiotics that are commonly used for selection of stably transfected cell lines. In this paper, we describe this unusual phenomenon and our initial attempts to understand its underlying mechanism. Our results provide the basis for a screenable cellular phenotype to assay the activity of neurotoxic forms of PrP, and they suggest several possible mechanisms by which these molecules could produce their pathological effects in vivo.
EXPERIMENTAL PROCEDURES
Reagents
G418 and Zeocin were obtained from Invitrogen, and hygromycin B and bleomycin were from Sigma, all as sulfate salts. Unless otherwise noted, all other reagents were from Sigma.
Transfected Cell Lines
HEK293 cells (ATCC CRL-1573) were grown in α-minimum Eagle's medium/Dulbecco's modified Eagle's medium (1:1) containing 10% fetal bovine serum, 2 mm glutamine, non-essential amino acids, and penicillin/streptomycin. Chinese hamster ovary (CHO) cells were grown in α-minimum Eagle's medium supplemented with 7% fetal bovine serum, non-essential amino acids, and penicillin/streptomycin.
To prepare stably transfected HEK cell clones, cells were transfected using Lipofectamine 2000 (Invitrogen) with pcDNA3.1(+) Hygro expression plasmids (Invitrogen) containing either no insert or cDNAs encoding WT or ΔCR mouse PrP (13). After selection in 200 μg/ml hygromycin B for 10–14 days, individual clones were isolated and maintained in 50 μg/ml hygromycin. For most experiments, at least two independent clones were tested with similar results.
HEK and CHO cells were transiently transfected with pcDNA3.1(+) Hygro or pcDNA3.1(+) Neo expression plasmids (Invitrogen) containing either no insert or cDNAs encoding WT or ΔCR mouse PrP using Lipofectamine 2000 and were used for experiments 24 h after transfection.
Neural Stem Cells
We utilized the procedure of Louis and Reynolds (17), with minor modifications. Brains dissected from embryonic day 13.5 transgenic mouse embryos were triturated in 5 ml of NeuroCult neural stem cell (NSC) basal medium containing NeuroCult NSC proliferation supplement (StemCell Technologies, Vancouver, Canada) along with 20 ng/ml epidermal growth factor. The formation of neurospheres was monitored daily. For differentiation of neurospheres, a 0.1–1-ml suspension containing ∼30–40 mature neurospheres was pipetted into each well of an 8-well chamber slide or a 24-well plate containing NeuroCult NSC basal medium with NeuroCult NSC differentiation supplement (StemCell Technologies) along with 10 μg/ml retinoic acid. Cells differentiated for 7 days were treated with Zeocin or G418 and were stained by terminal deoxynucleotidyltransferase dUTP nick end labeling (TUNEL) (see below) to assess cell death.
Cell Viability Assay
Cells were plated at >90% confluence in medium lacking hygromycin in 24-well plates and were treated with drugs for the indicated times at 37 °C. Medium was removed, and cells were incubated with 1 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) in PBS for 30 min at 37 °C. MTT was removed, and cells were resuspended in 500 μl of DMSO or isopropyl alcohol, and A570 was measured in a plate reader (Bio-Rad). Data were expressed as a percentage of A570 values in untreated cells.
TUNEL
Cells plated on glass coverslips were fixed with 4% paraformaldehyde for 20 min, rinsed twice with PBS, permeabilized with 0.5% Triton X-100 in PBS for 10 min, and then stained using a TMR Red in situ cell death detection kit, according to the manufacturer's directions (Roche Applied Science). Cell nuclei were counterstained with DAPI. Cells were mounted with Gel/Mount (Biomeda, Foster City, CA) and imaged on a Nikon TE2000E2 inverted fluorescence microscope. The number of TUNEL-positive cells as a percentage of DAPI-positive cells was determined in five fields for each sample group.
Western Blots
For detection of PrP, cells were lysed on ice for 10 min in Triton-DOC buffer (0.5% Triton X-100, 0.5% sodium deoxycholate, 150 mm NaCl, 50 mm Tris-HCl (pH 7.5) plus protease inhibitors). Lysates were centrifuged at 16,000 × g for 10 min to remove debris prior to analysis by SDS-PAGE. In some cases, proteins were enzymatically deglycosylated with PNGase F according to the manufacturer's directions (New England Biolabs, Beverly, MA). For detection of γ-H2AX, cells were lysed directly in 1× SDS-PAGE sample buffer (2% SDS, 10% glycerol, 100 mm Tris-HCl (pH 6.8), 0.002% bromphenol blue, 100 mm dithiothreitol) (150 μl/well of a 24-well plate), boiled at 95 °C for 10 min, and then frozen before use.
Following SDS-PAGE and electroblotting, blots were incubated with antibodies directed against PrP (6D11 (18), 8H4 (19), or SA65 (20)) or against the phosphorylated form of H2AX (Biolegend, San Diego, CA). Blots were visualized with ECL or with the Odyssey fluorescent imaging system (Li-Cor, Lincoln, NE). ECL signals were quantitated from scanned x-ray films using ImageJ (National Institutes of Health).
Lentiviral Transduction
Recombinant lentiviruses were created at the Hope Center Viral Vectors Core at Washington University. cDNAs encoding mouse PrP (WT and ΔCR) or enhanced green fluorescent protein were cloned into the transfer vector, pRRLsinCMV (21). Recombinant virus was generated by co-transfection of 293T packaging cells with the transfer vector, along with the plasmids pMD-Lg, pCMV-G, and RSV-REV (22). High titer (>109 plaque-forming units/ml), purified virus was applied directly to cells in tissue culture medium at a multiplicity of infection of 100 overnight at 37 °C. The next day, the medium was changed, and cells were incubated in the absence or presence of drugs before assay by MTT or Western blot as described above. Expression of the enhanced green fluorescent protein virus was verified by fluorescence microscopy.
Protein Synthesis Assay
HEK cells were preincubated in medium lacking cysteine and methionine for 30 min. EasyTagEXPRESS 35S-protein labeling mix (PerkinElmer Life Sciences) was added at a concentration of 200 μCi/ml, and cells were incubated for 60 min. Cells were then lysed in Triton-DOC buffer, and proteins were analyzed by SDS-PAGE followed by quantitation of radiolabeled bands using a Storm PhosphorImager (GE Healthcare). Multiple regions of the each lane were quantitated separately and yielded similar results. [35S]Methionine incorporation was normalized to the amount of total protein in each sample, assayed using a BCA kit (Pierce).
Immunofluorescence Staining
Cells were plated on glass coverslips in 24-well plates and grown to ∼50% confluence. For detection of PrP on the cell surface, living cells were stained with SA65 antibody (1:1,000) in Opti-MEM (Invitrogen) at 4 °C. Cells were then fixed and stained with secondary antibody as described below.
For detection of γ-H2AX, cells were fixed with 4% paraformaldehyde in PBS for 20 min, permeabilized with 0.5% Triton X-100 in PBS for 10 min, treated with block buffer (PBS plus 2% goat serum) for 30 min, stained with γ-H2AX antibody (Biolegend) (1:2,000) in block buffer for 60 min, and then stained with Alexa Fluor anti-mouse IgG secondary antibody (Invitrogen) in block buffer for 60 min. Cells were counterstained with 1 μg/ml DAPI for 5 min and then mounted in Gel/Mount. Cells were photographed on a Nikon TE2000E2 inverted fluorescent microscope equipped with a CCD camera, and images were processed using MetaMorph software (Molecular Devices, Sunnyvale, CA).
Semiautomated Microscopy
The staining procedure was identical to that described above, except that that cells were grown to ∼50% confluence directly in 24-well plates without coverslips, and plates were stored covered in PBS at 4 °C after staining. Data were acquired at the High-Throughput Screening Facility at Washington University, which is equipped with a Molecular Devices Image Express Micro High Content Imager. Images (9 fields/well; ∼1,000 cells total) were taken at ×20 magnification at 100 ms for H2AX (fluorescein isothiocyanate channel) and 70 ms for DAPI. Data were analyzed using multiwavelength cell scoring software (MetaExpress, Molecular Devices) to count both DAPI-positive cells and γ-H2AX-positive cells. The threshold was set to 50 gray levels above background, and minimum and maximum cell widths were 10 and 20 nm. The number of γ-H2AX-positive cells as a percentage of the number of DAPI-positive cells was calculated for each well.
