Abstract
Sonic hedgehog (Shh) and its main receptor Patched (Ptc) are implicated in both neural development and tumorigenesis1, 2. Beside the classic morphogen activity of Shh, Shh is also a survival factor3, 4. Along this line, Ptc has been shown to function as a dependence receptor, inducing apoptosis in the absence of Shh, while its pro-apoptotic activity is blocked in Shh presence5. Here we show that, in the absence of its ligand, Ptc interacts with the adaptor protein DRAL/FHL2. DRAL/FHL2 is required for the pro-apoptotic activity of Ptc both in immortalized cells and during neural tube development in chick embryo. We demonstrate that, in the absence of Shh, Ptc recruits a protein complex that includes DRAL, the CARD containing domain proteins TUCAN or NALP1 and the apical caspase-9. Ptc triggers caspase-9 activation and enhances cell death via a caspase-9-dependent mechanism. Thus, we propose that, upon absence of its ligand Shh, the dependence receptor Ptc serves as the anchor for a caspase-activating complex that includes DRAL, a CARD domain containing protein and caspase-9.
Keywords: Adaptor Proteins, Signal Transducing; genetics; metabolism; Animals; Apoptosis; physiology; Apoptosis Regulatory Proteins; genetics; metabolism; CARD Signaling Adaptor Proteins; genetics; metabolism; Caspase 9; metabolism; Cell Line; Chick Embryo; Hedgehog Proteins; genetics; metabolism; Homeodomain Proteins; genetics; metabolism; Humans; Multiprotein Complexes; metabolism; Muscle Proteins; genetics; metabolism; Neoplasm Proteins; genetics; metabolism; RNA, Small Interfering; genetics; metabolism; Receptors, Cell Surface; genetics; metabolism; Signal Transduction; physiology; Transcription Factors; genetics; metabolism; Two-Hybrid System Techniques
Dependence receptors now number more than a dozen that include DCC 6, UNC5H, some integrins, neogenin, p75NTR, RET, ALK, EPHA4 and TrkC 7–9, 10. They share several features in common (see reviews 7, 8): (i) these receptors create cellular states of dependence on specific ligands by inducing programmed cell death in the absence of their respective ligands, while the presence of their ligands inhibits the pro-apoptotic activity of these receptors; (ii) these receptors are typically involved in both neural development and neoplasia; and (iii) apoptosis induction requires a caspase-cleavage site in the intracytoplasmic domain.
Ptc fulfills all of these criteria as a candidate dependence receptor. The expression of Ptc induces apoptosis, and this is suppressed by its ligand, Shh. Beside its key role during development, Ptc is a tumour suppressor and mutations of Ptc are associated with neoplasia, especially basal cell carcinomas 11 and medulloblastomas 12. Moreover, apoptosis induction by Ptc requires the caspase-cleavage site at Asp1392 5.
However the mechanism by which the withdrawal of trophic ligands from their respective dependence receptors leads to apoptosis is in large part unknown. Previous studies on DCC have suggested that some of these dependence receptors may recruit a caspase-activating complex 13. This complex comprising the initiator/apical caspase-9 is independent of the two main apoptosis-related-caspase-activating complexes named, the DISC and the apoptosome 14. Yet, the precise identification of such an apoptosis-induced dependence receptor complex has been unsuccessful so far.
In an attempt to identify the downstream pro-apoptotic signaling of Ptc, a two-hybrid screen was performed using the pro-apoptotic domain of Ptc 1165–1392 5 as bait and a human embryonic brain cDNA library as prey (Fig. 1a). Among the 7 putative Ptc interactors (see list in Supplementary Fig. 1a), we focused our attention on DRAL/FHL2. Indeed, DRAL is a multifunctional protein that consists of four complete and one half LIM protein-protein interaction domains 15. DRAL participates in cellular processes that include regulation of gene expression, cytoarchitecture, cell adhesion, cell mobility, signal transduction and cell survival 15. Interestingly, DRAL over-expression appears to be pro-apoptotic in a variety of cells even though the mechanism for DRAL-induced apoptosis is unknown 16. DRAL has also been shown to interact with a CARD domain containing protein named TUCAN 17 which was proposed to act as an adaptor protein for caspase-1 18 or caspase-9 19.
Figure 1. The pro-apoptotic domain of Ptc interacts with DRAL.
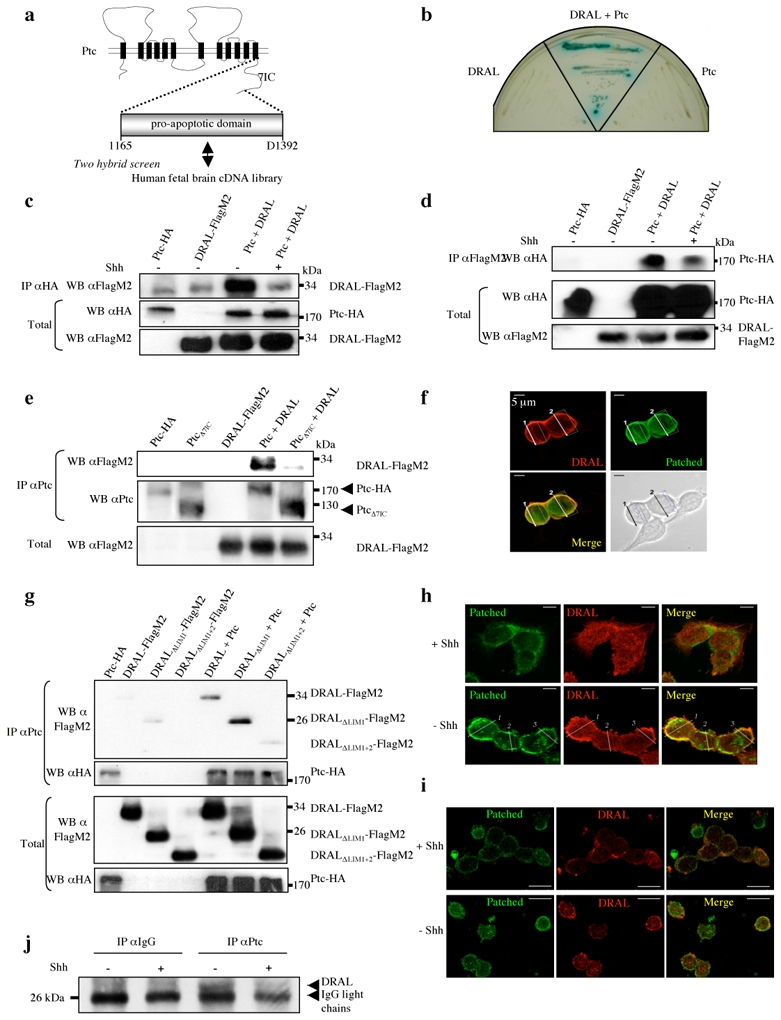
