Abstract
In many cell divisions, the position of the spindle apparatus is coordinated with polarity signals at the cell cortex so that copies of the genome are delivered to regions of the cell that are designated for differential inheritance by the two progeny. To coordinate spindle position with cell polarity, the spindle interfaces with elements on the cortex, where molecular motors often produce the forces that power displacement. Here we describe the molecular pathways by which cortical motors translocate the spindle in budding yeast, where the mechanisms are understood relatively well, and we compare these pathways to spindle positioning processes in metazoan systems, where the molecular details are less well understood.
Keywords: spindle, cytoplasmic microtubules, myosin, kinesin, dynein
1. Positioning the spindle within a dividing cell
The position of the mitotic spindle during cell division is rarely, perhaps never, random. Many eukaryotic cells undergo divisions in which the distribution of the duplicated genomes must be reconciled with an asymmetric feature of the dividing cell. This asymmetry is particularly critical during cell differentiation events that accompany development in the embryo and tissue homeostasis in the adult. In these scenarios, spindle movement is directed by crosstalk between the cell periphery and the microtubule cytoskeleton, and the power for spindle movement comes from molecular motors.
1.1 Spindle position and polarity in metazoan tissue organization
The organization of metazoan tissues is often dependent on the positioning of spindles in response to cues from the microenvironment. One example is neurogenesis in the Drosophila melanogaster embryo, where a program of spindle movements helps to generate the progenitors of the nervous system. Epithelial cells within the neuroectoderm may either maintain an epidermal fate by dividing symmetrically along the plane of the epithelium; or they may delaminate from the epithelium to become neuroblasts, which then divide asymmetrically to produce differentiating ganglion mother cells. During symmetric division, adherens junctions between epithelial cells maintain spindle orientation along the planar axis [1]. In contrast, delaminated neuroblasts shift the orientation of the spindle perpendicular to the planar axis, and maintain this orientation during subsequent asymmetric divisions through interactions with adjacent epithelial cells [2, 3]. Mutations that affect spindle orientation and position have increased numbers of neuroblasts, suggesting that differentiation has failed [4]. Another example is the stratified epithelium of the mammalian epidermis. Here, proliferative basal cells replenish the population of outer barrier cells by dividing perpendicular to the basement membrane [5]. This polarity depends on adherens junctions between basal cells and also interactions with the basement membrane; disruption of either structure leads to mis-aligned divisions and loss of epidermal organization [5].
Cells cultured in vitro can retain the ability to coordinate spindle orientation with external cues, providing insight into the underlying mechanisms. Epithelial cells grown to confluence on synthetic substrate orient their spindles parallel to plane of the monolayer [6]. Isolated epithelial cells lack cues from cell-cell contacts, but the cells still alter their spindle position in response to substrate interactions. This phenomenon is exemplified by cells grown on micropatterns of extracellular matrix, which stimulate spindle orientation toward regions of stronger cell adhesion in a manner dependent on the organization of the cortical actin cytoskeleton [7, 8].
Intestinal epithelial cells in 3-D culture form cysts by establishing an apical domain during the first cell division. The cells divide perpendicular to the apical-basal axis in subsequent divisions, resulting in a sheet of cells organized around a single fluid-filled lumen [9]. The small GTPase Cdc42, which has diverse roles in regulating cell polarity and actin organization, coordinates spindle orientation with the location of the apical domain in this context [9]. Together, these studies suggest a common theme where adhesive interactions with the external microenvironment stimulates the reorganization of the actin cytoskeleton, which in turn directs the position of the spindle.
1.2 Generating polarity to guide spindle position in single cells
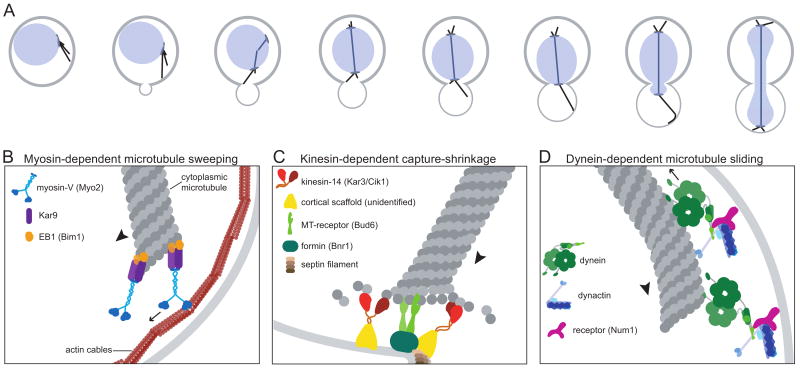
Spindle positioning programs can also be guided by intrinsic polarity cues. In single cells that divide asymmetrically, polarity is derived either from pre-existing landmarks or from stochastic symmetry-breaking events. Every cell division is asymmetric in the budding yeast, Saccharomyces cerevisiae. Daughter cells emerge from a site on the mother cell specified by local enrichment of Cdc42 activity, which stimulates the reorganization of the actin network to fuel polarized growth and transport into the bud [10]. The contents of the mother cell that are necessary for survival of the daughter must be transported through the mother-bud junction, termed the bud neck. One copy of the genome is provided to the daughter by delivering one end of the spindle and a portion of the nucleus into the bud (the nuclear envelope remains intact throughout the yeast cell cycle, therefore the spindle is contained within the nucleus; Fig.1A). The translocation of the spindle and nucleus through the bud neck requires that the spindle is first oriented along the mother-bud axis, and then one end is drawn into the bud. Only after the spindle is properly positioned are the two cells separated, by the processes of cytokinesis and septation at the bud neck.