RESULTS
Stable Expression of ΔCR PrP Has Minimal Effect on Cell Viability
To establish a cell culture system for analyzing the toxicity of ΔCR PrP, we stably transfected HEK293 cells with empty vector or with vector encoding WT or ΔCR PrP. The expression vector (pcDNA3.1(+) Hygro) encodes resistance to the antibiotic, hygromycin. We grew cells for 10–14 days in the presence of hygromycin B (200 μg/ml) and selected several antibiotic-resistant clones in which WT and ΔCR PrP were expressed at similar levels (supplemental Fig. S1A, lanes 2 and 3). The endogenous level of PrP in HEK cells was undetectable by Western blotting (supplemental Fig. S1A, lane 1). Both WT and ΔCR PrP migrated primarily as diglycosylated species with molecular sizes of 32–34 kDa. These forms were converted to unglycosylated species of 26 kDa (WT) and 24 kDa (ΔCR) by treatment with the N-glycosidase PNGase F (supplemental Fig. S1A, lanes 5 and 6). Immunostaining of unpermeabilized cells confirmed our previously published observation (16) that ΔCR PrP is present on the cell surface in a pattern that is indistinguishable from that of WT PrP (supplemental Fig. S1B).
We noted that, during selection of stable clones after transfection, we often recovered fewer colonies of cells expressing ΔCR PrP compared with cells expressing WT PrP or vector (data not shown). This observation suggested that ΔCR PrP may be detrimental to long term cell survival during the selection process. However, when we compared cell death in clones expressing matched levels of ΔCR or WT PrP, or vector alone, using two different assays, TUNEL (supplemental Fig. S1C) and propidium iodide staining (data not shown), we observed only a small detrimental effect of the ΔCR mutant (1.9 ± 0.7% TUNEL-positive cells for ΔCR compared with 0.9 ± 0.3% for WT and 0.8 ± 0.3% for vector; p < 0.01).
Screen for Drugs That Exacerbate ΔCR PrP Toxicity
Because there was minimal effect of ΔCR PrP on the base-line viability of HEK cells, we sought conditions that would selectively accentuate the toxicity of the mutant protein. We tested 24 different drugs affecting multiple cellular targets and pathways, including antibiotics that interfere with protein synthesis on ribosomes, genotoxic agents, oxidative stressors, inhibitors of protein folding and trafficking, and inhibitors of protein kinases and phosphatases (Table 1). Cells were exposed to each drug for 3 days, in some cases at two different concentrations, and cell viability was measured by an MTT assay. Our objective was to identify compounds that selectively impaired the viability of HEK cells expressing ΔCR PrP without significantly affecting the viability of cells expressing WT PrP.
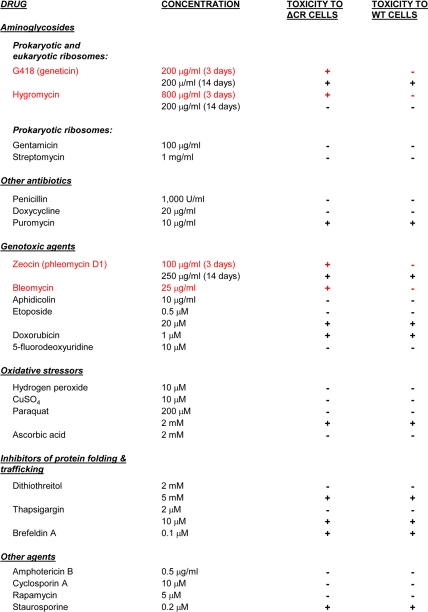
TABLE 1.
Drug sensitivity of HEK cells expressing WT or ΔCR PrP
Stably transfected HEK cells expressing WT or ΔCR PrP from a vector (pcDNA3.1(+) Hygro) encoding hygromycin resistance were treated with drugs at the indicated concentrations for 3 days (unless indicated otherwise) and then assayed for viability by MTT reduction. A positive toxic effect was defined as a reduction in viability of >50% in drug-treated cells compared with untreated cells. G418, hygromycin, Zeocin, and bleomycin (all shown in red) were selectively toxic to ΔCR but not WT cells under the indicated conditions. Other drugs had no effect on either cell type or were toxic to both. Vector-transfected cells behaved identically to cells expressing WT PrP in these assays (not shown).
Under the conditions tested, some of the compounds had no effect on either ΔCR or WT cells, whereas others were toxic to both cell lines. However, representatives of two classes of drugs showed a potent and selective toxicity toward cells expressing ΔCR PrP after 3 days of treatment. The first drug we identified, G418 (Geneticin) (supplemental Fig. S2), belongs to the family of aminoglycoside antibiotics that interfere with protein synthesis via binding to specific sites on ribosomal subunits (23). The second drug, Zeocin (a formulation of phleomycin D1; Invitrogen) (supplemental Fig. S2), is a genotoxic antibiotic that causes cell death by inducing DNA- and RNA- strand breaks (24, 25). Both drugs are commonly used for selection of stably transfected mammalian cell lines. Under typical selection conditions, these drugs kill cells lacking antibiotic resistance genes over a time course of 10–14 days (Table 1). In contrast, we observed substantial toxicity (>50% loss of viability) to cells expressing ΔCR PrP over a much shorter time period (3 days) (Table 1) (see below).
Characterization of Cellular Hypersensitivity to G418, Zeocin, and Related Compounds
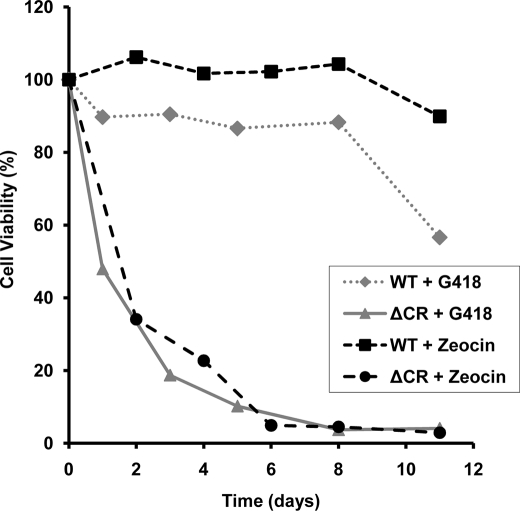
We performed an experiment to analyze in more detail the time course of the toxic effect G418 and Zeocin. We found that at 400 and 250 μg/ml, respectively, G418 and Zeocin caused 50% loss of viability of ΔCR PrP-expressing cells within 1–2 days (Fig. 1). In contrast, vector-transfected cells or cells expressing WT PrP displayed >80% viability for at least 8 days, and viability did not drop below 50% until 12–14 days (Fig. 1) (data not shown).
FIGURE 1.
HEK cells expressing ΔCR PrP are rapidly killed by G418 and Zeocin. Cells expressing ΔCR or WT PrP were treated with 400 μg/ml G418 or 250 μg/ml Zeocin for the indicated times, and viability was assayed by MTT reduction. Cell viability is expressed as the A570 value of treated cells as a percentage of the value for untreated cells. Data points are the mean values of duplicate wells from a single experiment and are representative of at least three similar experiments.
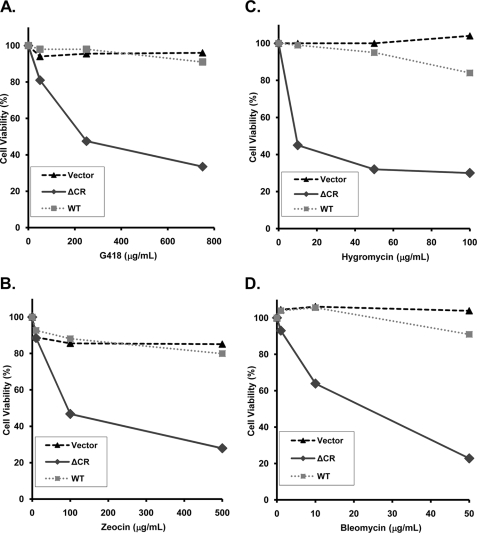
We performed a dose-response experiment to determine the effective concentration of antibiotic (EC50) that caused 50% loss of cell viability after 3 days as measured by MTT reduction (Fig. 2, A and B). The EC50 values for G418 and Zeocin were found to be 230 and 90 μg/ml, respectively, in cells expressing ΔCR PrP. During this time period, cells expressing WT PrP or transfected with empty vector maintained >90% viability.
FIGURE 2.
HEK cells expressing ΔCR PrP are hypersensitive to G418 (A), Zeocin (B), hygromycin (C), and bleomycin (D). Cells expressing WT or ΔCR PrP or transfected with empty vector were treated with the indicated concentrations of drug for 3 days, and viability was assayed by MTT reduction. Cell viability is expressed as the A570 value of treated cells as a percentage of the value for untreated cells. Data points are the mean values of duplicate wells from a single experiment and are representative of at least three similar experiments. Cells in A, B, and D were stably transfected using the expression vector, pcDNA3.1(+) Hygro; cells in C were transiently transfected using the expression vector, pcDNA3.1(+) Neo.
As an alternative to the MTT assay for measuring cellular toxicity in response to drug treatment, we used TUNEL staining to score the number of cells with fragmented DNA (supplemental Fig. S3A). Using this more sensitive read-out, we detected an effect of G418 and Zeocin on ΔCR PrP-expressing cells at concentrations as low as 50 and 10 μg/ml, respectively, after 3 days of treatment, concentrations that had no effect on WT or vector cells (supplemental Fig. S3, B and C).