(a) A two-hybrid screen with the pro-apoptotic domain of Ptc-1. (b) The Ptc-DRAL interaction was confirmed by direct two-hybrid. AH109 yeasts transformed with either a mock Gal4BD plasmid and a Gal4AD plasmid fused with DRAL (DRAL) or a mock Gal4AD plasmid and a Gal4BD plasmid fused with the Ptc pro-apoptotic domain (Ptc). (c) Co-immunoprecipitations were performed on HEK293T cells transiently expressing Ptc-HA (Ptc) or DRAL-FlagM2 (DRAL) or Ptc-HA and DRAL-FlagM2 (Ptc + DRAL) in the absence (−) or presence (+) of Shh (300 ng/ml). Pull-down with anti-HA antibody was used to immunoprecipitate Ptc (IP αHA) and DRAL was revealed by Western blot by using anti-FlagM2 antibody. Western blot on lysates before pull down are shown (Total). (d) Same experiment as in (c) but in the opposite direction: DRAL pull-down was done with anti-FlagM2 antibody (IP αFlagM2) and specific Ptc binding was revealed by using anti-HA antibody. Full blots are shown in Suppl. Figure 7a. (e) DRAL co-immunoprecipitation with Ptc but not with PtcΔ7IC. Pull-down of Ptc on lysates from cells expressing DRAL-FlagM2 (DRAL) and Ptc-HA (Ptc) or PtcΔ7IC (PtcΔ7IC). DRAL was revealed by Western blot by using anti-FlagM2 antibody. (f) Co-localization of Ptc and DRAL in transiently transfected HEK293T cells. Cells were stained with anti-Ptc (Patched, green) and anti-DRAL (DRAL, red) antibodies. A representative field is presented from 3 independent experiments. Superimposed photograph (Merge) is shown. (g) Co-immunoprecipitation of Ptc with DRAL deletion mutants was performed as described in (c) on cells expressing Ptc-HA and DRAL-FlagM2, DRALΔLim1-FlagM2, or DRALΔLIM1+2-FlagM2. DRAL presence was revealed with anti-FlagM2 antibody. (h,i) Co-localization of endogenous Ptc and DRAL in Daoy medulloblastoma cells (h) or in primary cultured cerebellar granule neurons (i) performed in the presence or absence of Shh. Cells were stained as described in (f). Superimposed photographs (Merge) are shown. (j) Immunoprecipitation on mice cortex of E16.5 embryos incubated in absence (−) or presence (+) of Shh was performed using Ptc antibody (IP α-Ptc) or IgG as control (IP α-IgG), and DRAL was detected by immunoblot by using anti-DRAL antibody.
To further validate the interaction between Ptc pro-apoptotic domain and DRAL, a targeted two-hybrid was performed. As shown in Fig. 1b, Ptc 1165–1392 and DRAL interact in yeast. To monitor whether this interaction could be observed in mammalian cells, co-immunoprecipitation studies were performed on HEK293T cells co-transfected with tagged Ptc and DRAL. As shown in Fig. 1c, full-length DRAL is pulled down by full-length Ptc. When Shh was added to the culture medium, Ptc/DRAL interaction was strongly decreased (Fig. 1c). Similar interaction data were observed when the reverse immunoprecipitation was performed (Fig. 1d). Moreover, as expected from the two-hybrid screen, the 7th intracellular region of Ptc is sufficient to interact with DRAL (Suppl. Fig. 1b), while deletion of the 7th intracellular region of Ptc that includes the pro-apoptotic domain prevents the interaction of Ptc with DRAL (Fig. 1e).
Confocal analysis of the two co-expressed full-length proteins shows a co-localization of both Ptc and DRAL in HEK293T cells (Fig. 1f and Suppl. Fig. 1c). In an attempt to define the region of DRAL important for the Ptc/DRAL interaction, we analyzed whether DRAL lacking LIM domains 1 and 2 could be pulled down by Ptc. While deletion of the LIM1 domain had no effect on Ptc/DRAL interaction, deletion of both LIM1 and LIM2 domains or deletion of the LIM2 domain only strongly decrease the ability of DRAL to interact with Ptc (Fig. 1g and Suppl. Fig 1d). Thus, both in yeast and in mammalian cells, Ptc interacts via its 7th intracellular domain with DRAL through a region including the DRAL LIM2 domain.
To address whether this interaction may be observed not only with ectopically expressed proteins but also with endogenous proteins, Ptc/DRAL co-localization was analyzed in Daoy medulloblastoma cell line in the absence or presence of Shh. While in the presence of Shh, DRAL was mainly cytoplasmic and failed to co-localize with Ptc, in the absence of Shh, DRAL and Ptc clearly co-localize at the membrane of these cells (Fig. 1h and Suppl. Fig. 1e). Similar results were obtained with primary cultured cerebellar granule cells (CGN) (Fig. 1i and Suppl. Fig. 1f). In vivo interaction of Ptc and DRAL was then addressed by co-immunoprecipitation in mouse developing brain extract. Semi-dissociated E16.5 brains were either left untreated or exposed to Shh and then further processed for Ptc immunoprecipitation. In the absence of Shh treatment, anti-Ptc antibody pulls down DRAL, while Ptc/DRAL association was not observed with an unrelated isotypic antibody confirming the specificity of the interaction. Addition of Shh inhibits the association of DRAL with Ptc (Fig. 1j). Thus, Ptc directly interacts with DRAL both in cell culture and in the developing brain and this interaction is prevented by Shh.
As a first assay to monitor the functional role of the Ptc/DRAL interaction, Ptc-induced cell death was assessed by Ptc transient expression in HEK293T cells 5 upon DRAL gene silencing by siRNA. DRAL siRNA dramatically reduced endogenous DRAL expression in HEK293T cells while the control siRNA did not (inset Fig. 2a). Correspondingly, Ptc-induced cell death was reduced when DRAL protein was down-regulated by siRNA (Fig. 2a). In contrast to Ptc, over-expression of the classic cell death inducer Bax triggered HEK293T cell death to a similar extent in the presence or absence of DRAL siRNA. Thus, in immortalized cells, Ptc requires DRAL to trigger cell death.
Figure 2. DRAL is required for Ptc pro-apoptotic activity.