Figure 1.
Models for spindle displacement by cortical motors. (A) Sequence of spindle positioning events during cell division in budding yeast. First, the plus ends of one set of microtubules are delivered to the bud. Second, microtubules that are attached to the bud neck shorten, drawing the spindle and nucleus toward the bud. Finally, microtubules are pulled along the bud cortex, toward the distal bud tip. This pulls the spindle and nucleus into the bud neck, and positions the anaphase spindle across the nascent site of cytokinesis. (B) Myosin-V transports microtubule plus ends along actin cables toward the bud. Arrow indicates the direction of myosin-V motility, determined by the polarity of actin cables that are nucleated at the bud tip and bud neck. Arrowhead indicates the force applied to the microtubule. (C) Capture-shrinkage at the bud neck. Microtubule plus ends are captured by the forming-interacting protein, Bud6, and are then induced to shorten by a mechanism that is likely to involve the depolymerase activity of kinesin-14. (D) Microtubules are pulled along the bud cortex by the minus-end directed motor dynein and its activator, dynactin. The dynein-dynactin complex is anchored to the cortex by Num1.
The first mitotic division during Caenorhabitis elegans development also relies on reorganization of the cortical actin cytoskeleton to direct the positioning of the spindle. Prior to division, the cortex of the one-cell embryo is covered with a meshwork of actin filaments under symmetrically distributed contractile force generated by the type-II non-muscle myosin motor and its activator, the small GTPase Rho [11-13]. Symmetry is broken by dampening contractility at a region of the cortex, and this appears to be cued by the location of sperm entry. The site of polarization correlates with the location and function of the sperm centrosomes and depends on the inhibition of Rho by the paternally-derived RhoGAP, CYK-4 [14-16]. Upon polarization, Rho and its activator, the RhoGEF ECT-2, are lost from this region, and the actomyosin network retracts away [12, 13]. Cdc42 subsequently promotes reorganization of the actomyosin network on the anterior half of the cell cortex, and translates this polarity into the differential recruitment of the PAR proteins [13, 17-19]. PAR protein polarity establishes anterior-posterior domains with signaling properties that guide the orientation of the spindle along the long axis of the cell and subsequently displace the spindle toward the posterior end in order to produce a larger and smaller daughter cell after cytokinesis [20-22].
1.3 The role of cytoplasmic microtubules
In each of the examples above, the spindle responds to cortical polarity through the action of the cytoplasmic, or astral, set of microtubules. Whereas the spindle microtubules separate the two copies of the genome, the cytoplasmic microtubules mediate interactions between the spindle and the cell periphery. The minus ends of cytoplasmic microtubules are anchored to microtubule-organizing centers (MTOCs), and are thereby tethered to the two poles of the spindle. The dynamic plus ends project out from the spindle and into the cytosol, where they undergo cycles of polymerization and depolymerization via dynamic instability. This behavior allows cytoplasmic microtubules to survey the cell cortex for binding partners. Upon binding to the cortex, force applied to cytoplasmic microtubules is transmitted to the spindle, producing displacement. Molecular motors generate this force in various ways.
2. Motors that generate force at the cell cortex
Molecular motors displace the microtubule network by applying tension to cytoplasmic microtubules at contact sites on the cell cortex. Time-lapse imaging of GFP-labeled tubulin in budding yeast has been useful for understanding how microtubules interact with the cortex. Yeast have only a few cytoplasmic microtubules at each SPB/MTOC, which simplifies resolution and allows for correlation between spindle movement and the behavior of individual microtubules. The displacement of the spindle and nucleus is accompanied by three classes of interactions between microtubules and the cell cortex[23-25]; Fig.1B-D). In the first class, growing microtubule plus ends collide with the cortex and then sweep along it while maintaining end-on contact. As the plus end moves across the cortex, the microtubule acts as a lever, applying torque to the microtubule network, which produces rotational movement of the MTOC. Thus, sweeping interactions are effective for concentrating microtubule plus ends at specific regions of the cortex and orienting the microtubule network toward these sites. In the second class of interactions, microtubule plus ends are attached, or captured, at fixed sites on the cell cortex, and they maintain end-on contact with these sites as they depolymerize from their plus ends. The shortening of the microtubule draws the MTOC and the attached microtubule network toward the site of cortical capture, pulling the spindle toward and into the neck. In the third class of interactions, microtubules exhibit lateral sliding along the cortex, maintaining contact with the cortex along their sides. The sliding of individual microtubules powers the movement of the MTOC and microtubule network toward the site of interaction on the cortex, drawing the spindle into the neck.
