Abstract
The RNA processing exosome complex was originally defined as an evolutionarily conserved multisubunit complex of ribonucleases responsible for the processing and/or turnover of stable RNAs. The exosome complex is also involved in the surveillance of mRNAs in both the nucleus and the cytoplasm, including nonsense-mediated decay (NMD) targets. The detailed mechanisms for how individual exosome subunits participate in each of these RNA metabolic pathways remains unclear. Here, we use RNAi to deplete exosome subunits, the exonucleases Rrp6 and Dis3, and an exosome cofactor in Drosophila melanogaster S2 tissue culture cells and assay the effects on global mRNA levels using gene expression microarrays. Consistent with the RNA degradative activities ascribed to the exosome, most mRNAs are increased. Notably, these stabilized mRNAs possess 3′ untranslated regions that are longer than the representative transcriptomic average. Moreover, our results reveal substantial differences in the pools of affected mRNAs for each depleted subunit. For example, ∼25% of the affected transcripts in Rrp6 depleted cells represent NMD substrates. While the affected mRNAs were dissimilar, they encode proteins that function in similar cellular pathways. We conclude that individual exosome subunits are largely functionally independent at the transcript level, but are interdependent on a transcriptomic level.
Keywords: Dis3, Rrp6, core exosome, RNase, mRNA turnover, NMD, UTR
INTRODUCTION
The RNA processing exosome was initially characterized as a multisubunit complex important for the 3′ → 5′ processing and degradation of many types of RNAs, including mRNA, rRNA, snRNA, snoRNA, and tRNA precursors in both the nucleus and the cytoplasm (Mitchell et al. 1997; Kadaba et al. 2004; Houseley et al. 2006; Schmid and Jensen 2008). Exosome subunits have also been implicated in the degradation of poly(A+) RNAs in the nucleus (DRN) and other specialized forms of nuclear mRNA turnover (Bousquet-Antonelli et al. 2000; Hilleren et al. 2001; Das et al. 2003; Houseley et al. 2006; Schmid and Jensen 2008). The complex is also involved in the destabilization of intergenic transcripts such as cryptic unstable transcripts (CUTs), upstream noncoding transcripts (UNTs), and promoter upstream transcripts (PROMPTs) (Wyers et al. 2005; Chekanova et al. 2007; Preker et al. 2008; Neil et al. 2009). In the cytoplasm, multiple exosome subunits have been shown to degrade transcripts targeted by both the siRNA and nonsense-mediated decay (NMD) machineries, although the mechanisms by which the transcripts are degraded vary by organism and system (Lejeune et al. 2003; Gatfield and Izaurralde 2004). Exosome subunits have also been linked to the degradation of mRNAs with specialized elements in their 3′ untranslated regions (UTRs) (Chen et al. 2001). Specifically, exosome subunits, in conjunction with cofactors, are important for the decay of the AU-rich element (ARE) containing c-fos and tumor necrosis factor (TNF) mRNAs (Chen et al. 2001). Exosome subunits have also been linked to the surveillance of other UTR elements, as evidenced by the phosphoglycerate kinase mRNA (Blattner and Clayton 1995; Colasante et al. 2007). Despite great progress in understanding the nature and scope of the RNA metabolic pathways and functions of individual exosome subunits, we are only beginning to comprehend how these subunits assemble and function as active complexes in vivo.
Strides toward understanding exosome subunit assembly and complex architecture have been made with in vitro reconstructions of archaeal and eukaryotic exosome complexes. Archaeal complexes have a hexameric ring of alternating RNase PH-domain subunits (Rrp41/Rrp42) topped with a cap of S1/KH-domain subunits (Rrp4, Cls4) (Buttner et al. 2005; Lorentzen et al. 2005; Liu et al. 2006). As with archaea, the human exosome complex has a cap consisting of three S1/KH-domain subunits (Csl4, Rrp4, and Rrp40), which rests upon a ring of six RNase PH-domain subunits (Ski6/Rrp41, Rrp42, Rrp43, Rrp45, Rrp46, and Mtr3). Further, the presence of all subunits was required to form this stable core complex in vitro (Liu et al. 2006). This observation is supported by work showing that certain subunits are codepleted when other subunits are targeted by RNAi, thus potentially destabilizing the core complex (Estevez et al. 2003; van Dijk et al. 2007). This nine-subunit core complex has been proposed to serve as a scaffold for two additional polypeptides, Dis3 and Rrp6, RNase II/R and RNase D homologs, respectively. The reconstructed core had limited RNase activity in vitro that increased when the remaining two polypeptides were added (Liu et al. 2006). Although it was initially shown that multiple subunits within the complex were catalytically active, recent studies have argued that this activity is predominantly, if not exclusively found in Dis3 and Rrp6 (Mitchell et al. 1997; van Hoof and Parker 1999; Liu et al. 2006; Dziembowski et al. 2007). In Drosophila, two distinct, yet functional exosome complexes, differing by the in- or exclusion of Rrp45, have been purified from S2 cells (Andrulis et al. 2002; Forler et al. 2003). As Drosophila also lacks an obvious Rrp43 homolog, Drosophila exosome complexes are also distinct from those of other eukaryotes.
Currently, all functions and contributions of individual subunits to distinct RNA processing and turnover events are thought to occur only in the context of the stoichiometric core complex outlined above. However, biochemical, cell biological, bioinformatic, and genetic evidence from recent work indicates that certain proteins, especially Rrp6, can function independent of other exosome subunits (Callahan and Butler 2008; Graham et al. 2009b) and form subcomplexes (Graham et al. 2006, 2009a). On a transcriptomic level, previous microarray experiments also show that many unique mRNAs are stabilized in rrp41-1, rrp6Δ, and rrp47Δ yeast strains (Houalla et al. 2006). This observation was confirmed and extended upon with tiling arrays using RNA harvested from depletions (Rrp4, Rrp41) or an exosome subunit null mutant (csl4-2) in plants (Chekanova et al. 2007). These observations present a puzzle. Namely, do exosome subunits always function as core complexes in vivo? Or can subunits work as subcomplexes, independently, or a combination of all of those possibilities?