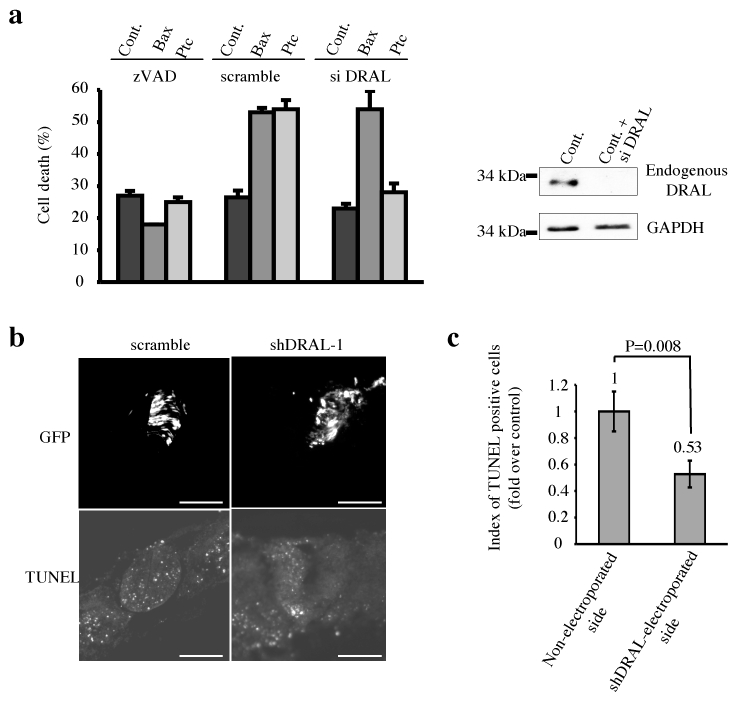
(a) HEK293T cells were transiently transfected with empty vector, siRNA scramble (scramble) or siRNA DRAL (si DRAL). 48 hours after siRNA transfection, cells were transfected a second time with empty vector (Cont.), Bax encoding vector (Bax) or Ptc encoding vector (Ptc) in the absence (−) or presence of zVAD-fmk (zVAD). Cell death was analyzed 24 hours after the second transfection by using trypan blue exclusion as described in 5. Standard deviations are indicated. siRNA DRAL efficiency on endogenous DRAL expression is shown by Western blotting using anti-DRAL antibody. Anti-GAPDH immunoblot is shown as a control of specificity and loading. (b) DRAL function in Ptc-induced apoptosis was also addressed in vivo in a context of absence of Shh created by removal of the caudal part of the Hensen node by microsurgery at E1.5 in the chick embryo in ovo. Before the microsurgery, the left side of the neural tube was electroporated with GFP and shDRAL-1 or scramble. Electroporation efficiency is shown on the upper picture (GFP) and TUNEL staining of the same section is shown on the lower picture (TUNEL). Scale bar : 100 μm. (c) Index of TUNEL positive cells is represented as the ratio between the number of cells stained in shDRAL-1 condition (shDRAL-1-electroporated side) to the number of cells stained in the control condition (Non-electroporated side) counted over 15 sections. Error bars indicate SEM. U test was performed (p=0.008). Note that DRAL shRNA electroporation alone or forced expression of DRAL had no significant effect on cell death (Suppl. Fig. 2ef).
We next analyzed whether Ptc-induced cell death requires DRAL in vivo. While it has been extensively demonstrated that Shh acts as a morphogen during nervous system development 1, several works have shown that Shh may also act as a survival factor 4, 20. Consistent with this, Le Douarin and colleagues have shown that the experimental withdrawal of Shh in chick embryos led to massive cell death of neuroepithelial cells in the developing neural tube 3, 20. We have demonstrated that this survival activity is due to the inhibition by Shh of Ptc pro-apoptotic activity 5. While in initial studies, we and others have used surgical procedure to prevent Shh production by the notochord and floor plate, we first assessed here whether a similar effect could be observed by the ectopic expression of HHIP1, a protein known to inhibit Shh signaling by competitively titrating this cue 21. The left side of the developing neural tube of HH11–13 chick embryos was co-electropored in ovo by GFP with either control or HHIP1 expression construct. As shown in Suppl. Fig. 2ab, while GFP was detected after electroporation, a significant increase in apoptosis, measured by TUNEL staining, was observed in HHIP1 electroporated neural tube compared to mock electroporated embryos. To further control that this HHIP1-dependent cell death was related to the loss of Shh inhibition of Ptc-induced apoptosis, a dominant-negative mutant of Ptc 5 was co-electroporated with HHIP1. In agreement with our previous data 5, Ptc dominant-negative mutant inhibits HHIP1-induced apoptosis (Suppl. Fig. 2ab). Thus, HHIP1 electroporation triggers Ptc-induced apoptosis in vivo. We then assessed the importance of DRAL in Ptc-induced cell death by co-electroporating HHIP1 with a DRAL shRNA-expressing construct that is effective at inhibiting chick DRAL expression (Suppl. Fig. 2c). While scrambled shRNA failed to suppress HHIP1-associated cell death, DRAL shRNA significantly inhibited apoptosis triggered by HHIP1 (Suppl. Fig. 2ab). Moreover, similar inhibition of HHIP1-induced apoptosis by DRAL shRNA was observed when electroporation of HHIP1 was performed more ventrally via the use of an electrode located below the neural tube (Suppl. Fig. 2d). It is however intriguing to note that HHIP1-electroporations were associated with cell death occurring mainly in the middle third and the dorsal third of the neural tube but only modestly in the 1/3 ventral neural tube. The discrepancy between this somehow dorsal cell death and the fact that higher Shh level is detected ventrally, raised the possible limitation of the Shh titration by HHIP1. However, similar cell death inhibition upon DRAL shRNA electroporation was observed when Shh was deprived using the surgical procedure described by Le Douarin and colleagues 3, 5 (Fig. 2bc). Thus altogether these data support the view that cell death induced upon inhibition of Shh in the developing neural tube requires DRAL.
DRAL has been shown to interact with TUCAN, a CARD domain containing protein initially described to bind caspase-9 19. Because caspase-9 has been implicated in the pro-apoptotic activity of many dependence receptors 13, 9, we explored whether TUCAN may be part of a Ptc/DRAL complex that could allow the recruitment of caspase-9, using co-immunoprecipitation experiments performed with HEK293 cells. As shown in Fig. 3a, Ptc immunoprecipitation resulted in the pull-down of DRAL and TUCAN and caspase-9. Similarly, TUCAN immunoprecipitation pulled down Ptc and caspase-9 (Fig. 3b). Interestingly, Shh presence strongly inhibited the interaction between TUCAN and Ptc while also decreasing the interaction between TUCAN and caspase-9 (Fig. 3ab). Thus, in the absence of the dependence ligand Shh, Ptc allows the recruitment of a complex that includes DRAL, TUCAN and caspase-9.
Figure 3. Ptc/DRAL serves as a platform to recruit TUCAN or NALP1 and caspase-9.