2.1 Myosin V – transporting microtubule ends along actin cables
The sweeping of microtubule ends along the cell cortex depends on cooperation between cytoplasmic microtubules and the cortical actin network (Fig.1B). In this mechanism, actin filaments are the substrate for motility, and microtubule ends are cargo. Budding yeast formins nucleate cables of actin filaments at the bud tip and neck; these cables cascade into the mother, creating bud-directed tracks for type-V myosins [26]. Other cargoes of this transport network include endoplasmic reticulum, mitochondria, secretory vesicles, mRNAs, and various proteins that contribute to the fitness of the daughter cell. Mutations and pharmacological perturbations that disrupt actin cables impair the delivery of microtubule ends to the bud, and delay the alignment of the spindle along the mother-bud axis [27-30]. Mutations in the type-V myosin, Myo2, elicit similar defects [30, 31]. Notably, reducing the velocity of Myo2 by truncating its lever arm also reduces the velocity of microtubule sweeping, providing strong evidence that Myo2 is the motor responsible for this process [32]. Consistent with this observation, GFP-labeled Myo2 is frequently detected at microtubule plus ends that are not in contact with the cortex [32]. It is not clear whether this represents a pool of Myo2 that associates with the microtubule end prior to its arrival at the cortex, or whether it is a remnant at the plus end, which remains after the microtubule disengages from the cortex. In either case, the observation reveals the presence of a motor complex at the plus end that is competent for motility along actin filaments.
Microtubule ends associate with Myo2 via a pathway involving the APC homologue Kar9 and the EB1 homologue Bim1, which are two of the numerous so-called “+TIP” proteins found near the plus end of the microtubule. Kar9 acts as an adapter by interacting simultaneously with Myo2 and Bim1 [29, 31-34]. Deletion of BIM1 abolishes the localization of Kar9 to cytoplasmic microtubules, suggesting that Kar9 may only associate with microtubules when in complex with Bim1 [29, 33, 34].
The localization of Kar9 at plus ends is important for proper function. Dynamically growing plus ends have a greater probability of colliding with the cortex and therefore encountering cortical Myo2 and/or actin cables than do the sides of microtubules. Consistent with this notion, the activity of the microtubule-sweeping mechanism correlates with the accumulation of Kar9 at plus ends. Kar9 localizes to the SPB during the G1 phase of the cell cycle, but moves out to the plus end during S phase and G2, when microtubule-sweeping is active [35]. The transfer of Kar9 from SPBs to plus ends is enhanced by the kinesin Kip2 and involves the plus end tracking proteins Bik1/CLIP-170 and Stu2/XMAP215 [35-37]. Fluorescence recovery after photobleaching (FRAP) experiments suggest that plus end-associated Kar9 is derived from the pool at the SPB, and not vice versa [38].
One potentially critical feature of Kar9 activity is its biased localization to microtubules emanating from one of the two SPBs. This selective recruitment is initiated at the SPB and requires the phosphorylation of Kar9 by the yeast CDK1/Cdc28 and several B-type cyclins [35, 38, 39]. Mutations that disrupt cyclin/CDK1 activity or phosphorylation sites on Kar9 allow localization to both sets of microtubules [37-39]. Under these conditions, microtubules from either SPB may be oriented toward the bud [36, 38]. Phospho-mimetic mutations rescue Kar9 asymmetry and spindle orientation, suggesting that phosphorylation by cyclin/CDK1 allows Kar9 to recognize a pre-exisiting asymmetry between the two SPBs [37]. The basis of this asymmetry is not yet clear. GFP-labeled Myo2 also exhibits asymmetric localization to the ends of microtubules emanating from one SPB, and this depends on Kar9 [32]. Thus, the selective recruitment of Kar9 to a specific set of microtubule plus ends appears to “license” those microtubules for myosin-dependent transport, thereby ensuring that only one end of the spindle is oriented toward the bud.
Although the microtubule-sweeping mechanism is well-characterized in budding yeast, it remains to be seen whether similar mechanisms contribute to spindle orientation in other organisms. The Bim1-Kar9 complex is regarded as the functional homologue of the EB1-APC complex, which connects microtubule ends to cortical polarity in animal cells [1, 40, 41]; however, EB1-APC is not known to transport microtubule ends along the cortex via myosin-V motors. Such a mechanism may not be necessary in cells that experience a greater frequency of microtubule-cortex interactions than do budding yeast cells, because this would increase the probability that microtubule ends will encounter docking sites on the cortex. For example, the C. elegans zygote possesses 100-fold more cytoplasmic microtubules than do budding yeast cells, and as many as 200 of these microtubules are in contact with the cortex at any single time point during mitosis [42]. In this scenario, spindle orientation may be accomplished by the stochastic capture of microtubule ends by alternative force generators that are confined to specific subdomains of the cortex.
2.2 Microtubule-depolymerizing kinesins – capture-shrinkage at the cortex
Tension can also be generated at the cortex by receptor complexes that couple microtubule attachment with depolymerization, known as capture-shrinkage (Fig.1C). Here, growing microtubule ends are captured by receptors and then undergo catastrophe - the transition to a depolymerization state. During depolymerization, the receptor remains processively bound to the shortening microtubule, resulting in the movement of the minus end and the attached microtubule network toward the cortical contact site. This model recalls capture-shrinkage mechanisms that produce chromosome movements by tethering kinetochores to depolymerizing microtubule ends (see review in this issue). In both scenarios, the force that produces movement may be derived from microtubule depolymerization itself; and therefore the motor would consist of the force generator (the depolymerizing microtubule end) and the receptor that tethers the cargo (the cell cortex or kinetochore) to the microtubule. In this mechanism, the motor protein may act only as a tether, not as a motor per se.