To better characterize the functions of individual exosome subunits in the context of the core in vivo, we carried out a nearly comprehensive microarray study by using RNAi to deplete exosome subunits in Drosophila S2 cells. Roughly 80% of the affected transcripts were increased when compared with a GFP dsRNA-treated control. In general, the affected transcripts had long UTRs, and known exosome targets, including NMD transcripts, were enriched in our data set. At the level of individual transcripts, the experiments yielded distinct profiles when different subunits were depleted; however, the pathways in which those altered transcripts function were similar. Our data show an unanticipated complexity for exosome subunit mediated RNA metabolism and suggest a more dynamic interplay between and among exosome subunits within the exosome complex or subcomplexes.
RESULTS
Depletion of core exosome subunits does not affect cell proliferation in S2 tissue culture cells
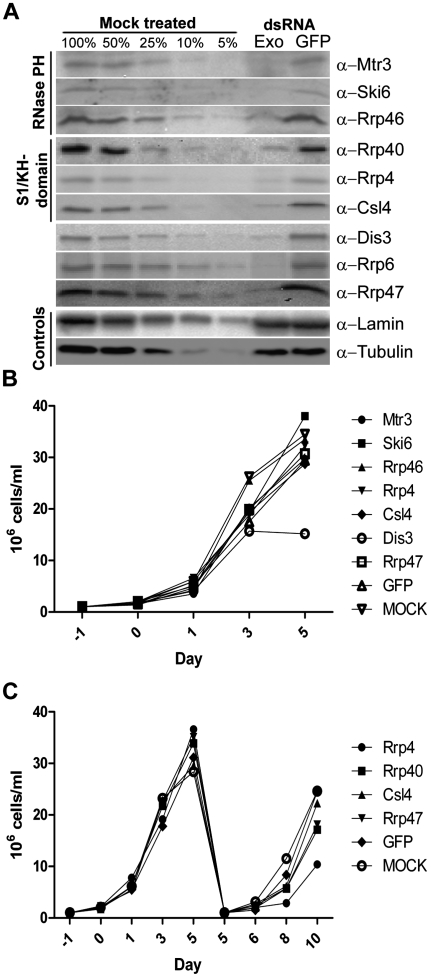
We previously demonstrated that Rrp6 is required for cell proliferation, whereas Rrp40 is not (Graham et al. 2009b). To determine whether other exosome subunits were important for viability we used RNAi to deplete the remaining subunits in Drosophila S2 tissue culture cells. Depletion was verified by Western blotting and percent depletion was calculated by comparing to a loading standard, Lamin Dm0 (Fig. 1A). Only cells in which subunits were verifiably depleted to below 10% of endogenous levels were used for further analysis. As mentioned above, Drosophila lacks an obvious Rrp43 homolog and Rrp42 and Rrp45 did not verifiably deplete to below 10%, and hence, these subunits are not included in this study.
FIGURE 1.
Depletion of most exosome subunits does not affect cell proliferation. (A) S2 cells were treated with exosome subunit specific dsRNAs as described in Materials and Methods. Effectiveness of dsRNA treatment was determined by Western blotting and depletion was calculated by comparing bands with a loading standard (Lamin Dm0). Representative blots for cell proliferation experiments are shown for each depleted exosome subunit. (B) During dsRNA treatment, cells were counted by hemacytometer at designated times. (C) S2 cells were depleted and counted as above. On day five, a portion of the cells were transferred to fresh flasks and treated with dsRNA for an additional 5 d. The graphs represent the average of three independent depletions for each subunit tested.
Most dsRNA-treated cells continued to proliferate, except Dis3 depletion elicited diminished cell proliferation (Fig. 1B,C; Supplemental Fig. S1). Rrp4-depleted cells grew slowly when the dsRNA treatment was extended to 10 d, but these cells proliferated normally when the dsRNA treatment was discontinued after 5 d (cf. Fig. 1C and Supplemental Fig. S1). These data indicate that most core exosome subunits, and hence, by inference, the exosome core, are not essential in S2 cells (Fig. 1B,C; Supplemental Fig. S1). We verified the functional efficacy of exosome subunit depletions by assaying 5.8S rRNA proccessing in dsRNA treated cells (Supplemental Fig. S2). The split structure of Dipteran 5.8S rRNA (Supplemental Fig. S2A) is distinct from most other eukaryotes and yields a multiple species of “mature” 5.8S rRNA in adult flies, plus a distinct pattern of processing intermediates in exosome subunit depleted cells (Supplemental Fig. S2B,C; Pavlakis et al. 1979; Shimada 1992; Fox 1997).
Earlier work in trypanosomes has shown that depleting certain exosome subunits results in the partial proteasome-mediated destabilization of other exosome subunits (Estevez et al. 2003). To determine if this was the case with the Drosophila complex, we examined the stability of a large number of exosome subunits following dsRNA treatment (Supplemental Fig. S3). The depletion of certain subunits resulted in one or more codepleted subunit(s) being reduced to ∼25%–40% of normal levels as judged by Western blotting (Supplemental Fig. S3). In general, depletion of RNase PH-domain subunits (Mtr3, Ski6, and Rrp46) caused the codepletion of one or more additional subunits. However, targeting the S1/KH-domain subunits (Rrp40, Rrp4, and Csl4) resulted in no codepletion (Supplemental Fig. S3). This result was unexpected, as in vitro reconstructions of the eukaryotic complex depended upon the presence of an intact S1/KH-domain cap (Liu et al. 2006). Thus, we do not obtain a broad destabilization of subunits following depletion, as might be expected for complete exosome complex disassembly (Estevez et al. 2003).
Genome-wide investigation of mRNAs regulated by exosome subunits in S2 cells
To understand better the contributions of individual exosome subunits to exosome complex function in vivo, we performed microarray analyses using RNA isolated from exosome subunit-depleted (below 10% of normal levels) S2 cells. RNA isolated from cells depleted of the exosome subunits Mtr3, Ski6, Rrp46, Rrp40, Rrp4, Csl4, Dis3, Rrp6, and the exosome cofactor Rrp47 was assayed using Drosophila_2 microarrays (Affymetrix). We limited nonspecifically affected transcripts in four ways and defined each depleted exosome subunit microarray profile (desMAP) as containing only reproducibly changed transcripts as detailed in the Materials and Methods.