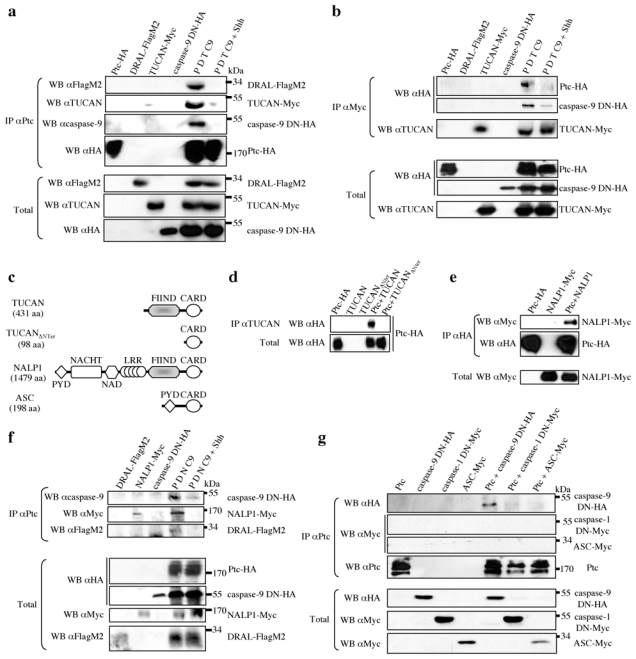
(a) Ptc pull-down (IP αPtc) on HEK293-EcrShh cells transfected with either Ptc (Ptc- HA), DRAL (DRAL-FlagM2), TUCAN (TUCAN-Myc) or caspase-9 DN (caspase-9 DN-HA) alone or the four constructs together in the absence (P D T C9) or presence (P D T C9 + Shh) of Shh (induction with Muristerone A plus recombinant Shh). DRAL, TUCAN and caspase-9 interaction with Ptc were revealed by Western blot by using anti-FlagM2, anti-Myc and anti-caspase-9 antibodies respectively. Full blots are shown in Suppl. Figure 7b. (b) TUCAN immunoprecipitation (IP αMyc) from HEK293T cells transfected with Ptc-HA, DRAL-FlagM2, TUCAN-Myc or caspase-9 DN-HA alone or the four constructs together in the absence (P D T C9) or presence (P D T C9 + Shh) of recombinant Shh; Ptc and caspase-9 were revealed after the pull-down by Western blotting using anti-HA antibody; TUCAN was revealed with anti-TUCAN antibody. Total: Western blots on lysate before pull-down. (c) Schematic representations of TUCAN, TUCAN mutant deleted of its N-Terminal domain, NALP1 and ASC. Note the domain of homology FIIND between TUCAN and NALP1 in their N-Terminal part (grey box). (d) TUCAN immunoprecipitation (using anti-TUCAN with an epitope in the CARD domain) from HEK293T cells transfected with empty vector and Ptc (Ptc-HA), TUCAN (TUCAN) or TUCAN deleted of its N-terminal part vectors (TUCANΔNter) or transfected with Ptc and either TUCAN (Ptc + TUCAN) or TUCAN deleted of its N-terminal region (Ptc + TUCANΔNter). Ptc was revealed in the pull-down by Western blot by using anti-HA antibody. (e) Ptc pull-down (IP αHA) on HEK293T cells transfected with empty vector and Ptc (Ptc-HA) or NALP1 vectors (NALP1-Myc) or transfected with Ptc and NALP1 (Ptc + NALP1). NALP1 pull-down with Ptc was revealed by anti-Myc Western blot. (f) Co-immunoprecipitations as in (a) except that TUCAN vector was replaced by NALP1 vector (NALP1-Myc). (g) Ptc pull-down (IP αPtc) on HEK293T cells transfected with Ptc vector (Ptc), caspase-9 DN (caspase-9 DN-HA), caspase-1 DN (caspase-1 DN-Myc) or ASC (ASC-Myc) alone or in combination with Ptc (Ptc + caspase-9 DN-HA; Ptc + caspase-1 DN-Myc; Ptc + ASC-Myc). Caspase-9, caspase-1 or ASC within the Ptc pull-down were detected using anti-HA or anti-Myc antibodies. Full blots are shown in Suppl. Figure 7c.
TUCAN is composed of two main domains: a FIIND domain located in the N-terminal region and a CARD domain at the C-terminus which is known to interact with caspases (see scheme Fig. 3c). Of interest, deletion of the N-terminal region of TUCAN is sufficient to abrogate the interaction of Ptc/DRAL with TUCAN (Fig. 3d). This N-terminal domain is closely related to a domain found in NALP1 (66% homology in human), another CARD domain-containing protein known to recruit caspases, that has been implicated in both cell death regulation but also in inflammation 22, 23 (Fig 3c). Using a similar co-immunoprecipitation approach, NALP1 was also pulled-down with Ptc, while this interaction was strongly reduced upon presence of Shh (Fig. 3ef). As a control for specificity, ASC, a protein related to NALP1, failed to interact with Ptc (Fig. 3cg). Interestingly, in HEK293T cells, while TUCAN siRNA, which was efficient in decreasing TUCAN protein levels (Fig. 4a), was not sufficient alone to fully and reproducibly inhibit Ptc-induced cell death (not shown), the combination of TUCAN and NALP1 siRNA completely inhibited cell death observed upon Ptc transfection (Fig. 4bc). Thus, NALP1 or TUCAN behave as adaptor proteins of the Ptc/DRAL complex that includes caspase- 9 and that appears to be required for Ptc-induced cell death. Even though TUCAN and NALPs have been implicated in NF-κB regulation, a classic survival pathway 22, 24, we were able to exclude a putative link between NF-κB pathway and the regulation of Ptc pro-apoptotic activity (Suppl. Fig. 4abc). Moreover, because TUCAN and NALP1 has been shown to also interact with caspase-1 18, 22, 25, we next investigated the specificity of the caspase-9 recruitment to Ptc. While Ptc interacts with caspase-9 upon expression in HEK293T cells, Ptc failed to pull down caspase-1 (Fig. 3g). Thus, Ptc/DRAL represents an anchor for a specific TUCAN/NALP1-caspase-9 containing complex.
Figure 4. TUCAN/NALP1 and caspase-9 are required for Ptc-induced apoptosis.