To extend these results to another cell type, we used TUNEL staining to assay the effect of G418 and Zeocin on transiently transfected CHO cells (supplemental Fig. S4A). We observed that both drugs increased TUNEL staining to a greater extent in cells expressing ΔCR PrP compared with cells expressing WT PrP or transfected with empty vector (supplemental Fig. S4, B and C). Similar results were obtained (using either MTT reduction or TUNEL staining as read-outs) in two other cell lines, MDCK and N2a (data not shown). Collectively, these results demonstrate that the expression of ΔCR PrP sensitizes multiple cell types, both stably and transiently transfected, to G418- and Zeocin-induced toxicity.
We also analyzed the effects of several drugs related to G418 and Zeocin. First, we tested hygromycin, an aminoglycoside antibiotic that is structurally similar to G418 (supplemental Fig. S2) and that also causes translational misreading at the A site of the small ribosomal subunit as a result of drug binding to specific nucleotides in rRNA (23). For this experiment, we utilized HEK cells transiently transfected with a vector (pcDNA3.1(+) Neo) encoding resistance to G418, rather than the stable HEK cell lines that carried a hygromycin resistance gene. Transiently transfected HEK cells expressing ΔCR PrP displayed an EC50 of 40 μg/ml hygromycin, whereas cells expressing WT PrP or empty vector showed >90% viability with up to 100 μg/ml drug (Fig. 2C). Thus, ΔCR PrP sensitizes cells to hygromycin as well as G418.
Interestingly, stable HEK cells lines expressing ΔCR PrP (but not those expressing WT PrP) from the hygromycin resistance vector were also susceptible to killing by hygromycin although with a higher EC50 (∼800 μg/ml after 3 days of treatment; Table 1). This result suggests that enzymatic detoxification of hygromycin by the vector-encoded resistance gene can be overcome by sufficiently high concentrations of the drug, such that ΔCR PrP-expressing cells are selectively killed. It is likely that this effect contributes to the lower recovery of stable clones expressing ΔCR PrP compared with those expressing WT PrP or vector (see above).
As controls, we also tested two aminoglycosides (streptomycin and gentamicin) that are specific for prokaryotic ribosomes and that have no effect on protein synthesis on eukaryotic ribosomes (23, 26). These two drugs had no effect on the viability of HEK cells expressing ΔCR or WT PrP or transfected with empty vector, even at concentrations well above their bacteriostatic levels (100 μg/ml for gentamicin and 1,000 μg/ml for streptomycin) (Table 1).
We also tested bleomycin, a genotoxic, copper-containing glycopeptide that is structurally related to Zeocin (supplemental Fig. S2) and that is used clinically as a chemotherapeutic agent (24). Bleomycin also showed a robust differential toxicity to cells expressing ΔCR PrP compared with control cells (Fig. 2D). The EC50 for bleomycin was 25 μg/ml. Other DNA-damaging agents with different chemical structures and modes of action (including aphidicolin, etoposide, doxorubicin, and 5-fluorodeoxyuridine) were not differentially toxic to ΔCR and WT PrP-expressing cells in this assay (Table 1).
Co-expression of WT PrP Suppresses Drug Hypersensitivity in Cells Expressing ΔCR PrP
Co-expression of WT PrP dramatically suppresses the toxicity of ΔCR PrP and other N-terminally deleted forms of PrP when expressed in transgenic mice (9, 10, 13). To investigate whether an analogous phenomenon was present in cultured cells, we tested whether introduction of WT PrP ameliorated the drug hypersensitivity induced by expression of ΔCR PrP in HEK cells.
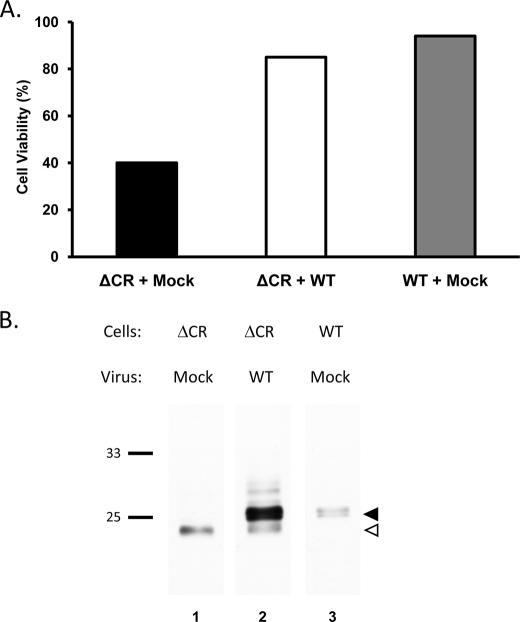
To achieve high level expression of WT PrP, which is necessary to rescue the neurodegenerative phenotype of Tg(ΔCR) mice (13), we transduced HEK cells stably expressing ΔCR PrP with a recombinant lentivirus encoding WT PrP. Transduced cells were treated for 2 days with 500 μg/ml Zeocin, after which cell viability was assayed by MTT reduction. As expected, mock-transduced cells expressing ΔCR PrP showed a marked loss of viability (Fig. 3A, left bar). In contrast, ΔCR PrP-expressing cells transduced with the lentivirus encoding WT PrP maintained normal viability (Fig. 3A, middle bar), comparable with mock-transduced cells stably expressing WT PrP (Fig. 3A, right bar).
FIGURE 3.
Co-expression of WT PrP suppresses drug hypersensitivity induced by ΔCR PrP. A, HEK cells expressing ΔCR or WT PrP were mock-transduced, or transduced with a recombinant lentivirus encoding WT PrP. After 18 h, cells were incubated with Zeocin (500 μg/ml) for 2 days and then assayed for viability by MTT reduction. Cell viability is expressed as the A570 value of drug-treated cells as a percentage of the value for untreated cells. B, HEK cells expressing ΔCR or WT PrP were mock-transduced or transduced with a recombinant lentivirus encoding WT PrP. After 3 days, cell lysates were prepared, and proteins were enzymatically deglycosylated with PNGase F, followed by Western blotting for PrP using 6D11 antibody. Bands corresponding to WT and ΔCR PrP are indicated by black and white arrowheads, respectively. Molecular size markers are given in kDa.
Western blots were performed on parallel cultures to assess the relative levels of WT and ΔCR PrP (Fig. 3B). Samples were enzymatically deglycosylated with PNGase to facilitate resolution of WT and ΔCR PrP based on the ∼2-kDa difference in Mr between the two forms. We observed that in cultures co-expressing both proteins, WT PrP was present at levels ∼6-fold higher than ΔCR PrP (Fig. 3B, lane 2, black and white arrowheads). Importantly, the level of ΔCR PrP was not altered by co-expression of WT PrP (Fig. 3B, compare lanes 1 and 2, white arrowhead), eliminating the possibility that the rescue effect produced by WT PrP is due to a reduction in the amount of ΔCR PrP. Expression of WT PrP at lower levels (by reducing the multiplicity of the lentiviral infection below 100) produced a less complete suppression of toxicity (data not shown), recapitulating the dose-dependent effect of WT PrP rescue in Tg(ΔCR) mice (13).
We also observed that transient transfection of a plasmid encoding ΔCR PrP produced less G418 toxicity in HEK cells stably expressing WT PrP compared with cells expressing empty vector (data not shown). Using this procedure, however, it was more difficult to control the relative expression levels of the two proteins.
Other Neurotoxic PrP Deletion Mutants and Dpl Sensitize Cells to Drug Treatment
PrP mutants harboring several other deletions encompassing the CR region, as well as Dpl, produce a spontaneous neurodegenerative phenotype in mice that is ameliorated by co-expression of WT PrP (9, 10, 12). We investigated whether these neurotoxic molecules, like ΔCR PrP, sensitized cultured cells to the effects of G418. We created stably transfected HEK cell lines expressing Δ32–134 PrP, Doppel, or yellow fluorescent protein (as an additional negative control) using the hygromycin resistance plasmid. Based on Western blotting with a PrP-specific antibody, the expression level of the Δ32–134 construct was similar to that of WT PrP and ΔCR PrP in the previously employed cell lines (Fig. 4B).
FIGURE 4.
Δ32–134 PrP and Doppel also sensitize HEK cells to G418 toxicity but are less potent than ΔCR PrP. A, stably transfected HEK cells expressing yellow fluorescent protein (YFP), WT PrP, ΔCR PrP, Δ32–134 PrP, or Doppel were treated with the indicated concentrations of drug for 5 days, and viability was assayed by MTT reduction. Cell viability is expressed as the A570 value of treated cells as a percentage of the value for untreated cells. Data points are the mean values of duplicate wells from a single experiment and are representative of at least three similar experiments. B, the expression levels of WT PrP, ΔCR PrP, Δ32–134 PrP, and Doppel were evaluated by Western blotting using anti-PrP antibody 6D11 (lanes 1–4) or an anti-Doppel antibody (lane 5). V, vector-transfected cells. Molecular size markers are given in kDa.
