Abstract
Natural killer (NK) cells are crucial in the control of cytomegalovirus infections in mice and humans. Here we show that the viral UL141 gene product has an immunomodulatory function that is associated with low-passage strains of human cytomegalovirus. UL141 mediated efficient protection of cells against killing by a wide range of human NK cell populations, including interferon-α-stimulated bulk cultures, polyclonal NK cell lines and most NK cell clones tested. Evasion of NK cell killing was mediated by UL141 blocking surface expression of CD155, which was previously identified as a ligand for NK cell-activating receptors CD226 (DNAM-1) and CD96 (TACTILE). The breadth of the UL141-mediated effect indicates that CD155 has a key role in regulating NK cell function.
Human cytomegalovirus (HCMV) is a ubiquitous human herpesvirus normally associated with subclinical primary infection followed by lifelong asymptomatic carriage during which the innate and adaptive immune responses act in concert to limit the consequences of infection. However, HCMV infections can cause severe disease in people whose immune systems are immature (such as those with congenital cytomegalic inclusion disease) or compromised (such as transplant recipients or patients with AIDS). The interplay between the virus and the immune system is central to our understanding of HCMV pathogenesis. People with defects in natural killer (NK) cell function are extremely sensitive to herpesvirus infections and to HCMV in particular1.
NK cell cytotoxicity is regulated by a fine balance of signals received by activating and inhibitory receptors. Interactions between endogenous human leukocyte antigen (HLA) class I molecules and NK cell inhibitory receptors normally dominate over signals received from activating receptors. The HCMV genome encodes four proteins that act in concert to downregulate HLA class I expression and thus would be predicted to render HCMV-infected cells susceptible to NK cell recognition2. However, responses of individual NK cell clones and lines to fibroblasts infected with HCMV vary greatly, ranging from activation to inhibition3. The mechanisms that determine whether activation or inhibition dominates during HCMV infection remain unclear, but the resistance of HCMV-infected cells to a proportion of NK cells can be attributed to specific virus genome–encoded products that mediate NK cell–evasion functions. A peptide derived from the leader sequence of HCMV gpUL140 stimulates cell surface expression of the nonclassical HLA class I molecule HLA-E independent of the transporters associated with antigen processing; HLA-E binds the NK cell inhibitory receptor complex CD94-NKG2A to suppress cytotoxicity mediated by CD94+NKG2A+ NK cells4,5. Several human UL16-binding proteins (ULBPs) were identified and named for their affinity for the HCMV protein UL16. ULBP1–ULBP3, major histocompatibility complex class I–related chain A (MICA) and MICB each bind the activating receptor NKG2D to stimulate NK cell functions. The protein gpUL16 acts by sequestering MICB, ULBP1 and ULBP2 in the endoplasmic reticulum. By impeding cell surface expression of these NKG2D ligands, UL16 suppresses NK cell recognition6-8. UL18 has long been known to be an HLA class I homolog with affinity for the inhibitory leukocyte immunoglobulin-like receptor 1 (also called immunoglobulin-like transcript 2), but its function in HCMV evasion of NK cell recognition remains controversial9.
HCMV has the largest genome (236 kilobases (kb)) of any characterized human virus, and most of its genes are nonessential for replication in fibroblasts in vitro10. Extensive in vitro passage of laboratory strains results in an accumulation of genetic defects, the most notable being the 15-kb and 13-kb deletions of strains AD169 and Towne, respectively, affecting one end of the long unique (UL) region commonly referred to as the UL/b′ sequence11. When assessed as potential vaccine candidates, both strains showed reduced virulence in vivo12. We were interested in the observation that cells infected with HCMV clinical isolates or the Toledo strain, which has been less extensively cultured in vitro, routinely provided substantially more protection against NK cell–mediated cytolysis than could be achieved with high-passage laboratory isolates5,13. The UL/b′ sequence deleted from strain AD169 is predicted to contain 23 genes that include many with potential immunomodulatory functions14-16. In a systematic search, we have identified and characterized an extremely efficient HCMV NK evasion function that mapped within the UL/b′ sequence to UL141.
RESULTS
NK cell protection associated with HCMV UL/b′
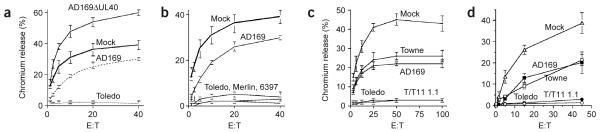
We initially investigated the susceptibility of cells infected with different HCMV strains to NK cell–mediated cytolysis with the transformed NK cell clone NKL as effectors against allogeneic primary human fetal foreskin fibroblasts or primary skin fibroblast targets. Consistent with previous findings, more protection against NK cell–mediated cytolysis was achieved by infection with the laboratory strain AD169 than with mock-infected cells, whereas infection with an HCMV UL40 deletion mutant (AD169ΔUL40) enhanced killing5 (Fig. 1a). However, nearly complete NK cell inhibition was achieved after infection with HCMV clinical isolates (6397 or Merlin) or the Toledo strain (Fig. 1a,b). Because certain NK cell receptors are sensitive to interactions with allogeneic HLA class I molecules, we also set up assays with matched NK cell effectors and skin fibroblasts derived from the same volunteer donors. We used three different types of NK cell effectors: bulk cultures depleted of T cells and activated with interferon-α (IFN-α), representing the broad NK cell response (D#NKb; in these designations, ‘#’ indicates the donor number); polyclonal NK cell lines (D#NKp); and NK cell clones derived from single-cell sorting (D#NKc). With NKL (Fig. 1c) and bulk cultures from three separate donors (Fig. 1d and Table 1), minimal killing was detectable against strain Toledo–infected cells, whereas cells infected with strain AD169 or strain Towne showed intermediate killing or killing comparable to that of mock-infected cells. To further characterize this enhanced protection, we tested a panel of 15 NK cell clones. Of these, nine clones capable of killing mock-infected targets (more than 10% killing) showed less killing against targets infected with strain Toledo than against those infected with strain AD169 (Table 1). Indeed, eight clones showed more than 10% killing against targets infected with strain AD169 compared with only two clones showing similar killing efficiency against cells infected with strain Toledo. Thus, results from NK cell bulk cultures, polyclonal lines and individual NK cell clones are all consistent with strain Toledo exhibiting an additional strong inhibitory function effective against most NK cells.
Figure 1.