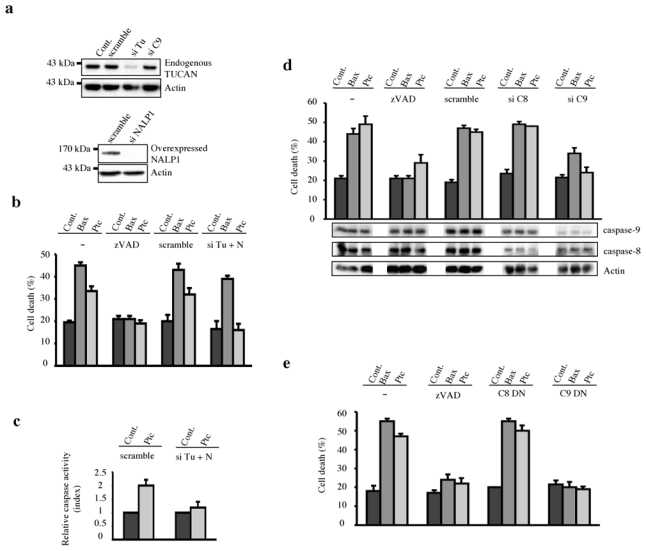
(a) siRNA TUCAN efficiency and specificity is shown by TUCAN Western blotting on endogenous TUCAN protein of HEK293T cells treated with scramble siRNA (scramble), siRNA TUCAN (si Tu) or siRNA caspase-9 (si C9). The same was done with NALP1 but siRNA NALP1 efficiency was assessed on HEK293T cells overexpressing NALP1. Anti-Actin immunoblot is shown as a control of specificity and loading. Full scanned blot is shown in Suppl. Figure 7d. (b) HEK293T cells were transfected with empty vector (Cont.), Bax or Ptc either without treatment (−) or in the presence of zVAD-fmk (zVAD), siRNA scramble (scramble), siRNA TUCAN plus siRNA NALP1 (si Tu + N) and cell death was analyzed by trypan blue exclusion assay. (c) Ptc-induced caspase activation is inhibited by a combination of siRNA TUCAN plus siRNA NALP1 as measured by relative caspase-3 activity. HEK293T cells were transfected with mock vector pcDNA3 (Cont.) or with Ptc in the presence of siRNA scramble (scramble) or siRNA TUCAN plus siRNA NALP1 (si Tu + N). Index of relative caspase activity is presented as the ratio between the caspase activity of the sample and that measured in HEK293T cells transfected with pcDNA3. Cell death was also compared in primary cultured cerebellar granule neurons from either CBA/J or C57Bl/6/J genetic background when deprived in Shh. While primary neurons death occurred in the CBA/J culture, no induction of cell death was detected in the C57Bl/6/J culture (Suppl. Fig.3). (d) HEK293T cells were transfected with empty vector (Cont.), Bax or Ptc either without treatment (−) or in the presence of zVAD-fmk (zVAD), siRNA scramble (scramble), siRNA caspase-8 (si C8) or siRNA caspase-9 (si C9) and cell death was analyzed by trypan blue exclusion assay. siRNA caspase-9 and caspase-8 efficiencies are shown by Western blotting using anti-caspase- 9 and anti-caspase-8 antibodies. Anti-Actin Western blot is shown as a control of specificity and loading. (e) HEK293T cells were co-transfected with empty vector (Cont.), Bax or Ptc with either empty vector (−), caspase-8 dominant-negative (C8 DN) or caspase-9 dominant-negative (C9 DN). Cell death was analyzed as in (d). For all experiments, error bars are s.d. n=3.
To assess the importance of this DRAL/TUCAN-NALP1/caspase-9 complex in Ptc-mediated cell killing, we next assessed whether Ptc-induced apoptosis requires caspase-9. As shown in Fig. 4d, in HEK293T cells, while Ptc-induced cell death (and Bax-induced cell death as a control) was not modulated by caspase-8 siRNA, it was strongly inhibited by caspase-9 siRNA (while each siRNA significantly reduces the levels of caspase-8 and caspase-9 protein expression respectively (Fig. 4d inset)). Similar results were obtained when dominant-negative mutants for caspase-8 and caspase-9 were used instead of siRNAs (Fig. 4e) or when using chemical caspase inhibitors (not shown). To address further the requirement of caspase-9 in a biological system where endogenous Ptc induces apoptosis upon Shh deprivation, developing neural tubes of HH11–13 chick embryos were co-electroporated in ovo by GFP and a HHIP1 expression construct together with a dominant-negative mutant for caspase-9. While HHIP1 electroporation triggers apoptosis, co-electroporation with the dominantnegative caspase-9 mutant strongly inhibited this Ptc-mediated cell death process (Suppl. Fig. 5ab).
Given that Ptc interacts with caspase-9 (through a DRAL/TUCAN-NALP1 complex) and that Ptc requires caspase-9 to trigger apoptosis both in immortalized cells in culture and in the developing neural tube in vivo, we hypothesized that unliganded Ptc activates caspase-9. To investigate this possibility, we analyzed the activity of caspase-9 by measurement of LEHD-AFC substrate cleavage in HEK293 cells transfected with Ptc. As shown in Fig. 5a, Ptc triggers caspase-9 activity while the presence of Shh strongly decreases Ptc-induced caspase-9 activity. We next analyzed whether this activation is associated with the formation of large caspase-9-containing complexes, by analogy to other initiator caspase-activating complexes 26. To this end, gel-exclusion chromatography analysis of caspase-9 was performed using lysates from control and Ptc-expressing HEK293 cells. As shown in Fig. 5b, while in absence of Ptc, caspase-9 is detected at small exclusion size (50–100kDa), the presence of Ptc triggers a shift of caspase-9 toward the higher molecular weight fractions, such that caspase-9 is also detected in the 1000–2000kDa size fraction. As a negative control, Ptc deleted to its pro-apoptotic domain (i.e., deleted of the 7th intracellular domain) failed to trigger this shift toward larger fractions (Fig. 5b). Moreover, the presence of caspase-9 in higher fractions observed upon Ptc expression is inhibited in presence of Shh (Fig. 5b). Interestingly, in the absence of Shh, when caspase-9 is detected in the high molecular weight fractions, caspase-9 is present in a complex containing Ptc and TUCAN, as shown by co-immunoprecipitation experiments (Fig. 5c). Moreover, when endogenous caspase-9 is pulled down by Ptc, the caspase-9 recruited in the Ptc complex is proteolytically processed (Suppl. Fig. 6), thus further supporting that (in the absence of Shh) Ptc induces a large molecular weight complex that includes DRAL and TUCAN (or/and NALP1) and that results in caspase-9 activation.
Figure 5. Ptc pro-apoptotic complex recruits and activates caspase-9.