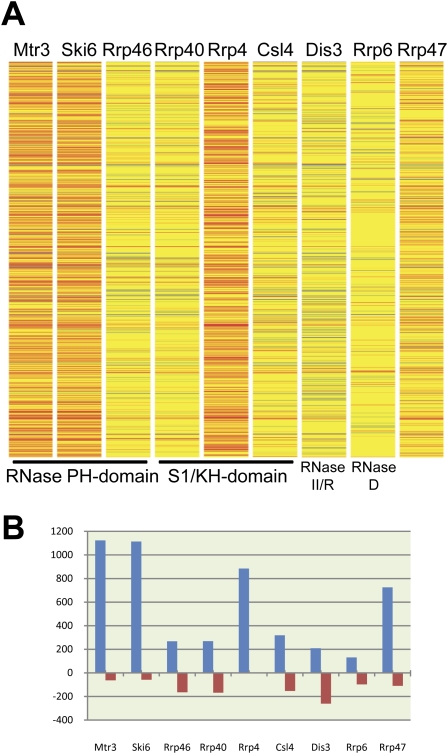
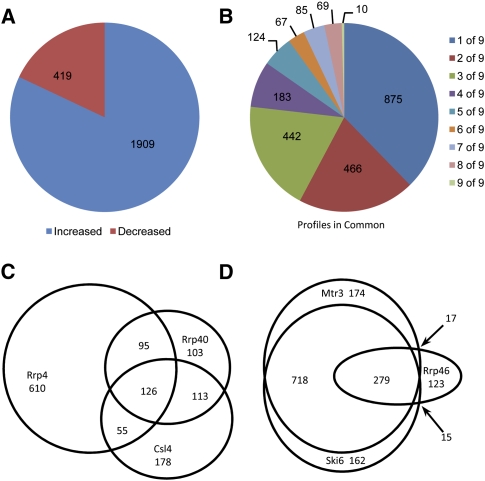
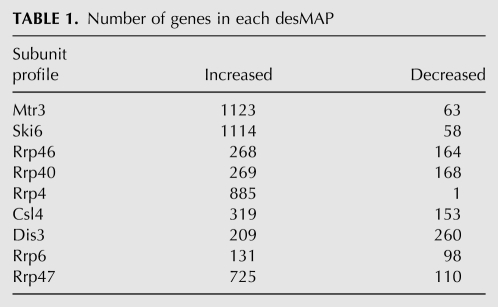
The heat map (Fig. 2A) depicts the desMAPs of all depleted exosome subunits. The changed transcripts vary greatly in number, identity, and the degree of change among desMAPs; however, several desMAPs have many transcripts in common (Figs. 2,3; Tables 1, 2; Supplemental Tables S1–S7). Depletion of Mtr3, Ski6, Rrp4, and the exosome cofactor Rrp47 affected the largest number of transcripts (Fig. 2B; Table 1). In contrast, depletion of Rrp6, the subunit commonly used to define the nuclear functions of the exosome, altered the fewest (Fig. 2B; Table 1). All affected transcripts in each desMAP are listed by subunit and fold change in Supplemental Table S1 and by subunit and gene description in Supplemental Table S2. For all exosome depletions, 6117 transcripts (5043 increased and 1074 decreased) were identified as reproducibly changed. When transcripts present in multiple desMAPs were accounted for, a total of 2328 (1909, or 82%, increased and 419 decreased), out of the 6117, transcripts were identified (Fig. 3A; Supplemental Table S3). These transcripts were then grouped based upon the number of desMAPs in which they occurred. A total of 875 (∼37%) of the affected transcripts were found only in a single desMAP, with 1783 (∼76%) being present in three or fewer desMAPs (Fig. 3B; Supplemental Table S3). Remarkably, only 0.4% of unique transcripts (10 total, all increased) were identified in all nine desMAPs (Fig. 3B; Supplemental Table S3). Even when the data were weighted to account for transcripts that appeared in multiple desMAPs, only ∼1.5% of transcripts were present in all desMAPs, while nearly 80% were found in three or fewer desMAPs (Supplemental Fig. S4; Supplemental Table S3).
FIGURE 2.
Microarray analysis of exosome subunit depleted cells reveals distinct transcriptomic profiles. (A) Heat map showing changes in mRNA levels by subunit. Unchanged mRNAs (fold change less than ±2) are shown in yellow, decreases (≤twofold change) are shown in shades of blue, marginal increases (two- to 3.5-fold changes) in shades of orange, and finally large increases (>3.5-fold change) in shades of red. (B) The number of altered transcripts in each desMAP.
FIGURE 3.
Characterization of depleted exosome subunit microarray profiles (desMAPs). (A) Number of increased and decreased transcripts in the pooled data set. (B) Distribution of transcripts in multiple desMAPs. Comparison of transcripts altered in (C) S1/KH-domain or (D) RNase PH-domain subunits.
TABLE 1.
Number of genes in each desMAP
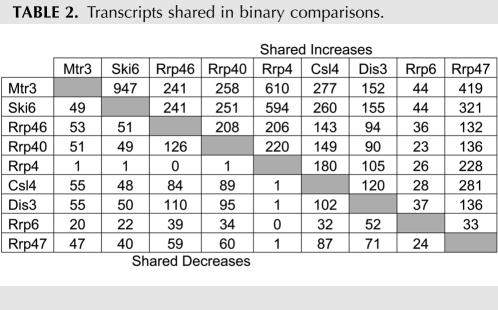
TABLE 2.
Transcripts shared in binary comparisons.