Resistance to NK cell attack maps to the HCMV UL/b′ sequence. (a,b) Human fetal foreskin fibroblasts (HFFFs) were mock infected (Mock) or were infected for 72 h with HCMV strains AD169, Toledo, Merlin or 6397 or a strain AD169ΔUL40 deletion mutant (multiplicity of infection, 10). NKL cells were used as effectors in allogeneic chromium release assays. (c) HFFFs were infected with strains AD169, Towne, Toledo or the recombinant virus Towne/Tol11 1.1 (T/T11 1.1) and NKLs were used as effectors in allogeneic NK cell assays. (d) Autologous assay with D9SF targets (primary skin fibroblast from donor 9) infected as described in c with IFN-α-activated bulk cultures derived from D9 PBMCs (D9NKb) as effectors. Results are mean ± s.d. of triplicate or quadruplicate cultures and are representative of at least three and up to eight independent experiments. E:T, effector:target ratio.
Table 1. Susceptibility of autologous HCMV-infected cells to NK cell–mediated lysis.
| NK cell | Target | NK:target | Mock | + AD169 | + Toledo | P |
|---|---|---|---|---|---|---|
| Primary T cell–depleted PBMC bulk cultures | ||||||
| D3NKb.1 | D3SF | 20:1 | 40 ± 6 | 21 ± 1 | 1 ± 1 | 0.00005 |
| D3NKb.2 | D3SF | 30:1 | 10 ± 1 | 13 ± 1 | 1 ± 1 | 0.0002 |
| D7NKb.1 | D7SF | 18:1 | 20 ± 2 | 18 ± 3 | 1 ± 4 | 0.0005 |
| D7NKb.2 | D7SF | 20:1 | 10 ± 2 | 16 ± 2 | 0 ± 1 | 0.002 |
| D7NKb.3 | D7SF | 25:1 | 56 ± 6 | 37 ± 2 | 4 ± 2 | 0.000002 |
| D7NKb.4 | D7SF | 25:1 | ND | 33 ± 1 | 0 ± 1 | 0.0000001 |
| D9NKb.1 | D9SF | 45:1 | 40 ± 5 | 20 ± 5 | 3 ± 1 | 0.01 |
| D9NKb.2 | D9SF | 39:1 | 40 ± 4 | 14 ± 1 | 2 ± 1 | 0.00008 |
| NK clones | ||||||
| D3NKc5E11 | D3SF | 46:1 | 1 ± 1 | 1 ± 1 | 2 ± 1 | NS |
| D3NKc5E11 | D3SF | 40:1 | 1 ± 1 | 15 ± 3 | 4 ± 1 | 0.03 |
| D3NKc5F8 | D3SF | 20:1 | 1 ± 1 | 1 ± 2 | 1 ± 1 | NS |
| D8NKc1C10 | D8SF | 58:1 | 48 ± 2 | 30 ± 2 | 13 ± 2 | 0.0003 |
| D8NKc1E11 | D8SF | 21:1 | 29 ± 2 | 10 ± 3 | 4 ± 1 | NS |
| D8NKc1F11 | D8SF | 44:1 | 24 ± 2 | 15 ± 2 | 3 ± 1 | 0.0008 |
| D8NKc2F3 | D8SF | 25:1 | 38 ± 3 | 13 ± 3 | 4 ± 1 | 0.009 |
| D8NKc2G9 | D8SF | 5:1 | 2 ± 1 | 3 ± 1 | 3 ± 1 | NS |
| D8NKc3C8 | D8SF | 26:1 | 8 ± 1 | 5 ± 1 | 3 ± 1 | NS |
| D8NKc3D9 | D8SF | 11:1 | 11 ± 1 | 15 ± 1 | 8 ± 1 | NS |
| D8NKc3E9 | D8SF | 32:1 | 35 ± 1 | 25 ± 2 | 9 ± 1 | 0.03 |
| D8NKc4F3 | D8SF | 25:1 | 48 ± 2 | 25 ± 1 | 10 ± 2 | 0.04 |
| D8NKc4E10 | D8SF | 44:1 | 13 ± 3 | 8 ± 1 | 6 ± 1 | NS |
| D8NKc5E7 | D8SF | 14:1 | 7 ± 3 | 2 ± 1 | 2 ± 1 | NS |
| D8NKc5C11 | D8SF | 63:1 | 36 ± 2 | 17 ± 2 | 13 ± 1 | NS |
Data are presented as percent chromium release ± s.e.m. D#SF, primary skin fibroblast; the number (#) designates the donor. ND, not done. P values compare killing of strain AD169–infected targets versus strain Toledo–infected targets (t-tests, assuming unequal variance)
NS, not significant (P > 0.05).
One or more NK cell–evasion function(s) must have been lost from the high-passage HCMV laboratory strains either through the deletion of the UL/b′ sequence or as a consequence of mutation elsewhere on their genomes. HCMV Towne/Tol11 1.1 corresponds to a recombinant strain Towne virus in which the UL/b′ from strain Toledo has been inserted17. The addition of this Toledo sequence in the strain Towne genome was sufficient to confer protection from NK cell killing comparable to that achieved by infection with strain Toledo alone (Fig. 1c,d). Thus, the 13-kb UL/b′ transferred from strain Toledo may contain one or more gene(s) that inhibit(s) NK cell recognition.
UL141 and NK cell–evasion function
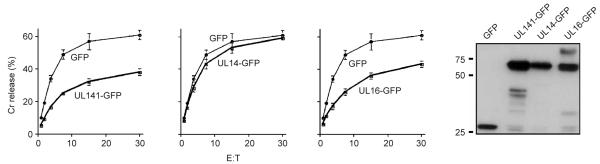
To screen for a NK cell evasion function, we transiently transfected human embryonic kidney (HEK) 293 cell lines with plasmids encoding open reading frames (ORFs) UL133–UL150. The ORFs cloned were as defined in the original analysis of the strain Toledo UL/b′ sequence11. The ORFs were expressed as green fluorescent protein (GFP) fusion proteins to track expression. Although none of this panel of transfected cells showed resistance to NK cell cytolysis, the assay was restricted by the use of a single NK cell line, DEL5. Sequencing of the UL133–UL150 PCR products identified a small number of errors in the original strain Toledo UL/b′ sequence that affected gene usage. A reannotated, revised version of the strain Toledo UL/b′ sequence was therefore generated18 (GenBank accession number AY446871). We cloned the corrected version of the strain Toledo UL141 ORF11,14 as a GFP fusion construct and sorted the transfected HEK 293 cell line for GFP expression, expanded the population in vitro and used these in assays within 3 weeks. Using NKL cells, HEK 293 cells expressing the UL141-GFP fusion protein showed resistance to NK cell–mediated cytolysis (Fig. 2). A UL16-GFP fusion protein was also capable of eliciting protection compared with UL14-GFP or control cells expressing GFP alone. UL16 is a recognized HCMV NK cell–evasion gene product6, whereas UL14 is an uncharacterized homolog of UL141. The GFP fusion proteins had comparable expression in the sorted cells used in the assay (Fig. 2). UL141 was thus identified as potentially encoding a product that mediates NK cell–evasion function.