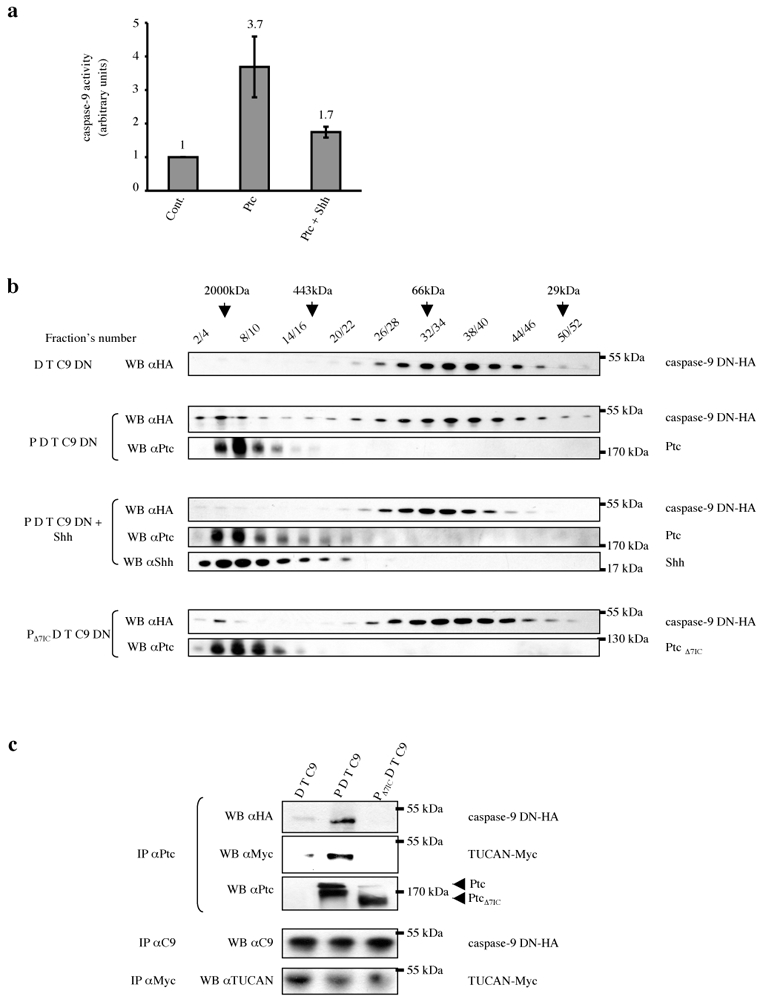
(a) HEK293-EcRShh cells were transfected with either empty vector (Cont.) or Ptc encoding vector in the absence (Ptc) or in the presence (Ptc + Shh) of Shh (induction with Muristerone A plus addition of 300 ng/ml of recombinant Shh). The cell lysates were subjected to caspase-9 activity assay over time using LEHD-AFC substrate of caspase-9. Error bars indicate s.d. n=4. (b) HEK293-EcRShh cells were transfected with DRAL-FlagM2, TUCAN-Myc and caspase-9 DN-HA vectors with either empty vector (D T C9) or Patched vector in the absence of Shh (P D T C9) or in the presence of Shh (P D T C9 + Shh; induction by Muristerone A plus recombinant Shh) or Ptc deleted of its last intracellular domain (PΔ7IC D T C9). Cell lysates were separated by gel filtration chromatography. Fractions were then analyzed by Western blotting for caspase-9 (WB αHA), Ptc (WB αPtc) and Shh (WB αShh). The fractions’ numbers and the fractions in which the molecular mass markers were eluted are shown. (c) The 20 first fractions were pooled and a pull-down of Ptc was performed (IP αPtc). Anti-HA and anti-Myc immunoblots were then performed and they show that caspase-9 and TUCAN interact with Ptc in these high molecular weight fractions. The relative quantity of proteins of interest (caspase-9 and TUCAN) in cell lysate of each condition was checked by immunoprecipitation (IP αC9 and IP αMyc respectively) and Western blotting with anti-caspase-9 and anti-TUCAN respectively.
Our data support the view that cell death initiation by the Ptc dependence receptor occurs through the recruitment of a protein complex that results in the activation of caspase-9. In this regard, caspase-9 is an initiator or apical protease involved in the control of a downstream cascade of caspase activation that leads to apoptosis. Initiator caspases are typically activated through formation of large multi-protein complexes (reviewed in 27). Among the known caspase-activating complexes of mammals are the apoptosome (Apaf-1; cytochrome c; caspase-9), the DISC (Death Inducing Signaling Complex) (Fas; FADD; caspase-8) 14, the PIDDosome (PIDD; RAIDD; caspase-2) 28 and the inflammasome (NALPs; ASC; caspase-1, caspase-5) 22. Here we report the existence of a fifth caspase-activating complex that signals downstream of the dependence receptor Ptc. Future work will define whether this “dependosome” is a common platform for dependence receptor death signaling as suggested by the fact that DCC has been shown to interact with and to require caspase- 9 to trigger cell death 13.
In addition to the adaptor protein DRAL, the Ptc-based activating complex also requires either TUCAN or NALP1. While TUCAN directly binds to pro-caspase-9 via their CARD-domains, NALP1 has been reported to indirectly bind via association with the pro-caspase-9-binding protein Apaf-129. Whether Apaf-1 can also be recruited to the Ptc-dependence receptor complex remains to be investigated. Similarly, it will be interesting to unravel the stoichiometry of the Ptc complex that activates caspase-9 since the observation that caspase-9 is detected in a 1000–2000kDa complex in Ptc-expressing cells suggests the formation of a multimeric complex, similar to other caspase-activating platforms.
Methods
Site directed mutagenesis and plasmid constructs
The different constructs used in this study are detailed in Supplementary Methods. The pcDNA3-Ptc-7ICmyr construct was obtained by PCR on the pcDNA3-Ptc-7IC plasmid with the primers Ptc-myr7IC-F and Ptc-7IC-HA-R (given in Suppl. Table1). Ptc mutant with the deletion of its last intracellular domain (pRK5-PtcΔ7IC) was obtained by directed mutagenesis via Quikchange strategy (Stratagene) on pRK5-Ptc with the primers given in Suppl. Table1 (mPtc1-Δ7IC-F and mPtc1-Δ7IC-R). For two-hybrid screen, the coding sequence of Ptc-7IC from pcDNA3-Ptc-7IC was amplified by PCR using the primers mPtc1-Eco-7IC-F and mPtc1-Pst-7IC-R (given in Suppl. Table1) and then inserted into pGBKT7 plasmid by digestion through EcoRI-PstI. By directed mutagenesis via Quikchange strategy (Stratagene) on the resulting plasmid, a stop amino acid was generated in position 1393, in order to obtain the pGBKT7-GAL4 DNA binding domain/Ptc 1165–1392 fusion plasmid (for primers sequence mPtc1-1393StopQC-F and mPtc1-1393StopQC-R, see Suppl. Table1). The construct pcDNA3-DRAL- 3xFlagM2ΔLIM2 -deleted only of its LIM2 domain- was obtained by directed mutagenesis via Quikchange strategy (Stratagene) using the primers DralΔLim2-F and DralΔLim2-R (see Suppl. Table1) on the plasmid pcDNA3-DRAL-3xFlagM2 as matrix. A pcDNA3 plasmid encoding HA-tagged DRAL from chicken was generated by PCR on cDNAs of one day chick embryo by using the primers Chick-DRAL-F and Chick-DRAL-R given in Suppl. Table1. This plasmid was used as matrix to amplify by PCR DRAL-encoding sequence using the primers pCAGGS-Chick-FRAL-XhoI-F and pCAGGS-Chick-FRAL-BglII- R (see Supp. Table1). The PCR product was inserted into pCAGGS vector. The pcDNA3-TUCANΔNTer plasmid was obtained by PCR using the primers TUCAN ΔNTerm- F and TUCAN ΔNTerm-Myc-R (see Suppl. Table1) on the pcDNA3-TUCAN-Myc plasmid. For the in ovo chick electroporation, most constructs were based in pCAGGS which was also used as empty vector. pCAGGS-caspase-9 dominant negative has been constructed by subcloning from pcDNA3-caspase-9 dominant negative 13 into pCAGGS vector. Primers to construct pSilencer1.0-U6 containing small interfering hairpins of DRAL mRNA were designed thanks to Promega siRNA Designer program.