We next examined the degree of overlap between individual desMAPs. We cataloged the transcripts that were present in each pairwise comparison (Table 2, increased [top], decreased [bottom]). All overlapping transcripts for each possible binary comparison are detailed in Supplemental Table S4 and the subtables therein. The desMAPs of Mtr3 and Ski6 shared the most (∼85%) transcripts, while the desMAPs of Rrp6 and Rrp4 had the fewest common (∼16%) transcripts, of which nearly a third were altered in opposite directions (Table 2; Supplemental Table S4). The desMAPs of Rrp6 and Dis3 shared the fewest transcripts and are the most divergent from the desMAPs of other exosome subunits (Table 2; Supplemental Table S4). These data show a large and unexpected variation in the transcriptomic profiles of individual exosome subunits and cofactors.
Comparison of transcript profiles based on exosome subunit homologies
We wanted to determine whether certain transcripts were shared among subunits with similar domain structures. The three S1/KH-domain subunits (Rrp40, Rrp4, and Csl4) had desMAPs that were between 20% (Rrp4 versus Csl4) and 50% (Csl4 versus Rrp40) similar (Fig. 3C; Supplemental Fig. S5A–C; Supplemental Table S5). Of the transcripts affected by depleting these three subunits, ∼70% were present only in one S1/KH-domain desMAP, just over 20% in two, with the remaining 10% being present in all three desMAPs (Fig. 3C; Supplemental Fig. S5A–C; Supplemental Table S5). We also compared the desMAPs yielded by depleting the RNase PH-domain containing exosome subunits (Mtr3, Ski6, and Rrp46). In this analysis, ∼30% of transcripts were found in only one RNase PH-domain desMAP, 50% were present in two of three desMAPs, and ∼19% were present in all three RNase PH-domain subunit desMAPs (Fig. 3D; Supplemental Fig. S5D-F; Supplemental Table S6).
These results show that even structurally similar subunits affect distinct sets of mRNAs. We also examined the transcripts from the structurally distinct subunits (Dis3 and Rrp6) and an exosome cofactor (Rrp47). The desMAPs of these three polypeptides were more distinct than those of the S1/KH- and RNase PH-domain subunits (Figs. 2, 3; Table 2; Supplemental Fig. S6). Examined in entirety, the desMAPs of all subunits were distinct, but the desMAPs of RNase PH-domain subunits were more similar to one another than other exosome subunits.
Transcripts affected by exosome subunit depletion have longer than average UTRs
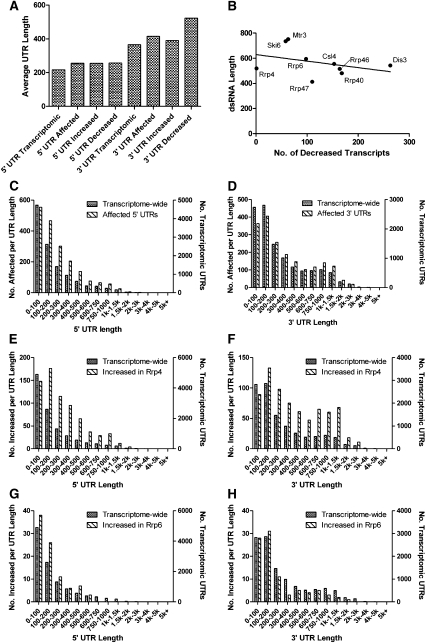
We next wanted to determine if the mRNAs identified by our arrays shared sequence and/or regulatory elements. Since exosome subunits have been linked to the surveillance of transcripts with specialized UTR elements, such as the AREs of c-fos and TNF mRNAs (Chen et al. 2001), we reasoned that transcripts with longer UTRs, and therefore a higher likelihood of containing a UTR element, would be more prevalent in our desMAPs. The 5′ and 3′ UTR libraries for D. melanogaster were downloaded from Flymine. In cases where UTRs of multiple lengths were annotated for a single transcript, the average of available UTR sizes was used. In total, 304 and 290 of the transcripts in our desMAPs either lacked annotated 5′ or 3′ UTRs, respectively, or lacked UTRs entirely; these transcripts were excluded from further analysis. When the average 5′ and 3′ UTR lengths of the affected genes were determined, ∼255 nt and ∼415 nt, respectively, they were significantly (P < 0.0001 for both) longer than the transcriptome-wide average, ∼216 for 5′ UTRs and ∼365 for 3′ UTRs (Fig. 4A). Further, in the case of 3′ UTRs these differences were more substantial in decreased transcripts (522 average 3′ UTR length, P < 0.0001) than in increased transcripts (390 average, P = 0.0212) (Fig. 4A). The same bias was not seen with 5′ UTRs, as the average length of both increased and decreased transcripts had very similar 5′ UTR lengths (Fig. 4A).
FIGURE 4.
The UTRs of affected genes are longer than predicted by genome-wide averages. (A) The average 5′ and 3′ UTR lengths of affected genes in each desMAP were compared with genome-wide average UTR lengths. (B) The length of the dsRNA used to deplete each exosome subunit was plotted opposite the number of decreased transcripts in each desMAP. The distribution of 5′ (C) and 3′ (D) UTR lengths in all desMAPs, left y-axes, plotted opposite the transcriptome-wide distribution of UTR lengths, right y-axes. The 5′ and 3′ UTR length profiles of transcripts increased in the desMAPs of (E,F) Rrp4 and (G,H) Rrp6 with axes as established above.
Several studies have indicated that RNAi treatments can result in the degradation of nontargeted mRNAs via off-target effects (Jackson et al. 2003, 2006; Jackson and Linsley 2004). To determine whether the dsRNAs used to deplete the exosome subunits could produce off-target effects, we first cross-analyzed the lengths of the dsRNAs and the number of affected transcripts in each desMAP. While Mtr3 and Ski6 have both the longest dsRNAs and the largest desMAPs (Table 1; Supplemental Fig. S7) the dsRNAs used to deplete Rrp4 and Rrp47, the next two largest desMAPs by transcript number, used average length (Rrp4) or the shortest dsRNAs (Rrp47). Further, of the four subunits with the largest number of affected transcripts over 87% (>95% for Mtr3, Ski6, and Rrp4) are increased where the preference for longer 3′ UTRs was less pronounced than in decreased transcripts (Fig. 4A; Supplemental Fig. S7). Since longer 3′ UTRs were more prevalent in decreased transcripts (Fig. 4A), we next plotted the number of decreased transcripts per desMAP versus the length of the dsRNA used in that depletion. There is a slight negative (Fig. 4B, trendline) bias against longer dsRNAs producing more decreased transcripts. Collectively, these metrics suggest that the correlation between exosome subunit depletion and increased UTR lengths is specific, and in most cases does not result from off-target effects.