Figure 2.
UL141 induces protection against NK cell attack. Cytotoxicity assay with NKLs as the effectors against stable HEK 293 cell lines expressing UL141-GFP, UL14-GFP or UL16-GFP fusion proteins or GFP alone. To ensure homogeneous expression, we sorted cell lines on the basis of GFP fluorescence before the assay. Far right, immunoblot with a GFP-specific rabbit polyclonal antibody, indicating fusion proteins were being expressed by the transfected cell lines. Left margin, molecular sizes (in kDa). Data are representative of three experiments.
Inhibition of a wide range of NK cells by gpUL141
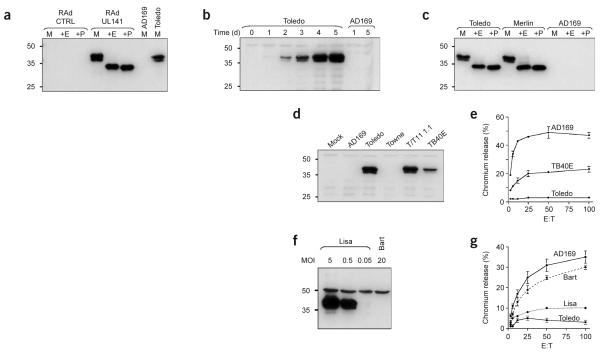
We constructed replication-deficient adenovirus vectors (RAds) containing the complete gene encoding UL141 alone (RAdUL141) or in combination with GFP (RAdUL141-GFP) to provide efficient expression of glycoprotein UL141 (gpUL141) in fibroblast targets (Fig. 3a). In NK cell assays, RAdUL141-GFP-infected cells showed notably reduced cytolysis compared with that of control cultures exposed to infection with an equivalent adenovirus recombinant encoding GFP alone (Table 2). We readily found inhibition with NKL cells (data not shown), NK cell bulk cultures from two different volunteers and five of six NK cell clones regardless of whether targets were allogeneic or autologous (Table 2). We further tested the specificity of gpUL141 by investigating its effects on bulk cultures depleted of CD94+ NK cells; UL40 inhibits a specific subset of CD94+NKG2A+ NK cells by the upregulation of HLA-E4,5. We noted gpUL141 inhibited complete and CD94-depleted NK cell bulk cultures with similar efficiency (Table 2) and had no differential effect on CD94hi and CD94lo NK cell clones (data not shown). Indeed, gpUL141 inhibited 67% (82 of 123) of the NK cell clones tested in an autologous setting by a margin of more than 10% in NK cell cytolysis assays, whereas three clones were stimulated (Table 3). The gpUL141-inhibited killing was thus independent of CD94 expression and was readily detected with freshly isolated NK cell bulk cultures and most NK cell clones.
Figure 3.
Characterization of gpUL141 expression. Cells were infected with recombinant adenoviruses or various HCMV strains. GpUL141 expression was detected with the M550 series of mAbs. (a) Expression of gpUL141, after infection of HFFFs with RAdUL141. The protein gpUL141 is sensitive to both Endo H (+E) and peptide-N-glycosidase F (+P). M, mock digest. (b) Time course of gpUL141 expression through the lytic replication cycle of HCMV strains Toledo and AD169. (c) Endo H (+E) and peptide-N-glycosidase F (+P) sensitivity of gpUL141 expressed from strain Toledo or Merlin. M, mock digest. (d) Immunoblot of the expression of gpUL141 in HFFFs infected with various HCMV strains (above lanes). (e) NK cell cytolysis assay with NKL as effectors, comparing the susceptibility of HFFFs after infection with strains TB40E, Toledo and AD169. (f) Immunoblot of strain TB40E that contained two viruses: UL141-replete (Lisa) and a virus without UL141 expression (Bart). Immunoblot with the M550 mAbs shows expression of gpUL141 in HFFFs infected with TB40E-Lisa or TB40E-Bart. MOI, multiplicity of infection. (g) NK cell cytolysis assay comparing the sensitivity to killing with NKL shown by cells infected with TB40E-Bart (dashed line) or TB40E-Lisa (dotted line). NK cell cytotoxicity results are mean ± sd. Data are one representative experiment of three. (a–d,f) Left margin, molecular sizes (in kDa).
Table 2. Expression of gpUL141 increases resistance to NK cell–mediated lysis.
| NK cell | Target | NK:target | Rad-GFP | RAdUL141-GFP | P |
|---|---|---|---|---|---|
| Fresh IFN-α-stimulated whole PBMCs | |||||
| D3PBMC | HFFF | 25:1 | 35 ± 1 | 20 ± 2 | 0.0006 |
| Primary T cell–depleted PBMC bulk cultures | |||||
| D3NKb1 | D3SF@ | 20:1 | 30 ± 2 | 13 ± 5 | 0.01 |
| D3NKb2 | D3SF@ | 12:1 | 17 ± 5 | 11 ± 1 | NS |
| D3NKb2 | HFFF | 12:1 | 22 ± 8 | 8 ± 2 | NS |
| D7NKb1 | D7SF@ | 18:1 | 20 ± 2 | 8 ± 2 | 0.0007 |
| Primary T cell– and CD94+ cell–depleted BMC bulk cultures | |||||
| D3NKb1(ΔCD94) | D3SF@ | 40:1 | 31 ± 3 | 11 ± 2 | 0.0008 |
| D7NKb1(ΔCD94) | D7SF@ | 22:1 | 16 ± 4 | 7 ± 3 | 0.01 |
| Expanded primary polyclonal NK lines | |||||
| D7NKp3.2 | D7SF@ | 40:1 | 18 ± 4 | 3 ± 3 | 0.009 |
| D7NKp3.2 | HFFF | 40:1 | 20 ± 3 | 8 ± 2 | 0.004 |
| NK clones | |||||
| D8NKc1C10 | D8SF@ | 8:1 | 21 ± 1 | 7 ± 1 | 0.0005 |
| D8NKc1E11 | D8SF@ | 40:1 | 18 ± 1 | 16 ± 2 | NS |
| D8NKc1F11 | D8SF@ | 32:1 | 19 ± 1 | 9 ± 1 | 0.002 |
| D8NKc3C8 | D8SF@ | 35:1 | 24 ± 1 | 8 ± 1 | 0.0003 |
| D8NKc4E10 | D8SF@ | 23:1 | 30 ± 3 | 11 ± 2 | 0.002 |
| D8NKc5C11 | D8SF@ | 66:1 | 36 ± 3 | 23 ± 3 | 0.02 |
Data are presented as percent chromium release ± s.e.m. @, autologous targets; D#NKb(ΔCD94), bulk cultures depleted of CD94+ NK cells (the number (#) indicates donor number). P values compare killing of RAd-GFP–-infected targets versus RAd-UL141–GFP–infected targets (t-test, assuming unequal variance)
NS, not significant (P > 0.05).