We found that the cell lines expressing Δ32–134 PrP and Dpl both showed significantly greater sensitivity to G418 compared with cell lines expressing WT PrP or yellow fluorescent protein (Fig. 4). However, the sensitizing effects of Δ32–134 PrP and Dpl were quantitatively less than those of ΔCR PrP. Thus, higher concentrations of G418 were required to kill cells expressing Δ32–134 PrP and Dpl compared with cells expressing ΔCR PrP. Based on a 5-day drug treatment, the corresponding EC50 values were 750 and 1000 μg/ml for Δ32–134 and Dpl, respectively, compared with 50 μg/ml for ΔCR (Fig. 4). Cells expressing WT PrP or yellow fluorescent protein remained >80% viable for this time period at G418 concentrations of up to 1,200 μg/ml. We also observed that HEK cells expressing Δ32–134 PrP and Dpl were rescued from G418 toxicity by lentiviral introduction of WT PrP, but this effect required lower levels of WT PrP than for cells expressing ΔCR PrP (data not shown).
The lower potency of Δ32–134 PrP and Dpl in the HEK cell assay parallels their lower neurotoxic potential in transgenic mice compared with ΔCR PrP (see “Discussion”). Taken together, these data demonstrate a striking correspondence between the effects of deleted forms of PrP and Dpl in transgenic mice and in the drug sensitivity assay.
Differentiated Neural Stem Cells Expressing ΔCR PrP Are Hypersensitive to Drugs
We wished to test whether the drug hypersensitivity phenomenon we observed in HEK and other transformed cell lines also applied to neuronal cells. Tg(ΔCR) mice on the Prn-p0/0 background, which exhibit the strongest neurodegenerative phenotype, die within 1 week of birth, making it difficult to recover neonatal animals for culturing cerebellar granule neurons, the primary cell type that dies in vivo (13). We therefore turned to lines of NSCs derived from transgenic mice at embryonic day 13.5 (17). In the presence of epidermal growth factor, these cells can be propagated in an undifferentiated, proliferative state in which they form spheroid bodies (neurospheres), measuring 100–200 μm in diameter and composed of ∼10,000 cells. When epidermal growth factor is removed and retinoic acid is added, NSCs differentiate along three major lineages, giving rise, after ∼7 days, to mixed cultures of neurons, astrocytes, and oligodendrocytes.
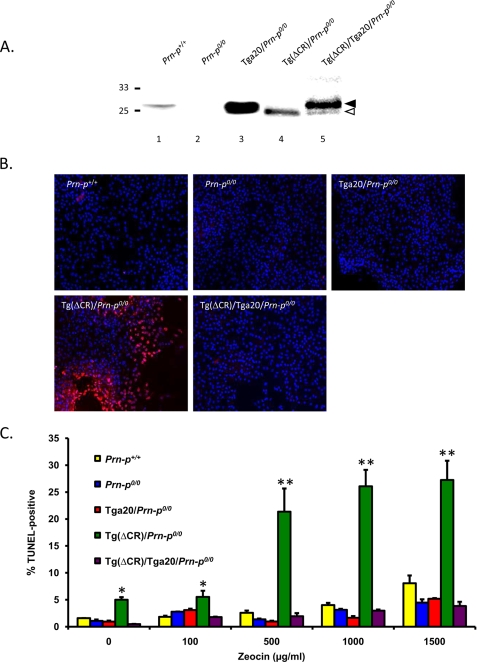
We prepared NSCs from the following five mouse lines and propagated them as neurospheres: Tg(ΔCR)/Prn-p0/0, Tg(ΔCR)/Tga20/Prn-p0/0, Tga20/Prn-p0/0, Prn-p0/0, and Prn-p+/+. After differentiation of the cultures, we confirmed the presence of three major cell types by immunostaining with lineage-specific markers (MAP-2 or NF-H for neurons, glial fibrillary acidic protein for astrocytes, and myelin basic protein for oligodendrocytes) (data not shown). Relative expression levels of PrP in NSC cultures were comparable with those in brain tissue from the corresponding mouse line (Fig. 5A) (data not shown). Detailed characterization of these NSC cultures will be subject of a forthcoming paper.6
FIGURE 5.
Differentiated NSCs expressing ΔCR PrP are hypersensitive to drugs. A, PrP expression in differentiated NSC clones of the indicated genotypes was analyzed by Western blotting using 8H4 antibody. Samples were enzymatically deglycosylated with PNGase F. Bands corresponding to WT and ΔCR PrP are indicated by black and white arrowheads, respectively. B, NSCs dissected at embryonic day 13.5 mouse embryos of the indicated genotypes were cultured as neurospheres and differentiated for 7 days in the presence of retinoic acid. Differentiated NSCs were treated for 72 h with Zeocin (500 μg/ml) and were then stained by TUNEL (red) to reveal fragmented DNA and with DAPI (blue) to reveal nuclei. C, differentiated NSCs (two independent clones for each genotype) were treated for 72 h with the indicated concentrations of Zeocin. The number of TUNEL-positive cells, expressed as a percentage of the number of DAPI-stained cells, was determined in five fields for each sample group. The bars show means ± S.E. (n = 3 independent experiments). The number of TUNEL-positive cells was significantly higher in Tg(ΔCR)/Prn-p0/0 cells than in control cells at all drug concentrations (*, p < 0.05; **, p < 0.01).
Differentiated NSC cultures were treated with G418 or Zeocin, and cell death was scored by TUNEL staining (Fig. 5, B and C). In the absence of drug treatment, we detected a small but significantly higher percentage of TUNEL-positive cells in NSCs derived from Tg(ΔCR)/Prn-p0/0 mice, as compared with NSCs from mice of the other genotypes (5% versus ∼1%, p < 0.05). However, the percentage of TUNEL-positive cells from Tg(ΔCR)/Prn-p0/0 mice was dramatically increased by treatment with Zeocin (500–1,500 μg/ml; Fig. 5, B and C) or G418 (data not shown). Importantly, both the spontaneous toxicity and the hypersensitivity to drugs were completely abolished by overexpression of WT PrP from the Tga20 transgene (ΔCR/Tga20/Prn-p0/0 genotype). This result parallels the situation in vivo, where the presence of the Tga20 transgene dramatically prolongs the life span of Tg(ΔCR) mice (13). In cultures from control mice (Tga20/Prn-p0/0, Prn-p0/0, and Prn-p+/+), the percentage of TUNEL-positive cells after drug treatment remained at <3%. Some of the dying cells in Tg(ΔCR)/Prn-p0/0 cultures are likely to be neurons, based on staining for MAP-2 or NF-H (not shown), but we do not yet know whether other cell types are also affected. Consistent with the results obtained in HEK293 and CHO cells, the NSC experiments demonstrate that, in differentiated neurons derived from mouse brain, expression of ΔCR PrP induces a low level of spontaneous cell death that is dramatically accentuated by exposure to G418 and Zeocin and that is suppressed by co-expression of WT PrP.
Drug Toxicity Is Induced Very Rapidly in Cells Expressing ΔCR PrP
One hypothesis to explain the drug-sensitizing effect of ΔCR PrP is that the mutant protein somehow facilitates intracellular accumulation of the drugs. In this case, expression of ΔCR PrP would be expected to accelerate the action of the drugs on their known intracellular targets: ribosomes (causing inhibition of protein synthesis) and DNA/RNA (inducing double strand breaks).
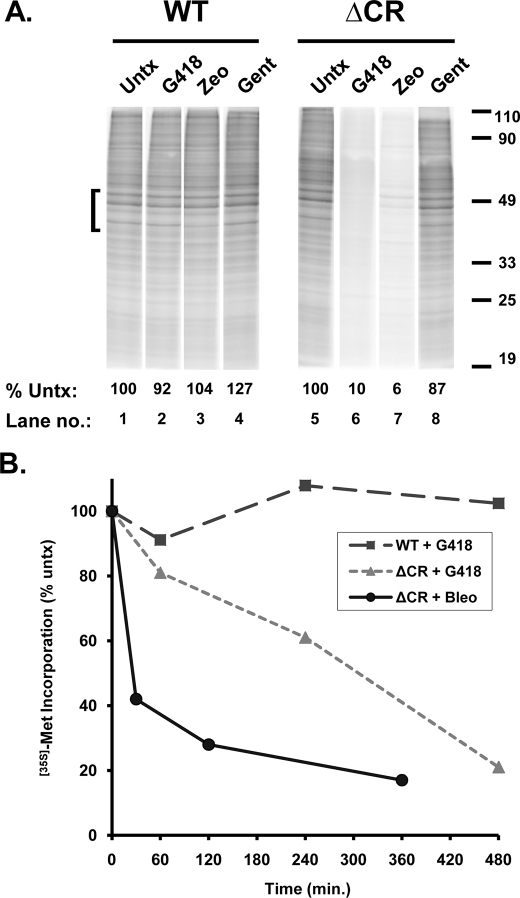
To test this prediction, we first measured the effect of G418 and Zeocin on levels of protein synthesis in stably transfected HEK cells expressing WT or ΔCR PrP. Both drugs are known to inhibit protein synthesis, G418 as a result of binding to sites on ribosomal RNA (23) and Zeocin (like other bleomycin-type compounds) as a result of its ability to cleave rRNA and tRNA (25). Cells were treated with drugs overnight (at a dose that does not affect cell viability) and then labeled with [35S]methionine in the presence of the drug to determine bulk protein synthesis rates.