Two key features of this mechanism are how capture sites are defined on the cortex and how microtubule depolymerization is exploited for motility. In yeast, capture-shrinkage events are biased toward the bud, drawing the pre-anaphase spindle toward the bud neck [23, 25]. This result indicates that although microtubule plus ends contact sites across the cell cortex, force-producing depolymerization events are limited to contacts at the bud neck and bud cortex. The establishment of capture-shrinkage sites requires the ring of septin filaments around the bud neck, which provides a scaffold for various signaling and transport pathways [43]. Another key component of this process is Bud6, a formin-binding protein that localizes to the bud neck and bud cortex and mediates microtubule capture at these sites [44-46]. While this mechanism does not require Kar9 for microtubule capture or depolymerization, the Myo2-Kar9-dependent sweeping mechanism enhances the efficiency of capture by delivering microtubule plus ends to the bud neck and bud [43, 45].
The component(s) of the capture-shrinkage mechanism that couple microtubule depolymerization and spindle movement toward the neck are poorly understood; however, kinesin motors that induce depolymerization are attractive candidates. Kar3 is a member of the kinesin-14 family of motors that was originally identified in a screen for mutations that disrupt nuclear fusion (karyogamy) during the mating of haploid yeast cells [47]. In addition, Kar3 has important roles in spindle function and positioning, which are differentially regulated by its light chains, Vik1 and Cik1 [48-50]. Loss-of-function alleles of kar3 exhibit longer cytoplasmic microtubules and are defective for positioning the pre-anaphase spindle at the bud neck; consistent with a disruption of the capture-shrinkage mechanism [48, 51]. cik1 mutants exhibit similar spindle positioning defects [48]. In vitro, Kar3-Cik1 complexes exhibit minus-end-directed motility in microtubule gliding assays, and depolymerization activity at plus ends [52]. Studies of Kar3 in mating yeast demonstrate how these activities may generate tension on cytoplasmic microtubules. During mating, activation of a G protein-coupled pheromone receptor triggers polarized growth, or schmooing. Cytoplasmic microtubules are delivered to this site by the Bim1-Kar9-Myo2 complex [29, 32, 53-55]. Kar3-Cik1 localizes to the plus ends of these microtubules, and mediates the formation of stable attachments with the Gα-subunit, Gpa1, at the schmoo cortex [56, 57]. Microtubules then shorten, bringing the SPB and nucleus move toward the site of polarization [56]. Null mutations of kar3 disrupt the attachment of cytoplasmic microtubules to the schmoo cortex; and mutations that specifically disrupt the ATPase activity of Kar3, which are predicted to lock the motor in a rigor state to its microtubule substrate, allow microtubules to attach to the cortex but not shorten [56]. These results support a dual function model for Kar3, wherein the kinesin first mediates the tethering of microtubule ends to cortical receptors, and then induces depolymerization via its ATPase activity.
Kip3, a member of the kinesin-8 family, may also be involved in the capture-shrinkage mechanism in budding yeast. Kip3 promotes depolymerization in vitro, and kip3 mutant cells exhibit long microtubules and defective positioning of the pre-anaphase spindle [58]. Nevertheless, capture-shrinkage events are still apparent in these mutants, indicating that Kip3 is not necessary for this process. Instead, the role of this kinesin may be to prevent continued microtubule growth from pushing the spindle away from the bud and back into the mother.
Depolymerizing microtubules may also provide the force for spindle movement in the C. elegans zygote. Elegant studies using laser microbeams to fragment the spindle or centrosomes during mitosis demonstrate that pulling force is applied to cytoplasmic microtubules from a limited number of sites on the cortex, with a greater magnitude of force generated at the posterior end of the cell [59, 60]. Imaging of microtubules at the cortex shows both end-on and lateral microtubule contacts during force generation. The appearance of end-on contacts is generally followed quickly by depolymerization away from the cortex. Microtubule ends exhibit greater dwell time at the posterior cortex; and this asymmetry depends on the PAR proteins [61]. During spindle movement, the number of end-on contacts transiently increases in the front and decreases in the rear [42]. Spindle movement can be inhibited by the microtubule-stabilizing drug, taxol, or by tubulin mutations that suppress microtubule dynamics [62]. Together these results in the C. elegans zygote are consistent with a capture-shrinkage mechanism, but confirming this model in vivo is challenging. The simultaneous contact of many microtubules with the cortex confounds the determination of the relevance of individual interactions. Furthermore, the presence of transient end-on interactions and lateral sliding interactions along the cortex raises the question of which type of interaction has the primary causative role in force production. A role for microtubule sliding has also been proposed in this system [63]. Distinguishing between these models will require the identification and disruption of factors that uniquely support either interaction. A role for depolymerizing kinesins in moving the spindle of the C. elegans zygote has not been established; however, the microtubule motor dynein is required for spindle orientation and displacement. The involvement of dynein suggests that sliding may be involved, based on the mechanisms of force generation by dynein in budding yeast, discussed next.