Table 3. Effect of UL141 expression on cytotoxicity mediated by individual NK cell clones derived from multiple donors.
| Donor | UL141 expression inhibited lysisa |
UL141 expression increased lysisa |
UL141 expression, no changeb |
|---|---|---|---|
| D3 | 4 of 16 | 0 of 16 | 12 of 16 |
| D7 | 21 of 24 | 0 of 24 | 3 of 24 |
| D8 | 9 of 16 | 1 of 16 | 6 of 16 |
| D9 | 48 of 67 | 2 of 67 | 17 of 67 |
| Total | 82 of 123 | 3 of 123 | 38 of 123 |
Data represent autologous assays; each event represents a single experiment with an individual clone.
Increase or decrease in cytolysis of more than 10% chromium release.
Increase or decrease in cytolysis of less than 10% chromium release.
Characterization of the UL141 gene product
UL141 is highly conserved between HCMV isolates, encoding a protein containing a potential signal peptide, a hydrophobic transmembrane domain and three potential N-linked glycosylations sites (Supplementary Fig. 1 online). A protein fold-recognition program indicates that gpUL141 as defined originally10 contains immunoglobulin-like regions19. Analysis of the updated sequence14,18 indicated the presence of an immunoglobulin-like β-sandwich domain at residues 15–114 in the mature protein (Supplementary Fig. 2 online). We expressed a truncated, secreted form of gpUL141, in which the predicted C-terminal cytoplasmic and transmembrane domains were replaced with an affinity tag (Strep-tag). This protein was expressed in abundance with a replication-deficient adenovirus vector and purified with a Streptactin Macroprep column (Supplementary Fig. 3 online). We determined the N-terminal amino acid sequence of purified soluble gpUL141 to identify the cleavage site for signal peptidase. The gpUL141 sequence started with the D residue specified by codon 37 in the ORF14. We also used purified, secreted gpUL141 as an immunogen to generate specific monoclonal antibodies. During productive HCMV infection, the UL141 protein could be readily detected at 24 h after infection and continued to accumulate into the late phase of the replicative cycle (Fig. 3b and Supplementary Fig. 4 online). We detected gpUL141 as a glycoprotein doublet with apparent molecular masses of 37 kDa and 40 kDa that after digestion with endo-N-acetylglucoseaminidase H (Endo H) or peptide-N-glycosidase F yielded a single 34-kDa product (Fig. 3a,c), consistent with the predicted molecular mass of 34.7 kDa (302 residues). Sensitivity to Endo H treatment was consistent with retention of gpUL141 in the endoplasmic reticulum, and this result was supported by immunocytochemistry (Supplementary Fig. 5 online). Expression of gpUL141 was readily detected with clinical HCMV isolate Merlin, strain Toledo or the recombinant HCMV Towne/Tol11 1.1 but not with the laboratory strains AD169 or Towne, which lack the UL/b′ nucleotide sequence (Fig. 3c,d).
UL141 frameshift mutation in HCMV strain TB40E
During screening of gpUL141 expression with different strains of HCMV, the amount of gpUL141 detected with the passaged strain TB40E-infected cells was much lower than with any other virus (Fig. 3d). Furthermore, infection with strain TB40E elicited intermediate NK cell resistance relative to infection with strains AD169 and Toledo (Fig. 3e). TB40E contained two viral species: a virus containing an intact copy of the UL141 gene (defined as the clonal strain TB40E-Lisa here) and a natural mutant thereof (called TB40E-Bart here). We cloned these separately by plaque purification (Fig. 3f). DNA sequence analysis showed that strain TB40E-Lisa and TB40E-Bart were identical throughout UL/b′, except that TB40E-Bart had a two–base pair (bp) frameshift insertion at codon 63 of UL141 and a 1,347-bp deletion removing all of UL144 and all but the six C-terminal codons of UL145 (Supplementary Fig. 1 online). Cells infected with strain TB40E-Bart (UL141−) were reproducibly more sensitive to NK cell–mediated lysis than were those infected with strain TB40E-Lisa or Toledo using NKL, D7NKb or D9NKb as effectors against allogeneic or autologous targets (Fig. 3g and data not shown). Infection with TB40E-Bart resulted in values for NKL killing close to those noted with strain AD169 (Fig. 3g). These results are consistent with gpUL141 providing a chief contribution to the differences in NK cell inhibition noted between strains Toledo and AD169. However, functions for UL144 or UL145 cannot be excluded in experiments comparing TB40E-Bart and TB40E-Lisa.
GpUL141 downregulates cell surface expression of CD155
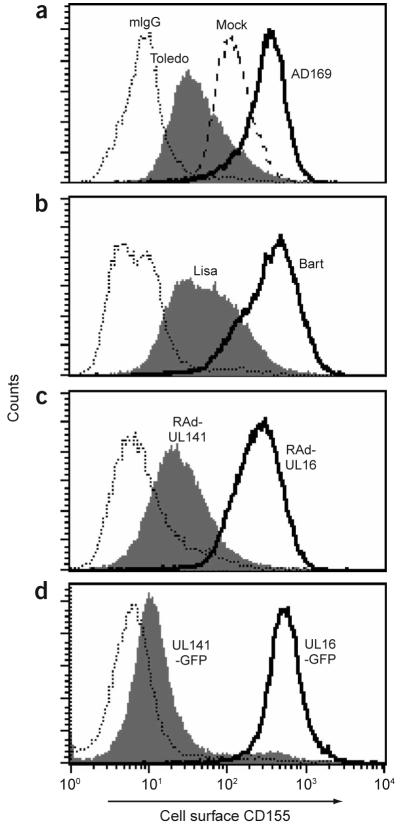
We attempted to identify the molecular target of gpUL141 by screening a broad range of previously identified NK cell receptor ligands by flow cytometry on UL141-transfected or RAdUL141-infected cell lines. The protein gpUL141 did not alter cell surface expression of HLA class I, MICA, MICB or a range of integrins or other adhesion molecules or the binding of NKp30-Fc and NKp46-Fc fusion proteins (data not shown). CD155 (also called poliovirus receptor or nectin-like molecule 5) and CD112 (also called nectin-2) are both cell surface ligands for the NK cell–activating receptors CD226 (also called DNAX accessory molecule 1) and CD96 (also called T cell–activation increased late expression)20,21. Infection with strains Toledo or TB40E-Lisa efficiently downregulated CD155 expression, as determined by the D171 monoclonal antibody (mAb), whereas infection with strain AD169 or TB40E-Bart (both UL141−) upregulated CD155 (Fig. 4). The NK cell cytotoxicity of strain TB40E-Bart-infected cells was partially inhibited by blocking with a mAb to CD155 (Supplementary Fig. 6 online). Furthermore, infection with adenovirus or transfection with a plasmid driving expression of gpUL141 down-regulated cell surface CD155, demonstrating that expression of gpUL141 or a UL141-GFP fusion protein was sufficient and necessary for this effect (Fig. 4). In contrast, HCMV gpUL16, UL16-GFP and the two additional ORFs deleted in TB40E-Bart (UL144 and UL145) had no effect on CD155 expression (Fig. 4 and Supplementary Fig. 7 online). UL141 did not affect CD112 expression on human fibroblasts or HEK 293 cells (data not shown). GpUL141 thus acts specifically to downregulate cell surface expression of the NK cell–activating receptor CD155. Soluble tagged gpUL141 inhibited binding of the D171 mAb to CD155 (Supplementary Fig. 8 online), suggesting a direct interaction between CD155 and gpUL141.