Importantly, the rate of protein synthesis in the absence of drug treatment was identical in WT and ΔCR cells, implying that ΔCR PrP does not cause a base-line metabolic impairment of cells (Fig. 6A, lanes 1 and 5). We observed a dramatic inhibition of protein synthesis specifically in ΔCR cells treated with G418 or Zeocin (Fig. 6A, lanes 6 and 7), whereas WT cells were unaffected (Fig. 6A, lanes 2 and 3). As a negative control, gentamicin, which is inactive against eukaryotic ribosomes (26), had no effect on protein synthesis in either kind of cell (Fig. 6A, lanes 4 and 8). We found that hygromycin at high doses (>500 μg/ml) also reduced protein synthesis by >50% in hygromycin-resistant ΔCR cells while leaving WT cells unaffected (not shown), consistent with the ability of high drug concentrations to overcome enzymatic detoxification and reduce viability in these cells (see above).
FIGURE 6.
G418 and Zeocin rapidly inhibit protein synthesis in cells expressing ΔCR but not WT PrP. A, HEK cells expressing either WT or ΔCR PrP were either untreated (Untx) or incubated for 20 h with 200 μg/ml either G418, Zeocin (Zeo), or gentamicin (Gent). Cells were then labeled with [35S]methionine for 60 min and lysed, and total cellular proteins were analyzed by SDS-PAGE and autoradiography. The amount of [35S]methionine incorporated into protein was quantitated by PhosphorImager analysis of the region of each lane indicated by the bracket. The numbers below each lane indicate the amount of radioactivity incorporated, expressed as a percentage of that in untreated control cells (lanes 1 and 5). B, HEK cells expressing WT or ΔCR PrP were treated with G418 (500 μg/ml) or bleomycin (50 μg/ml) for the indicated times, after which they were labeled with [35S]methionine for 60 min, and protein synthesis was quantitated as in A. The amount of radioactivity incorporated at each time point is expressed as a percentage of the amount incorporated prior to the start of drug treatment (at 0 min). Results from one of several representative experiments are shown.
We performed a time course to determine how quickly the drugs affected protein synthetic rates. G418 (500 μg/ml) significantly reduced protein synthesis in ΔCR cells within 4 h (T50 = 295 min) (Fig. 6B). Bleomycin (50 μg/ml) produced an even more rapid reduction (T50 = 30 min) (Fig. 6B). Additional experiments showed that a dose of 1,000–1,500 μg/ml of G418 or Zeocin for 24 h was required to diminish protein synthesis rates below 50% in WT or vector-transfected cells, whereas 20–50 μg/ml was sufficient in ΔCR cells (data not shown). Taken together, these results imply that the drugs reach effective intracellular concentrations in ΔCR cells within a very short time.
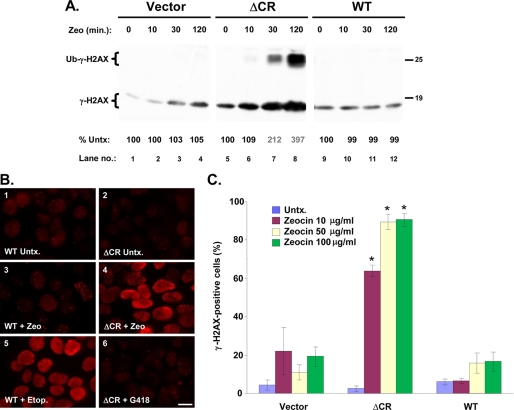
To further narrow the time window for drug toxicity, we assessed DNA damage induced by Zeocin, which, like other bleomycin-type chemotherapeutics, produces DNA double strand breaks (24). We took advantage of a sensitive assay based on phosphorylation of the non-replication-dependent histone variant, H2AX, which is recruited to sites of DNA double-strand breaks and is then rapidly phosphorylated (27). We treated HEK cells with Zeocin and detected phosphorylated H2AX (γ-H2AX) by Western blotting with a phospho-specific antibody. Within 30 min of Zeocin treatment, the amount of γ-H2AX in ΔCR cells had increased by ∼200%, and at 120 min it had reached ∼400% of starting values (Fig. 7A, lanes 7 and 8). Cells expressing WT PrP or empty vector showed minimal changes in γ-H2AX levels, demonstrating the specificity of the effect (Fig. 7A, lanes 1–4 and 9–12). The response of ΔCR cells was specific for drugs that induce DNA damage, because H2AX phosphorylation was not altered by treatment with G418, even at 1,000 μg/ml for 24 h, a condition that caused substantial death of these cells (data not shown). This latter result suggests that the observed increase in γ-H2AX is a direct reflection of Zeocin accumulation in the cell, rather than an indirect result of impaired cellular viability.
FIGURE 7.
Zeocin rapidly induces DNA damage in cells expressing ΔCR PrP. A, HEK cells stably transfected with empty vector or with vector encoding ΔCR or WT PrP were incubated with Zeocin (500 μg/ml) for the indicated times, after which cells were lysed, and γ-H2AX was detected by Western blotting. When large amounts of γ-H2AX are present, a monoubiquitinated form becomes visible (Ub-γ-H2AX; lanes 7 and 8). The numbers below each lane indicate the amount of γ-H2AX, expressed as a percentage of that in untreated control cells. B, HEK cells expressing WT or ΔCR PrP were untreated (Untx.) (panels 1 and 2) or were incubated for 60 min with Zeocin (100 μg/ml) (panels 3 and 4), etoposide (50 μm) (panel 5), or G418 (1,000 μg/ml) (panel 6). Cells were then fixed and stained with an antibody to γ-H2AX antibody and viewed by fluorescence microscopy. Scale bar in panel 6, 5 μm. C, HEK cells expressing vector, WT PrP, or ΔCR PrP were left untreated (untx) or were incubated for 60 min with the indicated concentrations of Zeocin. Cells were then stained for γ-H2AX and analyzed by semiautomated microscopy (see “Experimental Procedures”). The bars show the mean percentage ± S.E. of γ-H2AX-positive cells (nine separate fields, 1,000 total cells/well) from two independent experiments. *, values that are significantly different from corresponding values for vector and WT (p < 10−8).
We found that lentiviral overexpression of WT PrP suppressed Zeocin stimulation of H2AX phosphorylation in ΔCR cells (supplemental Fig. S5), paralleling the ability of the WT protein to rescue Zeocin-induced cell death in these cells (Fig. 3).
The rapid effect of Zeocin on H2AX phosphorylation was also observed by immunofluorescence staining. The majority of ΔCR cells treated with Zeocin showed robust nuclear staining for γ-H2AX within 60 min, whereas little or no staining was seen in untreated ΔCR cells or in WT cells either with or without drug (Fig. 7B, panels 1–4). As a control, both WT and ΔCR cells showed intense γ-H2AX staining after treatment with etoposide, a DNA-damaging agent that is structurally and mechanistically distinct from Zeocin (it is a topoisomerase II inhibitor) and that is equally toxic to both cell types (Fig. 7B, panel 5, and Table 1). As a negative control, γ-H2AX staining was not increased by treatment of ΔCR cells with G418 (Fig. 7B, panel 6), again demonstrating that the Zeocin effect depends specifically on DNA damage rather than on generalized cellular toxicity.
As a prelude to using drug hypersensitivity as a screenable cellular phenotype for ΔCR PrP activity, we adapted the γ-H2AX assay to a medium throughput format by semiautomated microscopy. As little as 10 μg/ml Zeocin for 60 min was sufficient to induce the appearance of γ-H2AX in a large percentage of ΔCR cells while showing no effect on WT or vector-transfected cells (Fig. 7C).
Taken together, our results suggest that the drugs under study rapidly accumulate in ΔCR cells and cause cell death by their known mechanisms of action: inhibition of protein synthesis on ribosomes (G418 and Zeocin) or induction of DNA/RNA damage (Zeocin).