2.3 Dynein – microtubule sliding
In the third mechanism that operates in budding yeast, microtubules slide along the cortex, drawing the spindle and nucleus into the bud neck (Fig.1D). Sliding events are initiated by the collision of a growing plus end with the cortex. The microtubule then curves along the cell cortex, maintaining contact along its side while moving toward its distal plus end.
Microtubule sliding requires the dynein motor. Cytoplasmic dynein is a large, multi-subunit complex necessary for microtubule-based processes in nearly all eukaryotes. Loss-of-function mutations in yeast dynein abrogate sliding events and delay the movement of the nucleus and spindle into the bud, consistent with the notion that sliding is powered by dynein motility [25, 64-67]. In dynein mutant cells, cytoplasmic microtubules are longer and exhibit a decrease in the frequency of dynamic end-on interactions [23]. This suggests that dynein may influence microtubule dynamics and/or the stability of microtubule-cortex interactions as part of its function.
Several lines of evidence indicate that microtubule sliding is driven by the minus end-directed motility of dynein motors at the cortex. Fluorescently labeled components of the dynein complex appear as punctae along microtubules and at stationary foci on the cortex, and mutations that abolish the cortical localization of dynein also abolish microtubule sliding [68-70]. These results suggest that dynein must associate with a cortical receptor in order to produce force. The model of dynein-driven sliding is also supported by observations on free cytoplasmic microtubules, which have detached from the SPB and then appear to slide along the cortex [25]. Two-color experiments in which α-tubulin and the minus end-capping γ-tubulin complex are differentially labeled demonstrate that free microtubules slide in a plus end-first orientation, consistent with cortical dynein stepping toward the minus end [71]. Moreover, the movement of fiducial fluorescent speckles indicates that free microtubules slide at velocities similar to those observed for purified yeast dynein in in vitro microtubule gliding assays [25, 72]. Together these data argue that dynein generates the force for sliding at the microtubule-cortex interface.
Genetic screens in yeast have identified a number of genes that are required for the dynein mechanism. Nearly all of the genes identified in these screens encode dynein regulators that are conserved across species. Based on this conservation, yeast has proven to be a powerful system for identifying the molecular contribution of these components to dynein function (reviewed in [73]). Current evidence supports a three-step model for activating microtubule sliding. First, the dynein motor complex is targeted to the plus ends of dynamic cytoplasmic microtubules. This requires Bik1/CLIP170 and the kinesin Kip2, along with the LIS1 and NudEL homologues, Pac1 and Ndl1 [68, 74-77]. By hitchhiking with these proteins to the plus end, dynein at the plus end may then probe the cortex in search of its cortical receptor - the PH domain-containing protein, Num1 [78-81].
Second, upon encountering Num1, dynein is anchored to the cortex through a mechanism that requires the dynactin complex. Dynactin is broadly conserved and required for perhaps every function of dynein in cells [82]. Yeast dynactin co-localizes with dynein on microtubules, and dynactin appears to transfer from plus ends to the cortex along with dynein [70]. In dynactin mutants, dynein accumulates at plus ends but is not found at the cortex [68, 74]. This may be analogous to the role of dynactin in linking dynein to vesicular cargoes; only in this case, the cortex is effectively the cargo.
Third and finally, with dynein bridging the microtubule and the cortex, the motor is activated and walks toward the minus end, sliding the microtubule past the anchor site. The timing of this last step is critical; activation prior to cortical docking would promote the streaming of dynein away from the plus end. The mechanism for activating dynein at the cortex is not understood; however, it is tempting to speculate that the activator may reside at the cortex, awaiting the arrival of dynein and the microtubule end.
The role of dynein in generating force for spindle movement is conserved in many eukaryotes. Dynein contributes to several types of spindle movement in the C. elegans zygote. Depletion of the dynein heavy chain subunit by RNAi delays the alignment of the spindle along the long axis of the cell [83]. In addition, either loss-of-function mutations of dynein heavy chain or RNAi-depletion of dynein components reduces the pulling forces that are applied to the spindle poles during mitosis [62, 84-87]. Similar observations have been made in Drosophila neuroblasts, where mutations that disrupt the function of Lis1 or the dynactin component, p150glued, exhibit defects in spindle alignment and subsequent movements [88]. Studies in these models have been particularly useful for uncovering the signaling pathways may translate cortical polarity into asymmetric force generation by dynein. For more information on this subject, the reader is directed to recent reviews [89, 90].
Given that dynein-dependent spindle movement coincides with microtubule sliding in the yeast model and with microtubule shortening in C. elegans, an important question is whether the mechanism of force generation differs in these two contexts. Purified dynein motors exhibit step-wise motility predominantly directed towards the minus ends of microtubules, in vitro [72]. To our knowledge, there is no biochemical evidence that dynein acts as a microtubule depolymerase. It is possible that the minus-end directed motility of dynein is coupled with a separate depolymerase module in some cells. This notion is reminiscent of the mechanism that produces movement of the MTOC and nucleus during meiosis in the fission yeast, Schizosaccharomyces pombe. Here, dynein-dependent microtubule sliding pulls the MTOC and nucleus back-and-forth between the two ends of the elongated cell [91, 92]. During nuclear movement, the leading microtubule extends to the opposite end of the cell, and that microtubule shortens from its plus end-cortex interface as the MTOC and nucleus move forward [93]. Laser cutting experiments demonstrate that depolymerization at the plus end accompanies but is not necessary for force generation. When the leading microtubule is cut in half, the microtubule that remains attached to the MTOC and nucleus continues to slide forward at a similar speed [92]. Interestingly, dynein mutants exhibit reduced depolymerization rates at the cell ends, suggesting that depolymerization is enhanced by dynein or perhaps by compressive force of the sliding microtubule plus end colliding with the cortex [93]. Thus, sliding and depolymerization appear to be components of a common dynein mechanism. Whereas the role of sliding is to produce force for motility, depolymerization may relieve steric constraints as the microtubule approaches the cortex.