Certain exosome subunits prefer mRNAs with longer UTRs
Based upon the observation that our depletions favored transcripts with longer than average UTRs, we wanted to determine if UTRs of particular lengths were favored in certain desMAPs. The transcriptome-wide distribution of different UTR lengths was determined for both 5′ and 3′ UTRs (Fig. 4C,D, right y-axes) and graphed opposite the distribution of UTR lengths of mRNAs in our microarray data set (Fig. 4C,D, left y-axes). The mRNAs increased in the desMAP of Rrp4 have longer 5′ and 3′ UTRs, while those increased in the desMAP of Rrp6 tend to have shorter UTRs (Fig. 4E–H). The UTR length distributions of the remaining subunits fall between these two extremes and are graphed in Supplemental Figures S8–S11. mRNAs with 5′ UTRs of 100–1000 nucleotides (nt) are enriched to varying degrees in most desMAPs (Supplemental Figs. S8, S9). When 3′ UTRs were examined, the predominant pattern showed that mRNAs with UTR lengths between 100–400 nt and 600–1500 nt were more prevalent in our data set (Supplemental Figs. S10, S11). This preference for longer UTRs suggests that AREs and/or additional UTR elements play a role in exosome-mediated mRNA surveillance.
NMD targets are differentially represented in exosome subunit depletions
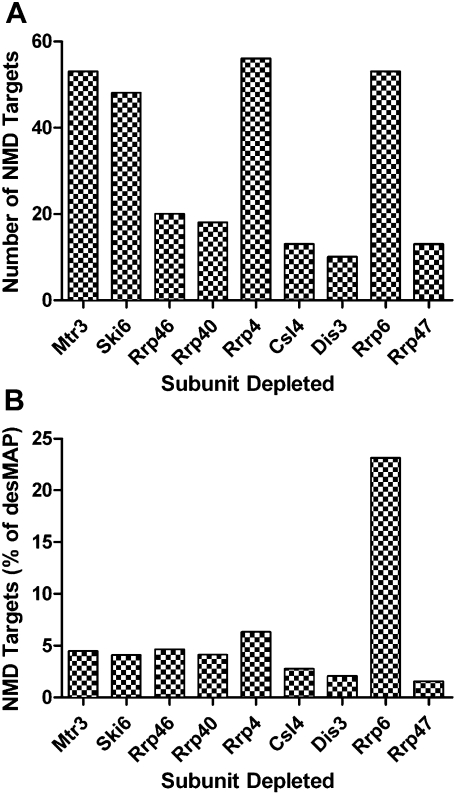
Although elements outside the coding sequence (longer UTRs) play a role in exosome-mediated mRNA surveillance, elements within the coding sequence are also known to serve this function. In this regard, we searched our microarray data to see if mRNAs with premature termination codons (i.e., NMD targets), whose turnover has been attributed to the exosome complex, were over-represented in our data set (Gatfield and Izaurralde 2004; Rehwinkel et al. 2005). We identified 40% of 185 mRNAs that are known Drosophila NMD targets in our pooled array data set (Supplemental Table S7) (Rehwinkel et al. 2005). These NMD targets were then mapped to individual desMAPs (Fig. 5A). The number of NMD-targeted transcripts varied by desMAP, but, in general, NMD targets constituted between 1.5% and 6.3% of affected transcripts in most desMAPs (Figs. 4B, 5A). The exception was the desMAP of Rrp6, where 23% of the transcripts were NMD targets (Fig. 5B). The data suggest a core exosome independent role for Rrp6 in mRNA surveillance.
FIGURE 5.
NMD targeted transcripts are differentially represented in desMAPs. (A) The numbers of known NMD targeted transcripts in each desMAP. (B) Prevalence of NMD transcripts, as percent of altered transcripts, in each desMAP.
Genes affected by exosome subunit depletions work in similar pathways
We wanted to determine whether the proteins encoded by the mRNAs affected by exosome subunit depletion had roles in similar biological pathways. We thus grouped the genes that were affected by exosome subunit depletion based upon the functions and interactions of the proteins they encoded. Increased and decreased genes were grouped and analyzed separately. A P-value was assigned to the detected pathways, depending upon the number of components within the individual pathway and their prevalence in our desMAPs. Specific and statistically significant pathways (P ≤ 0.05) were retained for further analysis; however, significant but overly general pathways (e.g., nucleus, cytoplasm, and molecular function) were winnowed. Related pathways were grouped into larger categories (e.g., mitosis and spindle were grouped into cell cycle). Certain pathways were present in multiple groups (e.g., protein ubiquitination was both protein modification and turnover), while other pathways did not fit into any group (e.g., male courtship behavior). The prevalence of a group of pathways was calculated by determining what percentage of total pathways for that desMAP resided in each subgroup.