Drug Toxicity Is Blocked by Antibodies to PrP and by Elevated Extracellular [K+]
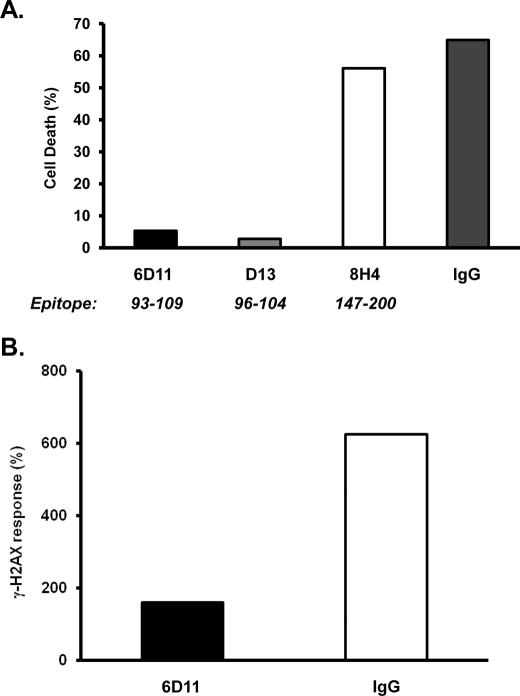
If ΔCR PrP sensitizes cells to drug toxicity by physically interacting with molecular targets on the cell surface, antibodies to specific regions of the PrP sequence might be expected to inhibit this effect. To test this prediction, we assessed the effects of several different PrP monoclonal antibodies on Zeocin-induced toxicity in ΔCR PrP-expressing cells. We found that two antibodies (6D11 and D13), directed against the 93–104 region of PrP (18, 28), dramatically reduced cell death induced by Zeocin. This effect was apparent using either the MTT viability assay (Fig. 8A) or the H2AX phosphorylation assay (Fig. 8B). In contrast, several other antibodies had no effect, including 8H4 (directed against PrP residues 147–200) (19), 3F4 (directed against residues 108–111 (29), which are missing in ΔCR PrP), and nonspecific IgG (Fig. 8, A and B) (data not shown). These results indicate that antibody binding to a specific region of PrP (residues 93–104) interferes with its ability to confer drug sensitivity on cells.
FIGURE 8.
Antibodies to the 93–104 region of ΔCR PrP inhibit drug sensitization. A, HEK cells expressing ΔCR PrP were pretreated with antibodies (5 μg/ml) for 60 min, after which Zeocin was added at a final concentration of 500 μg/ml. Forty-eight hours later, cells were assayed for viability by MTT reduction. Cell death (1 − A570) is plotted as a percentage of the value for non-drug-treated cells. B, HEK cells expressing ΔCR PrP were pretreated with antibodies for 60 min, after which Zeocin was added to a final concentration of 500 μg/ml. Sixty minutes later, cells were lysed and analyzed for γ-H2AX as described in the legend to supplemental Fig. S5B. The bars show means for duplicate wells.
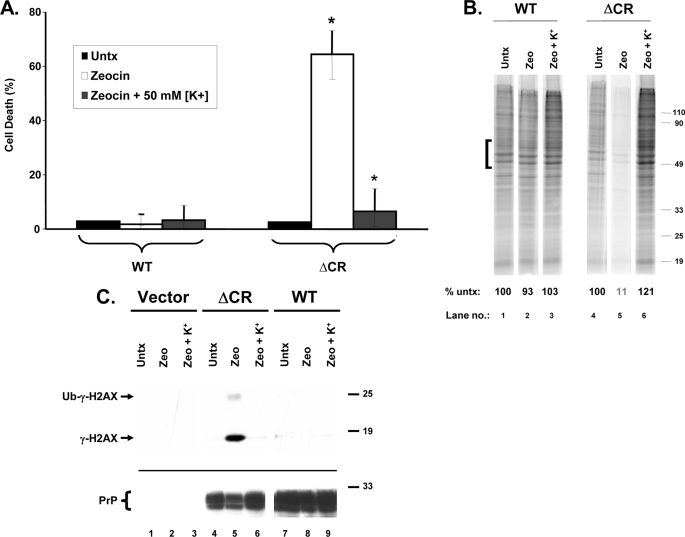
Because aminoglycosides and Zeocin/bleomycin are cationic, their entry into cells may depend on an electrochemical gradient (30, 31). If so, dissipating this gradient via membrane depolarization may reduce drug uptake and thereby suppress toxicity in ΔCR cells. To test this prediction, we incubated cells in elevated extracellular K+ (50 mm), which depolarizes the resting membrane potential, during Zeocin treatment. We found that this manipulation completely blocked the toxic effect of Zeocin, as measured by three different read-outs, including cell viability, [35S]methionine incorporation, and H2AX phosphorylation (Fig. 9, A–C). Elevated [K+] had no effect on any of these parameters in the absence of Zeocin and did not substantially alter the levels of PrP protein (Fig. 9C) (data not shown). Similar results were obtained with G418 and bleomycin and with cells expressing Δ32–134 PrP (data not shown).
FIGURE 9.
ΔCR PrP-induced drug toxicity is blocked by elevated extracellular [K+]. A, HEK cells expressing WT or ΔCR PrP were treated for 48 h with Zeocin (500 μg/ml), either in normal extracellular [K+] (5 mm) or in elevated extracellular [K+] (50 mm). Cell viability was then assayed by MTT reduction. Cell death (1 − A570) is plotted as a percentage of the value for non-drug-treated cells (Untx) in 5 mm [K+]. The bars represent means ± S.E. from three independent experiments. The values indicated by the asterisks are significantly different from each other (p < 0.02). B, HEK cells expressing WT or ΔCR PrP were untreated (lanes 1 and 4) or were incubated for 20 h with Zeocin (1,000 μg/ml) (lanes 2, 3, 5, and 6), either in 5 mm [K+] (lanes 1, 2, 4, and 5) or in 50 mm [K+] (lanes 3 and 6). Cells were then labeled with [35S]methionine and analyzed by SDS-PAGE as described in the legend to Fig. 6. The numbers below each lane indicate the amount of radioactivity incorporated in the region indicated by the bracket, expressed as a percentage of that in untreated control cells (lanes 1 and 4). C, HEK cells expressing vector, ΔCR PrP, or WT PrP were incubated for 24 h with Zeocin (100 μg/ml), either in 5 mm [K+] (lanes 1, 2, 4, 5, 7, and 8) or in 50 mm [K+] (lanes 3, 6, and 9). Cells were lysed and analyzed for γ-H2AX by Western blotting (top). PrP levels were verified by reprobing the blot with 6D11 antibody (bottom).
DISCUSSION
Transgenic mice expressing several different deletions within the flexible, N-terminal tail of PrP display a spontaneous neurodegenerative phenotype that is dose-dependently suppressed by co-expression of WT PrP (9, 10, 13). The shortest of these deletions, ΔCR, which is missing a highly conserved block of 21 amino acids (residues 105–125), is the most neurotoxic and produces a neonatal lethal phenotype in transgenic mice (13).
In an attempt to develop a cell culture model of ΔCR PrP toxicity, we made the surprising observation that expression of this protein renders a variety of transformed cell lines, as well as differentiated neural stem cells derived from transgenic mice, hypersensitive to the lethal effects of two structurally and mechanistically distinct classes of drugs that are commonly used for selection of transfected cells: aminoglycoside antibiotics (including G418 and hygromycin) and DNA-cleaving chemotherapeutic agents (including phleomycin (Zeocin) and bleomycin). Importantly, WT PrP does not possess this activity and indeed is able to suppress the drug-sensitizing effect of ΔCR PrP when the two proteins are expressed together.
This report describes the characteristics of this unusual phenomenon and provides insight into its underlying mechanism. Our results have important implications for understanding how the normal function of PrPC can be subverted to produce neurotoxic effects, and they lay the groundwork for developing cellular screening assays for identification of potentially novel anti-prion therapeutics.
Drug Hypersensitivity in Cultured Cells Mimics Essential Features of ΔCR PrP Toxicity in Vivo
Several observations demonstrate a striking parallel between the effects of ΔCR and other PrP deletion mutants in cultured cells and in transgenic mice. Most importantly, the drug-sensitizing effect of ΔCR PrP is suppressed by co-expression of WT PrP, similar to rescue of the neurodegenerative phenotype by WT PrP in transgenic mice. This result suggests that drug hypersensitivity does not represent a nonspecific toxic effect of ΔCR PrP but is instead related to a specific alteration in the cellular activity of PrPC that can be reversed by co-expression of the WT protein.
Second, another PrP deletion mutant (Δ32–134) as well as Dpl, both of which are neurotoxic in transgenic mice (9, 11), also confers drug hypersensitivity, and this effect is suppressible by co-expression of WT PrP. Moreover, there is a correlation between the potency of these molecules in vivo and strength of their drug-sensitizing effects in vitro, with ΔCR PrP displaying the greatest toxic potential in both settings. We have begun to perform a structure-activity analysis of PrP deletion mutants spanning the CR region and have found that Δ94–134, which is neurotoxic in transgenic mice (10), also sensitizes HEK cells to G418 toxicity.5 In contrast, deletion mutants (e.g. Δ23–31) that do not encompass the CR region lack drug-sensitizing activity,5 arguing for the specificity of the effect.