Cell cultures, transfection procedures, reagents and immunoblots
Transient transfection of Human Embryonic Kidney 293T cells (HEK293T) and Human Embryonic Kidney 293 cells expressing Muristerone A-inducible Shh (HEK293-EcRShh) were performed as previously described 5. Daoy cells (ATCC, #HTB-186) were grown in 10% DMEM media on poly-L-lysine (Sigma) coated round coverslip. Primary cultured cerebellar granule neurons (CGNs) were prepared from P6-aged mice (NMRI). 5e1 hybridoma cells producing a Shh blocking antibody (Developmental Studies Hybridoma Bank), were maintained in 20% DMEM media. Immunoblots were performed as described previously 6 using different source and dilution of antibodies described in Suppl. Methods. Recombinant Shh-N was from R&D systems. To induce Shh production by HEK293-EcRShh cells, Muristerone A (A.G. Scientific, inc) was used (1 μM).
siRNA transfection
For siRNA experiments, cells were transfected with 60 pmols siRNA total for a single siRNA transfection, or with 120 pmols siRNA total for double siRNA transfection. DRAL, caspase-8, caspase-9, NALP1, scramble siRNAs were from Santa Cruz Biotechnology; TUCAN siRNA was from Dharmacon.
Yeast two-hybrid screen and co-immunoprecipitation
Matchmaker two-hybrid system III (Clontech) was used according to the manufacturer’s instructions using the DNA binding domain GAL4 fused to Ptc 1165–1392 as a bait and the GAL4 transcriptional activation domain AD fused to a human fetal brain cDNA library (Clontech) as prey. In cellulo co-immunoprecipitation were carried out on HEK293T or HEK293-EcRShh cells transfected with various tagged constructs as described previously 13. For experiment requiring Shh, cells were treated or not with Muristerone A when plated and at the time of transfection, and recombinant Shh was added at a final concentration of 300 ng/ml 24 hours before harvesting the cells. In vivo immunoprecipitation were performed in OF-1 mice cortex embryos. Two cortices from E16.5 embryos were dissected out, cut in small species and triturated. Shh-treated cortices were incubated 40 minutes at 37°C in conditioned supernatant of HEK293- EcRShh-induced cells in which recombinant Shh-N (R&D systems) was also added at 900 ng per cortex. Non-treated cortices were incubated in the same condition in supernatant of non-induced HEK293-EcRShh cells and without addition of recombinant Shh-N. Dissociated cortices were further processed for immunoprecipitation using μMACS Protein G MicroBeads system (Miltenyi Biotec).
Co-localization studies
After transfection or treatment described in the supplementary materials, HEK293T, Daoy or CGNs cells were fixed in 4% paraformaldehyde for 30 minutes, washed and permeabilized with Triton 0.2 % in PBS 30 minutes. The coverslips were mounted in crystal/mount (Biomeda) and photographed with a Zeiss Axioplan2 LSM510 confocal microscope. Profiles were created using Axiovision 4.0V4.6.1.0 program.
Gel filtration
Gel filtration were performed as described in supplementary materials using size-exclusion chromatography on High Prep ™ Sephacryl ™ S300 (HR 16/60) columns using an FPLC protein purification system (Biologic HR, Biorad). Proteins were separated with the lysis buffer. Columns were calibrated with protein standards (29 kDa- 2000 kDa) according to the manufacturer’s instructions (Sigma). Aliquots of the fractions were analyzed on Criterion XT Precast gels (Bis-Tris 4–12%; Biorad) and Western blotting. Aliquots of the 20th first fractions were also pooled and used for co-immunoprecipitation experiments as described for in cellulo co-immunoprecipitation.
Cell death analysis and caspase assays
Cell death was analyzed 24h after transfection using trypan blue staining procedures as described previously 5. Cell death was also analyzed using Toxilight assay according to the manufacturer’s instructions (Lonza). For caspase-9 activity fluorometric assay, HEK293-EcRShh cells were treated twice (when seeded and when transfected) with Muristerone A for the condition with Shh, and recombinant Shh-N was added (300 ng/ml) 1 hour before harvesting the cells. Caspase-9 activity was then measured by using the caspase-9 fluorometric assay kit (BioVision). Caspase-3 activity was measured by using the caspase-3 fluorometric assay kit (BioVision) as described previously 5.
Chick in ovo experiments and TUNEL staining on chick embryos
Cell death analysis in the developing chick neural tube was performed using fertilized chicken eggs obtained from a local farm and incubated at 38°C. Embryos were staged according to Hamburger and Hamilton (HH). Chick embryos were electroporated with Sigma purified plasmid DNA (4.5 μg/μl) with 50 ng/ml Fast Green. For floor plate electroporation, chick embryos of stage HH10–11 were injected with plasmid DNA into the lumen of the neural tube and electroporated with specifically designed electrodes. In each condition, Green Fluorescent Protein (GFP) was co-electroporated to check the electroporation efficiency. Chick microsurgery was done as described in 5, except that the electroporation performed before microsurgery was done in only one side of the neural tube. Detection and quantification of apoptosis by TUNEL was performed using the in situ cell TUNEL detection kit from Roche as previously described 3. For each embryo, TUNEL-positive cells of electroporated and non-electroporated side of the neural tube were added for more than six sections with adequate GFP expression. The ratio between the number of TUNEL-positive cells from the electroporated side to the non-electroporated side was then calculated. For each condition, results of at least five embryos were used to calculate a mean of the ratio. A U test was done to evaluate if the difference between various conditions was significant. Fluorescent TUNEL was done as described in 30.
NF-kB luciferase reporter assays
Analysis of NF-kB activation was performed according classic luciferase reporter assays as described in the Supplementary Methods.