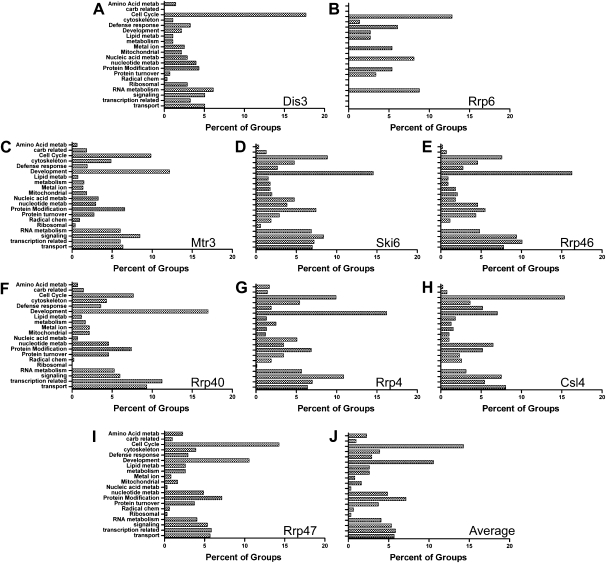
When the groupings for increased transcripts were plotted, the patterns of Rrp6 and Dis3 were different from the average pattern of other exosome subunits (cf. Fig. 6A,B,J). Cell cycle-related pathways were the most prominent in these two subunits, but Dis3 had comparatively few development-related transripts (cf. Fig. 6A,B,J). Also, the group pattern of mRNAs in Rrp6-depleted cells lacked many groups that were present in other exosome subunit depletions (cf. Fig. 6B,J). The group patterns of the remaining exosome subunits were very similar to one another (Fig. 6C–J). For those seven subunits (Mtr3, Ski6, Rrp46, Rrp40, Rrp4, Csl4, and Rrp47) development- and cell cycle-related transcripts were generally the most prominent classes; signaling-, transcription-, transport-, and protein-modification-related were also enriched to a similar extent (Figs. 6C–J). These similar patterns were surprising, since the desMAPs of individual subunits were quite different both by the number and the identity of the affected transcripts (Figs. 2B, 3; Supplemental Figs. S4–S7; Tables 1, 2). Further, these similarities were only apparent when examining increased transcripts, as a parallel analysis yielded no consensus pattern for the groupings of decreased transcripts (Supplemental Fig. S12). We surmise that although individual exosome subunits process and degrade distinct mRNAs, those mRNAs perform functionally interdependent roles in cellular pathways.
FIGURE 6.
Grouped analysis of cellular processes disrupted by transcripts increased in exosome subunit depletions. The cellular processes affected by the transcripts increased in each desMAP were identified by using Pathway Studio 5.0 (Ariadne). Pathways were grouped by similarity and patterns are shown as percentage of each group as a part of the whole profile. The patterns of (A) Dis3, (B) Rrp6, (C) Mtr3, (D) Ski6, (E) Rrp46, (F) Rrp40, (G) Rrp4, (H) Csl4, (I) Rrp47, and the (J) average pattern for all pathway groupings of increased transcripts are shown.
DISCUSSION
In this study we sought to identify and characterize mRNAs that were surveyed by the subunits that comprise and associate with the exosome complex in vivo. The current understanding of exosome complex function predicts that the transcript profiles of all exosome subunits should be very similar; however, both here and in other studies, different transcriptomic profiles were observed (Houalla et al. 2006; Chekanova et al. 2007). Our microarray data show that depletion of individual exosome subunits stabilizes distinct sets of mRNAs that vary greatly in both the size of the transcript pool and the identity of the transcripts affected. Validating their specificity, the arrays identified 74 NMD targeted transcripts, which are known exosome substrates. In general, the transcripts affected by exosome subunit depletion have larger than average 5′ and 3′ UTRs and encoded proteins that were involved in similar biological pathways.
In contrast to work in yeast showing that exosome subunit genes are essential, our data show that most exosome subunits are not required for cell proliferation in Drosophila S2 cells (Graham et al. 2009b). Only Rrp6 and Dis3 are essential in S2 cells, and their roles in cell proliferation are likely independent of other exosome subunits (Graham et al. 2009b). Our data agree with earlier observations showing that depletion of one exosome subunit codepletes one or more additional subunits (Estevez et al. 2003). However, depletion of individual S1/KH-domain subunits does not destabilize other exosome subunits, suggesting that the RNase PH-domain subunit ring may remain intact, and/or that individual S1/KH-domain subunits may be structurally redundant, as observed in archaea (Buttner et al. 2005). These partial (∼25%–40% remaining) codepletions make interpretation of the resulting data more complicated; however, closer examination of the desMAPs alleviates some of these concerns. The desMAPs of several codepleted pairs of subunits, such as Mtr3 and Csl4 (where Mtr3 depletion elicits Csl4 codepletion), are still substantially different. This may suggest that the codepleted subunits are not reduced below the threshold to produce effects on mRNA turnover and processing. It is also possible that the pools of exosome subunits remaining after depletion are sufficient to perform essential functions and thus allow the cells to proliferate. Nonetheless, given the dissimilarity of desMAPs and the reproducibility of the data, we suggest our findings to be of sufficient quality to determine the roles of subunits in the turnover of distinct classes of mRNAs.
Previously published microarray studies show heterogeneity and little transcriptomic overlap when individual exosome subunits are mutated or depleted (Houalla et al. 2006; Chekanova et al. 2007). Our microarray studies in Drosophila confirm and extend upon those previous works in yeast and plants and show that depletion of a limited number of individual exosome subunits affects different pools of mRNAs. We find the similarities among the available microarray studies noteworthy. First, the majority (∼80%) of the affected transcripts are increased in all three microarray studies (Houalla et al. 2006; Chekanova et al. 2007). Second, consistent with our study, the array profile of rrp6Δ yeast cells was the most divergent when compared with the profiles of other subunits (Houalla et al. 2006). Third, a small fraction of affected transcripts were altered in the same manner in three yeast strains, and furthermore, many transcripts were found in the profiles of only one or two of the three subunits tested (Houalla et al. 2006). Fourth, tiling microarray results from plants show that certain mRNAs were affected in a different manner depending on which subunit was depleted (Chekanova et al. 2007). Finally, the overall number and location of gene regions affected by exosome subunit RNAi depletion in plants was dependent on which subunit was depleted (Chekanova et al. 2007). Together, these microarray studies from three organisms (S. cerevisiae, A. thaliana, and D. melanogaster) and genetic studies in yeast demonstrate that depleting or mutating individual subunits has different effects on the processing and/or turnover of individual RNAs.