Finally, the drug-sensitizing effect of ΔCR PrP is seen in differentiated neural stem cells derived from Tg(ΔCR) mice, cultures of which include neurons, astrocytes, and oligodendrocytes. This result demonstrates that the drug effect is operative in the same cell types that show pathological changes in vivo and is not peculiar to transformed cell lines. Moreover, it is unrelated to transfection or to overexpression (the NSCs express ΔCR PrP at ∼0.5 times endogenous PrP levels). Taken together, these observations suggest that there is a mechanistic connection between the drug sensitivity produced by PrP deletion mutants and Dpl in cultured cells and the neurotoxic activity of these molecules in transgenic mice.
What Is the Cellular Basis for Drug Hypersensitivity?
A striking result of our study is the extremely rapid effect of the two classes of drugs, even at relatively low concentrations, on cells expressing ΔCR PrP. Inhibition of protein synthesis in response to G418 was detectable within several h, and DNA damage in response to Zeocin was detectable within 30 min. These results imply that the drugs reach effective intracellular concentrations in ΔCR cells within a very short time, long before any reduction in cellular viability is apparent (at >24 h). Taken together, our data suggest that ΔCR PrP sensitizes cells to drug toxicity by enhancing intracellular drug accumulation. Our attempts to demonstrate this point experimentally using fluorescent or radioactive versions of aminoglycosides have proven to be problematic due to high levels of nonspecific binding as well as uptake into endocytic compartments that are not relevant to cytoplasmic or nuclear accumulation.
One alternative explanation we considered is that ΔCR PrP-expressing cells have a base-line metabolic impairment and that stresses like protein synthesis inhibition or DNA damage “tip the cell over the edge,” triggering cell death. Arguing against this scenario, cells expressing ΔCR PrP show little alteration in base-line viability, growth rate, protein synthetic rate, or morphology in the absence of drug treatment. In addition, a number of other drugs were equally toxic to both WT and ΔCR cells (Table 1), suggesting that G418, hygromycin, Zeocin, and bleomycin possess specific features that account for their differential effect on ΔCR cells. Finally, drug treatment for up to 10 days had no effect on cells expressing WT PrP or empty vector, implying that toxic levels of the drugs are not reached until well after the time period during which ΔCR cells die (at 1–3 days).
How might ΔCR PrP facilitate intracellular drug accumulation? One possibility is that the protein enhances drug influx. Aminoglycosides are known to enter mammalian cells via endocytic pathways as well as via permeation of certain classes of cation-selective channels (30–33). There is also evidence that aminoglycosides as well as bleomycin are transported into yeast cells down an electrochemical gradient via membrane carriers or permeases (34–36). It is possible that ΔCR PrP modulates the activity of these or other drug uptake pathways. Our observation that elevated extracellular [K+] (which causes membrane depolarization) blocks the effect of Zeocin on ΔCR cells suggests involvement of an electrochemical gradient in drug accumulation and would be consistent with participation of an ion channel, transporter, or exchanger. Interestingly, the ability of WT PrP to suppress drug sensitivity induced by ΔCR PrP implies that the WT protein dampens whatever drug influx pathways are activated by the mutant protein, perhaps by competition for binding to the relevant channels or transporters.
An alternative possibility is that ΔCR PrP reduces drug efflux, for example via inhibition of multidrug resistance (MDR) pumps. In fact, there is evidence that PrP can modulate MDR-related pathways (37). Arguing against this idea, however, is our observation that MDR functional activity is normal in ΔCR cells,5 as assayed by efflux of doxorubicin, a substrate for several MDR isoforms (38). In addition, bleomycin, which is toxic to ΔCR cells, is not a substrate for MDR pathways (39). Whatever influx or efflux pathways are affected by ΔCR PrP are likely to be ubiquitous, because we have found that the mutant protein sensitizes a variety of cell types to drug toxicity.
Why is the sensitizing effect of ΔCR PrP apparently restricted to two classes of structurally and mechanistically unrelated drugs? G418 and hygromycin are positively charged, aminoglycoside antibiotics (Mr 496 and 528, respectively; supplemental Fig. S2) that interfere with protein synthesis on ribosomes (23). In contrast, Zeocin (phleomycin D1) and bleomycin are cationic, copper-containing glycopeptides (Mr 1,428 and 1,416, respectively; supplemental Fig. S2) that cause cleavage of DNA and RNA (24, 25). Mammalian cells have a relatively low permeability to each of these drugs, which is a key to their utility as selection agents; drugs that accumulated to toxic levels too quickly would not be completely inactivated by the corresponding resistance gene. The four active drugs are all positively charged, and it is possible that they share a common cellular uptake pathway, for example via cation-selective channels or transporters. In contrast, lipophilic drugs, such as cycloheximide, doxorubicin, or etoposide, which readily accumulate intracellularly, were rapidly toxic to both ΔCR and WT cells (Table 1).
We found that the drug-sensitizing effect of ΔCR PrP could be inhibited by treatment with anti-PrP antibodies (Fig. 8). Interestingly, the two antibodies with inhibitory effect are both directed against residues 93–104, just N-terminal to the ΔCR deletion. This result suggests that the activity of ΔCR PrP may depend on specific interactions between this region of the protein and target molecules on the cell surface. This region has been previously implicated in several properties of PrPC, including conformational conversion to PrPSc (40), apoptosis induced by antibody cross-linking (41), and binding of Alzheimer Aβ oligomers (42).
Applications of the Drug Sensitivity Assay
There have been several previous reports of spontaneous cell death induced in cultured neurons by expression of deleted forms of PrP and Dpl, although the effects observed have been relatively modest (43, 44). These results are consistent with our own observation that expression of ΔCR PrP has only a minor affect on the base-line viability of cultured cells (either transformed cell lines or differentiated neural stem cells). Taken together, these experiments suggest that factors present in the brain milieu are required for maximal toxicity of these molecules and that these factors are missing in existing cell culture systems.
The drug hypersensitivity phenomenon described here provides a robust cellular assay to investigate the biological activity of ΔCR PrP and related neurotoxic molecules. The strong parallels between the effects of these molecules in the drug assay and in transgenic mice make it likely that similar cellular mechanisms are operative in both settings. Thus, further studies of how ΔCR PrP accentuates drug accumulation are likely to provide important insights into the neurotoxic action of this protein in vivo. For example, such studies could lead to identification of the ion channels, pumps, or other molecular targets of ΔCR PrP. Abnormal activity of these target molecules, resulting in enhanced ion permeability, could contribute to the death of neurons expressing ΔCR PrP. In addition, the drug assay provides a rapid read-out for mutational analyses of PrP cytotoxic and cytoprotective activities, without the need to construct transgenic mice for each mutant. Finally, in the assay format described here (Fig. 7C), drug hypersensitivity can be used as a screenable phenotype for identification of RNA interference sequences or pharmacological compounds that inhibit ΔCR PrP toxicity. Although ΔCR PrP is an artificial molecule, it is likely to act by subverting a normal physiological function of PrPC, similar to what has been postulated for PrPSc. Thus, insights derived from the study of ΔCR PrP and related molecules will have applicability to understanding the pathogenesis of prion diseases and to working out effective therapies for these disorders.
Supplementary Material
Acknowledgments
We thank Aimin Li for help with DNA cloning, Heather Christensen and Nada Husic for assistance with lentiviral preparation, and Bill Nolan at the High-Throughput Screening Facility for help with semiautomated microscopy. We also acknowledge Laura Westergard for participation in the initial phase of the project. We thank Richard Kascsak, Man-Sun Sy, and Gianluigi Zanusso for providing anti-PrP antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grants NS052526 and NS040975 (to D. A. H.). This work was also supported by Department of Defense Grant DAMD17-03-1-0531 (to R. S. S.) and Telethon Foundation Grants GFP04007 (to E. B.), TCR08002 (to V. B.), and TCR08005 (to R. C.). The Hope Center Viral Vectors Core is supported by National Institutes of Health Neuroscience Blueprint Core Grant P30 NS057105 (to Washington University).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
H. M. Christensen and D. A. Harris, unpublished data.
E. Biasini, J. A. Turnbaugh, and D. A. Harris, manuscript in preparation.
- WT
- wild type
- CHO
- Chinese hamster ovary
- TUNEL
- terminal deoxynucleotidyltransferase dUTP nick end labeling
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PBS
- phosphate-buffered saline
- DAPI
- 4′,6-diamidino-2-phenylindole
- PNGase
- peptide:N-glycanase
- NSC
- neural stem cell
- MDR
- multidrug resistance.