Supplementary Material
Acknowledgments
We thank J. Briscoe and N. Le Douarin for advice and reagents, B.W. Schafer, C. Sardet and R. Sadoul for reagents, and K. Cywinska for her technical work on immunoprecipitation experiments. We thank L. Kremer from the Protein tools service at the Centro Nacional de Biotecnologia of Madrid for 5e1 hybridoma cell line. This work was supported by the Ligue Contre le Cancer, the Agence Nationale de la Recherche, the Institut National du Cancer, the Rhône-Alpes Region, the Centre National de la Recherche Scientifique, the EU grants Hermione and APO-SYS and the NIH. F.M. was supported by a Rhône-Alpes Region and an Association pour la Recherche sur le Cancer fellowships.
References
- 1.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 2.Murone M, Rosenthal A, de Sauvage FJ. Sonic hedgehog signaling by the patched-smoothened receptor complex. Curr Biol. 1999;9:76–84. doi: 10.1016/s0960-9822(99)80018-9. [DOI] [PubMed] [Google Scholar]
- 3.Charrier JB, Lapointe F, Le Douarin NM, Teillet MA. Anti-apoptotic role of Sonic hedgehog protein at the early stages of nervous system organogenesis. Development. 2001;128:4011–4020. doi: 10.1242/dev.128.20.4011. [DOI] [PubMed] [Google Scholar]
- 4.Litingtung Y, Chiang C. Specification of ventral neuron types is mediated by an antagonistic interaction between Shh and Gli3. Nat Neurosci. 2000;3:979–985. doi: 10.1038/79916. [DOI] [PubMed] [Google Scholar]
- 5.Thibert C, et al. Inhibition of neuroepithelial patched-induced apoptosis by sonic hedgehog. Science. 2003;301:843–846. doi: 10.1126/science.1085405. [DOI] [PubMed] [Google Scholar]
- 6.Mehlen P, et al. The DCC gene product induces apoptosis by a mechanism requiring receptor proteolysis. Nature. 1998;395:801–804. doi: 10.1038/27441. [DOI] [PubMed] [Google Scholar]
- 7.Bredesen DE, Mehlen P, Rabizadeh S. Apoptosis and dependence receptors: a molecular basis for cellular addiction. Physiol Rev. 2004;84:411–430. doi: 10.1152/physrev.00027.2003. [DOI] [PubMed] [Google Scholar]
- 8.Mehlen P, Bredesen DE. The dependence receptor hypothesis. Apoptosis. 2004;9:37–49. doi: 10.1023/B:APPT.0000012120.66221.b2. [DOI] [PubMed] [Google Scholar]
- 9.Tauszig-Delamasure S, et al. The TrkC receptor induces apoptosis when the dependence receptor notion meets the neurotrophin paradigm. Proc Natl Acad Sci U S A. 2007;104:13361–13366. doi: 10.1073/pnas.0701243104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furne C, et al. EphrinB3 is an Anti-apoptotic Ligand that Inhibits the Dependence Receptor Functions of EphA4 Receptors during adult neurogenesis. BBA Biol Mol Cell. 2008 doi: 10.1016/j.bbamcr.2008.09.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn H, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 12.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 13.Forcet C, et al. The dependence receptor DCC (deleted in colorectal cancer) defines an alternative mechanism for caspase activation. Proc Natl Acad Sci U S A. 2001;98:3416–3421. doi: 10.1073/pnas.051378298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmermann KC, Green DR. How cells die: apoptosis pathways. J Allergy Clin Immunol. 2001;108:S99–103. doi: 10.1067/mai.2001.117819. [DOI] [PubMed] [Google Scholar]
- 15.Johannessen M, Moller S, Hansen T, Moens U, Van Ghelue M. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell Mol Life Sci. 2006;63:268–284. doi: 10.1007/s00018-005-5438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholl FA, McLoughlin P, Ehler E, de Giovanni C, Schafer BW. DRAL is a p53-responsive gene whose four and a half LIM domain protein product induces apoptosis. J Cell Biol. 2000;151:495–506. doi: 10.1083/jcb.151.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stilo R, et al. TUCAN/CARDINAL and DRAL participate in a common pathway for modulation of NF-kappaB activation. FEBS Lett. 2002;521:165–169. doi: 10.1016/s0014-5793(02)02869-7. [DOI] [PubMed] [Google Scholar]
- 18.Razmara M, et al. CARD-8 protein, a new CARD family member that regulates caspase-1 activation and apoptosis. J Biol Chem. 2002;277:13952–13958. doi: 10.1074/jbc.M107811200. [DOI] [PubMed] [Google Scholar]
- 19.Pathan N, et al. TUCAN, an antiapoptotic caspase-associated recruitment domain family protein overexpressed in cancer. J Biol Chem. 2001;276:32220–32229. doi: 10.1074/jbc.M100433200. [DOI] [PubMed] [Google Scholar]
- 20.Charrier JB, Teillet MA, Lapointe F, Le Douarin NM. Defining subregions of Hensen’s node essential for caudalward movement, midline development and cell survival. Development. 1999;126:4771–4783. doi: 10.1242/dev.126.21.4771. [DOI] [PubMed] [Google Scholar]
- 21.Stamataki D, Ulloa F, Tsoni SV, Mynett A, Briscoe J. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev. 2005;19:626–641. doi: 10.1101/gad.325905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tschopp J, Martinon F, Burns K. NALPs: a novel protein family involved in inflammation. Nat Rev Mol Cell Biol. 2003;4:95–104. doi: 10.1038/nrm1019. [DOI] [PubMed] [Google Scholar]
- 23.Liu F, et al. Expression of NALP1 in cerebellar granule neurons stimulates apoptosis. Cell Signal. 2004;16:1013–1021. doi: 10.1016/j.cellsig.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Kinoshita T, Wang Y, Hasegawa M, Imamura R, Suda T. PYPAF3, a PYRIN-containing APAF-1-like protein, is a feedback regulator of caspase-1- dependent interleukin-1beta secretion. J Biol Chem. 2005;280:21720–21725. doi: 10.1074/jbc.M410057200. [DOI] [PubMed] [Google Scholar]
- 25.Faustin B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–724. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 26.Boatright KM, et al. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 27.Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 28.Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- 29.Chu ZL, et al. A novel enhancer of the Apaf1 apoptosome involved in cytochrome c-dependent caspase activation and apoptosis. J Biol Chem. 2001;276:9239–9245. doi: 10.1074/jbc.M006309200. [DOI] [PubMed] [Google Scholar]
- 30.Furne C, Rama N, Corset V, Chedotal A, Mehlen P. Netrin-1 is a survival factor during commissural neuron navigation. Proc Natl Acad Sci U S A. 2008;105:14465–14470. doi: 10.1073/pnas.0803645105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