In addition to similar effects at the transcriptomic level, our data are likewise consistent with effects observed for mRNA classes known to be exosome subunit substrates. For example, NMD targets are differentially represented in the profiles of exosome subunit depleted cells, especially that of Rrp6. In agreement with this result, Rrp6 has been shown to be involved in the turnover of NMD targeted transcripts in human cells and is both nuclear and cytoplasmic in both human and S2 cells (Lejeune et al. 2003; Graham et al. 2006, 2009a). Further, Rrp6 has recently been shown to have RNA processing functions that can occur independently of other exosome subunits in yeast (Callahan and Butler 2008). These results provide further evidence that Rrp6 may function in a core-exosome independent manner on certain substrates.
When the UTRs of the affected transcripts were examined, in general, they had longer than average 5′ and 3′ UTRs. The prevalence of mRNAs with longer UTRs is expected since ARE containing mRNAs are known exosome substrates, and longer UTRs have a greater probability of containing one or more AREs or other regulatory elements (Chen et al. 2001; Lin et al. 2007). These regulatory elements could be sequences that target transcripts to certain exosome subunits. In support of this, human PM-Scl-75/Rrp45, Oip2/Rrp43, and Rrp41/Ski6 have been shown to bind ARE-like RNAs in a sequence-specific manner in vitro (Anderson et al. 2006). Further, other RNA sequence elements can be present in other types of RNA, which may explain how the exosome complex is recruited to and processes or degrades other classes of RNAs. Detailed analysis of the UTR sequences of mRNAs identified by our microarrays could elucidate the identity of one or more of the putative regulatory elements.
To date, the data from multiple microarray studies (including this one) and associated experiments show that distinct RNAs are affected by exosome subunit depletion or mutation (Houalla et al. 2006; Chekanova et al. 2007; Preker et al. 2008). These data suggest that individual exosome subunits have roles in RNA processing that are either independent of the exosome complex, or that they serve specific roles within the core complex. These results are seemingly at odds with genetic and crystallographic data suggesting that all exosome subunits are required for complex integrity and the processing and/or turnover of RNAs (Mitchell et al. 1997; Allmang et al. 1999a,b, 2000; Estevez et al. 2003; Liu et al. 2006). However, targeted studies with Dis3 and Rrp6 have established that certain exosome subunits can function without associating with the entire complex (Houalla et al. 2006; Schneider et al. 2007; Callahan and Butler 2008). Further, other subunits such as the RNase PH-domain subunits mentioned above preferentially interact with certain RNA sequences, suggesting that they may serve as specificity modules for the exosome complex or work in a complex independent manner (Anderson et al. 2006; Houalla et al. 2006; Chekanova et al. 2007). Our grouped pathway data could resolve this paradox. While the individual transcripts affected by exosome subunit depletion in our microarrays are quite different, analysis of their biological functions show that the transcript sets are related. These data suggest interactions between and among the proteins encoded by exosome subunit surveyed mRNAs. These interactions may explain how the surveillance of distinct RNAs by different subunits yields very similar outcomes at the genetic and cellular level.
Our transcriptomic study represents an important advance in our understanding of the gene-specific and transcriptome-wide roles for individual exosome subunits in vivo. These data suggest that exosome subunits have specialized roles within the exosome complex independent from or within the context of exosome subcomplexes. Moreover, we identify cellular pathways that are regulated by distinct exosome subunits and provide compelling evidence for both overlapping and independent functions for Rrp6, Dis3, Rrp47, and other core exosome subunits. Future gene- or pathway-directed analysis will yield additional insights into the structure and function of the exosome core and its cofactors in living cells.
MATERIALS AND METHODS
Antibodies and S2 cell culture
Polyclonal exosome subunit-specific primary antibodies and HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) were described earlier (Graham et al. 2009b). D. melanogaster embryonic S2 cell lines were grown in Hyclone HyQ-CCM3 media at 27°C.
Double-stranded RNA preparation and RNA interference
dsRNA was prepared as described earlier (Graham et al. 2009b). T7 primer sequences are listed in Supplemental Table S8. Cells were grown in 25 cm2 flasks, treated with dsRNAs (30 μg/mL) on days 0, 1, and 3, resuspended by pipetting each time dsRNA was added, counted as earlier, and harvested on day 5 (Graham et al. 2009b). A portion of each culture was reserved for Western blotting as described previously (Graham et al. 2006). For extended cell-counting experiments, equal amounts of Day 5 dsRNA-treated cells were seeded into fresh 25 cm2 flasks with or without continued dsRNA treatment. Depletion of the targeted protein was verified via Western blotting and comparison to a loading standard (Lamin Dm0 or α-Tubulin) in Mock and dsGFP-treated cells. The remaining cells were pelleted and RNA was harvested using TRIzol Reagent (Invitrogen) according to the manufacturer's instructions.
Northern blotting
Samples (10 μg) of total RNA from independent dsRNA treatments were separated using 6% denaturing polyacrylamide gels. The separated RNAs were electroblotted onto Hybond-N (GE Healthcare) membranes and probed with 32P-labeled DNA oligonucleotide probes (Hu et al. 2009).
Microarray analysis
Once depletion of the subunits were verified by Western blotting, 50 μg samples of total RNA from independent, duplicate RNAi depletions (RNAi targeted protein reduced to <10% of normal levels as judged by Western blotting) were sent to the Gene Expression and Genotyping Core Facility, Case Comprehensive Cancer Center (http://gegf.chosencraft.com/web/). Samples were hybridized using Genechip Drosophila_2 microarrays (Affymetrix), washed, and data acquired (GeneChip Fluidics Station 450, GeneChip Scanner 3000, GeneChip Command Console Software Version 1.1) according to Affymetrix guidelines. The resulting data were analyzed using GCOS (Affymetrix), Microsoft Access, NetAffx (Affymetrix), and Genespring GX 10 (Agilent) as described earlier (Graham et al. 2009b). The raw and normalized data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE17874 (Edgar et al. 2002). (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17874).