Figure 4.
UL141-mediated downregulation of cell surface CD155 detected by flow cytometry. Flow cytometry with mAb D171 of cell surface CD155 on mock-infected cells or cells after infection with HCMV strains AD169 (UL141−) or Toledo (a), after infection with TB40E-Bart (UL141−) or TB40E-Lisa (b), after infection with adenovirus vectors encoding UL141 or UL16 (c), and in continuous cell lines expressing UL141-GFP or UL16-GFP fusion proteins (d). Dotted lines, staining with an isotype-matched control antibody. mIgG, mouse IgG.
We analyzed the effect of gpUL141 on CD155 expression with immunofluorescence studies of permeabilized cells. We used adenovirus and HCMV recombinants that also contained an unfused GFP gene as an infection tracker. Similarly, in transfected cells, specific fluorescence of UL141-GFP or UL16-GFP fusion proteins identified expressing cell populations. When expressed with an adenovirus vector, HCMV infection or DNA transfection, UL141 correlated exclusively with a loss of CD155 staining when detected with mAb D171. In contrast, UL141 had no obvious effect on the signal when we used the alternative 5D1 mAb to CD155 (Fig. 5). The disparate results obtained with the two CD155 mAbs were resolved in immunoblot experiments, which demonstrated that gpUL141 was compatible with CD155 expression. However, gpUL141 blocked CD155 maturation from an Endo H–sensitive, intracellular 69-kDa form to an Endo H–resistant cell surface 70- to 80-kDa species22 (Fig. 6). The D171 mAb is conformation specific23 and seemed to not recognize the immature 69-kDa CD155 form that accumulated when gpUL141 was present. In contrast, the 5D1 mAb was capable of recognizing immature forms24. With HCMV strains encoding UL141, mature CD155 was absent, whereas the immature Endo H–sensitive CD155 was synthesized in abundance. With HCMV strains lacking UL141, the mature 75-kDa form of CD155 was present, and an additional slower migrating and Endo H–resistant CD155 species was detected (Fig. 6). As CD155 has been associated with cell-cell interactions, we tested whether UL141 affected adhesion between NK cells and their targets, but noted only marginal differences (Supplementary Fig. 9 online). When expressed alone or in the context of a productive HCMV infection, gpUL141 thus acts to retain CD155 as an immature form associated with the endoplasmic reticulum to inhibit its cell surface expression and therefore prevent its interaction with an activating ligand on NK cells.
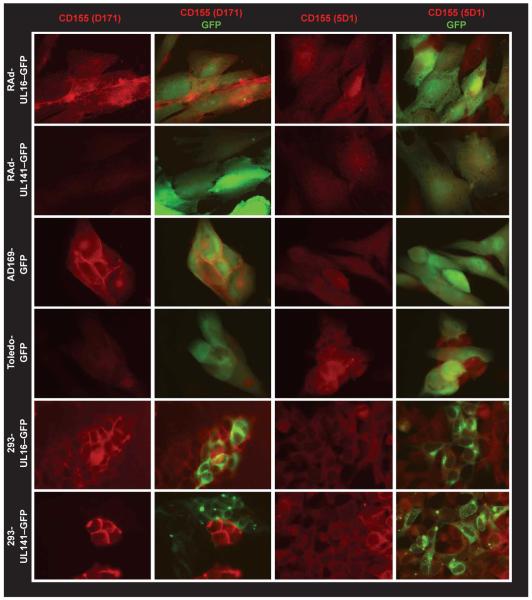
Figure 5.
UL141-mediated downregulation of cell surface CD155 detected by fluorescence microscopy. CD155 expression monitored by staining with the monoclonal antibodies D171 or 5D1 in the presence or absence of UL141. The gpUL141 was expressed with an adenovirus vector (RAdUL141-GFP), infection with strain Toledo-GFP in HFFFs or in UL141-GFP–transfected HEK 293 cells. Similarly, gpUL16 was expressed with an adenovirus vector (RAdUL16-GFP) or in UL16-GFP-transfected cells. HCMV strain AD169 is UL141 negative. HCMV and adenovirus vectors contain an intact GFP gene to identify infected cells. In HEK 293 cells, gpUL16-GFP and gpUL141-GFP fusion proteins traffick to the endoplasmic reticulum and concentrate in intracytoplasmic foci.
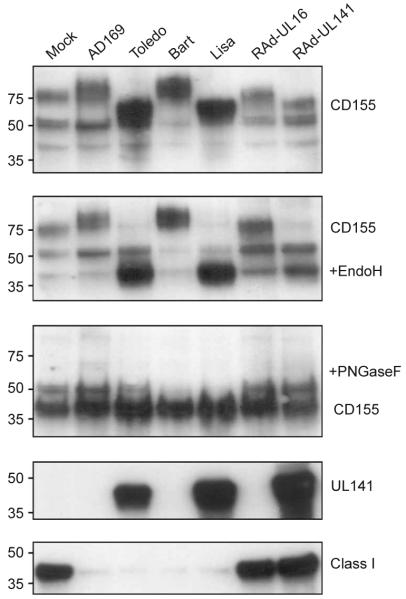
Figure 6.
UL141 inhibits maturation of CD155. Immunoblots of extracts of cells infected with viruses with the 5D1 mAb to CD155, the gpUL141-specific M550 mAbs or the HC-10 mAb to HLA class I. The sensitivity of the CD155 protein to Endo H and peptide-N-glycosidase F (PNGaseF) digestion is analyzed in parallel. HLA class I (Class I) downregulation is a surrogate marker of efficient HCMV infection. Left margin, molecular sizes (in kDa).