3. Concluding Remarks
Here we have discussed how molecular motors may alter the orientation and position the mitotic spindle during cell division. The fact that a simple organism such as budding yeast appears to employ three evolutionarily distinct classes of motors in this process raises the question of what advantages such diversification might confer. Multiple mechanisms could increase the robustness of spindle positioning through compensatory functions. Consistent with this notion, mutations that disrupt individual spindle positioning mechanisms in budding yeast do not impair viability; however, simultaneous disruption of multiple pathways results in severe growth defects [29, 48, 54, 94]. In addition, each mechanism may provide unique functions that improve the efficiency of spindle positioning. For instance, the microtubule-sweeping mechanism is likely to be more sensitive to actin-based cell polarity, whereas the microtubule-sliding mechanism may allow for greater force generation by providing more binding sites for motors along the sides of the microtubule. An important test of these models will be the identification of distinct mechanisms dedicated to specific modes of spindle movement in higher eukaryotes.
Acknowledgments
The writing of this review and research in our lab on this topic was supported by NIH R01 GM47337. We are grateful to Melissa D. Stuchell-Brereton for critical reading of this manuscript.
Abbreviations used
- MTOC
microtubule organizing center
- SPB
spindle pole body
- CLIP-170
cytoplasmic linker protein-170
- EB1
end-binding protein 1
- GFP
green fluorescent protein
- PH
pleckstrin homology
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lu B, Roegiers F, Jan LY, Jan YN. Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature. 2001;409:522–525. doi: 10.1038/35054077. [DOI] [PubMed] [Google Scholar]
- 2.Rebollo E, Roldan M, Gonzalez C. Spindle alignment is achieved without rotation after the first cell cycle in Drosophila embryonic neuroblasts. Development. 2009 doi: 10.1242/dev.041822. [DOI] [PubMed] [Google Scholar]
- 3.Siegrist SE, Doe CQ. Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development. 2006;133:529–536. doi: 10.1242/dev.02211. [DOI] [PubMed] [Google Scholar]
- 4.Cabernard C, Doe CQ. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev Cell. 2009;17:134–141. doi: 10.1016/j.devcel.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinsch S, Karsenti E. Orientation of spindle axis and distribution of plasma membrane proteins during cell division in polarized MDCKII cells. J Cell Biol. 1994;126:1509–1526. doi: 10.1083/jcb.126.6.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thery M, Racine V, Pepin A, Piel M, Chen Y, Sibarita JB, Bornens M. The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol. 2005;7:947–953. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- 8.Thery M, Jimenez-Dalmaroni A, Racine V, Bornens M, Julicher F. Experimental and theoretical study of mitotic spindle orientation. Nature. 2007;447:493–496. doi: 10.1038/nature05786. [DOI] [PubMed] [Google Scholar]
- 9.Jaffe AB, Kaji N, Durgan J, Hall A. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. J Cell Biol. 2008;183:625–633. doi: 10.1083/jcb.200807121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lew DJ. Yeast polarity: negative feedback shifts the focus. Curr Biol. 2005;15:R994–6. doi: 10.1016/j.cub.2005.11.048. [DOI] [PubMed] [Google Scholar]
- 11.Strome S. Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode Caenorhabditis elegans. J Cell Biol. 1986;103:2241–2252. doi: 10.1083/jcb.103.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Motegi F, Sugimoto A. Sequential functioning of the ECT-2 RhoGEF, RHO-1 and CDC-42 establishes cell polarity in Caenorhabditis elegans embryos. Nat Cell Biol. 2006;8:978–985. doi: 10.1038/ncb1459. [DOI] [PubMed] [Google Scholar]
- 14.Goldstein B, Hird SN. Specification of the anteroposterior axis in Caenorhabditis elegans. Development. 1996;122:1467–1474. doi: 10.1242/dev.122.5.1467. [DOI] [PubMed] [Google Scholar]
- 15.Cowan CR, Hyman AA. Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature. 2004;431:92–96. doi: 10.1038/nature02825. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins N, Saam JR, Mango SE. CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science. 2006;313:1298–1301. doi: 10.1126/science.1130291. [DOI] [PubMed] [Google Scholar]
- 17.Schonegg S, Hyman AA. CDC-42 and RHO-1 coordinate acto-myosin contractility and PAR protein localization during polarity establishment in C. elegans embryos. Development. 2006;133:3507–3516. doi: 10.1242/dev.02527. [DOI] [PubMed] [Google Scholar]
- 18.Gotta M, Abraham MC, Ahringer J. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr Biol. 2001;11:482–488. doi: 10.1016/s0960-9822(01)00142-7. [DOI] [PubMed] [Google Scholar]
- 19.Aceto D, Beers M, Kemphues KJ. Interaction of PAR-6 with CDC-42 is required for maintenance but not establishment of PAR asymmetry in C. elegans. Dev Biol. 2006;299:386–397. doi: 10.1016/j.ydbio.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng NN, Kirby CM, Kemphues KJ. Control of cleavage spindle orientation in Caenorhabditis elegans: the role of the genes par-2 and par-3. Genetics. 1995;139:549–559. doi: 10.1093/genetics/139.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyman AA, White JG. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. J Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- 23.Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- 27.Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller RK, Matheos D, Rose MD. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J Cell Biol. 1999;144:963–975. doi: 10.1083/jcb.144.5.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee L, Tirnauer JS, Li J, Schuyler SC, Liu JY, Pellman D. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 2000;287:2260–2262. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- 30.Beach DL, Thibodeaux J, Maddox P, Yeh E, Bloom K. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr Biol. 2000;10:1497–1506. doi: 10.1016/s0960-9822(00)00837-x. [DOI] [PubMed] [Google Scholar]