Nonspecific transcripts were limited in the following ways. First, RNA harvested from GFP dsRNA-treated cells was used as our baseline sample. This was to account for RNAs whose expression was altered simply by dsRNA treatment. Second, GCOS software (Affymetrix) assigned P-values to changed transcripts based on 14 probe set pairs per transcript per array (GFP and exosome subunit depleted). Only transcripts with change P-values of <0.005 (increases) or >0.995 (decreases) were retained. Third, transcripts with fold changes below ±2-fold were excluded from further analysis. Finally, all arrays were performed using RNA harvested from independent duplicate exosome subunit depletions and compared against arrays using RNA harvested from independent duplicate GFP dsRNA treated controls. This regime yielded four total comparisons. For the purposes of this study, a reproducibly changed transcript's expression is altered at least ±2-fold (in the same direction, with a P-value threshold of 0.005) in at least three of four independent exosome subunits depleted to GFP array comparisons. Proportional Venn and Euler diagrams were adapted from the following website: http://www.cs.kent.ac.uk/people/staff/pjr/EulerVennCircles/EulerVennApplet.html.
UTR analysis
All data for both 3′ and 5′ UTRs were downloaded from Flymine at http://www.flymine.org/release-18.0/begin.do. Final 5′ and 3′ UTR lengths of genes with multiple annotated UTRs were determined by averaging all of the 5′ or 3′ UTRs reported for that particular gene. The 5′ and 3′ UTR lengths of affected genes, both individually and in aggregate, were determined by matching gene identifiers with the averaged Flymine UTR datasets.
NMD analysis
Drosophila NMD targets were identified in a previously published work (Rehwinkel et al. 2005). Probe and gene identifiers from the two different arrays (Drosgenome1 and Drosophila_ 2; this study) were uploaded into NetAffx (Affymetrix) http://www.affymetrix.com/analysis/index.affx to standardize nomenclatures.
Grouped pathway analysis
Increased and decreased gene lists for each subunit were separately imported into Pathway Studio 5.0 (Ariadne). Pathway Studio 5.0 (Ariadne) then was directed to search for all connected and related proteins and biological pathways. Significant associations (P < 0.05) were grouped by similarity and tallied as percent of total significant pathways.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank the Coller Laboratory, especially Wenqian Hu, for technical assistance. We also express gratitude to Dr. Paul Fisher for the Lamin antibody; and Alan Tartakoff, Donal Luse, Jonatha Gott, and members of the Andrulis laboratory for discussions and comments on the manuscript. This work is supported by grant GM072820 from the NIH (to E.D.A). E.D.A. is a Mount Sinai Health Care Foundation Scholar.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1906710.
REFERENCES
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999a;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes & Dev. 1999b;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JR, Mukherjee D, Muthukumaraswamy K, Moraes KC, Wilusz CJ, Wilusz J. Sequence-specific RNA binding mediated by the RNase PH domain of components of the exosome. RNA. 2006;12:1810–1816. doi: 10.1261/rna.144606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- Blattner J, Clayton CE. The 3′-untranslated regions from the Trypanosoma brucei phosphoglycerate kinase-encoding genes mediate developmental regulation. Gene. 1995;162:153–156. doi: 10.1016/0378-1119(95)00366-e. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- Buttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008;36:6645–6655. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Colasante C, Robles A, Li CH, Schwede A, Benz C, Voncken F, Guilbride DL, Clayton C. Regulated expression of glycosomal phosphoglycerate kinase in Trypanosoma brucei. Mol Biochem Parasitol. 2007;151:193–204. doi: 10.1016/j.molbiopara.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Das B, Butler JS, Sherman F. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:5502–5515. doi: 10.1128/MCB.23.16.5502-5515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez AM, Lehner B, Sanderson CM, Ruppert T, Clayton C. The roles of intersubunit interactions in exosome stability. J Biol Chem. 2003;278:34943–34951. doi: 10.1074/jbc.M305333200. [DOI] [PubMed] [Google Scholar]
- Forler D, Kocher T, Rode M, Gentzel M, Izaurralde E, Wilm M. An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat Biotechnol. 2003;21:89–92. doi: 10.1038/nbt773. [DOI] [PubMed] [Google Scholar]
- Fox MG. Identification of a new ribosomal RNA in Drosophila melanogaster. Gene. 1997;194:291–295. doi: 10.1016/s0378-1119(97)00136-4. [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- Graham AC, Kiss DL, Andrulis ED. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol Biol Cell. 2006;17:1399–1409. doi: 10.1091/mbc.E05-08-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AC, Davis SM, Andrulis ED. Interdependent nucleocytoplasmic trafficking and interactions of Dis3 with Rrp6, the core exosome, and importin-α3. Traffic. 2009a;10:499–513. doi: 10.1111/j.1600-0854.2009.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AC, Davis SM, Andrulis ED. Core exosome-independent roles for Rrp6 in cell cycle progression. Mol Biol Cell. 2009b;20:2242–2253. doi: 10.1091/mbc.E08-08-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- Houalla R, Devaux F, Fatica A, Kufel J, Barrass D, Torchet C, Tollervey D. Microarray detection of novel nuclear RNA substrates for the exosome. Yeast. 2006;23:439–454. doi: 10.1002/yea.1369. [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Linsley PS. Noise amidst the silence: Off-target effects of siRNAs? Trends Genet. 2004;20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes & Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- Lin WJ, Duffy A, Chen CY. Localization of AU-rich element-containing mRNA in cytoplasmic granules containing exosome subunits. J Biol Chem. 2007;282:19958–19968. doi: 10.1074/jbc.M702281200. [DOI] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;451:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Pavlakis GN, Jordan BR, Wurst RM, Vournakis JN. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979;7:2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. The exosome: A multipurpose RNA-decay machine. Trends Biochem Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T. Distribution of split 5.8S ribosomal RNA in Diptera. Insect Mol Biol. 1992;1:45–49. doi: 10.1111/j.1365-2583.1993.tb00076.x. [DOI] [PubMed] [Google Scholar]
- van Dijk EL, Schilders G, Pruijn GJ. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA. 2007;13:1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Parker R. The exosome: A proteasome for RNA? Cell. 1999;99:347–350. doi: 10.1016/s0092-8674(00)81520-2. [DOI] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]