DISCUSSION
The HCMV genome shows an inherent instability during in vitro culture18,25. The spontaneous deletion of 13–15 kb from HCMV laboratory strains correlates with reduced virulence in vivo and increased susceptibility of HCMV-infected cells to NK cell–mediated cytolysis in vitro. Downregulation of surface HLA class I strips HCMV-infected cells of a key set of NK cell inhibitory ligands and thus renders them potentially vulnerable to NK cell attack. Should activating ligands become dominant, the balance would shift toward priming of NK cell killing. However, HCMV has responded to this strong selective pressure. Multiple NK cell inhibitory functions have been identified in studies with strain AD169: UL16 downregulates activating NK cell ligands MICB and ULBP1 and ULBP2, UL40 upregulates the inhibitory NK cell ligand HLA-E, and UL18 may itself act as an inhibitory NK cell ligand. However, infection with HCMV clinical isolates consistently bestowed substantially more effective protection from NK cell attack than did the highly passaged laboratory strain AD169, indicating that NK cell–evasion functions had been lost from laboratory strains. Here we have identified UL141 as a powerful NK cell evasion function that has a chief function in the enhanced protective effect noted with HCMV clinical isolates. Whether UL141 is the only gene in the UL/b′ region to inhibit NK cells cannot be concluded from these studies, as we used only a single NK cell line for our initial screening of the UL133–UL150 genes. Treatment of TB40E-Bart-infected targets, which showed disruptions in the UL141, UL144 and UL145 genes, with the D171 mAb to CD155 only partially blocked NK cell killing, indicating either an inefficient blocking of the CD155-CD226 interaction or an added function for either UL144 or UL145 in NK inhibition.
Regardless of this, UL141 was capable of suppressing killing of 67% of all NK cell clones tested, consistent with targeting of a principal NK cell–regulatory pathway by the gene. We calculated this value using a rigorous definition of inhibition (more than 10% reduction in specific target lysis). Insight into the broad NK cell inhibitory effect of UL141 came from the identification of its cellular target, CD155. CD155 is a ligand for the activating receptor CD226, which has been reported as being present on almost all NK cells20, and was expressed universally on all the NK cell bulk cultures and clones evaluated here (data not shown). UL141 showed specificity for CD155 in that it did not influence the expression of CD112 (nectin-2), another reported ligand for CD226. Furthermore, the UL141-related gene HCMV UL14 had no detectable effect on either CD112 or CD155 (data not shown). The efficiency with which UL141 inhibits NK cell function indicates the interaction of CD226 with CD155 has a major role in the regulation of NK cell function26.
Parallels exist between the action of UL141 and the HCMV-encoded NK cell inhibitor UL16. Both are proteins residing in the endoplasmic reticulum that act to promote intracellular retention of NK cell–activating ligands. HCMV contains many gene families thought to be generated through gene duplication, but the lack of sequence similarity between the UL16 and UL141 proteins suggests they may act through different mechanisms. Perhaps the most important functional distinction is that UL16 targets stress-induced NKG2D ligands, whereas UL141 targets a ligand that is constitutively expressed by a wide range of cell types. The function of CD155 as a constitutive activating ligand is likely to stimulate NK cell recognition if surface HLA class I is downregulated, as noted during HCMV infection as well as during other viral infections and cell transformation. UL141 suppressed NK cell function when expressed in either stably transfected cells or with the use of a replication-deficient adenovirus vector, neither of which downregulates HLA class I expression or upregulates CD155. In autologous assays, constitutive CD155 expression may therefore have been sufficient to stimulate NK cell killing of fibroblasts expressing physiological amounts of HLA class I. Such spontaneous killing may be accentuated by in vitro cell culture. In tissues, heterophilic interactions between membrane-bound CD155 and nectin-3 contribute to cell-cell and cell-matrix adhesion and restrict these proteins to adhesion junctions. During in vitro culture, virus infection27 or cell transformation, cell-cell and cell-matrix interactions become disrupted, potentially making CD155 available to interact with CD226 resulting in NK cell activation and cytolysis.
The function of CD226 is dependent on its forming a complex with the adhesion molecule LFA-1 on the effector cell. Activation of NK cell effectors through CD226 thus also indicates involvement in effector-target adhesion through the interaction of LFA-1 on effectors with the adhesion molecule ICAM-1 on the HCMV-infected target; it is well established that ICAM-1 is upregulated during HCMV infection27,28. Furthermore, CD155 interacts with another nectin family member, CD96 (TACTILE), to stimulate adhesion between NK cells and their targets and thus promote NK cell–mediated cytotoxicity21. Given those observations, we tested whether UL141 affected adhesion between NK cells and their targets but were unable to detect any changes. UL141 therefore seems to act mainly by impeding signaling through the activating receptor CD226 rather than by radically reducing adhesion between NK cell effectors and their targets.
The function of UL141 may yet prove to transcend its function in impeding NK cell recognition, as CD226 is also expressed on T cells, monocytes, megakaryocytes and B cell subsets20,29-31. The best characterized functions of CD155 are associated with its roles in cell-cell adhesion and cell motility. CD155 interacts with Tctex-1, a subunit of the dynein motor complex linked to endocytosis and retrograde transport31,32, is found at the leading edges of migrating cells33 and has been linked to fibroblast migration34 and monocyte transendothelial migration35. UL141 suppression of CD155 function has the potential to affect multiple cell processes and may thus have even more influence on the biological interaction of HCMV and its host.
METHODS
Cells
Human fetal foreskin fibroblasts, primary skin fibroblasts from volunteer donors, human alveolar epithelial carcinoma A549 cells, human embryonic retinoblast 911 cells and human embryonic kidney HEK 293 cells were all grown in DMEM supplemented with 10% FCS (Invitrogen). Primary human fibroblasts were preferred targets in NK cell assays because they support productive HCMV infection. Skin fibroblast cultures from biopsies were immortalized with human telomerase reverse transcriptase36. The established NK cell clone NKL and the methods used for generating NK cell lines and T cell–depleted, IFN-α-activated bulk cultures have been described5,37. NK cell cloning was accomplished by single-cell sorting with clones stimulated weekly with irradiated allogeneic peripheral blood mononuclear cells (PBMCs), mouse mAb OKT3 to CD3, and IL-2 (1,000 IU/ml). All NK cell clones were maintained in SCGM medium (Cellgro) supplemented with 5% human AB serum and were allowed to ‘rest’ from feeder stimulation for at least 1 week before use in assays. This project was approved by the Bro Taf Local Research Ethics Committee. Prior consent was obtained from all blood and skin biopsy donors.