REFERENCES
- 1.Aguzzi A., Baumann F., Bremer J. (2008) Annu. Rev. Neurosci. 31, 439–477 [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S. B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris D. A., True H. L. (2006) Neuron 50, 353–357 [DOI] [PubMed] [Google Scholar]
- 4.Brandner S., Isenmann S., Raeber A., Fischer M., Sailer A., Kobayashi Y., Marino S., Weissmann C., Aguzzi A. (1996) Nature 379, 339–343 [DOI] [PubMed] [Google Scholar]
- 5.Mallucci G., Dickinson A., Linehan J., Klöhn P. C., Brandner S., Collinge J. (2003) Science 302, 871–874 [DOI] [PubMed] [Google Scholar]
- 6.Chesebro B., Trifilo M., Race R., Meade-White K., Teng C., LaCasse R., Raymond L., Favara C., Baron G., Priola S., Caughey B., Masliah E., Oldstone M. (2005) Science 308, 1435–1439 [DOI] [PubMed] [Google Scholar]
- 7.Rambold A. S., Müller V., Ron U., Ben-Tal N., Winklhofer K. F., Tatzelt J. (2008) EMBO J. 27, 1974–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westergard L., Christensen H. M., Harris D. A. (2007) Biochim. Biophys. Acta 1772, 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shmerling D., Hegyi I., Fischer M., Blättler T., Brandner S., Götz J., Rülicke T., Flechsig E., Cozzio A., von Mering C., Hangartner C., Aguzzi A., Weissmann C. (1998) Cell 93, 203–214 [DOI] [PubMed] [Google Scholar]
- 10.Baumann F., Tolnay M., Brabeck C., Pahnke J., Kloz U., Niemann H. H., Heikenwalder M., Rülicke T., Bürkle A., Aguzzi A. (2007) EMBO J. 26, 538–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore R. C., Lee I. Y., Silverman G. L., Harrison P. M., Strome R., Heinrich C., Karunaratne A., Pasternak S. H., Chishti M. A., Liang Y., Mastrangelo P., Wang K., Smit A. F., Katamine S., Carlson G. A., Cohen F. E., Prusiner S. B., Melton D. W., Tremblay P., Hood L. E., Westaway D. (1999) J. Mol. Biol. 292, 797–817 [DOI] [PubMed] [Google Scholar]
- 12.Rossi D., Cozzio A., Flechsig E., Klein M. A., Rülicke T., Aguzzi A., Weissmann C. (2001) EMBO J. 20, 694–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li A., Christensen H. M., Stewart L. R., Roth K. A., Chiesa R., Harris D. A. (2007) EMBO J. 26, 548–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivera-Milla E., Oidtmann B., Panagiotidis C. H., Baier M., Sklaviadis T., Hoffmann R., Zhou Y., Solis G. P., Stuermer C. A., Málaga-Trillo E. (2006) FASEB J. 20, 317–319 [DOI] [PubMed] [Google Scholar]
- 15.Li A., Barmada S. J., Roth K. A., Harris D. A. (2007) J. Neurosci. 27, 852–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen H. M., Harris D. A. (2009) J. Neurochem. 108, 44–56 [DOI] [PubMed] [Google Scholar]
- 17.Louis S. A., Reynolds B. A. (2005) Methods Mol. Biol. 290, 265–280 [DOI] [PubMed] [Google Scholar]
- 18.Pankiewicz J., Prelli F., Sy M. S., Kascsak R. J., Kascsak R. B., Spinner D. S., Carp R. I., Meeker H. C., Sadowski M., Wisniewski T. (2006) Eur. J. Neurosci. 23, 2635–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zanusso G., Liu D., Ferrari S., Hegyi I., Yin X., Aguzzi A., Hornemann S., Liemann S., Glockshuber R., Manson J. C., Brown P., Petersen R. B., Gambetti P., Sy M. S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8812–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matucci A., Zanusso G., Gelati M., Farinazzo A., Fiorini M., Ferrari S., Andrighetto G., Cestari T., Caramelli M., Negro A., Morbin M., Chiesa R., Monaco S., Tridente G. (2005) Brain Res. Bull. 65, 155–162 [DOI] [PubMed] [Google Scholar]
- 21.Wolkowicz R., Nolan G. P., Curran M. A. (2004) Methods Mol. Biol. 246, 391–411 [DOI] [PubMed] [Google Scholar]
- 22.Li M., Rossi J. J. (2005) Methods Mol. Biol. 309, 261–272 [DOI] [PubMed] [Google Scholar]
- 23.Poehlsgaard J., Douthwaite S. (2005) Nat. Rev. Microbiol. 3, 870–881 [DOI] [PubMed] [Google Scholar]
- 24.Chen J., Stubbe J. (2005) Nat. Rev. Cancer 5, 102–112 [DOI] [PubMed] [Google Scholar]
- 25.Abraham A. T., Lin J. J., Newton D. L., Rybak S., Hecht S. M. (2003) Chem. Biol. 10, 45–52 [DOI] [PubMed] [Google Scholar]
- 26.Hobbie S. N., Kalapala S. K., Akshay S., Bruell C., Schmidt S., Dabow S., Vasella A., Sander P., Böttger E. C. (2007) Nucleic Acids Res. 35, 6086–6093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celeste A., Fernandez-Capetillo O., Kruhlak M. J., Pilch D. R., Staudt D. W., Lee A., Bonner R. F., Bonner W. M., Nussenzweig A. (2003) Nat. Cell Biol. 5, 675–679 [DOI] [PubMed] [Google Scholar]
- 28.Williamson R. A., Peretz D., Pinilla C., Ball H., Bastidas R. B., Rozenshteyn R., Houghten R. A., Prusiner S. B., Burton D. R. (1998) J. Virol. 72, 9413–9418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolton D. C., Seligman S. J., Bablanian G., Windsor D., Scala L. J., Kim K. S., Chen C. M., Kascsak R. J., Bendheim P. E. (1991) J. Virol. 65, 3667–3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myrdal S. E., Steyger P. S. (2005) Hear. Res. 204, 170–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcotti W., van Netten S. M., Kros C. J. (2005) J. Physiol. 567, 505–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myrdal S. E., Johnson K. C., Steyger P. S. (2005) Hear. Res. 204, 156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandoval R. M., Dunn K. W., Molitoris B. A. (2000) Am. J. Physiol. Renal Physiol. 279, F884–F890 [DOI] [PubMed] [Google Scholar]
- 34.Thornton G., Wilkinson C. R., Toone W. M., Jones N. (2005) Genes Cells 10, 941–951 [DOI] [PubMed] [Google Scholar]
- 35.Aouida M., Leduc A., Wang H., Ramotar D. (2004) Biochem. J. 384, 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aouida M., Pagé N., Leduc A., Peter M., Ramotar D. (2004) Cancer Res. 64, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 37.Du J., Pan Y., Shi Y., Guo C., Jin X., Sun L., Liu N., Qiao T., Fan D. (2005) Int. J. Cancer 113, 213–220 [DOI] [PubMed] [Google Scholar]
- 38.Sharom F. J. (2008) Pharmacogenomics 9, 105–127 [DOI] [PubMed] [Google Scholar]
- 39.Schurr E., Raymond M., Bell J. C., Gros P. (1989) Cancer Res. 49, 2729–2733 [PubMed] [Google Scholar]
- 40.Peretz D., Williamson R. A., Matsunaga Y., Serban H., Pinilla C., Bastidas R. B., Rozenshteyn R., James T. L., Houghten R. A., Cohen F. E., Prusiner S. B., Burton D. R. (1997) J. Mol. Biol. 273, 614–622 [DOI] [PubMed] [Google Scholar]
- 41.Solforosi L., Criado J. R., McGavern D. B., Wirz S., Sánchez-Alavez M., Sugama S., DeGiorgio L. A., Volpe B. T., Wiseman E., Abalos G., Masliah E., Gilden D., Oldstone M. B., Conti B., Williamson R. A. (2004) Science 303, 1514–1516 [DOI] [PubMed] [Google Scholar]
- 42.Laurén J., Gimbel D. A., Nygaard H. B., Gilbert J. W., Strittmatter S. M. (2009) Nature 457, 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drisaldi B., Coomaraswamy J., Mastrangelo P., Strome B., Yang J., Watts J. C., Chishti M. A., Marvi M., Windl O., Ahrens R., Major F., Sy M. S., Kretzschmar H., Fraser P. E., Mount H. T., Westaway D. (2004) J. Biol. Chem. 279, 55443–55454 [DOI] [PubMed] [Google Scholar]
- 44.Watts J. C., Drisaldi B., Ng V., Yang J., Strome B., Horne P., Sy M. S., Yoong L., Young R., Mastrangelo P., Bergeron C., Fraser P. E., Carlson G. A., Mount H. T., Schmitt-Ulms G., Westaway D. (2007) EMBO J. 26, 4038–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.