- 31.Yin H, Pruyne D, Huffaker TC, Bretscher A. Myosin V orientates the mitotic spindle in yeast. Nature. 2000;406:1013–1015. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
- 32.Hwang E, Kusch J, Barral Y, Huffaker TC. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J Cell Biol. 2003;161:483–488. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korinek WS, Copeland MJ, Chaudhuri A, Chant J. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 2000;287:2257–2259. doi: 10.1126/science.287.5461.2257. [DOI] [PubMed] [Google Scholar]
- 34.Miller RK, Cheng SC, Rose MD. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol Biol Cell. 2000;11:2949–2959. doi: 10.1091/mbc.11.9.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maekawa H, Usui T, Knop M, Schiebel E. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 2003;22:438–449. doi: 10.1093/emboj/cdg063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore JK, D'Silva S, Miller RK. The CLIP-170 homologue Bik1p promotes the phosphorylation and asymmetric localization of Kar9p. Mol Biol Cell. 2006;17:178–191. doi: 10.1091/mbc.E05-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore JK, Miller RK. The cyclin-dependent kinase Cdc28p regulates multiple aspects of Kar9p function in yeast. Mol Biol Cell. 2007;18:1187–1202. doi: 10.1091/mbc.E06-04-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–574. doi: 10.1016/s0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- 39.Maekawa H, Schiebel E. Cdk1-Clb4 controls the interaction of astral microtubule plus ends with subdomains of the daughter cell cortex. Genes Dev. 2004;18:1709–1724. doi: 10.1101/gad.298704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- 41.Reilein A, Nelson WJ. APC is a component of an organizing template for cortical microtubule networks. Nat Cell Biol. 2005;7:463–473. doi: 10.1038/ncb1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozlowski C, Srayko M, Nedelec F. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell. 2007;129:499–510. doi: 10.1016/j.cell.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 43.Kusch J, Meyer A, Snyder MP, Barral Y. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 2002;16:1627–1639. doi: 10.1101/gad.222602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segal M, Bloom K, Reed SI. Bud6 directs sequential microtubule interactions with the bud tip and bud neck during spindle morphogenesis in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:3689–3702. doi: 10.1091/mbc.11.11.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huisman SM, Bales OA, Bertrand M, Smeets MF, Reed SI, Segal M. Differential contribution of Bud6p and Kar9p to microtubule capture and spindle orientation in S. cerevisiae. J Cell Biol. 2004;167:231–244. doi: 10.1083/jcb.200407167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delgehyr N, Lopes CS, Moir CA, Huisman SM, Segal M. Dissecting the involvement of formins in Bud6p-mediated cortical capture of microtubules in S. cerevisiae. J Cell Sci. 2008;121:3803–3814. doi: 10.1242/jcs.036269. [DOI] [PubMed] [Google Scholar]
- 47.Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- 48.Cottingham FR, Gheber L, Miller DL, Hoyt MA. Novel roles for saccharomyces cerevisiae mitotic spindle motors. J Cell Biol. 1999;147:335–350. doi: 10.1083/jcb.147.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manning BD, Barrett JG, Wallace JA, Granok H, Snyder M. Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J Cell Biol. 1999;144:1219–1233. doi: 10.1083/jcb.144.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allingham JS, Sproul LR, Rayment I, Gilbert SP. Vik1 modulates microtubule-Kar3 interactions through a motor domain that lacks an active site. Cell. 2007;128:1161–1172. doi: 10.1016/j.cell.2006.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saunders W, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr Biol. 2005;15:1420–1427. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maddox P, Chin E, Mallavarapu A, Yeh E, Salmon ED, Bloom K. Microtubule dynamics from mating through the first zygotic division in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1999;144:977–987. doi: 10.1083/jcb.144.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller RK, Rose MD. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molk JN, Salmon ED, Bloom K. Nuclear congression is driven by cytoplasmic microtubule plus end interactions in S. cerevisiae. J Cell Biol. 2006;172:27–39. doi: 10.1083/jcb.200510032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maddox PS, Stemple JK, Satterwhite L, Salmon ED, Bloom K. The minus end-directed motor Kar3 is required for coupling dynamic microtubule plus ends to the cortical shmoo tip in budding yeast. Curr Biol. 2003;13:1423–1428. doi: 10.1016/s0960-9822(03)00547-5. [DOI] [PubMed] [Google Scholar]