Cell lines
UL14, UL16 and UL141 were amplified by PCR from HCMV genomic DNA and were inserted directly into the pcDNA3.1/CT-GFP-TOPO vector (Invitrogen) such that the genes were expressed as GFP fusion (C-terminal) proteins. The primers used were as follows: for strain Toledo UL141, 5′-ATCATGTGCCGCCGGGAGTCG-3′ (forward) and 5′-GCCTCTTCATCTTTCTAACACC-3′ (reverse), for strain AD169 UL16, 5′-CCGGCATGGAGCGTCGCCGA-3′ (forward) and 5′-GGTCCTCGGTGCGTAACCGCT-3′ (reverse); and for strain AD169 UL14, 5′-CCGGCATGGAGCGTCGCCGA-3′ (forward) and 5′-GGTCCTCGGTGCGTAACCGCT-3′ (reverse). The identity of all PCR-amplified sequences was confirmed by sequencing. HEK 293 cells were transfected with the cloned GFP fusion products and cell lines were selected with 2 mg/ml of geneticin (Invitrogen) and cell sorting. Purity of the cloned populations were more than 90%.
Virus
The HCMV strain AD169, deletion mutant AD169ΔUL40, strain Merlin, strain TB40E, strain Toledo encoding GFP (TOLΔβ2.7) and strain AD169 encoding GFP (RCMV288) have been described5,18,36, and strains Towne, Toledo and recombinant Towne/Tol11 1.1 were provided by E. Mocarski17 (Stanford University, Palo Alto, California). For NK cell cytolysis assays, fibroblasts were infected for 72 h with HCMV (10 plaque-forming units per cell). Replication-deficient adenovirus recombinants were constructed with the AdEasy-1 vector system provided by B. Vogelstein38 (Johns Hopkins Oncology Center, Baltimore, Maryland). HCMV genes were amplified by PCR, then were subcloned into the transfer vectors pShuttle-CMV or pAd-Track-CMV. The adenovirus recombinants, referred to by their transgenes in the text, have the following ‘formal’ designations: RAd592 has no transgene insert; RAd502 encodes GFP alone; RAd588 encodes both UL16 and GFP; RAd522 encodes both the complete strain Toledo UL141 and GFP genes; RAd587 encodes strain Toledo UL141 alone; RAd550 encodes GFP and strain Toledo UL141 with the transmembrane and cytoplasmic domains replaced with a streptactin-binding tag (Strep-tag; IBA). For RAd550, the Strep-tag was inserted by PCR amplification of the UL141 gene with the forward primer 5′-GGGGTACCATCATGTGCCGCCGGGAGTCG-3′ and the reverse primer 5′-CCGCTCGAGTTATTTTTCGAACTGCGGGTGGCTCCAAGCGCTCCGAGTGGCCCAGGGAGACATC-3′ (Invitrogen).
Protein analysis
Soluble tagged gpUL141 protein (designated P550) was prepared by infection of A549 cells for 72 h with RAd550 (multiplicity of infection, 10) in serum-free medium. Culture supernatant was collected and secreted P550 was purified on a Streptactin Macroprep column (IBA) according to the manufacturer’s instructions. N-terminal sequencing was done on an Applied Biosystems Procise Sequencer (LSUMC Core Laboratories, USA). Purified P550 was also used to generate mouse mAbs M550.2, M550.3 and M550.4 (all isotype IgG1) to gpUL141. A mixture of M550.2, M550.3 and M550.4 was used for immunoblots, and a combination of M550.3 and M550.4 was used for immunocytochemistry. Glycosylation was analyzed by digestion with Endo H or peptide-N-glycosidase F according to the manufacturer’s instructions (New England Biolabs).
Flow cytometry and immunohistochemistry
A FACSCalibur with CELLQuest PRO software (Becton Dickinson) was used for flow cytometry, and cell sorting was provided by the Cardiff University Central Biotechnology Service with a Cytomation MoFlo sorter. Before immunocytochemistry, cells were fixed with paraformaldehyde and were permeabilized with detergents. Fluorescence was detected and analyzed with a Leica DMIRBE microscope with Improvision Openlab software. Antibodies used included mouse mAb to CD155 (D171, ab3142; Abcam); another mouse mAb to CD155 (5D1; ref. 24); goat antibody to mouse IgG conjugated to AlexaFluor 594 (A-11020; Molecular Probes) or to AlexaFluor 647 (A-21237; Molecular Probes); mouse mAb to calnexin (MAB3126; Chemicon International); and concavalin A–AlexaFluor 488 (C-11252; Molecular Probes); the Zenon mouse IgG1 fluorescein labeling kit was also used (Z-25045; Molecular Probes).
Other reagents
Other antibodies and reagents used were as follows: rabbit anti-GFP (sc-8334; Santa-Cruz), goat anti-rabbit–horseradish peroxidase (170-6515; Bio-Rad), goat anti-mouse–horseradish peroxidase (170-6516; Bio-Rad), HC-10 mouse mAb to HLA class I heavy chain, Streptactin–horseradish peroxidase (2-1502-001; IBA), mouse mAb to Streptag (2-1507-001; IBA), the VYBRANT CFDA SE Cell Tracer Kit (V-12883, Molecular Probes) and the Cell Trace Far Red DDAO-SE kit (C-34553, Molecular Probes).
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Jones and D. Kipling for their cooperation with the telomerase immortalization of fibroblasts; V. Groh for MIC mAbs; O. Mandelboim for NKp30-Fc and NKp46-Fc proteins; and E. Mocarski, M. Wills and J. Sathish for discussions. Supported by the Wellcome Trust, Biotechnology and Biological Sciences Research Council and Medical Research Council.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Note: Supplementary information is available on the Nature Immunology website.
References
- 1.Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med. 1989;320:1731–1735. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 2.Karre K. How to recognize a foreign submarine. Immunol. Rev. 1997;155:5–9. doi: 10.1111/j.1600-065x.1997.tb00935.x. [DOI] [PubMed] [Google Scholar]
- 3.Carr WH, Little AM, Mocarski E, Parham P. NK cell-mediated lysis of autologous HCMV-infected skin fibroblasts is highly variable among NK cell clones and polyclonal NK cell lines. Clin. Immunol. 2002;105:126–140. doi: 10.1006/clim.2002.5273. [DOI] [PubMed] [Google Scholar]
- 4.Tomasec P, et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287:1031–1033. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 5.Wang EC, et al. UL40-mediated NK evasion during productive infection with human cytomegalovirus. Proc. Natl. Acad. Sci. USA. 2002;99:7570–7575. doi: 10.1073/pnas.112680099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosman D, et al. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 7.Kubin M, et al. ULBP1, 2, 3: novel MHC class I-related molecules that bind to human cytomegalovirus glycoprotein UL16, activate NK cells. Eur. J. Immunol. 2001;31:1428–1437. doi: 10.1002/1521-4141(200105)31:5<1428::AID-IMMU1428>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 8.Welte SA, et al. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur. J. Immunol. 2003;33:194–203. doi: 10.1002/immu.200390022. [DOI] [PubMed] [Google Scholar]
- 9.Leong CC, et al. Modulation of natural killer cell cytotoxicity in human cytomegalovirus infection: the role of endogenous class I major histocompatibility complex and a viral class I homolog. J. Exp. Med. 1998;187:1681–1687. doi: 10.1084/jem.187.10.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn W, et al. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA. 2003;100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cha TA, et al. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berstein DL, Plotkin SA. Cytomegalovirus vaccines. In: Levine MM, Kaper JB, Rappuloi R, Liu MA, Good MF, editors. New Generation Vaccines. Marcel Dekker; New York: 2004. pp. 649–659. [Google Scholar]
- 13.Cerboni C, et al. Human cytomegalovirus strain-dependent changes in NK cell recognition of infected fibroblasts. J. Immunol. 2000;164:4775–4782. doi: 10.4049/jimmunol.164.9.4775. [DOI] [PubMed] [Google Scholar]
- 14.Davison AJ, et al. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 2003;84:17–28. doi: 10.1099/vir.0.18606-0. [DOI] [PubMed] [Google Scholar]
- 15.Penfold ME, et al. Cytomegalovirus encodes a potent α chemokine. Proc. Natl. Acad. Sci. USA. 1999;96:9839–9844. doi: 10.1073/pnas.96.17.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benedict CA, et al. Cutting edge: a novel viral TNF receptor superfamily member in virulent strains of human cytomegalovirus. J. Immunol. 1999;162:6967–6970. [PubMed] [Google Scholar]
- 17.Mocarski ES, Kemble GW, Lyle JM, Greaves RF. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolan A, et al. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 2004;85:1301–1312. doi: 10.1099/vir.0.79888-0. [DOI] [PubMed] [Google Scholar]
- 19.Novotny J, Rigoutsos I, Coleman D, Shenk T. In silico structural and functional analysis of the human cytomegalovirus (HHV5) genome. J. Mol. Biol. 2001;310:1151–1166. doi: 10.1006/jmbi.2001.4798. [DOI] [PubMed] [Google Scholar]
- 20.Bottino C, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J. Exp. Med. 2003;198:557–567. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs A, Cella M, Giurisato E, Shaw AS, Colonna M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155) J. Immunol. 2004;172:3994–3998. doi: 10.4049/jimmunol.172.7.3994. [DOI] [PubMed] [Google Scholar]
- 22.Bernhardt G, Bibb JA, Bradley J, Wimmer E. Molecular characterization of the cellular receptor for poliovirus. Virology. 1994;199:105–113. doi: 10.1006/viro.1994.1102. [DOI] [PubMed] [Google Scholar]
- 23.Zibert A, Wimmer E. N glycosylation of the virus binding domain is not essential for function of the human poliovirus receptor. J. Virol. 1992;66:7368–7373. doi: 10.1128/jvi.66.12.7368-7373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoki J, Koike S, Ise I, Sato-Yoshida Y, Nomoto A. Amino acid residues on human poliovirus receptor involved in interaction with poliovirus. J. Biol. Chem. 1994;269:8431–8438. [PubMed] [Google Scholar]
- 25.Tahara-Hanaoka S, et al. Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112) Int. Immunol. 2004;16:533–538. doi: 10.1093/intimm/dxh059. [DOI] [PubMed] [Google Scholar]
- 26.Leis M, Marschall M, Stamminger T. Downregulation of the cellular adhesion molecule Thy-1 (CD90) by cytomegalovirus infection of human fibroblasts. J. Gen. Virol. 2004;85:1995–2000. doi: 10.1099/vir.0.79818-0. [DOI] [PubMed] [Google Scholar]
- 27.Scholz M, et al. Cytomegalovirus- and interferon-related effects on human endothelial cells. Cytomegalovirus infection reduces upregulation of HLA class II antigen expression after treatment with interferon-gamma. Hum. Immunol. 1992;35:230–238. doi: 10.1016/0198-8859(92)90004-7. [DOI] [PubMed] [Google Scholar]
- 28.Kojima H, et al. CD226 mediates platelet and megakaryocytic cell adhesion to vascular endothelial cells. J. Biol. Chem. 2003;278:36748–36753. doi: 10.1074/jbc.M300702200. [DOI] [PubMed] [Google Scholar]
- 29.Shibuya K, et al. CD226 (DNAM-1) is involved in lymphocyte function-associated antigen 1 costimulatory signal for naive T cell differentiation and proliferation. J. Exp. Med. 2003;198:1829–1839. doi: 10.1084/jem.20030958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohka S, et al. Receptor (CD155)-dependent endocytosis of poliovirus and retrograde axonal transport of the endosome. J. Virol. 2004;78:7186–7198. doi: 10.1128/JVI.78.13.7186-7198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mueller S, Cao X, Welker R, Wimmer E. Interaction of the poliovirus receptor CD155 with the dynein light chain Tctex-1 and its implication for poliovirus pathogenesis. J. Biol. Chem. 2002;277:7897–7904. doi: 10.1074/jbc.M111937200. [DOI] [PubMed] [Google Scholar]
- 32.Takai Y, Irie K, Shimizu K, Sakisaka T, Ikeda W. Nectins and nectin-like molecules: roles in cell adhesion, migration, and polarization. Cancer Sci. 2003;94:655–667. doi: 10.1111/j.1349-7006.2003.tb01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oda T, Ohka S, Nomoto A. Ligand stimulation of CD155α inhibits cell adhesion and enhances cell migration in fibroblasts. Biochem. Biophys. Res. Commun. 2004;319:1253–1264. doi: 10.1016/j.bbrc.2004.05.111. [DOI] [PubMed] [Google Scholar]
- 34.Reymond N, et al. DNAM-1 and PVR regulate monocyte migration through endothelial junctions. J. Exp. Med. 2004;199:1331–1341. doi: 10.1084/jem.20032206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McSharry BP, Jones CJ, Skinner JW, Kipling D, Wilkinson GW. Human telomerase reverse transcriptase-immortalized MRC-5 and HCA2 human fibroblasts are fully permissive for human cytomegalovirus. J. Gen. Virol. 2001;82:855–863. doi: 10.1099/0022-1317-82-4-855. [DOI] [PubMed] [Google Scholar]
- 36.Robertson MJ, et al. Characterization of a cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp. Hematol. 1996;24:406–415. [PubMed] [Google Scholar]
- 37.McSharry BP, Tomasec P, Neale ML, Wilkinson GW. The most abundantly transcribed human cytomegalovirus gene (β 2.7) is non-essential for growth in vitro. J. Gen. Virol. 2003;84:2511–2516. doi: 10.1099/vir.0.19298-0. [DOI] [PubMed] [Google Scholar]
- 38.He TC, et al. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.