- 57.Zaichick SV, Metodiev MV, Nelson SA, Durbrovskyi O, Draper E, Cooper JA, Stone DE. The mating-specific Galpha interacts with a kinesin-14 and regulates pheromone-induced nuclear migration in budding yeast. Mol Biol Cell. 2009;20:2820–2830. doi: 10.1091/mbc.E09-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta MLJ, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- 59.Grill SW, Gonczy P, Stelzer EH, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- 60.Grill SW, Howard J, Schaffer E, Stelzer EH, Hyman AA. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–521. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- 61.Labbe JC, Maddox PS, Salmon ED, Goldstein B. PAR proteins regulate microtubule dynamics at the cell cortex in C. elegans. Curr Biol. 2003;13:707–714. doi: 10.1016/s0960-9822(03)00251-3. [DOI] [PubMed] [Google Scholar]
- 62.Nguyen-Ngoc T, Afshar K, Gonczy P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol. 2007;9:1294–1302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- 63.Tsou MF, Ku W, Hayashi A, Rose LS. PAR-dependent and geometry-dependent mechanisms of spindle positioning. J Cell Biol. 2003;160:845–855. doi: 10.1083/jcb.200209079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li YY, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci U S A. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci U S A. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reck-Peterson SL, Vale RD. Molecular dissection of the roles of nucleotide binding and hydrolysis in dynein's AAA domains in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:1491–1495. doi: 10.1073/pnas.2637011100. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee WL, Kaiser MA, Cooper JA. The offloading model for dynein function: differential function of motor subunits. J Cell Biol. 2005;168:201–207. doi: 10.1083/jcb.200407036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moore JK, Li J, Cooper JA. Dynactin function in mitotic spindle positioning. Traffic. 2008;9:510–527. doi: 10.1111/j.1600-0854.2008.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moore JK, Sept D, Cooper JA. Neurodegeneration mutations in dynactin impair dynein-dependent nuclear migration. Proc Natl Acad Sci U S A. 2009;106:5147–5152. doi: 10.1073/pnas.0810828106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moore JK, Stuchell-Brereton MD, Cooper JA. Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell Motil Cytoskeleton. 2009;66:546–555. doi: 10.1002/cm.20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 75.Carvalho P, Gupta MLJ, Hoyt MA, Pellman D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev Cell. 2004;6:815–829. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 76.Caudron F, Andrieux A, Job D, Boscheron C. A new role for kinesin-directed transport of Bik1p (CLIP-170) in Saccharomyces cerevisiae. J Cell Sci. 2008;121:1506–1513. doi: 10.1242/jcs.023374. [DOI] [PubMed] [Google Scholar]
- 77.Li J, Lee WL, Cooper JA. NudEL targets dynein to microtubule ends through LIS1. Nat Cell Biol. 2005;7:686–690. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heil-Chapdelaine RA, Oberle JR, Cooper JA. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J Cell Biol. 2000;151:1337–1344. doi: 10.1083/jcb.151.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Farkasovsky M, Kuntzel H. Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J Cell Biol. 2001;152:251–262. doi: 10.1083/jcb.152.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Markus SM, Punch JJ, Lee WL. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr Biol. 2009;19:196–205. doi: 10.1016/j.cub.2008.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang X, Punch JJ, Lee WL. A CAAX motif can compensate for the PH domain of Num1 for cortical dynein attachment. Cell Cycle. 2009;8 doi: 10.4161/cc.8.19.9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 83.Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schmidt DJ, Rose DJ, Saxton WM, Strome S. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol Biol Cell. 2005;16:1200–1212. doi: 10.1091/mbc.E04-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Severson AF, Bowerman B. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans. J Cell Biol. 2003;161:21–26. doi: 10.1083/jcb.200210171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pecreaux J, Roper JC, Kruse K, Julicher F, Hyman AA, Grill SW, Howard J. Spindle oscillations during asymmetric cell division require a threshold number of active cortical force generators. Curr Biol. 2006;16:2111–2122. doi: 10.1016/j.cub.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 87.Couwenbergs C, Labbe JC, Goulding M, Marty T, Bowerman B, Gotta M. Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J Cell Biol. 2007;179:15–22. doi: 10.1083/jcb.200707085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Siller KH, Doe CQ. Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Dev Biol. 2008;319:1–9. doi: 10.1016/j.ydbio.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- 90.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 91.Yamamoto A, West RR, McIntosh JR, Hiraoka Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol. 1999;145:1233–1249. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vogel SK, Pavin N, Maghelli N, Julicher F, Tolic-Norrelykke IM. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 2009;7:e1000087. doi: 10.1371/journal.pbio.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamamoto A, Tsutsumi C, Kojima H, Oiwa K, Hiraoka Y. Dynamic behavior of microtubules during dynein-dependent nuclear migrations of meiotic prophase in fission yeast. Mol Biol Cell. 2001;12:3933–3946. doi: 10.1091/mbc.12.12.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]